Abstract
Purpose/Objective(s)
Clinical proton beam therapy has been based on the use of a generic relative biological effectiveness (RBE) of ~1.1. However, emerging data suggest that Fanconi Anemia (FA) and homologous recombination pathway defects may lead to a variable RBE, at least in-vitro. Here, we investigated the role of SLX4 (FANCP), which acts as a docking platform for the assembly of multiple structure-specific endonucleases, in the response to proton irradiation.
Methods and Materials
Isogenic cell pairs for the study of SLX4, XPF/ERCC1, MUS81, and SLX1 were irradiated at the mid-Spread-Out Bragg Peak of a clinical proton beam (linear energy transfer, 2.5 keV/μm) or with 250 kVp X-rays, and clonogenic survival fractions (SF) were determined. To estimate the RBE of protons relative to Cobalt-60 photons (Co60eq), we assigned a RBE(Co60Eq) of 1.1 to X-rays to correct the physical dose measured. Standard DNA repair foci assays were employed to monitor damage responses, and cell cycle distributions were assessed by flow cytometry. The PARP inhibitor olaparib was used for comparison.
Results
Loss of SLX4 function resulted in an enhanced proton RBE(Co60Eq) of 1.42 compared to 1.11 for wild-type cells (at SF=0.1, p<0.05) which correlated with increased persistent DNA double-strand breaks in cells in S/G2 phase. Genetic analysis identified the SLX4-binding partner MUS81 as a mediator of resistance to proton radiation. Both proton irradiation and olaparib treatment resulted in a similar prolonged accumulation of RAD51 foci in SLX4/MUS81-deficient cells, suggesting a common defect in the repair of DNA replication fork-associated damage.
Conclusions
A defect in the FA pathway at the level of SLX4 results in hypersensitivity to proton radiation, which is at least in part due to impaired MUS81-mediated processing of replication forks that stall at clustered DNA damages. In-vivo and clinical studies are needed to confirm these findings in human cancers.
Introduction
Clinical proton beam therapy has been based on the use of a generic relative biological effectiveness (RBE) of ~1.1 relative to Cobalt-60 reference radiation (Co60Eq) [1]. Clinical experience suggests that a proton RBE(Co60Eq) of 1.1 is likely reasonably accurate for normal tissues [2-4]. However, there remains very little data on proton RBE variations in human cancers which frequently harbor alterations in DNA repair pathways [5,6]. The exact frequencies of genetic/epigenetic defects in DNA damage response and repair genes in human cancers remain to be established but it has been postulated that alterations exist in most if not all human cancers [7].
Homologous recombination repair (HRR), which depends on BRCA1 and BRCA2, is a major pathway for the repair of DNA double-strand breaks [6]. HRR is also involved in the repair and restart of collapsed DNA replication forks where it converges with the Fanconi Anemia (FA) pathway of replication fork maintenance and repair to form a common FA/BRCA pathway [6,8]. Recent observations have implicated mutations in BRCA1 (FANCS) and the RAD51 (FANCR) recombinase in the pathogenesis of FA, further solidifying the notion of a single pathway [9,10]. Altogether, there are now at least 17 FA genes [8]. Of interest is a recently identified member, SLX4 (FANCP), which is involved in the assembly of several structure-specific endonucleases with functions in the incision of DNA lesions at stalled forks as well the resolution of HRR intermediates [11-14]. FA/BRCA defects are found in several human cancer types including lung cancer (up to 20-30%) and SLX4 mutations are seen in about 5% of lung squamous cell cancers [6,15,16]. To our knowledge, there have been no studies investigating the role of SLX4 in the cellular response to ionizing radiations.
We recently demonstrated a variable proton RBE in a diverse panel of human lung cancer cell lines, with the most sensitive cell lines yielding RBE(Co60Eq) estimates of 1.31-1.77 for a clonogenic survival fraction (SF) of 0.5 at the center of the Spread-Out Bragg Peak (SOBP) [5]. In several cell lines, the increased RBE was correlated with defects in the FA/BRCA pathway, and comparisons in isogenic models for FANCA, BRCA1, and FANCD2 confirmed these findings. Similarly, data from another group recently revealed the importance of RAD51 and BRCA2 (FANCD1) for the repair of proton induced DNA damage [17,18]. Together, these observations suggest that defects in the FA/BRCA pathway may render the affected tumor cells hypersensitive to the slightly more complex clustered DNA damages caused by protons compared to other low linear energy transfer (LET) radiations, resulting in a RBE(Co60Eq) > 1.1. In contrast, repair-proficient tumor (and normal) cells remove proton- and photon-induced damages almost equally well, reflected by a RBE of 1.1.
The specific repair protein requirements at sites of replication forks colliding with clustered DNA damages caused by protons remain to be defined. Here, we asked whether loss of SLX4, which is downstream of FANCD2 [19,20], results in an increased cellular sensitivity to proton radiation as used in the clinic.
Materials and Methods
Cell lines and culture
Immortalized human fibroblasts derived from a FA complementation group P patient with bi-allelic SLX4 mutations or derivates complemented with wild-type protein or dissociation-of-functions mutants were a kind gift from Dr. A Smogorzewska, Rockefeller University, New York, NY [11]. These mutants were deleted (Δ) for either one of three interaction domains: ΔXPF (MLR), ΔMUS81 (SAP), or ΔSLX1 (SBD). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich) with 1 μg/ml puromycin and 20% Bovine Growth Serum (BGS) (HyClone, South Logan, Utah). Immortalized mouse embryonic fibroblasts (MEF) derived from Mus81 wild-type (+/+) or deficient (−/−) mice were kindly provided Drs. R. Hakem and P. McPherson, University of Toronto, Toronto, Canada [21]. Cells were maintained in DMEM with 1 μg/ml puromycin. Chinese hamster ovary (CHO) cell lines AA8 and the ERCC1-mutant line UV20 were cultured in modified McCoy's 5a medium (Gibco, Carlsbad, CA). All cell lines were maintained in a humidified incubator at 37°C and 5% CO2, with 10% BGS, 20 mM HEPES, 1% Streptomycin-Penicillin and 2 mM L-Glutamine (Sigma-Aldrich, St. Louis, MO) supplemented to the medium. Cells were tested mycoplasma free (MycoAlert, Lonza, Walkersviller, MD) and used within 10 passages, with 2-3 passages per week, for all experiments after resuscitation.
Irradiation and dosimetry
Irradiation conditions for proton, X-ray, and Cs-137 γ-ray treatments, as well as dosimetric validation, were described in detail previously [5]. Briefly, cells were irradiated either at mid-SOBP using a clinical set-up with a dose rate of 1.7 Gy/min, by Cs-137 γ-rays delivered at 2.4 Gy/min, or by X-rays produced by a Siemens Stabilipan 2 X-ray generator, which was operated at 250 kVp and 12 mA with a half-value layer of 0.5 mm Cu. A dose rate of 1.6 Gy/min was defined at 50 cm SSD with full backscatter for the treatment of adherent cells. The X-ray unit was calibrated according to the AAPM's TG61 using a 0.6cc Farmer-type ionization chamber and electrometer. X-ray output was confirmed three times over the course of the past year and found to be constant.
Drug treatments
Olaparib was purchased from LC Laboratories (Woburn, MA, USA), and dissolved in Dimethyl Sulfoxide (DMSO, Sigma-Aldrich) to generate a stock concentration at 10 mM. It was aliquoted and stored at −70°C with protection from light for a maximum of 6 weeks. Cells were treated with olaparib at indicated concentrations in experiments without wash-off.
Clonogenic assay
Standard colony formation assays were performed as described in detail previously [5]. Briefly, exponentially growing cells were harvested, diluted to the appropriate densities in single cell suspension, and plated into flasks for attachment 8 hours prior to irradiation. Samples for irradiation with x-rays, γ-rays or protons were prepared in parallel for irradiation on the same day. Culture vessels were incubated for 8-24 days, and colonies with >50 viable cells were scored.
Flow cytometry
Standard protocol for cell cycle analysis using the DNA dye propidium iodide (Sigma-Aldrich) for flow cytometry was used. Cells in exponential growth were prepared for mock control, X-ray, or proton irradiation in parallel. Cells were collected and washed twice with phosphate-buffered saline (PBS). Single cell suspension in PBS was made and then fixed in 1% paraformaldehyde (PFA) and 80% ice-cold ethanol. Cells were kept in −20°C until analysis. Cells were also co-stained with γ-H2AX antibody (Millipore, Temecula, CA). Ethanol fixative was removed, and cells were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in PBS. Following blocking in 5% goat serum (Sigma-Aldrich) for 30 min, cells were incubated with γ-H2AX antibody for 2 hours at room temperature. After twice washing in PBS, cells were incubated in Alexa-488 conjugated secondary antibody (Invitrogen, Carlsbad, CA) for 1 hour. 1 mg/ml RNAse and 500 μg/ml propidium iodide (PI) were added, and then cells were analyzed using a LSRII flow cytometer (BD Biosciences, San Jose, CA, USA). Data were analyzed using FlowJo software (FlowJo, LLC; Ashland, OR). Cells were gated into subpopulations according to their cell cycle phase indicated by PI staining of DNA content. Gates in untreated cells were set to minimize baseline γ–H2AX signal. Background signal was subtracted from the increased signal observed in irradiated samples.
Immunofluorescence microscopy
Staining and quantification of DNA damage repair foci was performed as previously described [5,16]. Briefly, cells were plated on chamber slides with appropriate cell density to ensure subconfluence before fixing. They were fixed with 4% PFA for 10 min, permeabilized with 0.5% TritonX-100 for 15 min, and blocked with 5% goat serum in PBS 0.1% TritonX-100 for 30 min (all Sigma-Aldrich). Cells were then incubated with γ-H2AX mouse monoclonal antibody (Millipore, Temecula, CA) at 1:500 dilution, or RAD51 rabbit polyclonal antibody (Santa Cruz, Dallas, Texas) at 1:200 dilution followed by incubation with mouse or rabbit Alexa-488 conjugated secondary antibody (Invitrogen, Carlsbad, CA) at 1: 1,000, respectively. All slides were counterstained with DAPI (4',6-diamidino-2-phenylindole), examined and photographed by fluorescence microscopy (Olympus BX51, Center Valley, PA).
Statistical analysis
The data were analyzed by GraphPad Prism 6 (GraphPad Software, La Jolla, CA). Log transformed survival fractions were fitted using the Linear-Quadratic (LQ) formula with weighting to 1/Y2. RBE was defined as the ratio of the physical doses of reference radiation and protons that yield a clonogenic survival fraction (SF) of 0.1 or 0.5. Individual RBE values were taken from the LQ fitted survival curves in each independent repeat experiment and averaged to report a mean RBE. To estimate proton RBE values relative to Co60 γ-rays, proton RBE values measured with X-rays as reference in individual repeats were multiplied with a factor of 1.1 and then averaged. All statistical comparisons were performed with the student's T-test. P-values of ≤ 0.05 (two-sided) were considered statistically significant. All data were typically based on three independent repeat experiments.
Results
SLX4 mutation causes sensitivity to proton radiation
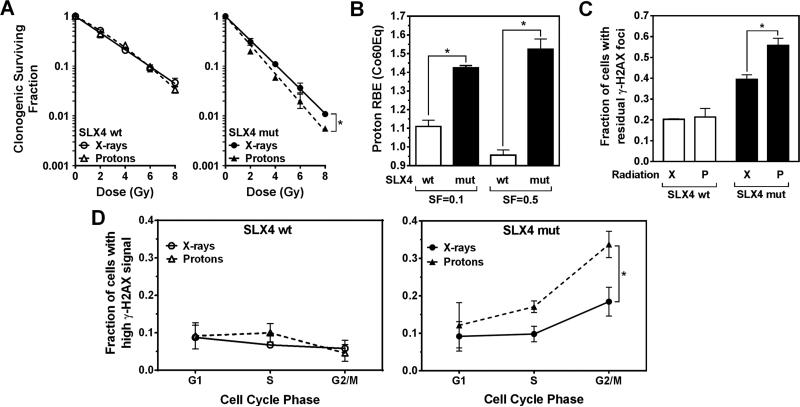
To study the role of SLX4 in determining cellular resistance to proton radiation, we employed a previously established experimental set-up where cells are irradiated at the mid-SOBP of a clinical proton beam (dose-averaged LET, 2.5 keV/μm) or with 250 kVp X-rays (~2.0 keV/μm) [5]. Cellular radiosensitivity was determined by standard colony formation assays. Reproducibility of prior results with this set-up was confirmed (Supplementary Fig. S1A). Human fibroblasts defective for SLX4 that have been derived from a FA-P patient displayed a greater X-ray sensitivity than fibroblasts complemented with wild-type SLX4, for example, SF2Gy of 0.31 versus 0.53, respectively (Supplementary Fig. S1B). Furthermore, SLX4-deficient cells were significantly more sensitive to protons than to X-rays (RBE(X-rayEq)= 1.29 for SF=0.1) in contrast to SLX4 wild-type cells (RBE=1.01) (Fig. 1A, Supplementary Fig. S2A).
Fig. 1.
Role of SLX4 for cellular resistance to proton radiation. (A) Clonogenic cell survival curves for immortalized human fibroblasts with bi-allelic SLX4 mutations (mut) or complemented with wild-type (wt) protein. Data points represent means of log transformed survival fractions +/− standard error (SE). (B) Estimated proton RBE values relative to Co60 γ-rays (Co60Eq). To convert the measured proton RBE(X-rayEq) in Panel A to RBE(Co60Eq), we used a multiplicator of 1.1, based on published data that orthovoltage X-rays have a RBE(Co60Eq) of 1.1-1.15 [22,23]. The purpose of this conversion is to report clinically relevant proton RBE values. We note that a 1.1 multiplier is an oversimplification as the RBE of X-rays compared to Co60 γ-rays may differ along the survival curve and between cell lines. SF, survival fraction. (C) Fraction of cells with ≥ 20 foci at 18 hours after 6 Gy irradiation with X-rays (X) or protons (P). Bars represent means +/− SE. (D) Fractions of cells with high γ-H2AX signal using flow cytometry 18 hours after 6 Gy irradiation. *, p≤0.05, paired T-test.
Because X-rays have a RBE(Co60Eq) of 1.1-1.15 [22,23], we adjusted the RBE(X-rayEq) values using a conservative multiplier of 1.1 to derive RBE(Co60Eq) values for protons. This revealed average RBE(Co60Eq) values of 1.42 and 1.1 at SF=0.1 for SLX4 deficient and wild-type cells, respectively (Fig. 1B). Direct comparison with Cs-137 γ-rays, which have LET and RBE values that are close to Co60 γ-rays, confirmed these estimates (Supplementary Fig. S2B).
Function of SLX4 in the repair of proton damage to DNA is mediated through MUS81
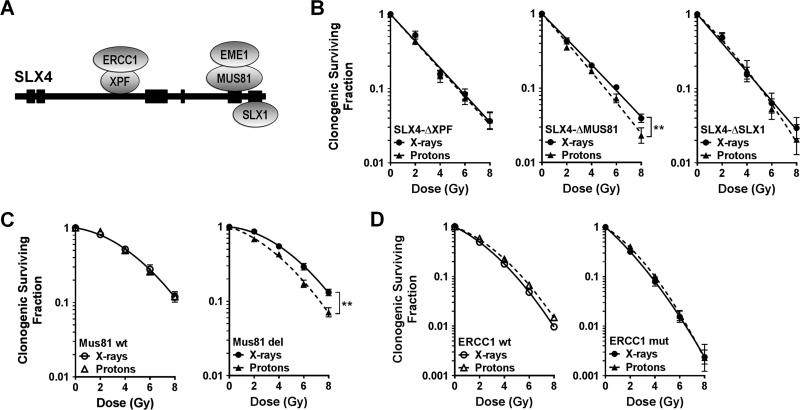
Next, we employed a panel of FA-P fibroblasts expressing dissociation-of-function SLX4 deletion mutants where binding either to XPF (ERCC4/FANCQ), MUS81, or SLX1 was abolished (see Materials and Methods) (Fig. 2A). Only the SLX4-ΔMUS81 mutant was associated with increased cellular sensitivity to protons, i.e., RBE(Co60Eq) of 1.26 at SF=0.1 (Fig. 2B), while binding of neither XPF nor SLX1 to SLX4 was required in the proton response. We confirmed these observations by employing MEFs either wild-type (RBE(Co60eq)=1.1) or null for Mus81 (RBE(Co60Eq)=1.29), as shown in Fig. 2C. In contrast, the absence in CHO cells of functional ERCC1, the binding partner of XPF, did not lead to increased sensitivity to protons compared to ERCC1 wild-type cells (Fig. 2D). Of note, all SLX4 dissociation-of-function mutants were more resistant to X-rays than cells lacking SLX4 function (Supplementary Figure S1B). Taken together, the data in Fig. 2 establish that the repair of proton radiation damage to DNA by SLX4 is at least in part mediated by MUS81, which is known to process stalled replication forks and resolve HRR intermediates [13,21,24].
Fig. 2.
Role of SLX4 associated endonuclease complexes for cellular resistance to protons. (A) Simplified schema of SLX4 protein illustrating binding of structure-specific endonucleases. (B) Clonogenic survival curves for SLX4-deficient fibroblasts complemented with dissociation-of-function mutants, analogous to Fig. 1A. (C) Clonogenic survival curves for MEFs either wild-type (wt) for Mus81 or with biallelic deletion (del). (D) Analogous survival curves for CHO cells wither wt or mutant (mut) for ERCC1. **, p≤0.01, paired T-test.
Similar phenotype of proton- and olaparib-treated SLX4/MUS81-deficient cells
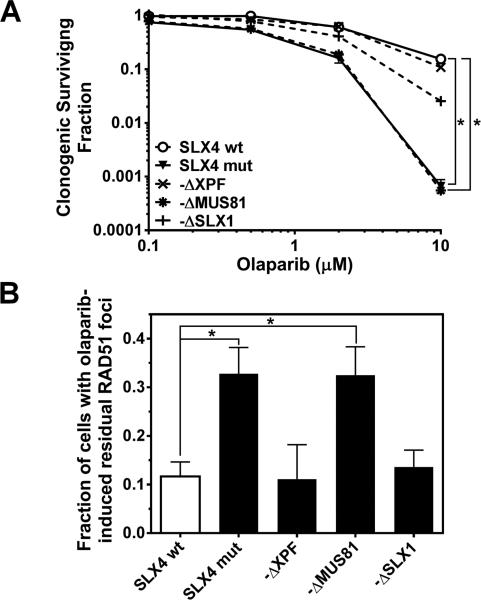
We previously reported that cells deficient in FANCD2, which is a binding partner of SLX4, unexpectedly displayed increased numbers of RAD51 foci at ≥18 hours following proton compared to X-irradiation [5]. Fig. 3A shows a similar observation made in SLX4 deficient cells. The observed increase in RAD51 foci was not due to a difference in cell cycle distribution between X- and proton-irradiated cells (Fig. 3B). When we evaluated our panel of SLX4 dissociation-of-function mutants we observed an analogous persistence of RAD51 foci only in cells expressing SLX4-ΔMUS81 (Fig. 3C), which correlated with an accumulation of DSB (Fig. 3D) and the previously found RBE increase (Fig. 2B). Interestingly, this phenotype mirrored the observed hypersensitivity of SLX4 deficient and SLX4-ΔMUS81 expressing cells to olaparib (Fig. 4A) and the corresponding over-accumulation of RAD51 foci (Fig. 4B). Together, these data suggest that HRR is either aberrant or is initiated properly but blocked at a late stage in proton-irradiated SLX4/MUS81 deficient cells, thereby leading to genotoxicity.
Fig. 3.
RAD51 foci formation in SLX4/MUS81 defective cells. (A) Left panel, representative images of subnuclear RAD51 foci in SLX4 cells with bi-allelic mutations (mut) or cells complemented with wild-type (wt) protein. Right panel, quantification of results showing the fraction of cells with at least 10 foci at 18 hours after 6 Gy irradiation with either X-rays (X) or protons (P). Foci in untreated cells were subtracted. Various foci number cut-offs to define a positive cell were explored (> 5, 10, 15, 20) and a cut-off of 10 provided the best discrimination consistent with published data ref. Bars represent means +/− SE. (B) Cell cycle distributions determined in parallel to the foci experiments in panel A. (C) Analysis of RAD51 foci formation in the SLX4 dissociation-of-function mutants analogous to panel A. (D) Parallel analysis of γ-H2AX foci formation analogous to panel 1C. *, p≤0.05, paired T-test.
Fig. 4.
PARP inhibitor phenotype of SLX4 mutant cell lines. (A) Clonogenic survival curves for SLX4 mutant (mut), wild-type (wt), and the dissociation-of function mutant cell lines treated with olaparib at the concentrations indicated. (B) Fraction of cells with at least 10 RAD51 foci after 32 hours of olaparib treatment (5 μM). Foci in untreated cells were subtracted. Bars represent mean +/− SE. *, p≤0.05, T-test.
Discussion
We report that SLX4, which acts as a docking platform for the assembly of multiple structure-specific endonucleases, is important for the cellular response to proton induced DNA damage (Fig. 1A,B). Through genetic analysis we show that the SLX4 binding partner MUS81 mediates or at least contributes to this function (Fig. 2A-C). MUS81 has been implicated in the processing of stalled replication forks following protracted treatment with the crosslinker mitomycin C and at sites of single-stranded DNA lesions [11,25]. In the latter scenario, this activity is likely needed to avoid the conversion of single-stranded lesions into DSB in cells treated with topoisomerase I or PARP inhibitors [11,24]. We hypothesize that clustered damages caused by protons are more complex than those produced by X-rays and contain protruding single-stranded lesions that induce fork reversal. Loss of MUS81 function will result in persistent DSB, either through fork collapse or failed repair of lesions within the damage cluster, as demonstrated by γ-H2AX foci accumulating in the S/G2 phases of the cell cycle (Fig. 1D). Subsequently, RAD51 filaments may accumulate either in an attempt to protect the stalled replication forks, as has been shown for example by Jasin [26], or to participate in futile HRR. This interpretation is supported by the observed similar phenotypes of proton- and olaparib-treated SLX4/MUS81-defective cells. Furthermore, we previously found that loss of FANCD2, which binds SLX4, resulted in a similar accumulation of DSB and RAD51 foci after proton irradiation and olaparib treatment [5,27].
Another possibility to consider is that the accumulation of RAD51 foci represents an impaired resolution of HRR intermediates given the reported Holliday junction resolvase function of MUS81/EME1 [12,13]. If proton induced damages require HRR for repair more frequently than X-ray induced lesions, for the same dose of radiation, then impaired resolvase activity will lead to persistent and genotoxic recombination intermediates. This may hold true for complete loss of SLX4 which will impair more than just one resolvase. However, loss of MUS81 alone may be compensated by other resolvases such as SLX1 and GEN1. Furthermore, there is no change in X-ray sensitivity upon loss of MUS81 activity (Fig. 2C, Supplementary Fig. S1B) [25], strongly arguing against a role of MUS81 in the resolution of HRR intermediates after irradiation. We, therefore, favor a role for MUS81 that is upstream of clustered damage repair. Thus the increased proton sensitivity observed here is likely not due to a bona fide HRR defect.
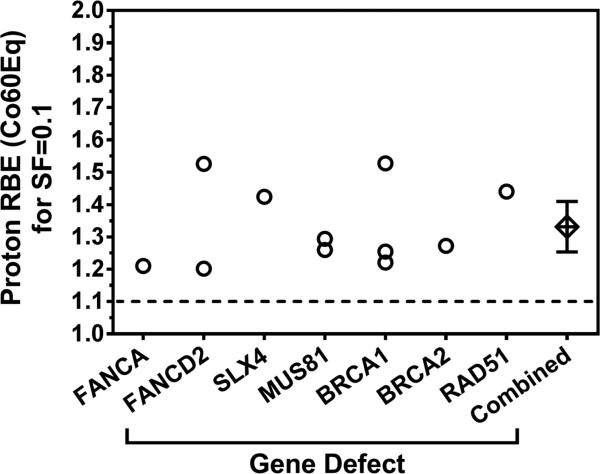
Our study adds to an emerging body of data that implicates a number of FA and HRR genes in the repair of proton damage to DNA. The estimated RBE(Co60Eq) values at SF=0.1 are in the range of 1.20-1.47 (mean, 1.33) at mid-SOBP, as shown in Fig. 5. For SF=0.5, where the estimates tend to be less robust but are more relevant for the clinically administered daily dose of ~ 2 Gy, we generally derive higher values depending on the shoulder region of the respective survival curves, such as 1.34-1.77 for BRCA1 loss and 1.52 for SLX4 (Fig. 1B) [5]. Of note, a recent study did not find increased proton sensitivity in HRR-deficient CHO cells [28]. In that report cells were trypsinized right after irradiation and seeded for colony formation, in contrast to studies by us and others [5,17,18]; however, more data are needed for a better understanding of these differences.
Fig. 5.
Compilation of published RBE values resulting from disruptions in the FA pathway and related genes. Data points are taken from this report and references [5] and [17]. Cumulative data point represents mean with 95% confidence intervals. SF, survival fraction
The clinical significance of the increased in-vitro RBE estimates is that, if they can be confirmed in appropriate in-vivo experiments, proton beam radiation for cancers with a defective FA or HRR pathway may lead to an enhanced therapeutic ratio. This would represent a novel biological benefit of protons in tumors, in addition to the known dosimetric advantage with regard to normal tissue sparing. We stress that in-vivo and clinical data are urgently needed to validate the RBE increases observed invitro and identify predictive biomarkers of increased sensitivity to fractionated proton radiation treatment in patients.
Supplementary Material
Summary.
Clinical proton beam therapy has been based on the use of a generic RBE =1.1. However, emerging evidence suggests that defective DNA repair as found in many cancers yields a variable RBE. We show that SLX4, which acts as a docking platform for the assembly of structure-specific endonucleases, is required for DNA repair and survival after proton irradiation. This function is at least partially mediated through MUS81. Findings support a “new biology” for proton radiation.
Acknowledgments
Support: Federal Share of program income earned by Massachusetts General Hospital on C06 CA059267, Proton Therapy Research and Treatment Center (T.S.H, J.A.E., H.P., H-M.L., K.D.H., H.W.); Massachusetts General Hospital Executive Committee of Research - Deliberative Interim Support Funding (H.W.); NCI Cancer Clinical Investigator Team Leadership Award through a supplement to P30CA006516 (T.S.H.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None
References
- 1.Paganetti H, Niemierko A, Ancukiewicz M, et al. Relative biological effectiveness (RBE) values for proton beam therapy. Int J Radiat Oncol Biol Phys. 2002;53:407–421. doi: 10.1016/s0360-3016(02)02754-2. [DOI] [PubMed] [Google Scholar]
- 2.Bush DA, Cheek G, Zaheer S, et al. High-dose hypofractionated proton beam radiation therapy is safe and effective for central and peripheral early-stage non-small cell lung cancer: results of a 12-year experience at Loma Linda University Medical Center. Int J Radiat Oncol Biol Phys. 2013;86:964–968. doi: 10.1016/j.ijrobp.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Sejpal S, Komaki R, Tsao A, et al. Early findings on toxicity of proton beam therapy with concurrent chemotherapy for nonsmall cell lung cancer. Cancer. 2011;117:3004–3013. doi: 10.1002/cncr.25848. [DOI] [PubMed] [Google Scholar]
- 4.Westover KD, Seco J, Adams JA, et al. Proton SBRT for medically inoperable stage I NSCLC. J Thorac Oncol. 2012;7:1021–1025. doi: 10.1097/JTO.0b013e31824de0bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q, Ghosh P, Magpayo N, et al. Lung Cancer Cell Line Screen Links Fanconi Anemia/BRCA Pathway Defects to Increased Relative Biological Effectiveness of Proton Radiation. Int J Radiat Oncol Biol Phys. 2015;91:1081–1090. doi: 10.1016/j.ijrobp.2014.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Willers H, Pfäffle HN, Zou L, et al. Targeting Homologous Recombination Repair in Cancer. In: Kelley MR, editor. DNA Repair in Cancer Therapy: Molecular Targets and Clinical Applications. Academic Press; Elsevier: 2012. pp. 119–16. [Google Scholar]
- 7.Jackson SP Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang AT Smogorzewska A. SnapShot: Fanconi anemia and associated proteins. Cell. 2015;160:354–354. e351. doi: 10.1016/j.cell.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Wang AT, Kim T, Wagner JE, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer SL, Tian L, Kahkonen M, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, Spitz GS, Veturi U, et al. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garner E, Kim Y, Lach FP, et al. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarbajna S, Davies D West SC. Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 2014;28:1124–1136. doi: 10.1101/gad.238303.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svendsen JM, Smogorzewska A, Sowa ME, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sousa FG, Matuo R, Tang SW, et al. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair (Amst) 2015;28:107–115. doi: 10.1016/j.dnarep.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birkelbach M, Ferraiolo N, Gheorghiu L, et al. Detection of Impaired Homologous Recombination Repair in NSCLC Cells and Tissues. J Thorac Oncol. 2013;8:279–286. doi: 10.1097/JTO.0b013e31827ecf83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fontana AO, Augsburger MA, Grosse N, et al. Differential DNA repair pathway choice in cancer cells after proton- and photon-irradiation. Radiother Oncol. 2015 doi: 10.1016/j.radonc.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Grosse N, Fontana AO, Hug EB, et al. Deficiency in homologous recombination renders Mammalian cells more sensitive to proton versus photon irradiation. Int J Radiat Oncol Biol Phys. 2014;88:175–181. doi: 10.1016/j.ijrobp.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto KN, Kobayashi S, Tsuda M, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein Douwel D, Boonen RA, Long DT, et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McPherson JP, Lemmers B, Chahwan R, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 22.Hall EJ. The relative biological efficiency of x rays generated at 220 kVp and gamma radiation from a cobalt 60 therapy unit. Br J Radiol. 1961;34:313–317. doi: 10.1259/0007-1285-34-401-313. [DOI] [PubMed] [Google Scholar]
- 23.Spadinger I Palcic B. The relative biological effectiveness of 60Co gamma-rays, 55 kVp X-rays, 250 kVp X-rays, and 11 MeV electrons at low doses. Int J Radiat Biol. 1992;61:345–353. doi: 10.1080/09553009214551031. [DOI] [PubMed] [Google Scholar]
- 24.Regairaz M, Zhang YW, Fu H, et al. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J Cell Biol. 2011;195:739–749. doi: 10.1083/jcb.201104003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dendouga N, Gao H, Moechars D, et al. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol Cell Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlacher K, Wu H Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfäffle HN, Wang M, Gheorghiu L, et al. EGFR-activating mutations correlate with a Fanconi anemia-like cellular phenotype that includes PARP inhibitor sensitivity. Cancer Res. 2013;73:6254–6263. doi: 10.1158/0008-5472.CAN-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerelchuluun A, Manabe E, Ishikawa T, et al. The major DNA repair pathway after both proton and carbon-ion radiation is NHEJ, but the HR pathway is more relevant in carbon ions. Radiat Res. 2015;183:345–356. doi: 10.1667/RR13904.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.