Abstract
Urinary proteomics has become one of the most attractive topics in disease biomarker discovery. Mass spectrometry (MS)-based proteomic analysis has advanced continuously and emerged as a prominent tool in the field of clinical bioanalysis. However, only few protein biomarkers have made their way to validation and clinical practice. Biomarker discovery is challenged by many clinical and analytical factors including, but not limited to, the complexity of urine and the wide dynamic range of endogenous proteins in the sample. This article highlights promising technologies and strategies in the MS-based biomarker discovery process, including study design, sample preparation, protein quantification, instrumental platforms, and bioinformatics. Different proteomics approaches are discussed, and progresses in maximizing urinary proteome coverage and standardization are emphasized in this review. MS-based urinary proteomics has great potential in the development of noninvasive diagnostic assays in the future, which will require collaborative efforts between analytical scientists, systems biologists, and clinicians.
Keywords: Biomarker, Mass spectrometry, Proteomics, Urine
1 Introduction
Urine is among the most valuable sample materials for disease biomarker discovery. Urine collection is simple, non-invasive, and available volume is relatively abundant compared to other biological fluids. It contains information from systemic circulation to local tissues via extracellular vesicles, proteins, and small molecules. Differential protein profiles and abundances are often observed for many diseases, due to an estimated 1,800,000 human proteoforms (including PTMs) from ~20,300 genes [1]. The proteome represents the entire profile of proteins expressed in a biological sample under a defined condition. Interest in the proteome is buttressed by the assertion that nucleic acid-level studies might often be too far removed from disease states and processes - where the transcriptome was shown to predict only about half of the variations observed in proteins [2]. Proteomics provides a reliable vantage point for cellular stress responses and other changes. The standardized definition of proteomic biomarker was proposed as “a specific peptide or protein that is associated with a specific condition, such as the onset, manifestation, or progression of a disease or a response to treatment” [3]. It is further suggested that biomarkers be defined solely by their association with a disease, barring further mechanistic evidence, and that extrapolation to similar, yet disparate diseases be avoided.
MS-based techniques allow a unique window into biological perturbations via global or targeted proteomics strategies; the scale, specificity, and discovery potential of which are unmatched by other currently available methods. MS-based methods have enjoyed technological gains largely due to the popularity and interest across many research genres, such as drug development, biomedical research, and toxicology. Common biomarker-related research foci include chemical proteomics or drug target identification, drug companion diagnostics, protein-protein interaction, drug metabolism, and markers of subacute exposure [4]. Characterization of the exposome, the biological record of a lifetime of subacute toxicant exposure, may be an integral part of future clinical bioanalysis [5]. Supporting the idea that a dynamic record of all exposures across a lifetime could prove useful is the concept that cancer and other diseases could potentially arise from exposure to combinations of individual benign agents [6]. While assessing biomarkers over time is promising, this monitoring goal is currently unattainable due to lack of information on disease etiology and associated costs. MS-based methods offer the greatest promise, now and into the future.
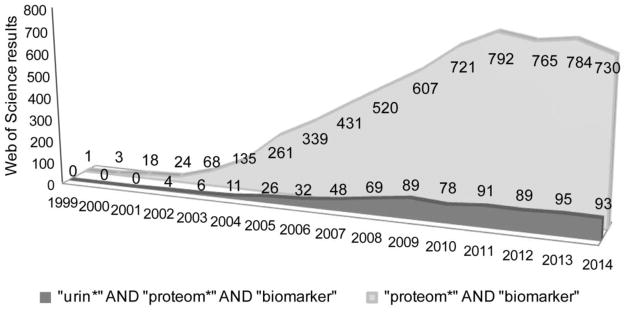
Despite the widespread application of protein biomarker discovery, to date few candidate biomarkers have been moved forward to validation, regulatory approval, and clinical use, and to our knowledge were discovered via immuno-based assays (Table 1) [7, 8]. Effective biomarker discovery has been challenged by many factors, including inherent biological complexity and variability (e.g. intra-individual variation [9] and disease marker promiscuity [10]), stability of analytical platforms, efficiency of data analysis, validation of biomarkers in the general population, and the expense and time needed to bring a test into the clinic. Biomarker and drug development projects operate on similar budget- and time- scales [11], however, suggesting the fruits of recent MS studies may still be ripening. The lack of immediate successes in this first wave of publications has led to some skepticism and might explain some of the decrease in proteomic biomarker publications of the last few years. As discussed by Mischak et al [12], publications found via “proteom*” AND “biomarker” on Web of Science are declining in frequency with a global maxima at 2011, while the inclusion of “urin*” results in a plateau rather than a decline (Fig. 1). This resistance to decline could be due to the unique promise of urine as a sample source for proteomic biomarker discovery. The ten most influential papers of this plateau period, related to urine protein biomarker discovery or validation, were found via search of Web of Science for “urin* proteo* biomarker”. Focusing on the top two primary studies of each year 2011–2015, a large proportion sought markers for kidney diseases [13–15] – some focusing on exosomes in kidney disease [16, 17]. Other diseases studied include bladder cancer [18], preeclampsia [19], type 2 diabetes [20], and prostate and other epithelial cancers where urine samples were used for biomarker studies [21]. A final category contains an influential method development study [9].
Table 1.
FDA-approved urine protein biomarkers (found via patent and literature searches)
| Protein | Clinical use | Disease |
|---|---|---|
| Nuclear matrix protein 22 (NMP-22, NuMA) [7, 8] | Diagnosis and monitoring | Bladder cancer |
| Fibrin/fibrinogen degradation products (FDP) [7] | Monitoring | Bladder cancer |
| Bladder tumor antigen (BTA) [7] | Monitoring | Bladder cancer |
| High molecular weight CEA and mucin [7] | Monitoring | Bladder cancer |
| Tissue inhibitor of metalloproteinase-2 (TIMP-2) [137] | Early detection | Acute kidney injury |
| Insulin-like growth factor binding protein 7 (IGFBP7) [137] | Early detection | Acute kidney injury |
Fig. 1.
Web of Science search results of the yearly trend in publishing frequency for general “proteom*” biomarker papers, and those including “urin*”
In this review we discuss the manifold expert suggestions and new technical developments in urine biomarker discovery. With the purpose of organizing the collective thought and streamlining the path to clinical success, major steps of the biomarker discovery process are highlighted including study design, sample collection and preparation, protein quantification, instrumental platforms, and data analysis.
2 Human urine composition and anatomical inputs
Urine solutes include salts, small molecules, soluble and insoluble proteins, extracellular vesicles, nucleic acids, and cells and cell debris. Urinary proteins include a soluble fraction (typically < 40 kDa) from plasma or secreted from urogenital epithelium or prostate (male), comprising about half of total urine proteins; a sediment protein fraction from sloughed epithelial cells of renal or urogenital origin comprising most of the other half; and an exosomal fraction from epithelial cells lining the urogenital tract or from immune cells or platelets in plasma, comprising the final few percent of total urine proteins [22]. Approximately 30% of the urinary proteome is expected to be of systemic origin, with the remainder from the urogenital tract [23]. A comparison of high quality proteomics data of disparate patient origins found about 26% of total urine protein identifications were also found in plasma after removing proteins known to be prostatic secretions, and only about 15% of total plasma proteins were also found in urine [24].
On average about 180 liters of plasma holding 13 ± 3 kilograms of protein is filtered by the kidneys daily [25]. The ratio of urine protein concentration to plasma protein concentration is also known as sieving coefficient. The sieving coefficient for albumin is roughly 6:10,000, meaning for every 10,000 units of albumin per milliliter of plasma, only 6 units end up in a milliliter of final urine [26]. From this sieving coefficient, if different plasma proteins vary in concentration from roughly 1010 to 100 pg/mL [27], and plasma proteins make up roughly 30% of total urine protein [23], concentrations of different urine proteins can be estimated from 107 pg/mL down to 10−3 pg/mL for low abundance proteins. Typically smaller proteins and albumin are nearly completely reabsorbed into plasma, leading to low daily total protein loss to urine of around 150 mg protein per day, 10 mg of which is albumin [22].
3 Study design of urinary protein biomarker discovery
3.1 Patient selection
Patient selection is critical and a starting point for successful biomarker discovery. Patient groups must be carefully chosen to avoid discovering artifacts of lurking variables (e.g., age, sex, similar diseases, etc.). Age is known to have a strong influence on the urinary proteome, generally attributed to kidney aging [28, 29]. Typical control groups are age- and sex-matched healthy volunteers. More strictly selected control groups are highly recommended, which recruit patients known to have a disease with a similar perturbation in biological function to the disease of interest. A good example of this is a recent study that sought biomarkers for prostate cancer and included patients with benign prostatic hyperplasia, patients with other uropathies, as well as a healthy control cohort [30]. Recruiting a related disease control group will bring a higher likelihood of discovering true unique markers with higher specificity to the disease of interest. The importance of this criterion is evidenced by a meta-analysis of 238 disease states, where more than 80% of the putative protein biomarkers (disease vs. control) are linked to multiple diseases [10]. The authors offer the idea of establishing broad-scale inter-disease network models to aid in the identification of distinct markers. In light of the frequency of candidate marker promiscuity, the elusive tissue-specific marker sounds ideal and the importance of tuning the often-suggested panel of markers to high selectivity resonates.
3.2 Statistical design
Key to successful biomarker discovery is a study design that avoids bias and is sufficiently-powered to find true differences above the noise of biological and technical variation. The importance of rigorous statistical design cannot be understated, yet detailed treatment of the subject is beyond the scope of this review. Topics explored in more appropriate depth elsewhere include: the determination of sample size while balancing the high cost of working with clinical samples [31–33], replication, randomization, blocking [34], and common areas for improvement and standardization [3]. As a quick heuristic, an experiment can be started by analyzing 12 controls and 12 cases, followed by calculation of the final sample size needed for the chosen level of test performance [35]. Guidelines for the standardized reporting of diagnostic accuracy studies (STARD [36]), tumor marker studies (REMARK [37]), and observational studies in biomedical research (STROBE [38]) can also provide an outline of critical aspects relevant to any biomarker study.
3.3 Orthogonal model systems to facilitate biomarker discovery
Working with an orthogonal model system could improve success in urine biomarker discovery. The genetic signature of a disease was shown to be robust across different tissues – more robust even than a single tissue’s signature is under different disease conditions [39]. This suggests similar markers could be found for a specific disease throughout the body. Orthogonal studies, using tissue or other biofluids samples, could potentially augment a urine biomarker study [40]. Other relevant orthogonal models, including cell culture (e.g., secretome analysis) and animal models (e.g., xenograft analysis), allow the manipulation of genetic and environmental conditions, reducing the biological variation and isolating the significant disease-related factors [41, 42].
4 Urine sample collection and preparation
4.1 Urine collection
The standardization of urine collection and storage protocols has been championed by the Human Urine Proteome Project (HKUPP) and the European Kidney and Urine Proteomics (EuroKUP) groups. The guidance is available online (http://www.hkupp.org) and is generally consistent with published reports. Briefly, mid-stream urine from the second morning void is collected into appropriate containers, centrifuged at 1000 g for 10 minutes, aliquoted, and stored at −80°C (avoiding unnecessary freeze-thaw cycles) [43–45]. Five percent sodium azide solution may be added as a biocide. Protease inhibitors are not recommended for untargeted urine proteomics since studies showed that they failed to increase protein identifications and may interfere with the subsequent digestion procedure [46, 47]. Protein can precipitate due to freezing, so care should be taken to avoid centrifugation after freezing and to fully resolubilize protein upon thawing via sonication, detergents, or addition of Tris buffer to pH~8 [44]. Urine samples need to be kept on ice during sample preparation.
4.2 Protein extraction
MS-based proteomics strategies are typically divided into three major groups: top-down (analysis of intact protein), middle-down (analysis of large peptides), and bottom-up (analysis of proteolytic peptides), each requiring different sample preparation methods. Bottom-up proteomics is the most widely-used approach in urine protein biomarker studies and methods relevant to this strategy are discussed here. Removal of small molecules in the urine is important as they can interfere with the bicinchoninic acid (BCA) assay, used to determine total protein concentration and normalize samples [48]. Small molecules can also bind and block ligand sites when using enrichment techniques like combinatorial peptide ligand library (CPLL) [49].
4.2.1 Protein precipitation
Protein precipitation techniques via different organic solvents are classic methods for protein extraction, such as methanol, ethanol, acetone, acetonitrile, and mixed solvents such as chloroform/methanol. Results of studies comparing solvent precipitation efficiency vary, likely due to different specific protocols and the inherent variation of samples from different disease states [46, 50]. The extraction efficiency of each solvent tends to favor certain protein classes and is affected by specific variables such as initial protein concentration and ionic strength in the urine [51]. Factors beyond extraction efficiency may need to be considered. For example, acetone precipitation was reported to cause a potential modification of peptides. About 5% of the time when glycine is the second amino acid in a peptide, a gas-phase modification due to acetone could make glycine take on the mass of proline, resulting in misidentification [52]. Overall, precipitation techniques should align with the experimental goals and be optimized within each lab, striking a balance between speed, cost, and performance.
4.2.2 Ultrafiltration
Ultrafiltration via molecular-weight cutoff (MWCO) spin columns with small pore sizes (e.g., 3 kDa and 10 kDa) is popular for straining and concentrating the protein fraction, removing metabolites, salts, detergents, and other small molecules, however there is concern over sample loss due to membrane adsorption [46]. Studies showed that optimizing elution buffers and adding salt or detergent modifiers can help minimize the sample loss in MWCO [53, 54]. Ultrafiltration devices are also useful for peptidomic studies; retaining larger proteins in a 20 kDa MWCO filter unit while allowing peptide analytes through, for example [13, 20].
4.2.3 MWCO filter-aided sample preparation
Filter-aided sample preparation (FASP) was developed to extract proteins from tissues and cell culture samples [55, 56]. Briefly, the protein fraction is concentrated in the membrane after MWCO. Protein reduction, alkylation, and digestion are performed on the membrane, and digested peptides are eluted and collected for subsequent analysis. Erde et al. [57] reported an enhanced FASP method incorporating alternative reagents to improve proteome coverage and sample recovery. Of great relevance here, a high-throughput quantitative urine proteomics study was recently reported, adapting FASP to a 96-well filter plate [58].
4.2.4 Dialysis and lyophilization
Dialysis followed by lyophilization can provide high recovery of protein samples. Unfortunately this technique is time and volume-consuming and can result in a sample mixture containing many small molecules and salts, whose interference in downstream analyses may preclude its utility [50].
Further work is needed to conclusively show which technique best balances efficiency and workflow simplicity in urine sample analyses. From our perspective, an ideal sample preparation method efficiently extracts protein from a sample, minimizes sample handling, and lends itself to automation - making the optimized FASP variations, especially in 96-well format, promising candidates for future urinary proteomics studies.
4.3 Maximizing urinary proteome coverage
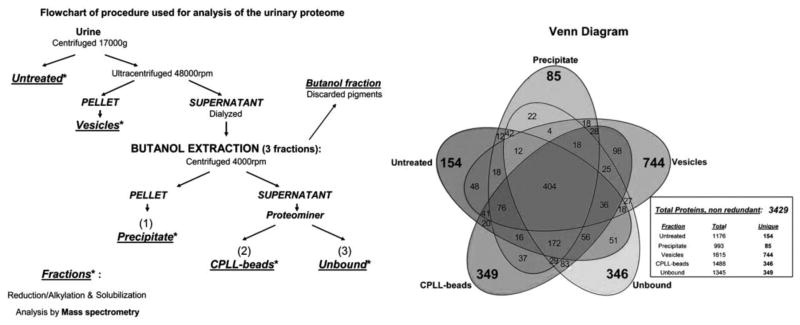
Deepening coverage of the urine proteome should help improve success in biomarker discovery. There is growing interest in exploring proteomes of ever-decreasing sample amounts with ever-increasing depth [46]. To this end a new subfield is emerging within proteomics working on the microgram-scale dubbed microproteomics [59]. Urine proteomics and microproteomics share a similar challenge – lack of robustness against sample loss, since the typically more interesting or important proteins are found at concentrations near or below the method detection limit. Beyond efficient extraction, much effort has gone into sample enhancement strategies to overcome this analytical barrier. Using some of these, Santucci et al. [49] recently reported a deep profiling of the urinome with the identification of nearly 3500 proteins in human urine (Fig. 2).
Fig. 2.
Flowchart of the multi-fractionation procedures for urinary proteomic analysis (left) and Venn diagram of the detected 3429 proteins (right), adapted and modified from Santucci et al. [49] with permission.
4.3.1 Affinity fractionation to reduce dynamic range in protein concentration
The dynamic range of protein concentrations in urine is around 10 orders of magnitude, similar to plasma [60]. The goal of affinity fractionation strategies is to reduce the dynamic range and increase protein IDs by bringing the low abundance proteins out from the shadow of high abundance proteins. These methods usually rely on adsorption to a functionalized solid phase of beads or column [61], such as immunodepletion, CPLL beads (ProteoMiner™), and functionalized magnetic beads (ClinProt™) [46, 49, 62]. Immunodepletion techniques do not seem to provide an increase in identifications when applied to urine [46, 63]. While ion exchange and immunodepletion strategies effectively removed high abundance targets like albumin, the lack of increase in identification number is attributed to remaining, untargeted high- or medium-abundance proteins, or to low-abundance proteins falling below the method’s limit of detection [63]. The ClinProt™ technique improves protein identification numbers via the same principle as off-line liquid chromatography: increasing sample fractions. CPLL beads have successfully increased protein coverage in urine by binding proteins to a library of millions of random hexapeptide ligands [49, 64, 65]. When a complex protein mixture is applied to this library, abundant proteins quickly saturate the sites of complementary ligands, with the excess flowing past. Low abundance proteins accumulate on their respective ligand sites and all are eluted with a decreased dynamic range. CPLL enhances detection of low-abundance proteins with fewer additional fractions than many other techniques for more efficient down-stream analysis, likely brings low abundance proteins up into the detectable range, and appears to be a highly desirable enrichment strategy.
4.3.2 Targeting specific solute compartments
Analyzing typically neglected sample fractions could lead to increased success in discovery experiments. Extracellular vesicles (40–5000 nm diameter), known as mediators of intercellular signaling and transportation, have emerged as potential sources of biomarkers, especially for urogenital cancers [41, 66]. Extracellular vesicles are categorized by size and origin as exosomes, microvesicles, and apoptotic bodies. Exosomes within urine could hold important proteins otherwise lost in traditional urine proteomics workflows [67, 68]. Interest in this solute compartment is strong, with the current highest-cited “urine proteom* biomarker” publication on Web of Science being a urine exosome study [69]. Separation techniques rely on size and density variations: ultrafiltration, density gradient centrifugation, and the gold standard differential ultracentrifugation [41]. Once separated, these can be extracted for protein similar to cell lysis [70].
5 Protein quantification for biomarker discovery
MS-based protein quantification is a key component in biomarker discovery, as biomarkers are usually selected via variations in protein or peptide concentrations between sample groups. Quantification techniques change as a marker progresses along the biomarker pipeline from discovery-phase relative quantification to validation-phase absolute quantification.
5.1 Relative quantification for the selection of candidate biomarkers
5.1.1 Label-free quantification
The identification of candidate protein biomarkers can be achieved by relative quantification through label-free or label-based approaches. Label-free quantification compares proteins or peptides between different LC-MS runs, usually through spectral counting and peak area or intensity measurements [71–74]. Since no labeling reagent is involved, the sample preparation is often simpler and relatively inexpensive. It offers greater dynamic range and proteome coverage compared to label-based methods [75]. Major hurdles of label-free approaches are the high dependence on instrument reproducibility between sample runs and low throughput, which requires more instrument time than label-based approaches.
5.1.2 Label-based quantification
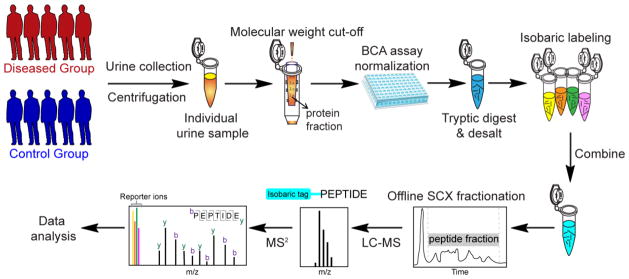
Stable isotope labeling enables accurate and simultaneous comparison of multiple samples for protein and peptide quantification. It can be categorized into mass difference labeling and isobaric labeling. Mass-difference labeling, such as formaldehyde dimethylation [76–78] introduces a fixed mass difference onto proteins or peptides by using heavy stable isotopes. It provides more accurate quantification than label-free methods, but is criticized for increasing mass spectral complexity, negatively influencing protein coverage and quantification. Isobaric labeling by iTRAQ [79, 80], TMT [81–84], and DiLeu [85–89] tags has multiplexing capability and has been applied to many quantitative proteomic studies. A typical workflow using isobaric labeling strategy is illustrated in Fig. 3. Isobaric tags do not increase the spectral complexity because of their balanced structure, including an isotopically-coded reporter, a balance group, and an amine reactive group. After labeling, samples are combined at equal ratios and each labeled sample produces a unique reporter ion upon MS2 fragmentation for quantification. Recent efforts have been made to increase the multiplexing capacity of labeling reagents, including 8-plex iTRAQ [90], 6-plex [91], 8-plex [92], and up to 54-plexTMT [93]. Robinson and coworkers [94] have developed combined precursor isotopic labeling and isobaric tagging (cPILOT), extending multiplexing capabilities to 12 and 16 samples. High-resolution FT-ICR and Orbitrap instruments have allowed the development of new tags, exploiting the subtle mass defects that arise due to variations in nuclear binding energy between different stable isotopes [95–97].
Fig. 3.
A representative workflow of quantitative urine proteomics using isobaric labeling. Urine samples are collected from human patients (control and disease groups) and centrifuged to remove particulates and cell debris. Protein fraction from each urine sample is obtained with molecular weight cut-off filter. Total protein concentration is measured with bicinchoninic acid (BCA) assay for normalization. After protein digestion and desalting, each sample is labeled by different channels of isobaric tags and combined after labeling reaction. Offline SCX LC is often performed to fractionate samples before LC-MS/MS analysis. The intensities of reporter ions are used for relative quantification.
DiLeu reagents, developed by our group, have shown comparable performance with commercial tags, with greatly reduced cost and improved reporter ions intensities. The development and performance of 4-plex [85], 8-plex [86] and 12 plex [87] DiLeu tags have been successfully demonstrated in quantitative proteomics and peptidomics analysis. Most recently, 4-plex DiLeu labels have been applied to large-scale human urine proteomics, enabling accurate relative quantification of urinary proteins in men with lower urinary tract symptoms [48].
5.2 Absolute quantification for biomarker verification
Untargeted proteomics analysis can generate a panel of candidate disease biomarkers, but further verification through targeted absolute quantification is necessary before clinical validations and applications. Verifying the list of candidate biomarkers is expensive and challenging, and is a major reason for the dearth of clinically-implemented biomarkers [98, 99]. Antibody-based immunoassays, particularly the ELISA, are currently the most common techniques for biomarker validation [100]. ELISA is highly sensitive and specific but is also limited by the availability of antibodies and the expense of assay development for new target analytes [101, 102].
MS-based absolute quantification has become a successful alternative to immunoassays, offering reduced costs, shortened lead-time, and greatly improved throughput. Targeted detection and quantification of candidate biomarkers is generally achieved by selected reaction monitoring (SRM), also referred to as multiple reaction monitoring (MRM). The most common method for absolute quantification uses spiked stable isotope-labeled peptide standards to construct calibration curve [21, 103–105]. This method is much faster than ELISA and enables the quantification of multiple peptides in one run. But typical commercial isotopically labeled peptide standards are relatively expensive and custom synthesis can be even more costly for this type of experiment [88, 106]. Another MS-based absolute quantification method is through multiplexed stable isotope labeling. The non-isobaric mTRAQ tags allow specific quantification of each version of the labeled peptide by unique MRM transitions, and have been applied to verify candidate biomarkers in various applications [107–109]. Recently, Greer et al. [88] developed novel isotopic N,N-dimethyl leucine (iDiLeu) reagents for absolute quantification of peptides and proteins. With the five-plexed iDiLeu tags, a calibration curve and peptide surrogate can be analyzed in one LC-MS run, demonstrating increased throughput and higher quantification accuracy compared with the traditional method using isotope-encoded peptide standards.
6 MS-based instrumental platforms
Due to the complexity of the urinary proteome, hyphenated MS platforms such as LC-MS [18, 110], CE-MS [111–113], and 2DE-MS [23, 114] have been extensively employed in protein biomarker discovery. As compared in more details in several reviews [23, 115], each platform has its own advantages and disadvantages. 2DE-MS can detect large molecules, but is time consuming and difficult to automate [115]. LC-MS provides sensitive and reproducible analysis of protein digests [46]. CE-MS requires low sample volume and allows fast and high resolution analyte separation. A comparative study of LC-MS/MS and CE-MS/MS analysis demonstrated complementary coverage of peptides in human urine [116]. Emerging techniques such as ion mobility separations and microfluidic devices also possess great potential [117, 118]. Newly developed high-accuracy, high-resolution instruments (e.g., Q-Exactive Orbitrap™ and Orbitrap Fusion™) allow greater sequence coverage and more accurate quantification of proteins.
New data acquisition methods also show promise in proteomics. Data-independent acquisition (DIA) approach was demonstrated to provide complementary, if not better, proteome coverage and dynamic range than the traditional data-dependent acquisition (DDA) [119]. This relatively new data acquisition mode requires the construction of reference spectral libraries and special data processing tools [120]. One application of DIA, known as sequential window acquisition of all theoretical spectra (SWATH), was shown to remain linear over four orders of magnitude, with an impressive limit of detection in the amol range [121].
7 Data analysis and bioinformatics
Proteomics studies typically employ a long series of statistical filters from the determination of peptide false detection rates to inferential tests to tests for enrichment of biological functions and other processes. Care must be taken to properly utilize sophisticated statistics, which ultimately determine the selection or disposal of candidate markers [122]. Given the complexity of the topic, in-depth coverage of bioinformatics is not possible here. Rigorous data analysis [35] and the promising application of machine learning to biomarker research is covered elsewhere [123]. Gene ontology (GO) tools are useful for interpreting the potential biological function and molecular process of a given protein, allowing a large protein dataset to be sorted into biological processes, cellular locations, and biochemical functions [124]. A number of online tools offer these enrichment analyses, including DAVID, PSEA-Quant analysis, and protein annotation through evolutionary relationship (PANTHER) [125, 126]. Data mining is another promising informatics approach that can expedite the validation phase, efficiently leading to new biomarkers [127]. Efforts to improve and standardize metadata included with primary data have been undertaken to aid in successful data mining [128].
8 Conclusions and outlook
Future success in biomarker discovery efforts will rely heavily on collaboration among clinicians, analytical chemists and bioinformaticians, and will incorporate the state-of-the-art tools and techniques highlighted here. Automation and standardization in analytical methodology and quality control will generate more reproducible results, allowing distant labs to pool data or work cooperatively on larger efforts, particularly on studies with very large sample sizes. Toward this end, Mischak et al. generated human urine standards as a potential mechanism for controlling inter- and intra-lab instrument variations [129].
As discussed earlier, the development of biomarkers and drugs occurs on a similar time scale. Ongoing clinical trials involving protein biomarker discovery or validation were found via clinicaltrials.gov (Table 2). Of these, four are centrally-focused on urine for protein biomarker discovery in urologic diseases and ten include urine along with orthogonal bodily fluids or tissue samples for biomarkers of diseases spanning neurology, cancer, and cardiology, among others. While urine-focused trials are still mostly undertaken for urologic diseases, there is interest in investigating urine as an orthogonal non-invasive sample material for studying urinary proteins of systemic origin.
Table 2.
Active protein biomarker discovery and validation trials involving urine analysis; via clinicaltrials.gov.
| Conditions | Enrollment | Identifier | Urine focus |
|---|---|---|---|
| Female Stress Urinary Incontinence | 40 | NCT02023502 | + |
| IgA Nephropathy | 200 | NCT01879514 | + |
| Diabetic Nephropathy, Retinopathy | 3500 | NCT02040441 | + |
| Acute Kidney Injury in Sepsis Patients | 150 | NCT01981993 | + |
| Ischemic Brain Injury, Stroke | 750 | NCT00983723 | − |
| Chronic Kidney Disease (CKDu) | 350 | NCT02226055 | − |
| Colorect., Esophag., Gastric, Panc. Cancer | 1000 | NCT00899626 | − |
| Lung Cancer | 2000 | NCT00898313 | − |
| Heart Transplantation | 482 | NCT00042614 | − |
| Duchenne Muscular Dystrophy (DMD) | 220 | NCT01380964 | − |
| Lung Transplant Rejection | 60 | NCT00558597 | − |
| Kidney Transplant Infection | 1000 | NCT01515605 | − |
| Urinary Incontinence | 10 | NCT01987336 | − |
| Hyperoxia | 40 | NCT02553668 | − |
Note: Many studies include urine orthogonally with serum, CSF, or tissue samples for biomarker discovery and are shown as (−) under “urine focus” where urine analysis provides complementary information. Those primarily analyzing urine for biomarkers are shown as (+) under “urine focus”.
Regulatory qualification is critical and will necessarily slow progress to clinical implementation. Efforts to standardize reporting [130, 131] and quality control in proteomics studies will streamline the qualification process [132]. An additional challenge will be sorting through the myriad unpursued discovery-phase reports to find promising classifiers for further validation, requiring collaboration with bioinformatics specialists.
Perhaps at some point in the future, urine proteomics will allow the identification of gradual onset of any disease, allowing the therapy of this future time to steer processes back to a normal state [133]. The concept has made the news thanks to one public effort to characterize the healthy human state: Google’s Baseline Study [134]. As discussed in the current paper, at least three deep interrogations of the healthy human “urinome” have been reported, providing deep proteome coverage [28, 49, 129]. Perhaps these efforts and more can be compiled to better characterize a healthy reference urinome.
MS analyses have brought proteomics research into the modern era and will continue to be the go-to technique for accurate and sensitive bioanalysis. Some within biomedical research assert that journals will soon require MS data and turn away results derived from immunoassays [135]. Continuing in this direction, some are proposing a shift away from the end goal of expensive immunoassays like ELISA as the ultimate assay for clinical application [136]. MS methodology is quite promising - it is the future of biomedical research and will be the premier source of important discoveries in modern biomedical research.
Acknowledgments
The authors would like to thank National Institutes of Health funding through Grants P20 DK097826; R01 DK093690; and U54 DK104310. S.T. acknowledges an NIH-supported Molecular and Environmental Toxicology Training Program Predoctoral Fellowship (grant number T32-ES007015). LL acknowledges an H. I. Romnes Faculty Research Fellowship and a Vilas Distinguished Achievement Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Abbreviations
- MWCO
Molecular weight cut-off
- FASP
Filter-aided sample preparation
- CPLL
Combinatorial peptide ligand library
- GO
Gene ontology
- AQUA
Absolute quantification
Footnotes
The authors have declared no conflict of interest.
References
- 1.Legrain P, Aebersold R, Archakov A, Bairoch A, et al. The human proteome project: current state and future direction. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009993. M111 009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mischak H, Allmaier G, Apweiler R, Attwood T, et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 4.Titz B, Elamin A, Martin F, Schneider T, et al. Proteomics for systems toxicology. Comput Struct Biotechnol J. 2014;11:73–90. doi: 10.1016/j.csbj.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rappaport SM. Biomarkers intersect with the exposome. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals. 2012;17:483–489. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodson WH, Lowe L, Carpenter DO, Gilbertson M, et al. Assessing the carcinogenic potential of low-dose exposures to chemical mixtures in the environment: the challenge ahead. Carcinogenesis. 2015;36:S254–S296. doi: 10.1093/carcin/bgv039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlou MP, Diamandis EP, Blasutig IM. The Long Journey of Cancer Biomarkers from the Bench to the Clinic. Clinical Chemistry. 2013;59:147–157. doi: 10.1373/clinchem.2012.184614. [DOI] [PubMed] [Google Scholar]
- 8.Fuzery AK, Levin J, Chan MM, Chan DW. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clinical proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 10.Dudley JT, Butte AJ. Identification of Discriminating Biomarkers for Human Disease Using Integrative Network Biology. Pacific Symposium on Biocomputing; 2009; 2009. pp. 27–38. [PMC free article] [PubMed] [Google Scholar]
- 11.Drabovich AP, Martinez-Morillo E, Diamandis EP. Toward an integrated pipeline for protein biomarker development. Biochim Biophys Acta. 2015;1854:677–686. doi: 10.1016/j.bbapap.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Mischak H, Vlahou A, Righetti PG, Calvete JJ. Putting value in biomarker research and reporting. Journal of Proteomics. 2014;96:A1–A3. doi: 10.1016/j.jprot.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Argiles A, Siwy J, Duranton F, Gayrard N, et al. CKD273, a New Proteomics Classifier Assessing CKD and Its Prognosis. Plos One. 2013;8 doi: 10.1371/journal.pone.0062837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zurbig P, Jerums G, Hovind P, MacIsaac RJ, et al. Urinary Proteomics for Early Diagnosis in Diabetic Nephropathy. Diabetes. 2012;61:3304–3313. doi: 10.2337/db12-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schanstra JP, Zurbig P, Alkhalaf A, Argiles A, et al. Diagnosis and Prediction of CKD Progression by Assessment of Urinary Peptides. Journal of the American Society of Nephrology. 2015;26:1999–2010. doi: 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zubiri I, Posada-Ayala M, Sanz-Maroto A, Calvo E, et al. Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. Journal of Proteomics. 2014;96:92–102. doi: 10.1016/j.jprot.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 17.Bourderioux M, Thao NK, Chhuon C, Jeanson L, et al. A New Workflow for Proteomic Analysis of Urinary Exosomes and Assessment in Cystinuria Patients. Journal of Proteome Research. 2015;14:567–577. doi: 10.1021/pr501003q. [DOI] [PubMed] [Google Scholar]
- 18.Chen YT, Chen HW, Domanski D, Smith DS, et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. Journal of Proteomics. 2012;75:3529–3545. doi: 10.1016/j.jprot.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Carty DM, Siwy J, Brennand JE, Zurbig P, et al. Urinary Proteomics for Prediction of Preeclampsia. Hypertension. 2011;57:561–U387. doi: 10.1161/HYPERTENSIONAHA.110.164285. [DOI] [PubMed] [Google Scholar]
- 20.Roscioni SS, de Zeeuw D, Hellemons ME, Mischak H, et al. A urinary peptide biomarker set predicts worsening of albuminuria in type 2 diabetes mellitus. Diabetologia. 2013;56:259–267. doi: 10.1007/s00125-012-2755-2. [DOI] [PubMed] [Google Scholar]
- 21.Shi TJ, Gao YQ, Quek SI, Fillmore TL, et al. A Highly Sensitive Targeted Mass Spectrometric Assay for Quantification of AGR2 Protein in Human Urine and Serum. Journal of Proteome Research. 2014;13:875–882. doi: 10.1021/pr400912c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barratt J, Topham P. Urine proteomics: the present and future of measuring urinary protein components in disease. CMAJ. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decramer S, de Peredo AG, Breuil B, Mischak H, et al. Urine in Clinical Proteomics. Molecular & Cellular Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Jia L, Zhang L, Shao C, Song E, et al. An attempt to understand kidney’s protein handling function by comparing plasma and urine proteomes. PLoS One. 2009;4:e5146. doi: 10.1371/journal.pone.0005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saritas T, Kuppe C, Moeller MJ. Progress and controversies in unraveling the glomerular filtration mechanism. Curr Opin Nephrol Hypertens. 2015;24:208–216. doi: 10.1097/MNH.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 26.Tojo A, Kinugasa S. Mechanisms of glomerular albumin filtration and tubular reabsorption. Int J Nephrol. 2012;2012:481520. doi: 10.1155/2012/481520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker ES, Liu T, Petyuk VA, Burnum-Johnson KE, et al. Mass spectrometry for translational proteomics: progress and clinical implications. Genome Med. 2012;4:63. doi: 10.1186/gm364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zurbig P, Decramer S, Dakna M, Jantos J, et al. The human urinary proteome reveals high similarity between kidney aging and chronic kidney disease. Proteomics. 2009;9:2108–2117. doi: 10.1002/pmic.200800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macoska JA, Kasina S, Campeau L, Andersson K-E. Geriatric Urology. Springer; 2014. pp. 13–35. [Google Scholar]
- 30.Adeola HA, Soares NC, Paccez JD, Kaestner L, et al. Discovery of novel candidate urinary protein biomarkers for prostate cancer in a multiethnic cohort of South African patients via label-free mass spectrometry. Proteomics Clin Appl. 2015;9:597–609. doi: 10.1002/prca.201400197. [DOI] [PubMed] [Google Scholar]
- 31.Maes E, Cho WC, Baggerman G. Translating clinical proteomics: the importance of study design. Expert Rev Proteomics. 2015;12:217–219. doi: 10.1586/14789450.2015.1041512. [DOI] [PubMed] [Google Scholar]
- 32.Cairns DA, Barrett JH, Billingham LJ, Stanley AJ, et al. Sample size determination in clinical proteomic profiling experiments using mass spectrometry for class comparison. Proteomics. 2009;9:74–86. doi: 10.1002/pmic.200800417. [DOI] [PubMed] [Google Scholar]
- 33.Levin Y. The role of statistical power analysis in quantitative proteomics. Proteomics. 2011;11:2565–2567. doi: 10.1002/pmic.201100033. [DOI] [PubMed] [Google Scholar]
- 34.Oberg AL, Vitek O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J Proteome Res. 2009;8:2144–2156. doi: 10.1021/pr8010099. [DOI] [PubMed] [Google Scholar]
- 35.Dakna M, Harris K, Kalousis A, Carpentier S, et al. Addressing the challenge of defining valid proteomic biomarkers and classifiers. BMC Bioinformatics. 2010;11:594. doi: 10.1186/1471-2105-11-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bossuyt PM. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Bmj. 2003;326:41–44. doi: 10.1136/bmj.326.7379.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McShane LM, Altman DG, Sauerbrei W, Taube SE, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 38.von Elm E, Altman DG, Egger M, Pocock SJ, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. The Lancet. 2007;370:1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 39.Dudley JT, Tibshirani R, Deshpande T, Butte AJ. Disease signatures are robust across tissues and experiments. Mol Syst Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee G, Singanamalli A, Wang H, Feldman MD, et al. Supervised multi-view canonical correlation analysis (sMVCCA): integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging. 2015;34:284–297. doi: 10.1109/TMI.2014.2355175. [DOI] [PubMed] [Google Scholar]
- 41.Nawaz M, Camussi G, Valadi H, Nazarenko I, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11:688–701. doi: 10.1038/nrurol.2014.301. [DOI] [PubMed] [Google Scholar]
- 42.Patel S, Ngounou Wetie AG, Darie CC, Clarkson BD. Cancer secretomes and their place in supplementing other hallmarks of cancer. Advances in experimental medicine and biology. 2014;806:409–442. doi: 10.1007/978-3-319-06068-2_20. [DOI] [PubMed] [Google Scholar]
- 43.Thomas CE, Sexton W, Benson K, Sutphen R, Koomen J. Urine collection and processing for protein biomarker discovery and quantification. Cancer Epidemiol Biomarkers Prev. 2010;19:953–959. doi: 10.1158/1055-9965.EPI-10-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson DH, Banks RE. Banking of clinical samples for proteomic biomarker studies: a consideration of logistical issues with a focus on pre-analytical variation. Proteomics Clin Appl. 2010;4:250–270. doi: 10.1002/prca.200900220. [DOI] [PubMed] [Google Scholar]
- 45.Lee RS, Monigatti F, Briscoe AC, Waldon Z, et al. Optimizing sample handling for urinary proteomics. Journal of proteome research. 2008;7:4022–4030. doi: 10.1021/pr800301h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afkarian M, Bhasin M, Dillon ST, Guerrero MC, et al. Optimizing a proteomics platform for urine biomarker discovery. Mol Cell Proteomics. 2010;9:2195–2204. doi: 10.1074/mcp.M110.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hepburn S, Cairns DA, Jackson D, Craven RA, et al. An analysis of the impact of pre-analytical factors on the urine proteome: Sample processing time, temperature, and proteolysis. Proteomics Clin Appl. 2015;9:507–521. doi: 10.1002/prca.201400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greer T, Hao L, Nechyporenko A, Lee S, et al. Custom 4-Plex DiLeu Isobaric Labels Enable Relative Quantification of Urinary Proteins in Men with Lower Urinary Tract Symptoms (LUTS) Plos One. 2015;10 doi: 10.1371/journal.pone.0135415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santucci L, Candiano G, Petretto A, Bruschi M, et al. From hundreds to thousands: Widening the normal human Urinome (1) Journal of Proteomics. 2015;112:53–62. doi: 10.1016/j.jprot.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 50.Olszowy P, Buszewski B. Urine sample preparation for proteomic analysis. Journal of Separation Science. 2014;37:2920–2928. doi: 10.1002/jssc.201400331. [DOI] [PubMed] [Google Scholar]
- 51.Feist P, Hummon AB. Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int J Mol Sci. 2015;16:3537–3563. doi: 10.3390/ijms16023537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpson DM, Beynon RJ. Acetone precipitation of proteins and the modification of peptides. J Proteome Res. 2010;9:444–450. doi: 10.1021/pr900806x. [DOI] [PubMed] [Google Scholar]
- 53.Cunningham R, Wang JX, Wellner D, Li LJ. Investigation and reduction of sub-microgram peptide loss using molecular weight cut-off fractionation prior to mass spectrometric analysis. J Mass Spectrom. 2012;47:1327–1332. doi: 10.1002/jms.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greening DW, Simpson RJ. A centrifugal ultrafiltration strategy for isolating the low-molecular weight (<or=25K) component of human plasma proteome. J Proteomics. 2010;73:637–648. doi: 10.1016/j.jprot.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 56.Wisniewski JR, Ostasiewicz P, Mann M. High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J Proteome Res. 2011;10:3040–3049. doi: 10.1021/pr200019m. [DOI] [PubMed] [Google Scholar]
- 57.Erde J, Loo RR, Loo JA. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res. 2014;13:1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Y, Suh MJ, Sikorski P, Kwon K, et al. Urine sample preparation in 96-well filter plates for quantitative clinical proteomics. Anal Chem. 2014;86:5470–5477. doi: 10.1021/ac5008317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutstein HB, MorriS JS, Annangudi SP, Sweedler JV. Microproteomics: Analysis of protein diversity in small samples. Mass Spectrometry Reviews. 2008;27:316–330. doi: 10.1002/mas.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson NL. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Molecular & Cellular Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 61.Millioni R, Tolin S, Puricelli L, Sbrignadello S, et al. High abundance proteins depletion vs low abundance proteins enrichment: comparison of methods to reduce the plasma proteome complexity. PLoS One. 2011;6:e19603. doi: 10.1371/journal.pone.0019603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiedler GM, Baumann S, Leichtle A, Oltmann A, et al. Standardized peptidome profiling of human urine by magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem. 2007;53:421–428. doi: 10.1373/clinchem.2006.077834. [DOI] [PubMed] [Google Scholar]
- 63.Filip S, Vougas K, Zoidakis J, Latosinska A, et al. Comparison of Depletion Strategies for the Enrichment of Low-Abundance Proteins in Urine. PLoS One. 2015;10:e0133773. doi: 10.1371/journal.pone.0133773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castagna A, Cecconi D, Sennels L, Rappsilber J, et al. Exploring the hidden human urinary proteome via ligand library beads. J Proteome Res. 2005;4:1917–1930. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 65.Candiano G, Dimuccio V, Bruschi M, Santucci L, et al. Combinatorial peptide ligand libraries for urine proteome analysis: investigation of different elution systems. Electrophoresis. 2009;30:2405–2411. doi: 10.1002/elps.200800762. [DOI] [PubMed] [Google Scholar]
- 66.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13:1572–1580. doi: 10.1002/pmic.201200285. [DOI] [PubMed] [Google Scholar]
- 68.Alvarez ML, Khosroheidari M, Ravi RK, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 69.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pitto M, Corbetta S, Raimondo F. Preparation of urinary exosomes: methodological issues for clinical proteomics. Methods in molecular biology (Clifton, NJ) 2015;1243:43–53. doi: 10.1007/978-1-4939-1872-0_3. [DOI] [PubMed] [Google Scholar]
- 71.Megger DA, Bracht T, Meyer HE, Sitek B. Label-free quantification in clinical proteomics. Biochim Biophys Acta. 2013;1834:1581–1590. doi: 10.1016/j.bbapap.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 72.Buchberger A, Yu Q, Li L. Advances in Mass Spectrometric Tools for Probing Neuropeptides. Annual review of analytical chemistry. 2015;8:485–509. doi: 10.1146/annurev-anchem-071114-040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adeola HA, Soares NC, Paccez JD, Kaestner L, et al. Discovery of novel candidate urinary protein biomarkers for prostate cancer in a multiethnic cohort of South African patients via label-free mass spectrometry. Proteomics Clinical Applications. 2015;9:597–609. doi: 10.1002/prca.201400197. [DOI] [PubMed] [Google Scholar]
- 74.Duan X, Young R, Straubinger RM, Page B, et al. A straightforward and highly efficient precipitation/on-pellet digestion procedure coupled with a long gradient nano-LC separation and Orbitrap mass spectrometry for label-free expression profiling of the swine heart mitochondrial proteome. Journal of proteome research. 2009;8:2838–2850. doi: 10.1021/pr900001t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sandin M, Chawade A, Levander F. Is label-free LC-MS/MS ready for biomarker discovery? Proteomics Clin Appl. 2015;9:289–294. doi: 10.1002/prca.201400202. [DOI] [PubMed] [Google Scholar]
- 76.Han DK, Eng J, Zhou HL, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nature Biotechnology. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji CJ, Guo N, Li L. Differential dimethyl labeling of N-termini of peptides after guanidination for proteome analysis. Journal Of Proteome Research. 2005;4:2099–2108. doi: 10.1021/pr050215d. [DOI] [PubMed] [Google Scholar]
- 78.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nature Protocols. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 79.Ross PL, Huang YLN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 80.Jing LH, Parker CE, Seo D, Hines MW, et al. Discovery of biomarker candidates for coronary artery disease from an APOE-knock out mouse model using iTRAQ-based multiplex quantitative proteomics. Proteomics. 2011;11:2763–2776. doi: 10.1002/pmic.201000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson A, Schafer J, Kuhn K, Kienle S, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 82.Sinclair J, Timms JF. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS. Methods. 2011;54:361–369. doi: 10.1016/j.ymeth.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 83.Hung CW, Tholey A. Tandem Mass Tag Protein Labeling for Top-Down Identification and Quantification. Analytical Chemistry. 2012;84:161–170. doi: 10.1021/ac202243r. [DOI] [PubMed] [Google Scholar]
- 84.Murphy JP, Everley RA, Coloff JL, Gygi SP. Combining Amine Metabolomics and Quantitative Proteomics of Cancer Cells Using Derivatization with Isobaric Tags. Analytical Chemistry. 2014;86:3585–3593. doi: 10.1021/ac500153a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiang F, Ye H, Chen RB, Fu Q, Li LJ. N,N-Dimethyl Leucines as Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics. Analytical Chemistry. 2010;82:2817–2825. doi: 10.1021/ac902778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frost DC, Greer T, Xiang F, Liang Z, Li L. Development and characterization of novel 8-plex DiLeu isobaric labels for quantitative proteomics and peptidomics. Rapid Commun Mass Spectrom. 2015;29:1115–1124. doi: 10.1002/rcm.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frost DC, Greer T, Li LJ. High-Resolution Enabled 12-Plex DiLeu Isobaric Tags for Quantitative Proteomics. Analytical Chemistry. 2015;87:1646–1654. doi: 10.1021/ac503276z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greer T, Lietz CB, Xiang F, Li LJ. Novel isotopic N,N-Dimethyl Leucine (iDiLeu) Reagents Enable Absolute Quantification of Peptides and Proteins Using a Standard Curve Approach. Journal of the American Society for Mass Spectrometry. 2015;26:107–119. doi: 10.1007/s13361-014-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hao L, Zhong XF, Greer T, Ye H, Li LJ. Relative quantification of amine-containing metabolites using isobaric N,N-dimethyl leucine (DiLeu) reagents via LC-ESI-MS/MS and CE-ESI-MS/MS. Analyst. 2015;140:467–475. doi: 10.1039/c4an01582g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choe L, D’Ascenzo M, Relkin NR, Pappin D, et al. 8-Plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rauniyar N, Gao BB, McClatchy DB, Yates JR. Comparison of Protein Expression Ratios Observed by Sixplex and Duplex TMT Labeling Method. Journal Of Proteome Research. 2013;12:1031–1039. doi: 10.1021/pr3008896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Werner T, Becher I, Sweetman G, Doce C, et al. High-Resolution Enabled TMT 8-plexing. Analytical Chemistry. 2012;84:7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- 93.Everley RA, Kunz RC, McAllister FE, Gygi SP. Increasing Throughput in Targeted Proteomics Assays: 54-Plex Quantitation in a Single Mass Spectrometry Run. Analytical Chemistry. 2013;85:5340–5346. doi: 10.1021/ac400845e. [DOI] [PubMed] [Google Scholar]
- 94.Robinson RAS, Evans AR. Enhanced Sample Multiplexing for Nitrotyrosine-Modified Proteins Using Combined Precursor Isotopic Labeling and Isobaric Tagging. Analytical Chemistry. 2012;84:4677–4686. doi: 10.1021/ac202000v. [DOI] [PubMed] [Google Scholar]
- 95.McAlister GC, Huttlin EL, Haas W, Ting L, et al. Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Analytical Chemistry. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hebert AS, Merrill AE, Bailey DJ, Still AJ, et al. Neutron-encoded mass signatures for multiplexed proteome quantification. Nature Methods. 2013;10:332-+. doi: 10.1038/nmeth.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hebert AS, Merrill AE, Stefely JA, Bailey DJ, et al. Amine-reactive Neutron-encoded Labels for Highly Plexed Proteomic Quantitation. Molecular & Cellular Proteomics. 2013;12:3360–3369. doi: 10.1074/mcp.M113.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnology. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 99.Huttenhain R, Malmstrom J, Picotti P, Aebersold R. Perspectives of targeted mass spectrometry for protein biomarker verification. Current Opinion In Chemical Biology. 2009;13:518–525. doi: 10.1016/j.cbpa.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zangar RC, Daly DS, White AM. ELISA microarray technology as a high-throughput system for cancer biomarker validation. Expert Rev Proteomic. 2006;3:37–44. doi: 10.1586/14789450.3.1.37. [DOI] [PubMed] [Google Scholar]
- 101.Makawita S, Diamandis EP. The Bottleneck in the Cancer Biomarker Pipeline and Protein Quantification through Mass Spectrometry-Based Approaches: Current Strategies for Candidate Verification. Clinical Chemistry. 2010;56:212–222. doi: 10.1373/clinchem.2009.127019. [DOI] [PubMed] [Google Scholar]
- 102.Addona TA, Shi X, Keshishian H, Mani DR, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nature Biotechnology. 2011;29:635–U119. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-MS/MS using protein cleavage and isotope dilution mass spectrometry. Journal Of Proteome Research. 2004;3:644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 104.Li WL, Nemirovskiy O, Fountain S, Mathews WR, Szekely-Klepser G. Clinical validation of an immunoaffinity LC-MS/MS assay for the quantification of a collagen type II neoepitope peptide: A biomarker of matrix metalloproteinase activity and osteoarthritis in human urine. Analytical Biochemistry. 2007;369:41–53. doi: 10.1016/j.ab.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 105.Simon R, Lemoine J, Fonbonne C, Jaffuel A, et al. Absolute quantification of podocin, a potential biomarker of glomerular injury in human urine, by liquid chromatography-multiple reaction monitoring cubed mass spectrometry. Journal of pharmaceutical and biomedical analysis. 2014;94:84–91. doi: 10.1016/j.jpba.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 106.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 107.DeSouza LV, Taylor AM, Li W, Minkoff MS, et al. Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. Journal Of Proteome Research. 2008;7:3525–3534. doi: 10.1021/pr800312m. [DOI] [PubMed] [Google Scholar]
- 108.Yoon JY, Yeom J, Lee H, Kim K, et al. High-throughput peptide quantification using mTRAQ reagent triplex. Bmc Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-S1-S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin HR, Zhang L, Xie LQ, Huang LY, et al. Hyperplex-MRM: A Hybrid Multiple Reaction Monitoring Method Using mTRAQ/iTRAQ Labeling for Multiplex Absolute Quantification of Human Colorectal Cancer Biomarker. Journal Of Proteome Research. 2013;12:3912–3919. doi: 10.1021/pr4005025. [DOI] [PubMed] [Google Scholar]
- 110.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nature Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 111.Mischak H, Delles C, Klein J, Schanstra JP. Urinary Proteomics Based on Capillary Electrophoresis-Coupled Mass Spectrometry in Kidney Disease: Discovery and Validation of Biomarkers, and Clinical Application. Adv Chronic Kidney D. 2010;17:493–506. doi: 10.1053/j.ackd.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 112.Coon JJ, Zurbig P, Dakna M, Dominicza AF, et al. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clinical Applications. 2008;2:964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pejchinovski M, Hrnjez D, Ramirez-Torres A, Bitsika V, et al. Capillary zone electrophoresis on-line coupled to mass spectrometry: A perspective application for clinical proteomics. Proteomics Clin Appl. 2015;9:453–468. doi: 10.1002/prca.201400113. [DOI] [PubMed] [Google Scholar]
- 114.Airoldi L, Magagnotti C, Iannuzzi AR, Marelli C, et al. Effects of cigarette smoking on the human urinary proteome. Biochem Bioph Res Co. 2009;381:397–402. doi: 10.1016/j.bbrc.2009.02.055. [DOI] [PubMed] [Google Scholar]
- 115.Fliser D, Novak J, Thongboonkerd V, Argiles A, et al. Advances in urinary proteome analysis and biomarker discovery. Journal of the American Society of Nephrology. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 116.Klein J, Papadopoulos T, Mischak H, Mullen W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis. 2014;35:1060–1064. doi: 10.1002/elps.201300327. [DOI] [PubMed] [Google Scholar]
- 117.Angel TE, Aryal UK, Hengel SM, Baker ES, et al. Mass spectrometry-based proteomics: existing capabilities and future directions. Chem Soc Rev. 2012;41:3912–3928. doi: 10.1039/c2cs15331a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thongboonkerd V, Songtawee N, Sritippayawan S. Urinary proteome profiling using microfluidic technology on a chip. Journal Of Proteome Research. 2007;6:2011–2018. doi: 10.1021/pr060586+. [DOI] [PubMed] [Google Scholar]
- 119.Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin Appl. 2015;9:307–321. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 120.Egertson JD, MacLean B, Johnson R, Xuan Y, MacCoss MJ. Multiplexed peptide analysis using data-independent acquisition and Skyline. Nat Protoc. 2015;10:887–903. doi: 10.1038/nprot.2015.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schubert OT, Gillet LC, Collins BC, Navarro P, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 122.Thiese MS, Arnold ZC, Walker SD. The misuse and abuse of statistics in biomedical research. Biochem Med (Zagreb) 2015;25:5–11. doi: 10.11613/BM.2015.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Swan AL, Mobasheri A, Allaway D, Liddell S, Bacardit J. Application of Machine Learning to Proteomics Data: Classification and Biomarker Identification in Postgenomics Biology. Omics-a Journal of Integrative Biology. 2013;17:595–610. doi: 10.1089/omi.2013.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carnielli CM, Winck FV, Paes Leme AF. Functional annotation and biological interpretation of proteomics data. Biochim Biophys Acta. 2015;1854:46–54. doi: 10.1016/j.bbapap.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 125.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Frantzi M, Bhat A, Latosinska A. Clinical proteomic biomarkers: relevant issues on study design & technical considerations in biomarker development. Clin Transl Med. 2014;3:7. doi: 10.1186/2001-1326-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Acosta-Martin AE, Lane L. Combining bioinformatics and MS-based proteomics: clinical implications. Expert Rev Proteomics. 2014;11:269–284. doi: 10.1586/14789450.2014.900446. [DOI] [PubMed] [Google Scholar]
- 128.Griss J, Perez-Riverol Y, Hermjakob H, Vizcaino JA. Identifying novel biomarkers through data mining-a realistic scenario? Proteomics Clin Appl. 2015;9:437–443. doi: 10.1002/prca.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mischak H, Kolch W, Aivaliotis M, Bouyssie D, et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteomics Clinical Applications. 2010;4:464–478. doi: 10.1002/prca.200900189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taylor CF, Paton NW, Lilley KS, Binz PA, et al. The minimum information about a proteomics experiement (MIAPE) Nature Biotechnology. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 131.Taylor CF, Binz PA, Aebersold R, Affolter M, et al. Guidelines for reporting the use of mass spectrometry in proteomics. Nature Biotechnology. 2008;26:860–861. doi: 10.1038/nbt0808-860. [DOI] [PubMed] [Google Scholar]
- 132.Gu Q, Yu LR. Proteomics quality and standard: From a regulatory perspective. Journal Of Proteomics. 2014;96:353–359. doi: 10.1016/j.jprot.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Anderson L. Six decades searching for meaning in the proteome. Journal of Proteomics. 2014;107:24–30. doi: 10.1016/j.jprot.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 134.Barr A. Wall Street Journal (Online) New York, N.Y: 2014. [Google Scholar]
- 135.Mapes M. Endocrine News. Endocrine Society; 2015. pp. 44–45. [Google Scholar]
- 136.Carr SA, Anderson L. Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory? Clin Chem. 2008;54:1749–1752. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Endre ZH, Pickering JW. Cell cycle arrest biomarkers win race for AKI diagnosis. Nat Rev Nephrol. 2014;10:685–686. doi: 10.1038/nrneph.2014.198. [DOI] [PubMed] [Google Scholar]