Summary
Csr is a conserved global regulatory system, which uses the sequence specific RNA binding protein CsrA to activate or repress gene expression by binding to mRNA and altering translation, stability and/or transcript elongation. In Escherichia coli, CsrA activity is regulated by two sRNAs, CsrB and CsrC, which bind to multiple CsrA dimers, thereby sequestering this protein away from its mRNA targets. Turnover of CsrB/C sRNAs is tightly regulated by a GGDEF-EAL domain protein, CsrD, which targets them for cleavage by RNase E. Here, we show that EIIAGlc of the glucose-specific PTS system is also required for the normal decay of these sRNAs and that it acts by binding to the EAL domain of CsrD. Only the unphosphorylated form of EIIAGlc bound to CsrD in vitro and was capable of activating CsrB/C turnover in vivo. Genetic studies confirmed that this mechanism couples CsrB/C sRNA decay to the availability of a preferred carbon source. These findings reveal a new physiological influence on the workings of the Csr system, a novel function for the EAL domain, and an important new way in which EIIAGlc shapes global regulatory circuitry in response to nutritional status.
Keywords: sRNA decay, Csr system, PTS, GGDEF-EAL protein, global regulation, CsrD
Introduction
The Csr (carbon storage regulator) or Rsm (repressor of stationary phase metabolites) system is present in diverse eubacteria, where it globally regulates metabolism, biofilm formation, motility, virulence, quorum sensing, and stress response systems (Romeo, 1998, Babitzke & Romeo, 2007, Romeo et al., 2013, Vakulskas et al., 2015). The RNA binding protein CsrA/RsmA of the Csr system regulates gene expression by interacting with sequences in mRNA, thus altering translation, mRNA stability, and/or transcript elongation. CsrA governs genes responsible for bacterial lifestyle transitions, repressing processes that are triggered upon entry into the stationary phase of growth and conferring stress resistance, while activating processes such as glycolysis, which support vigorous growth. In Escherichia coli, CsrA activity is mainly controlled by the noncoding sRNAs, CsrB and CsrC, which contain multiple CsrA binding sites, allowing them to antagonize CsrA activity by sequestering it away from lower affinity mRNA targets (Liu et al., 1997, Weilbacher et al., 2003). Fluctuations in CsrB/C levels play a central role in regulating Csr system and the bacterial lifestyle.
Multiple factors ensure appropriate expression of csrB/C. Transcription is activated by the BarA-UvrY two-component signal transduction system (TCS) in response to carboxylic acids (Suzuki et al., 2002, Chavez et al., 2010, Martínez et al., 2014). In a complex negative feedback loop, CsrA regulates csrB/C transcription by activating both the expression of the response regulator UvrY and its phosphorylation by the sensor-kinase BarA (Suzuki et al., 2006, Camacho et al., 2015, Suzuki et al., 2002). Amino acid starvation and other stresses activate csrB/C transcription via the stringent response components ppGpp and DksA (Edwards et al., 2011) and two DEAD-box RNA helicases, DeaD and SrmB, activate csrB/C transcription by distinct mechanisms (Vakulskas et al., 2014).
In contrast to synthesis, the decay of CsrB/C RNAs is not well understood. A specificity factor, CsrD, is necessary for degradation of CsrB/C by the housekeeping nucleases RNase E and PNPase (Suzuki et al., 2006). CsrD does not appear to be a nuclease, but renders CsrB/C susceptible to degradation by RNase E, thus affecting the expression of CsrA-regulated genes in a predictable fashion. CsrD contains GGDEF and EAL domains, which are often responsible for synthesis and degradation of the secondary messenger cyclic dimeric (3′→5′) GMP (c-di-GMP). However, biochemical and genetic studies indicated that CsrD displays no c-di-GMP synthetic or hydrolytic activity and that CsrD activity is not regulated by c-di-GMP in vivo. At present, the molecular mechanism of CsrD effects on CsrB/C degradation is unclear. CsrD bound nonspecifically to CsrB/C in vitro (Suzuki et al., 2006). Accordingly, one hypothesis is that CsrD evolved from the GGDEF-EAL domain family, becoming an RNA binding protein. How CsrD activity is regulated is another open question. CsrA weakly represses csrD expression in E. coli and Salmonella Typhimurium (Suzuki et al., 2006, Jonas et al., 2010), but does not seem to affect CsrB turnover in E. coli (Gudapaty et al., 2001), and no other factors are known to affect CsrD activity.
Glucose is the preferred carbon and energy source for E. coli and is taken up primarily by the glucose-specific phosphoenolpyruvate-dependent sugar-phosphotransferase system, PTS (Deutscher et al., 2006, Deutscher et al., 2014, Lengeler & Jahreis, 2009). This system consists of two cytoplasmic proteins, enzyme I (EI) and histidine phosphocarrier protein (HPr) that are used for transporting many sugars, and two glucose-specific proteins, enzyme IIAGlc (EIIAGlc) and the membrane-bound enzyme IIBCGlc (EIIBCGlc). Glucose uptake is coupled to its phosphorylation. The phosphoryl group is donated by PEP and transferred to glucose via a phosphorylation cascade formed by EI, HPr, EIIAGlc, and EIIBCGlc proteins. Thus, the phosphorylation state of the PTS proteins depends both on extracellular carbon availability and the metabolic state of the cell.
The glucose-PTS proteins also mediate regulatory functions (Gabor et al., 2011, Deutscher et al., 2014). EIIAGlc is a central regulator of carbon metabolism. Unphosphorylated EIIAGlc mediates inducer exclusion by binding to and inhibiting transporters of non-PTS sugars (Deutscher et al., 2014, Deutscher et al., 2006). It also inhibits metabolism of alternative carbon sources, e.g. by binding to glycerol kinase (Postma et al., 1984). In contrast, phosphorylated EIIAGlc (EIIAGlc-P) binds to and stimulates the activity of adenylate cyclase, which produces cAMP. This compound acts as a secondary messenger that binds to the cAMP receptor protein (Crp), forming a transcription factor (cAMP-Crp) that exerts global effects on the proteome, ensuring efficient resource utilization (Krin et al., 2002, Park et al., 2006, Bao & Duong, 2013). With the discovery of novel EIIAGlc binding partners in various species, EIIAGlc has been found to participate in chemotaxis (Neumann et al., 2012), respiration/fermentation (Koo et al., 2004), biofilm formation (Pickering et al., 2012) and virulence (Kim et al., 2010, Mazé et al., 2014). The EIIBCGlc protein also carries out a variety of regulatory functions (Lux et al., 1995, Nam et al., 2001, Tanaka et al., 2000, Lee et al., 2000).
In a screen for EIIAGlc binding partners in Vibrio cholerae, a CsrD homologue, MshH was identified (Pickering et al., 2012). While no function was assigned to this interaction, it was hypothesized that EIIAGlc might affect the decay of Csr sRNAs. Here, we present the results of a detailed investigation of the role of EIIAGlc in CsrB/C decay in E. coli. Unphosphorylated EIIAGlc binds specifically to the EAL domain of CsrD and stimulates CsrB/C turnover. We propose that this mechanism helps to increase the concentration of free CsrA when it is needed to support growth, and simultaneously poises the Csr system for rapid response to changing environmental conditions.
Results
EIIAGlc activates CsrB/C decay via CsrD
To start to investigate the possible connection between EIIAGlc and the Csr system in E. coli, we decided to determine if EIIAGlc participates in the degradation of CsrB/C in this species. To do so, we first determined the stability of CsrB/C in the presence or absence of EIIAGlc (Δcrr) after the addition of rifampicin to the exponentially growing cultures. Deletion of crr decreased CsrB and CsrC decay rates by about 5-fold and 3-fold, respectively (Figure 1A–D). Ectopic expression of crr complemented the crr defect, confirming that EIIAGlc somehow regulates CsrB/C decay (Figure 1A–D). As discussed below, a site-directed crr mutant allele, encoding an EIIAGlc protein that cannot be phosphorylated, H91A, also complemented the crr deletion (Figure 1A–D).
Figure 1.
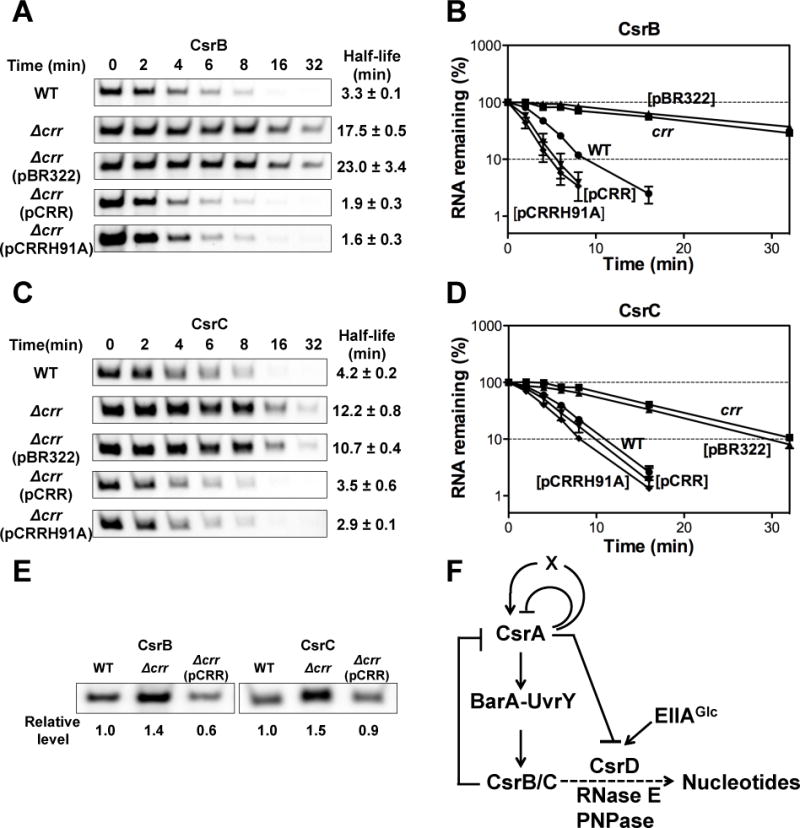
EIIAGlc affects CsrB/C decay rates and levels in E. coli.
A and C. Decay rates of CsrB/C were determined by Northern blotting of RNA extracted from exponential phase cultures (OD600 ~0.5) at various times following the addition of rifampicin. Culture identities: E. coli MG1655 (WT) or its Δcrr mutant with or without plasmid pBR322, pCRR or pCRRH91A (nonphosphorylatable EIIAGlc). The RNA half-lives were determined from the linear portions of their decay curves, shown in B and D. Standard deviations of values from two independent experiments are shown.
E. CsrB/C steady state levels determined by Northern blotting of RNA from exponentially growing cultures (OD600 ~0.5), as above.
F. A model for the Csr regulatory circuitry that includes EIIAGlc activation of CsrD-mediated CsrB/C decay. A broken line indicates an undefined mechanism(s).
As shown in Figure 1E, while EIIAGlc greatly stimulated CsrB/C decay, it modestly reduced the levels of CsrB/C in the cell. A similar observation was also made previously in a csrD mutant strain, and was shown to be the result of the Csr regulatory circuitry (Suzuki et al., 2006; Figure 1F). CsrA indirectly activates transcription of CsrB/C via the BarA-UvrY TCS (Suzuki et al., 2002). Thus, when CsrB/C decay is inhibited, these sRNAs accumulate and sequester CsrA, causing a decrease in their transcription and an attenuated effect on their levels.
We next performed an epistasis experiment to determine whether the effect of EIIAGlc on CsrB/C decay was dependent on CsrD. A Δcrr ΔcsrD double deletion strain was severely defective in CsrB/C decay (Figure 2A and B), as reported previously for the ΔcsrD mutant (Suzuki et al., 2006). While ectopic expression of crr in the Δcrr ΔcsrD strain failed to restore CsrB/C decay, ectopic overexpression of csrD enhanced CsrB/C decay rates to wild-type levels (Figure 2A and B). This finding suggested that CsrD functions downstream of EIIAGlc in CsrB/C turnover.
Figure 2.
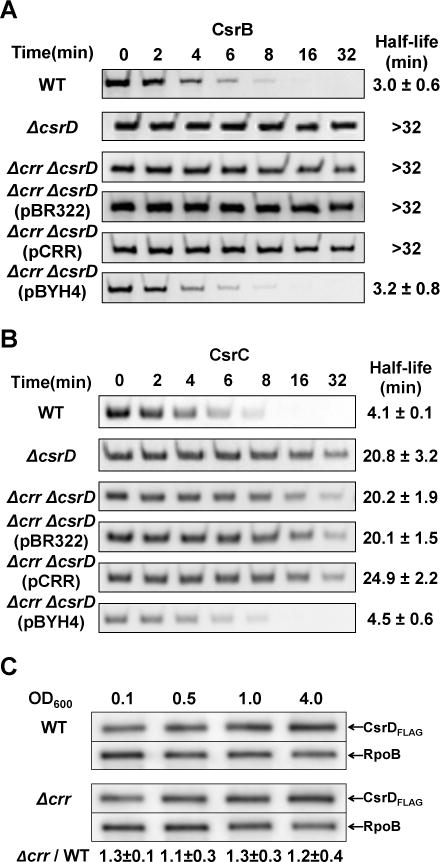
EIIAGlc (crr) activates CsrB/C decay via CsrD, but does not enhance cellular CsrD levels.
A and B. Decay rates of CsrB (A) and CsrC (B) were determined by Northern blotting of RNA from E. coli MG1655 (WT), ΔcsrD, Δcrr ΔcsrD strains with or without plasmid pBR322, pCRR or pBYH4 (expressing CsrD), as described in Figure 1.
C. Western blots depicting the effect of crr deletion on the level of CsrD protein. RpoB was used as loading control. CsrD protein levels in Δcrr relative to those in the wild-type strain (WT) are shown at the bottom. Standard derivations from triplicate experiments are indicated.
A possible explanation for the epistasis results is that EIIAGlc affects CsrD levels in the cell by altering its stability or synthesis. However, deletion of crr had no effect on the levels of a chromosomally encoded and biologically functional CsrD-FLAG protein (Figure 2C and Figure S1).
In vitro binding of EIIAGlc to CsrD requires the EAL domain
Because EIIAGlc regulates the activities of several proteins via direct binding, it was reasonable to speculate that EIIAGlc affects CsrB/C decay via direct binding to CsrD in E. coli. To test this idea, we examined the binding of EIIAGlc and CsrD in an in vitro binding assay or pull-down assay using His-tagged EIIAGlc and a soluble recombinant CsrD protein (CsrDΔTM) in which the N-terminal transmembrane domains of CsrD were replaced with a maltose binding protein (MBP) tag (Figure 3A). Previous studies showed that the transmembrane domains of CsrD were dispensable for its activity when the protein was ectopically expressed (Suzuki et al., 2006). In this assay, CsrD was mixed with Ni-NTA resin with or without His-tagged EIIAGlc. As shown in Figure 3B, the CsrDΔTM variant was retained by the Ni-NTA resin when EIIAGlc was bound to it, but remained in the unbound fraction when EIIAGlc was absent, indicating that EIIAGlc bound directly to CsrD.
Figure 3.
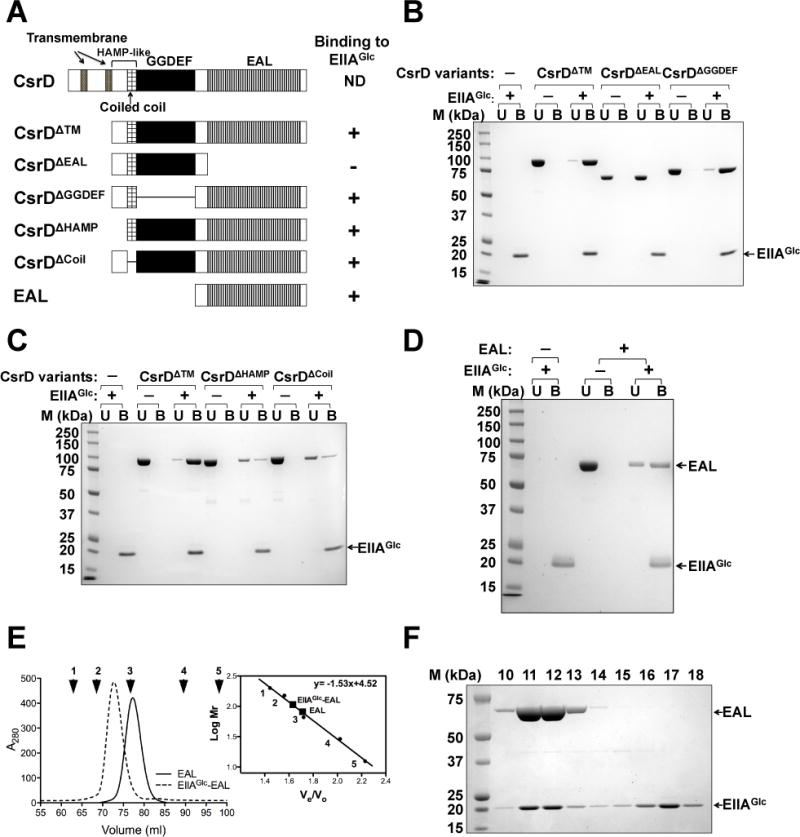
EIIAGlc interacts specifically with the EAL domain of CsrD.
A. CsrD variants used to identify the domain that binds to EIIAGlc.
B, C and D. In vitro assays for binding of EIIAGlc to CsrD variants depicted in panel A. Each reaction contained 8 μM of CsrD variants and 16 μM of EIIAGlc. U and B: unbound and bound proteins. Control reactions were performed in the absence of EIIAGlc to confirm that CsrD varients do not bind nonspecifically to Ni-NTA resin.
E. Gel filtration assay of EIIAGlc, EAL, and EIIAGlc-EAL mixture. Proteins were fractionated on a Superdex 200 column (HiLoad™ 16/60, 120 ml). The solid line corresponds to EAL domain alone; the dashed line corresponds to the EIIAGlc-EAL mixture. The chromatogram for EIIAGlc was not shown because this protein displays little absorbance at 280 nm. Arrows indicate elution volumes of molecular weight markers used to calibrate the column (Experimental procedures).
F. SDS-PAGE and Coomassie blue staining of proteins from gel filtration chromatography fractions of the EIIAGlc-EAL mixture shown in panel E. Fractions (3 ml) were collected starting at 40 ml.
To determine which domain of CsrD protein is involved in the interaction with EIIAGlc, we tested similar MBP fusions of CsrD lacking the EAL (CsrDΔEAL), GGDEF (CsrDΔGGDEF), or HAMP-like domain (CsrDΔHAMP and CsrDΔCoil), using the pull-down assays. While the other CsrD variants retained the ability to bind to EIIAGlc, CsrDΔEAL lost all detectable binding, suggesting that the EAL domain is involved in this interaction (Figure 3B and 3C). Moreover, the EAL domain alone bound to EIIAGlc in this assay (Figure 3D). These results indicated that EIIAGlc binds specifically to the EAL domain of CsrD.
To examine the binding reaction of EIIAGlc with CsrD in more detail, the size and composition of the EIIAGlc-EAL complex was analyzed by gel-filtration chromatography. The free EAL and EIIAGlc eluted at positions corresponding to sizes of their monomeric forms (EAL, 72kDa; EIIAGlc, 20 kDa) (Figure 3E and F). When EIIAGlc and EAL were mixed to allow binding, and fractionated on Superdex 200 (HiLoad™ 16/60, GE Healthcare), a new peak was observed at a position corresponding to a size of 98kDa, approximately that of a heterodimer of EIIAGlc-EAL (92kDa). To determine the ratio of EIIAGlc and EAL in the complex, column fractions corresponding to the presumptive EIIAGlc-EAL complex and the free EIIAGlc were analyzed by SDS-PAGE with Commassie blue staining and quantification of the stained proteins (Figure 3F). This experiment revealed the molar ratio of EAL bound to EIIAGlc in peak fractions (11 and 12) to be 1:1 suggesting that EIIAGlc binds to the EAL domain of CsrD in a one to one ratio.
In the pull down assay, the relative amount of CsrDΔTM or CsrDΔGGDEF bound by EIIAGlc was much greater than that of proteins that lacked the intact HAMP-like domain, CsrDΔHAMP or CsrDΔCoil (Figure 3B and C). The HAMP domain typically promotes dimerization or protein-protein interactions and plays important roles in signal transduction (Hulko et al., 2006). We used gel filtration assays to determine the in vitro oligomeric states of CsrD variants containing or lacking the HAMP-like domain. All CsrD variants containing the HAMP-like domain, CsrDΔTM, CsrDΔGGDEF and CsrDΔEAL, eluted at volumes consistent with their tetrameric forms (Figure S2 and Table S4). In contrast, the CsrD variants with a disrupted HAMP-like domain, CsrDΔHAMP and CsrDΔCoil, eluted as apparent monomers (Figure S2 and Table S4). The precise way in which tetramerization affected EIIAGlc binding by the CsrD variants was not further investigated.
EIIAGlc regulates CsrB/C decay in a phosphorylation-dependent manner
EIIAGlc typically modulates the activity of its binding partners in a phosphorylation-dependent manner. Accordingly, we tested the effect of the phosphorylation state of EIIAGlc on CsrB/C decay. We first investigated the impact of the unphosphorylated EIIAGlc on CsrB/C turnover using a site-directed mutant protein that could not be phosphorylated (EIIAGlc H91A). Plasmid complementation of the Δcrr strain with EIIAGlc H91A restored CsrB/C decay rates to slightly higher than in the wild-type strain (Figure 1A–D), demonstrating that phosphorylation of EIIAGlc was dispensable for activation of CsrB/C turnover.
Next, we performed pull-down assays to determine the effect of EIIAGlc phosphorylation on binding to CsrD. In these experiments, CsrD containing an N-terminal MBP tag was bound to amylose resin and then mixed with EIIAGlc in reactions that were designed to produce either the phosphorylated or unphosphorylated form of this protein. In one reaction, E. coli Hpr, EI and PEP were mixed to provide phosphorylated EIIAGlc. In the other reaction, pyruvate was added instead of PEP to maintain EIIAGlc in the unphosphorylated form. Strikingly, while most of the unphosphorylated EIIAGlc bound to CsrD, no binding was observed between the phosphorylated EIIAGlc and CsrD (Figure 4). These results were in agreement with the observation that EIIAGlc did not require phosphorylation for activation of CsrB/C turnover in vivo (Figure 1A–D), and indicated that the binding of unphosphorylated EIIAGlc to CsrD activates CsrB/C sRNA decay.
Figure 4.
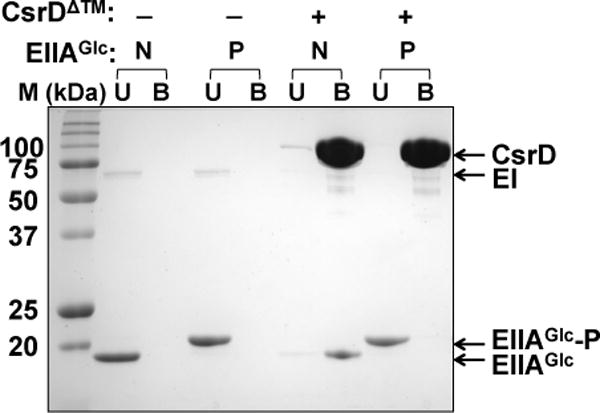
CsrD binds only to unphosphorylated EIIAGlc in pull-down assays.
MBP-tagged CsrD protein was bound to amylose resin and then mixed with EIIAGlc in the nonphosphorylated (N) or phosphorylated state (P) to permit binding reactions to occur. The reactions were processed as described in the Experimental procedures. Control reactions were performed without CsrD to test for nonspecific binding of the two forms of EIIAGlc to amylose resin. Note that the two forms of EIIAGlc were resolved from each other on 15% SDS-PAGE gel. U and B refer to proteins that were unbound vs. bound by the amylose resin.
cAMP-Crp modestly represses CsrB turnover
While the unphosphorylated EIIAGlc bound to CsrD in vitro and was able to activate CsrB/C decay in vivo, we wondered if the phosphorylated form of EIIAGlc might affect CsrB/C turnover via its important role in cAMP-Crp production (Krin et al., 2002, Park et al., 2006). Deletion of cyaA or crp modestly increased the CsrB decay rate by 2-fold (Figure 5A), while exhibiting weak or negligible effects on CsrC decay (Figure 5B). The increased decay rate of CsrB in the cyaA mutant was restored by exogenous cAMP (10 mM), confirming that cAMP-Crp somehow inhibits CsrB decay. Deletion of cyaA or crp in the Δcrr background had twofold effects on CsrB decay rates that were similar to those in the wild-type background, and deletion of crr had similar fivefold effects on CsrB decay in both the wild type strain and its isogenic crp and cyaA mutants (Figure 5A). These findings confirmed that the major effect of EIIAGlc on CsrB decay is mediated independently of cAMP-Crp. The modest effect of cAMP-Crp in the Δcrr background was likely due to basal adenylate cyclase activity in the crr mutant (Lévy et al., 1990, Feucht & Saier, 1980, Reddy & Kamireddi, 1998).
Figure 5.
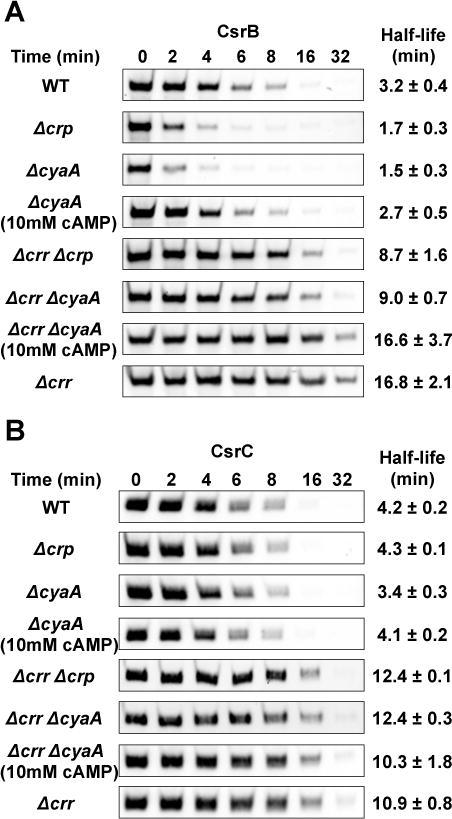
Effects of cAMP-Crp on CsrB/C decay.
A and B. Decay rates of CsrB/C were determined by Northern blotting of RNA from E. coli MG1655 (WT) and isogenic deletion mutants: cyaA, crp, cyaA, crr crp, crr cyaA, crr with or without added 10 mM cAMP, as described in Figure 1.
CsrB/C decay is regulated in response to carbon availability via the phosphorylation state of EIIAGlc
The phosphorylation state of EIIAGlc is determined by carbon sources that are taken up and metabolized (Hogema et al., 1998, Deutscher et al., 2014). Preferred carbon sources such as glucose lead to net dephosphorylation of PTS proteins, including EIIAGlc, whereas unfavorable carbon sources or carbon starvation conditions cause the accumulation of phosphorylated EIIAGlc. Because the unphosphorylated EIIAGlc bound to CsrD in vitro and promoted CsrB/C decay in vivo (Figures 1, 4), we expected that CsrB/C decay rates should be elevated in the presence of glucose. Consequently, we first examined CsrB/C decay in minimal medium supplemented with 0.2% glucose, glycerol or succinate. Both the phosphorylation state of EIIAGlc and CsrB/C decay rates responded predictably to these carbon sources; more rapid decay was observed in glucose compared to glycerol or succinate (Figure 6).
Figure 6.
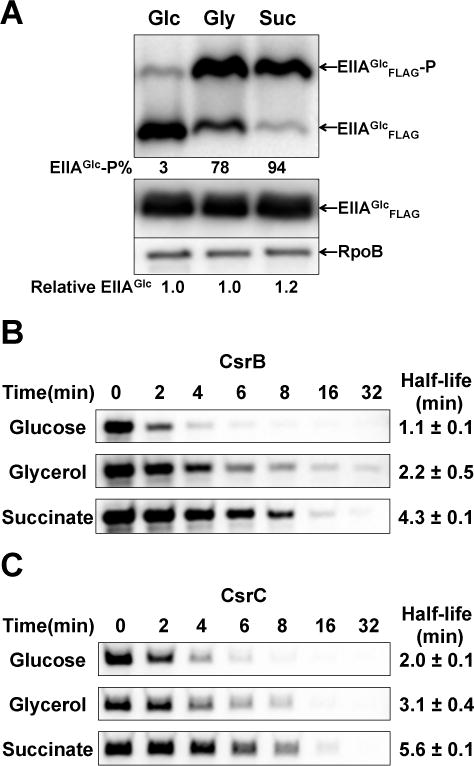
Effects of carbon sources on EIIAGlc phosphorylation and CsrB/C decay in minimal media.
A. Western blot depicting the phosphorylation state of EIIAGlc of E. coli MG1655 growing in minimal medium A supplemented with 0.2% glucose, glycerol or succinate. Exponential phase extracts were fractionated in gels with Phos-tag™ reagent and analyzed by Western blot. Relative level of total EIIAGlc was analyzed by western blot without using Phos-tag™ reagent. The percentage of phosphorylated (EIIAGlc-P) and unphosphorylated EIIAGlc (EIIAGlc) and relative total EIIAGlc protein levels are given.
B and C. Northern blot depicting CsrB/C decay rates in E. coli MG1655 exponentially growing in minimal medium A supplemented with 0.2% glucose, glycerol or succinate. The RNA half-lives were determined as shown in Figure 1.
To verify that the phosphorylation state of EIIAGlc determines the decay rates of CsrB/C in response to different carbon sources, we examined CsrB/C turnover in a strain that expresses the mutant EIIAGlc protein, H91A, which cannot be phosphorylated. Because the strain expressing H91A has a significant growth defect in minimal medium (data not shown) the WT and H91A strains were first grown in LB broth to exponential phase and then washed and inoculated into minimal medium lacking a carbon compound or containing 0.2% glucose or succinate. The phosphorylation state of EIIAGlc was determined from growth in LB (Figure 7A) and 10 min after inoculation into minimal media (Figure 7B). EIIAGlc phosphorylation in LB was ~40% and increased to ~90% in media with succinate or lacking a carbon source, while it decreased to 4% at 10 min after glucose exposure. Decay rates of CsrB/C were determined 10 min after inoculation (Figure 8). The decay rates of CsrB and CsrC in the wild-type strain (WT) were ~3.5 and 2.5-fold greater, respectively, in medium with glucose vs. no carbon source. A more modest, but reproducible difference (~2 fold) was observed for CsrB/C decay rates in glucose compared to succinate. These data support the observations described above, showing that CsrB/C decay rates vary in response to different carbon conditions, although the difference in decay between succinate and carbon-deficient media does not seem to be explained by EIIAGlc phosphorylation alone (Figure 8), as both conditions resulted in similar EIIAGlc-P levels (Figure 7B). Most importantly, CsrB/C decay in the H91A strain was rapid and virtually identical in all three media, confirming that the phosphorylation state of EIIAGlc determines CsrB/C decay in response to carbon substrate availability. The levels of CsrB/C RNAs (Figure 8E) were consistent with these decay rates, but as observed previously (Figure 1), the effects of turnover may be attenuated via the feedback loop of the Csr circuitry (Figure 1F; Suzuki et al., 2002; 2006).
Figure 7.
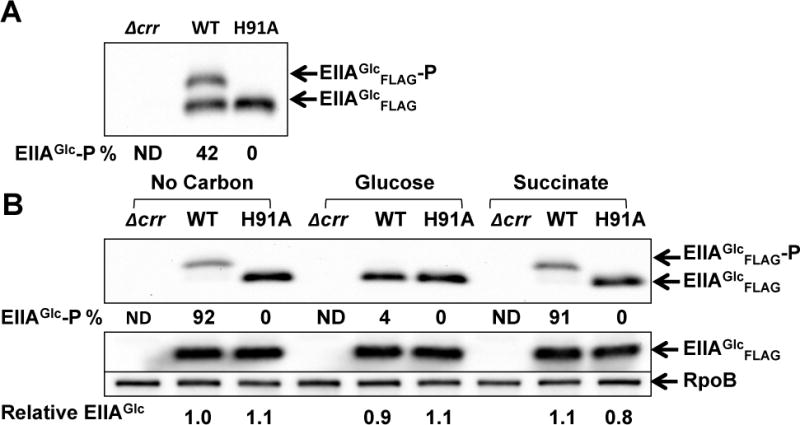
The phosphorylation state of EIIAGlc before and 10 min after shift from LB broth into minimal media.
A and B. Western blot depicting the phosphorylation state and relative level of EIIAGlc in E. coli MG1655 (WT), Δcrr and H91A (mutant strain that carrying a chromosomal EIIAGlcH91A point mutation). Extracts were prepared from cultures grown in LB broth (A) and at 10 min after reinoculation into minimal medium without carbon, with 0.2% glucose or succinate (B). The percentage of phosphorylated EIIAGlc relative to total EIIAGlc was determined as described in Figure 6.
Figure 8.
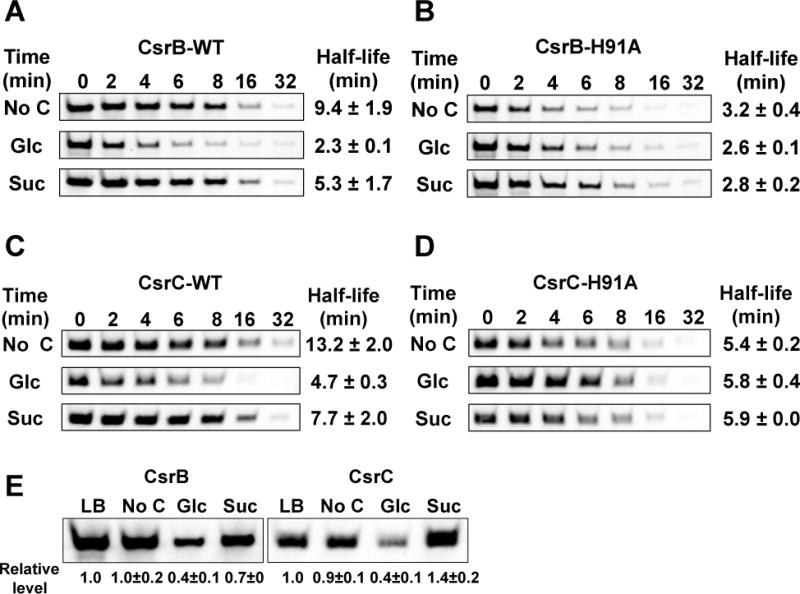
CsrB/C decay rates and levels after shift from LB to minimal media.
A, B, C and D. Decay rates of CsrB (A and B) and CsrC (C and D) determined by Northern blotting of RNA from E. coli MG1655 (WT) and mutant strain H91A (unphosphorylatable EIIAGlc). Strains were grown in LB broth to exponential phase, washed and reinoculated into M9 minimal medium without carbon or with 0.2% glucose or succinate. Rifampicin was added 10 min after inoculation into minimal media and RNA half-lives determined as in Figure 1.
E. CsrB/C steady state levels determined by Northern blotting of RNA from E. coli MG1655 (WT) in LB broth at exponential growth phase and 10 min after inoculation into minimal media.
EIIAGlc and MshH promote Csr sRNA decay in Vibrio cholerae
EIIAGlc, RNase E and CsrD orthologs are widespread in Enterobacteriaceae, Vibrionaceae, and Shewanellaceae species (Suzuki et al., 2006, Vakulskas et al., 2015, Comas et al., 2008), suggesting that a common mechanism may exist for Csr sRNA decay in members of these bacterial families. As a proof of principle, we tested the effects of EIIAGlc and the CsrD homolog, MshH, on decay of the V. cholerae sRNAs, CsrB, CsrC and CsrD (Lenz et al., 2005). The V. cholerae sRNAs exhibited longer half-lives compared to the E. coli sRNAs under our growth conditions (Figure S3). Nevertheless, deletion of mshH, crr or both genes greatly decreased CsrB and CsrD turnover. These effects were not apparent for CsrC, which was already extremely stable in the wild-type strain. This experiment demonstrated the potential of EIIAGlc and CsrD to activate the decay of Csr sRNAs in this important member of the Vibrionaceae family.
Discussion
Here, we identified a new regulatory function for EIIAGlc, in which it binds to the sRNA decay protein CsrD and stimulates CsrB/C decay when glucose is present. This mechanism should enhance the concentration of free CsrA, which activates glycolysis and represses gluconeogenesis, secondary metabolism, and stress resistance responses such as biofilm formation (Babitzke & Romeo, 2007, Vakulskas et al., 2015, Romeo et al., 2013). Because CsrA regulates lifestyle transitions in many bacterial species and interacts with hundreds of transcripts in E. coli (Babitzke & Romeo, 2007, Patterson-Fortin et al., 2013, Edwards et al., 2011, Vakulskas et al., 2015), we propose that this represents a particularly important role for EIIAGlc. A high rate of turnover can facilitate rapid changes in transcript levels. Therefore, EIIAGlc-CsrD interactions should not only allow CsrB/C decay rates to be reset in response to changing glucose availability, but may poise the Csr system for rapid responses to other cues or conditions when glucose is present. The V. cholerae CsrD ortholog protein MshH and EIIAGlc also activated the decay of CsrB and CsrD sRNAs (Figure S3). We suspect that the mechanism described for E. coli CsrB/C turnover operates in many species of Enterobacteriaceae, Vibrionaceae and Shewanellaceae.
The conclusion that only the unphosphorylated form of EIIAGlc is able to promote CsrB/C decay through binding interactions with CsrD was based on a combination of biochemical and genetic evidence. CsrD bound only to the unphosphorylated form of EIIAGlc in vitro (Figure 4). Furthermore, a non-phosphorylatable protein, EIIAGlc-H91A, sustained CsrB/C decay rates that were similar to or even greater than the wild-type protein (Figure 1A–D). The presence of glucose also caused net dephosphorylation of EIIAGlc and supported rapid decay of CsrB/C sRNAs relative to carbon starvation conditions or alternative carbon sources (Figure 6–8). Importantly, EIIAGlc-H91A supported high decay rates under all of these conditions, confirming that the effects of carbon availability on CsrB/C decay are mediated thorough altered phosphorylation of EIIAGlc (Figure 8). A previous study with MshH of V. cholerae concluded that both the phosphorylated and unphosphorylated forms of EIIAGlc bind to the CsrD homolog. This conclusion was based on the observation that in a two-hybrid assay, MshH interacted with mutant proteins designed to mimic the unphosphorylated (EIIAGlc-H91A) or the phosphorylated (EIIAGlc-H91D) forms of EIIAGlc (Pickering et al., 2012). However, we caution that EIIAGlc-H91D does not appear to mimic EIIAGlc-P. Another putative EIIAGlc-P mimic, EIIAGlc-H91E, was unable to activate adenylate cyclase (Reddy & Kamireddi, 1998). Similarly, effects of EIIANtr-P were not mimicked by replacing its phosphorylatable His residue with either Asp or Glu (Lüttmann et al., 2009). Finally, we constructed EIIAGlc-H91D in E. coli and found that it behaved similarly to the unphosphorylated EIIAGlc rather than EIIAGlc-P in CsrB/C decay (Figure S4).
The phosphorylated form of EIIAGlc activates cAMP synthesis by binding to adenylate cyclase. Because cAMP and Crp modestly repressed CsrB decay (Figure 5), we propose that EIIAGlc-P indirectly and modestly represses CsrB turnover, reinforcing the positive effect of unphosphorylated EIIAGlc on CsrB decay. Because a potential Crp binding site was predicted in the untranslated leader region of csrD (data not shown), we tested the possibility that Crp inhibits CsrB decay by controlling csrD expression. Weakly positive to negligible effects of Crp were observed on CsrD levels (Figure S5), which might contribute to the effect of Crp on CsrB decay.
Given that EIIAGlc acts via CsrD without altering its levels in the cell and that overexpression of csrD restored CsrB/C decay in a strain deleted for the EIIAGlc gene, crr (Figure 2), we propose that EIIAGlc functions as an allosteric activator of CsrD, perhaps similar to the role of EIIAGlc-P in activating adenylate cyclase (Saier, 1989, Park et al., 2006). Structural studies of EIIAGlc show that it possesses a concave face that allows it to interact with globular target proteins (Hurley et al., 1993, Chen et al., 2013, Wang et al., 2000, Cai et al., 2003). Other EIIAs seem unable to duplicate this function (Deutscher et al., 2006). We deleted the genes for five other EIIAs (fruB, mtlA, chbA, manX and ptsN) and found that none of them regulated CsrB/C decay (data not shown).
This study also expands our understanding of the functionality of the EAL domain. This is not the first report of a catalytically inactive EAL domain performing a regulatory role via protein-protein interactions (Römling et al., 2013). The enzymatically inactive EAL domain protein YdiV of E. coli binds to the transcription activator FlhD4C2 and prevents it from binding to target DNA (Li et al., 2012). Similarly, the EAL domain of the E. coli photoreceptor YcgF (BluF) binds to the MerR-like repressor YcgE (BluE) in the presence of blue light and prevents it from binding to DNA (Tschowri et al., 2009). Complex regulation exists for the EAL domain of FimX, which binds to c-di-GMP as well as the PilZ protein, and is required for biogenesis of the Type IV pilus (Guzzo et al., 2009). In all of these cases, the EAL domain-containing protein acts as a sensor that uses its EAL domain to transmit information to another protein. In contrast, the EAL domain of CsrD acts as a receiver, to detect signaling information from a sensory protein. We did not examine other EAL domain proteins for binding interactions with EIIAGlc. However, none of the binding partners in E. coli or the potential ones in V. cholerae contains an EAL domain except for CsrD/MshH (Deutscher et al., 2014, Pickering et al., 2012). The modular structure of CsrD suggests that the effect of EIIAGlc might be transmitted through other CsrD domains, such as the GGDEF domain, which is also necessary for CsrD activity (Suzuki et al., 2006), although this possibility has not been explored.
Our previous data suggested that CsrD might function as a CsrB/C binding protein, converting the sRNAs into substrates for RNase E degradation (Suzuki et al., 2006). However, EIIAGlc did not increase the binding affinity or specificity of CsrD for CsrB in electrophoretic mobility shift assays (Figure S6). Presently, we cannot distinguish whether (i) additional unknown factors or conditions are required to reconstitute the effect of EIIAGlc on CsrD RNA binding, (ii) EIIAGlc stimulates CsrD activity at a step in the decay pathway other than CsrB/C binding, or (iii) perhaps RNA binding by CsrD is an in vitro artifact and not part of its biological function.
While the circuitry surrounding the Csr system is extensive, its role in carbon and energy pathways is particularly wide-ranging and important (Edwards et al., 2011, Patterson-Fortin et al., 2013, Yang et al., 1996, Romeo, 1996, Romeo et al., 2013, Sabnis et al., 1995, Pernestig et al., 2003). Previous studies have shown that carboxylic acid-containing end products of carbon metabolism, such as acetate and formate, stimulate CsrB transcription via the BarA-UvrY TCS (Chavez et al., 2010). Thus, the synthesis pathway and the newly discovered turnover pathway for CsrB/C should mediate reinforcing positive effects on the levels of these sRNAs when preferred carbon resources have been expended and end products have accumulated. The resulting decrease in CsrA activity under this condition should promote the transition from glycolytic metabolism and active growth to gluconeogenesis, glycogen biosynthesis and the formation of a stress resistant phenotype. We caution that other regulators influence the expression of CsrB/C RNAs, e.g. ppGpp, DksA, DeaD and SrmB helicases, and impact the workings of this circuitry. An understanding of the combinatorial effects of all of these factors, and perhaps unknown ones, will require additional investigation.
Experimental procedures
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table S1. Bacterial strains were routinely grown in Luria-Bertani (LB) broth unless otherwise indicated. For synthetic minimal medium, minimal medium A (Hogema et al., 1998) and M9 minimal medium supplemented with indicated carbohydrates were used. When necessary, the following antibiotics were added to the growth media: ampicillin (100 μg ml−1), tetracycline (15 μg ml−1), kanamycin (50 μg ml−1), and chloramphenicol (25 μg ml−1). E. coli and V. cholerae strains were grown at 37°C and 27°C, respectively. Stationary phase cultures were routinely used to inoculate LB broth or minimal media unless otherwise indicated. For strains carrying cyaA deletion or crp disruption, exponentially growing cultures were used to inoculate LB broth supplemented with or without 10 mM cAMP to minimize the growth defect.
Construction of plasmids and mutant strains
The plasmids and related primers and restriction sites used in this study are listed in Table S2 and S3. The plasmid pCRR, used for complementation of a crr deletion strain, carries the crr gene under a mutant lacUV5 promoter, in which −8 A of the −10 hexamer consensus was replaced with −8 T for decreased promoter activity (Moyle et al., 1991). To generate pCRR, the crr gene was amplified from chromosomal DNA of E.coli MG1655 and ligated into vector pBR322. Plasmid pCRRH91A expressed the mutant crr gene, producing the protein EIIAGlcH91A. This plasmid was constructed similarly to pCRR except that the His91Ala (CAC to GCC) substitution was introduced by the megaprimer PCR procedure (Ke & Madison, 1997). Plasmid pBYH4 used for complementation of the csrD deletion strain was described previously (Suzuki et al., 2006). To construct plasmid pETCRR for expression of C-terminal His-tagged EIIAGlc, crr was amplified and cloned into plasmid pET24a. Plasmids overexpressing CsrD variants were generated by amplifying the coding regions corresponding to CsrDΔTM (residues 156–646), CsrDΔHAMP (residues 192–646), CsrDΔCoil (residues 156–199 and 220–656), CsrDΔGGDEF (residues 156–223 and 393–646), CsrDΔEAL (residues 156–385) and EAL (residues 393–646) by standard PCR or overlapping PCR (Urban et al., 1997), and cloning the resulting products into vector pmal-c5x, yielding N-terminally MBP-tagged CsrD variants.
E. coli gene deletions were created by the standard P1vir transduction procedure or the lambda Red system as described (Datsenko & Wanner, 2000). Chromosomal C-terminal FLAG-tagged fusions were generated using the phage lambda Red system as described (Datsenko & Wanner, 2000, Uzzau et al., 2001). The mutant strain H91A carrying a chromosomal point mutation of crr, producing EIIAGlc H91A protein, was constructed by using overlapping PCR mutagenesis and the pKOV gene replacement protocol as described (Urban et al., 1997, Link et al., 1997).
V. cholerae ∆crr and ∆mshH mutants were created in the C6706str2 wild-type background (Thelin & Taylor, 1996) by double homologous recombination as previously described (Haugo & Watnick, 2002, Pickering et al., 2012).
Expression and Purification of EIIAGlc and CsrD variants
EIIAGlc was overproduced in E. coli strain BL21(DE3) grown in 1L of M9 minimal medium supplemented with 0.8% glucose (w/v). Three hours after the induction with 1mM IPTG at OD600 ~ 0.6, cells were harvested and lysed using a French Press. After centrifugation (20,000 × g, 30 min, 4 °C), the soluble fraction of the lysate was applied to a HisTrap column (1ml, GE Healthcare) and eluted with a gradient of imidazole (20–500 mM). Eluted proteins were further purified by gel filtration chromatography (Superdex 75 10/300, GE Healthcare), dialyzed against dialysis buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 1 mM DTT, 10% glycerol) and stored for subsequent experiments.
Overproduction of CsrD variants was from E. coli BL21(DE3) strains containing the corresponding plasmids, which were grown in LB medium supplemented with 0.2% glucose (w/v). Cells were induced with 0.3 mM IPTG at OD600 ~ 0.6 and lysed as mentioned above. The soluble fraction of the lysate was applied to an MBPTrap column (1ml, GE Healthcare) and eluted with a gradient of 0–10 mM maltose. To obtain homogeneous proteins, eluted proteins were further purified by gel filtration chromatography (Superdex 200 10/300, GE Healthcare) and dialyzed against dialysis buffer.
Northern blotting
Northern blot analysis was performed as previously described, with minor modifications (Vakulskas et al., 2014). Bacterial culture were immediately stabilized by the addition of RNAprotect™ Bacteria Reagent (Qiagen) or 0.125 volumes of stop solution (10% phenol/90% ethanol). Total RNA was isolated using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Total cellular RNA (1–2 μg) was separated on denaturing 5% acrylamide/7 M urea gels and transferred to a positively charged nylon membrane (Roche Diagnostics) by electroblotting. The membrane was cross-linked using UV light and hybridized overnight at 68°C (CsrB/C of E. coli) or 70°C (CsrB/C/D of V. cholerae) using a DIG-labeled antisense RNA probe (Table S3), which was prepared with the DIG Northern Starter kit (Roche Diagnostics). Transcripts were detected using the DIG Northern Starter kit (Roche Diagnostics) according to the manufacturer’s instructions. Blots were imaged using the ChemiDoc XRS+ system (Bio-Rad) and RNAs were quantified using Quantity One image analysis software (Bio-Rad). Prior to hybridization, the rRNAs (16S and 23S) were stained by methylene blue, which served as loading controls for signal correction.
Western blotting
Western blot analysis was performed using standard laboratory protocols as described (Vakulskas et al., 2014). Briefly, total cellular proteins were separated by SDS-PAGE and transferred to 0.2 μm polyvinylidene difluoride membranes (Bio-Rad) by electroblotting. Blots for FLAG epitope-tagged proteins used the anti-FLAG M2 monoclonal antibody (Sigma) at 1:2,000 dilution. Blots for RpoB used anti-RpoB monoclonal antibodies (Neoclone) at 1:50,000 dilution. Western blots were detected using horseradish peroxidase-linked secondary antibodies and the SuperSignal West Femto Chemiluminescent Substrate (Pierce) as recommended by the manufacturer. Proteins were quantified using Quantity One image analysis software (Bio-Rad).
Protein Pull-down assays
Interaction between CsrD variants and EIIAGlc was assessed using purified His-tagged EIIAGlc protein to pull down MBP-tagged CsrD variants. In a 60 μl reaction, 8 μM EIIAGlc and 4 μM CsrD variants were incubated with 15 μl Ni-NTA resin (Qiagen) in binding buffer (50 mM MES pH 6.5, 1 mM DTT, 20 mM Imidazole) at 4°C for 1hour. Unbound proteins remaining in the supernatant solution were collected after brief centrifugation of the resin. The resin was washed three times with 1 ml washing buffer (50 mM MES pH 6.5, 1mM DTT, 60 mM Imidazole), and the bound proteins were eluted with 45 μl of elution buffer (25 mM Tris-HCl pH 8.0, 500 mM NaCl, and 500 mM Imidazole).
To determine the phosphorylation dependence of the interaction between EIIAGlc and CsrD, the MBP-tagged CsrDΔTM was used to pull down His-tagged EIIAGlc. His-tagged EIIAGlc (8 μM) was incubated in reactions containing EI (1 μM), Hpr (8 μM) and either 5 mM PEP or 5 mM pyruvate in 60 μl buffer (20mM Tris-HCl pH8.0, 2 mM MgCl2, 1 mM DTT) at room temperature for 10 min. These reactions were designed to produce the phosphorylated or unphosphorylated form of EIIAGlc, respectively. Reactions were dialyzed against the binding buffer (50mM MES pH6.5, 1mM DTT) with Slide-A-Lyzer dialysis cassette (Thermo) for 1 hour and subsequently incubated at 4°C for 1 hr with CsrDΔTM (95 μg) pre-bound to amylose resin. Unbound proteins were collected after brief centrifugation of the resin and bound proteins were eluted with 60 μl of elution buffer (20 mM Tris-HCl pH7.5, 200 mM NaCl, 10 mM maltose, 1 mM DTT) after extensive rinsing of the resin with binding buffer.
Unbound and bound proteins in the pull-down reactions were detected by SDS-PAGE (10% or 15%, as required) and Coomassie blue staining. Proteins were quantified using Quantity One image analysis software (Bio-Rad).
Determination of the EIIAGlc phosphorylation state
The phosphorylation state of EIIAGlc was determined as previously described (Hogema et al., 1998), with modifications. Briefly, 0.2 ml of bacterial culture (OD600 ~ 0.5) was treated with the addition of 20 μl of 10 M NaOH followed by 1 ml of ethanol and 180 μl of 3 M sodium acetate, pH 5.2. After chilling at −80°C for 2 hr, precipitates were collected by centrifugation, rinsed with 70% ethanol, and suspended in 100 μl of sample buffer (0.16 M Tris HCl, pH 7.5, 4% SDS, 20% glycerol, and 10% 2-mercaptoethanol). To achieve a good separation of the two forms of EIIAGlc, samples were fractionated on SDS-PAGE gels containing 50 μM of Phos-tag reagent as previously described (Vakulskas et al., 2014). Subsequently, gels were washed with Western transfer buffer (25 mM Tris, 192 mM Glycine, 20% methanol, and 0.1% SDS) containing 1 mM EDTA for 10 min, followed by a second wash with transfer buffer for 20 min. Western blot analysis was then performed as described above.
Gel filtration analysis of EIIAGlc in complex with the EAL domain or other CsrD variants
For analysis of the EIIAGlc-EAL complex, a 0.5 ml reaction containing 33 μM EAL, 78 μM EIIAGlc or both proteins were incubated at 4 °C for 30 min and subjected to gel filtration analysis using an AKTA-FPLC system (Superdex 200, HiLoad™ 16/60, 120ml, GE Healthcare), and subsequently eluted at 4°C with a flow rate of 1 ml/min in buffer containing 20 mM Tris HCl pH 7.5, 100 mM NaCl, 1mM DTT. For CsrD variants, 0.5 ml samples containing purified CsrDΔTM, CsrDΔEAL, CsrDΔHAMP, CsrDΔCoil, or CsrDΔGGDEF was separated on the same system. Fractions (3 ml) were collected and analyzed by SDS-PAGE and Coomassie blue staining. For EIIAGlc-EAL analysis, the column was pre-calibrated using 5 gel filtration molecular weight markers (1, sweet potato β-amylase, 200 kDa; 2, yeast alcohol dehydrogenase, 150 kDa; 3, bovine serum albumin, 66 kDa; 4, carbonic anhydrase from bovine erythrocytes, 29 kDa; 5, horse heart cytochrome C, 12.4 kDa), and blue dextran 2000. For CsrD variants analysis, the column was calibrated using 5 molecular weight markers (1, equine spleen apoferritin, 443 kDa; 2, sweet potato β-Amylase, 200 kDa; 3, alcohol yeast dehydrogenase,150 kDa; 4, bovine serum albumin, 66 kDa; 5, carbonic anhydrase from bovine erythrocytes, 29 kDa), and blue dextran 2000. The relative molecular masses of proteins or protein complexes were calculated by logarithmic interpolation from the standards.
Supplementary Material
Acknowledgments
We thank Dr. Alan Peterkofsky of Laboratory of Cell Biology, National Heart, Lung, and Blood Institute, National Institutes of Health, for providing purified EI and Hpr proteins. These studies were funded by National Institutes of Health grants R01GM059969, R01AI097116 and F32AI100322.
References
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Bao H, Duong F. Phosphatidylglycerol directs binding and inhibitory action of EIIAGlc protein on the maltose transporter. J Biol Chem. 2013;288:23666–23674. doi: 10.1074/jbc.M113.489567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Williams DC, Wang G, Lee BR, Peterkofsky A, Clore GM. Solution structure of the phosphoryl transfer complex between the signal-transducing protein IIAGlucose and the cytoplasmic domain of the glucose transporter IICBGlucose of the Escherichia coli glucose phosphotransferase system. J Biol Chem. 2003;278:25191–25206. doi: 10.1074/jbc.M302677200. [DOI] [PubMed] [Google Scholar]
- Camacho MI, Alvarez AF, Chavez RG, Romeo T, Merino E, Georgellis D. Effects of the global regulator CsrA on the BarA/UvrY two-component signaling system. J Bacteriol. 2015;197:983–991. doi: 10.1128/JB.02325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez RG, Alvarez AF, Romeo T, Georgellis D. The physiological stimulus for the BarA sensor kinase. J Bacteriol. 2010;192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Oldham ML, Davidson AL, Chen J. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature. 2013;499:364–368. doi: 10.1038/nature12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, González-Candelas F, Zúñiga M. Unraveling the evolutionary history of the phosphoryl-transfer chain of the phosphoenolpyruvate:phosphotransferase system through phylogenetic analyses and genome context. BMC Evol Biol. 2008;8:147. doi: 10.1186/1471-2148-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Aké FM, Derkaoui M, Zébré AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol. 2011;80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht BU, Saier MH. Fine control of adenylate cyclase by the phosphoenolpyruvate:sugar phosphotransferase systems in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980;141:603–610. doi: 10.1128/jb.141.2.603-610.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor E, Göhler AK, Kosfeld A, Staab A, Kremling A, Jahreis K. The phosphoenolpyruvate-dependent glucose-phosphotransferase system from Escherichia coli K-12 as the center of a network regulating carbohydrate flux in the cell. Eur J Cell Biol. 2011;90:711–720. doi: 10.1016/j.ejcb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo CR, Salinas RK, Andrade MO, Farah CS. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol. 2009;393:848–866. doi: 10.1016/j.jmb.2009.07.065. [DOI] [PubMed] [Google Scholar]
- Haugo AJ, Watnick PI. Vibrio cholerae CytR is a repressor of biofilm development. Mol Microbiol. 2002;45:471–483. doi: 10.1046/j.1365-2958.2002.03023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogema BM, Arents JC, Bader R, Eijkemans K, Yoshida H, Takahashi H, Aiba H, Postma PW. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Faber HR, Worthylake D, Meadow ND, Roseman S, Pettigrew DW, Remington SJ. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Ahmad I, Romeo T, Römling U, Melefors O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol. 2010;12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke SH, Madison EL. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Ryu Y, Koo BM, Lee NY, Chun SJ, Park SJ, Lee KH, Seok YJ. A mammalian insulysin homolog is regulated by enzyme IIAGlc of the glucose transport system in Vibrio vulnificus. FEBS Lett. 2010;584:4537–4544. doi: 10.1016/j.febslet.2010.10.035. [DOI] [PubMed] [Google Scholar]
- Koo BM, Yoon MJ, Lee CR, Nam TW, Choe YJ, Jaffe H, Peterkofsky A, Seok YJ. A novel fermentation/respiration switch protein regulated by enzyme IIAGlc in Escherichia coli. J Biol Chem. 2004;279:31613–31621. doi: 10.1074/jbc.M405048200. [DOI] [PubMed] [Google Scholar]
- Krin E, Sismeiro O, Danchin A, Bertin PN. The regulation of Enzyme IIA(Glc) expression controls adenylate cyclase activity in Escherichia coli. Microbiology. 2002;148:1553–1559. doi: 10.1099/00221287-148-5-1553. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Boos W, Bouché JP, Plumbridge J. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 2000;19:5353–5361. doi: 10.1093/emboj/19.20.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol. 2009;16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H, Xu S, Wang HW, Gu L. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 2012;40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF, Oakford L, Yüksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Lux R, Jahreis K, Bettenbrock K, Parkinson JS, Lengeler JW. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy S, Zeng GQ, Danchin A. Cyclic AMP synthesis in Escherichia coli strains bearing known deletions in the pts phosphotransferase operon. Gene. 1990;86:27–33. doi: 10.1016/0378-1119(90)90110-d. [DOI] [PubMed] [Google Scholar]
- Lüttmann D, Heermann R, Zimmer B, Hillmann A, Rampp IS, Jung K, Görke B. Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIANtr in Escherichia coli. Mol Microbiol. 2009;72:978–994. doi: 10.1111/j.1365-2958.2009.06704.x. [DOI] [PubMed] [Google Scholar]
- Martínez LC, Martínez-Flores I, Salgado H, Fernández-Mora M, Medina-Rivera A, Puente JL, Collado-Vides J, Bustamante VH. In silico identification and experimental characterization of regulatory elements controlling the expression of the Salmonella csrB and csrC genes. J Bacteriol. 2014;196:325–336. doi: 10.1128/JB.00806-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazé A, Glatter T, Bumann D. The central metabolism regulator EIIAGlc switches Salmonella from growth arrest to acute virulence through activation of virulence factor secretion. Cell Rep. 2014;7:1426–1433. doi: 10.1016/j.celrep.2014.04.022. [DOI] [PubMed] [Google Scholar]
- Moyle H, Waldburger C, Susskind MM. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam TW, Cho SH, Shin D, Kim JH, Jeong JY, Lee JH, Roe JH, Peterkofsky A, Kang SO, Ryu S, Seok YJ. The Escherichia coli glucose transporter enzyme IICBGlc recruits the global repressor Mlc. EMBO J. 2001;20:491–498. doi: 10.1093/emboj/20.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Grosse K, Sourjik V. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:12159–12164. doi: 10.1073/pnas.1205307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, Lee BR, Seok YJ, Peterkofsky A. In vitro reconstitution of catabolite repression in Escherichia coli. J Biol Chem. 2006;281:6448–6454. doi: 10.1074/jbc.M512672200. [DOI] [PubMed] [Google Scholar]
- Patterson-Fortin LM, Vakulskas CA, Yakhnin H, Babitzke P, Romeo T. Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J Mol Biol. 2013;425:3662–3677. doi: 10.1016/j.jmb.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernestig AK, Georgellis D, Romeo T, Suzuki K, Tomenius H, Normark S, Melefors O. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol. 2003;185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BS, Smith DR, Watnick PI. Glucose-specific enzyme IIA has unique binding partners in the vibrio cholerae biofilm. MBio. 2012;3:e00228–00212. doi: 10.1128/mBio.00228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma PW, Epstein W, Schuitema AR, Nelson SO. Interaction between IIIGlc of the phosphoenolpyruvate:sugar phosphotransferase system and glycerol kinase of Salmonella typhimurium. J Bacteriol. 1984;158:351–353. doi: 10.1128/jb.158.1.351-353.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Kamireddi M. Modulation of Escherichia coli adenylyl cyclase activity by catalytic-site mutants of protein IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1998;180:732–736. doi: 10.1128/jb.180.3.732-736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T. Post-transcriptional regulation of bacterial carbohydrate metabolism: evidence that the gene product CsrA is a global mRNA decay factor. Res Microbiol. 1996;147:505–512. doi: 10.1016/0923-2508(96)84004-6. [DOI] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol. 2013;15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis NA, Yang H, Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem. 1995;270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- Saier MH. Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol Rev. 1989;53:109–120. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kimata K, Aiba H. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 2000;19:5344–5352. doi: 10.1093/emboj/19.20.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschowri N, Busse S, Hengge R. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 2009;23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban A, Neukirchen S, Jaeger KE. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 1997;25:2227–2228. doi: 10.1093/nar/25.11.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci U S A. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas CA, Pannuri A, Cortés-Selva D, Zere TR, Ahmer BM, Babitzke P, Romeo T. Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol Microbiol. 2014;92:945–958. doi: 10.1111/mmi.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. Regulation of Bacterial Virulence by Csr (Rsm) Systems. Microbiol Mol Biol Rev. 2015;79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Louis JM, Sondej M, Seok YJ, Peterkofsky A, Clore GM. Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIAglucose of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. EMBO J. 2000;19:5635–5649. doi: 10.1093/emboj/19.21.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu MY, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


