Abstract
Protein glycosylation is an important and common post-translational modification. More than 50% of human proteins are believed to be glycosylated to modulate the functionality of proteins. Aberrant glycosylation has been correlated to several diseases, such as inflammatory skin diseases, diabetes mellitus, cardiovascular disorders, rheumatoid arthritis, Alzheimer’s and prion diseases, and cancer. Many approved cancer biomarkers are glycoproteins which are not highly abundant proteins. Therefore, effective qualitative and quantitative assessment of glycoproteins entails enrichment methods. This chapter summarizes glycoprotein enrichment methods, including lectin affinity, immunoaffinity, hydrazide chemistry, hydrophilic interaction liquid chromatography, and click chemistry. The use of these enrichment approaches in assessing the qualitative and quantitative changes of glycoproteins in different types of cancers are presented and discussed. This chapter highlights the importance of glycoprotein enrichment techniques for the identification and characterization of new reliable cancer biomarkers.
Keywords: cancer biomarkers, click-chemistry enrichment, glycoproteomics, glycoprotein enrichment, HILIC enrichment, hydrazide chemistry, immunoaffinity-based enrichment, LC-MS/MS, lectin chromatography
Glycoproteins as cancer biomarkers
Glycosylation is one of the most important and common post-translational modifications. More than 50% of human proteins have been reported to be glycosylated. It is involved in many physiological functions and biological pathways such as protein stabilization, maintaining tissue structures, turnover and activity, cell–cell adhesion and cell–matrix attachment [1]. N-linked glycosylation or O-linked glycosylation are typical types of protein glycosylations. Aberrant glycosylation has been observed in many diseases such as inflammatory skin diseases, diabetes mellitus, cardiovascular disorders, rheumatoid arthritis, Alzheimer’s and prion diseases and cancer [2,3]. In the case of cancer, a profound correlation between glycosylation and disease development and/or malignancy has been demonstrated. Accordingly, unequivocal identification and quantification of glycoproteins-associated with cancer provide an opportunity to discover reliable and sensitive cancer biomarkers that permit the detection of the disease at early stages.
Two ‘omics’ fields aimed at evaluating the glycosylation of glycoconjugates in biological systems have been developed, namely: glycomics and glycoproteomics. This chapter deals only with glycoproteomics. The focus of this field is evaluating the changes of glycosylated proteins, in terms of determining glycosylation site occupancy (or absence) in proteins. Therefore, glycoproteomics defines the microheterogeneity associated with the glycosylation sites of proteins. Also, macro-heterogeneity can be monitored with multiple glycosylation sites in a single peptide backbone, which can be produced as a result of incomplete enzymatic digestion.
Glycoproteins are typically subjected to enzymatic digestion and then, to LC-MS or LC-MS/MS analyses. Therefore, the choice of enzyme is one of the key factors for efficient characterization of glycopeptides/glycoproteins. Trypsin has been the most widely used enzyme. Tryptic glycopeptides ionize more efficiently than other proteolytic glycopeptides since tryptic glycopeptides possess C-terminus lysine or arginine residues. Recently, endoproteinase Lys-C or endoproteinase Glu-C have been incorporated into tryptic digestion to improve the cleavage efficiency and modulate the size of the generated peptides to promote effective LC-MS/MS analyses.
To study the microheterogeneity of glycopeptides, MS/MS techniques are required for better interpretation of glycan linkages to peptide backbone sequences. This also distinguishes many isobaric glycopeptides observed in MS. Collision-induced dissociation (CID) has been the routine dissociation method of choice in glycopeptides identification. The complementary glycan structures of glycopeptides are assessed from CID tandem MS. Higher-energy collision dissociation creates the diagnostic oxonium ions of glycopeptides at low m/z values, such as 138, 204, 366, etc. Electron transfer dissociation (ETD) has been recently applied for better identification of glycopeptides. This fragmentation technique permits the sequence of peptide backbone, since it prompts fragmentation of peptide backbone but not the glycan structure. Combining these different dissociation methods, such as CID-higher-energy collision dissociation or CID-ETD, enables comprehensive characterizations of glycopeptides. Employing such tandem MS techniques should facilitate the effective characterizations of glycopeptides/glycoproteins, which eventually results in the identification and validation of glycoprotein cancer biomarkers.
The focus of this chapter is highlighting the glycoproteomic changes associated with different type of cancers. These have been studied using LC-MS/MS mainly in conjunction with lectin affinity (LAC) enrichment, hydrazide chemistry-based enrichment, hydrophilic interaction liquid chromatographic enrichment (HILIC) or other enrichment approaches, such as click chemistry-based enrichment or immunoaffinity. First, the different methods employed to enrich glycoproteins are briefly described and introduced. Next, a summary of recent studies demonstrating the potential of using the enrichment methods in search of reliable and sensitive glycoproteins cancer biomarkers will be discussed and described.
Enrichment of glycoproteins: the technique of choice
Several analytical challenges are associated with LC-MS/MS-based glycoproteomics. First, glycoproteins are present in low abundances in biological systems. Also, LC-MS/MS analysis of glycopeptides is hampered by their microheterogeneity and low-ionization efficiencies in the presence of other peptides. Accordingly, glycoproteins/glycopeptides enrichment is currently considered the method of choice to overcome these challenges. Enrichment of glycoproteins also enhances their LC-MS/MS analysis by overcoming the concentration dynamic range that is commonly associated with proteomics. Lectin affinity chromatography and hydrazide chemistry-based approaches are currently considered the two commonly employed glycoprotein/glycopeptide enrichment techniques. Recently, HILIC is considered as a promising enrichment technique to capture glycopeptides. Other techniques such as click chemistry and immunoaffinity will be also introduced in this chapter.
Lectin affinity enrichment
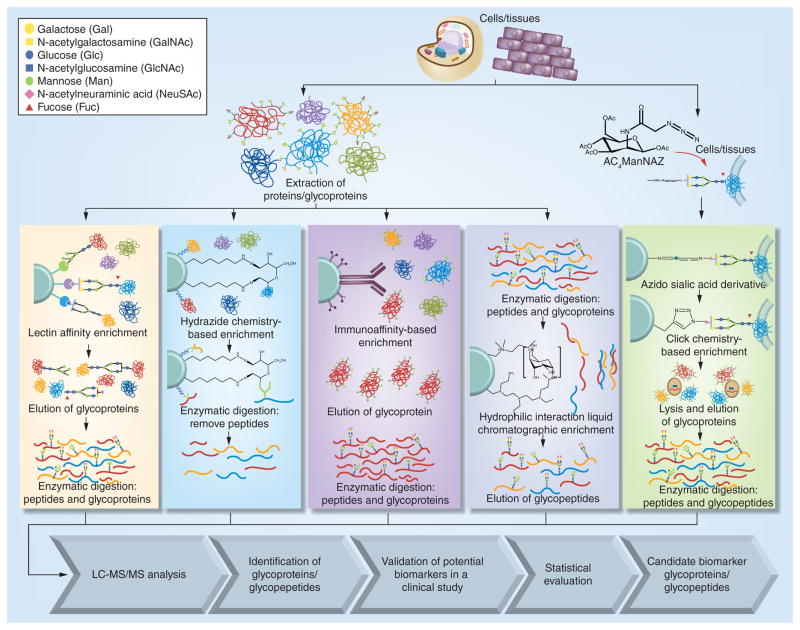
The different glycoproteomics strategies that will be discussed in this chapter are outlined in Figure 1. Lectin affinity enrichment is one of the mainly used techniques utilizing the specificity of lectins to a particular type of glycan residues or linkages [4]. The different types of commercially available lectins are listed in Table 1. Lectins are the glycoproteins that have specific affinity to a particular type of glycans. For example, concanavalin A (ConA) specifically binds to man-nose containing glycans, Sambucus nigra agglutinin binds to sialic acid containing glycans and wheat germ agglutinin (WGA) mainly interacts with GlcNAc residues of a glycan structure. Lectin affinity enrichment commonly employs a type of glycans to enrich specific glycan structures. However, a multilectin mixture has been used to capture and enrich a wide range of glycans. Lectin enrichment technique has been effectively applied to study glycoproteomic changes associated with different types of cancer, including lung, breast and liver.
Figure 1.
Workflow outlining glycoprotein enrichment and LC-MS/MS qualitative and quantitative characterization.
Table 1.
Commercially available lectin materials, abbreviation, sources and glycan specificity.
| Lectin | Abbreviation | Source† | Glycan specificity |
|---|---|---|---|
| Concanavalin A | Con A | Canavalia ensiformis (Jack Bean) seeds | αMan, αGlc |
| Galanthus nivalis agglutinin | GNL, GNA | Galanthus nivalis (Snowdrop) bulbs | αMan |
| Lens culinaris agglutinin | LCA, LcH | Lens culinaris (lentil) seeds | αMan, αGlc |
| Wheat Germ agglutinin | WGA | Triticum vulgaris (wheat germ) | GlcNAc |
| Sambucus nigra agglutinin | SNA, EBL | Sambucus nigra (Elderberry) bark | Neu5Acα6Gal/GalNAc |
| Aleuria aurantia lectin | AAL | Aleuria aurantia mushrooms | Fucα6GlcNAc |
| Maackia amurensis lectin I | MAL I, MAL | Maackia amurensis seeds | Galβ4GlcNAc |
| Jacalin lectin | Jacalin, AIL | Artocarpus integrifolia (Jackfruit) seeds | Galβ3GlcNAc |
| Peanut agglutinin | PNA | Arachis hypogaea peanuts | Galβ3GalNAc |
The information about source was obtained from [5].
Hydrazide chemistry-based enrichment
Enrichment of glycoproteins through hydrazide chemical reactions has been recently demonstrated [6]. Upon peroxidation, a secondary hydroxyl group of glycan residues is converted to an aldehyde group, which is able to covalently bind to hydrazide-functionalized beads as shown in Figure 1. Since the resulting hydra-zone bond is irreversible, another chemical reaction or additional enzyme is needed to release the glycopeptides from the beads. For example, PNGase F has been widely applied to release glycopeptides bound to beads. Since this enzyme cleaves N-glycosidic bond between glycans and N-glycosylation sites, the microheterogeneity of the glycosylation sites is lost through this approach. Hydrolysis is recently applied to release glycopeptides without sialic acid residue. Thus, partial structural studies of glycans are enabled connecting to their glycosylation sites. This procedure was recently applied to assess glycoprotein changes associated with lung, breast and prostate cancers as well as hepatocellular carcinoma.
Hydrophilic interaction liquid chromatographic enrichment
Hydrophilic interaction liquid chromatographic enrichment is considered an attractive enrichment technique to capture glycopeptides by virtue of the hydrophilicity of glycan moieties of glycopeptides. HILIC enrichment of glycopeptides is based on the hydrophilic interaction between the stationary phase and glycan moieties of glycopeptides as shown in Figure 1. Enrichment is attained at high content of organic solvent such as 80% acetonitrile while glycopeptides are eluted with increasing water content. HILIC is widely applicable since the sample preparation includes a broad range of stationary/mobile phase and solid phase extraction formats, suitable for different types of separation/detection including mass spectrometer. Also, HILIC enrichment is performed on glycopeptide, thus enabling the efficient removal of peptides after enzymatic digestion. Therefore, increasing the likelihood of acquiring tandem MS of glycopeptides, and thus facilitating more confident assignment of glycoprotein microheterogeneity. Recently, this technique was successfully applied to assess potential glycopeptide biomarkers of hepatocellular carcinoma as described below.
Click chemistry-based enrichment
Click chemistry was originally developed for selective detection of cell-surface glycoproteins. This enrichment is based on including a labeled monosaccharide into the glycan residues during cell growth. As shown in Figure 1, metabolic labeling inserts azide-modified glycan residues into the cell membrane. Click chemistry then occurs based on cooper-catalyzed azide-alkyne cycloaddition. By this way, the relatively high abundant cytosolic proteins are removed, enabling better characterization of cell-surface glycoproteins. This method was applied to identify cell-surface glycoprotein biomarkers for prostate cancer as described below.
Immunoaffinity glycoprotein enrichment
Certain glycoproteins are typically purified using antibodies. Antibodies can be expressed to recognize specific glycoproteins of interest. This method involves the use of an immunoaffinity column that contains the antibody or antibody-related agent, which has antigen-antibody specificity to a target glycoprotein. It enables detailed characterization of a target glycoprotein, pursuing site-specific occupancies or types of glycan residues. This method is dependent on the availability of antibodies capable of recognizing the glycoproteins of interest. Therefore, this method might be expensive. The utility of this method to characterize haptoglobin isolated from lung cancer samples is discussed in this chapter.
Defining glycoprotein biomarkers by enrichment techniques for different types of cancer
Lung cancer
Lung cancer is the leading cause of cancer deaths in the USA with an overall 5-year survival rate of 17% [7]. In 2013, 159,480 new cases and 228,190 deaths related to this disease were reported. Poor survival rate is highly associated with the late stages of this disease, although early treatment relatively improves patient survival. Therefore, the development of early stages diagnosis or prognosis is utterly needed.
Using multilectin affinity enrichment, the different N- and O-linked serum glycoproteins were monitored between healthy and lung cancer patients by Heo et al. [8]. To enrich a wide range of glycoproteins, several lectin materials were used in this study, including ConA, GNA and LCH for Man binding, MAL, SNA and WGA for sialic acid and GlcNAc bindings and AIL and PNA for Gal or GalNAc bindings (see Table 1 for the full names of the different lectin materials). According to this study, many identified glycoproteins exhibited an elevated level in lung cancer patients, such as haptoglobin, inter-α-trypsin inhibitor heavy chain 3 and 4, leucine-rich α-2-glycoprotiein, plasma kallikrein precursor and α-1-antitrypsin. The results of plasma kallikrein and inter-α-trypsin inhibitor heavy chain 3 were validated by western blot. Interestingly, none of the glycoproteins observed in this research exhibited downregulation.
On the other hand, Hongsachart et al. [9] focused on the different expression of only WGA bound glycoproteins in the blood serum of lung cancer patients. They initially removed the highly abundant haptoglobin glycoprotein to effectively enrich low-abundant glycoproteins. Out of 39 differentially expressed glycoproteins, 27 upregulated glycoproteins and 12 down-regulated glycoproteins were observed in lung cancer patients relative to matching control. They suggested that three upregulated glycoproteins (adiponectin, ceruloplasmin and glycosylphosphatidyl-inositol-80) and two downregulated glycoproteins (cyclin H and Fyn) could be potentially used as lung cancer biomarkers. These were validated by western blot analysis. Interestingly, α-1-antitrypsin has been reported in both studies by Heo et al. [8] and Hongsachart et al. [9], representing upregulation in lung cancer patients by 1.8- and 1.25-times, respectively. This glycoprotein is a member of serine protease inhibitor (serpin) superfamily proteins. The upregulation of this glycoprotein has been reported in many cancers since it is an acute phase glycoprotein, highly involved in inflammation, cancer and infection.
Hydrazide chemistry-based enrichment was also applied to define lung cancer glycoprotein biomarkers. As mentioned above, this method was recently used to define the occupied glycosylation sites but not the microheterogeneities of these sites. Zeng et al. [10] have reported that 38 deglycosylated peptides from 22 different glycoproteins were observed. These peptides exhibited statistically significant differences among lung cancer and disease free samples. In adenocarcinoma and squamous cell carcinoma, a peptide derived from isoform γ-B of fibrinogen γ-chain was observed as the most upregulated, while peptides derived from serotransferrin detected as the most downregulated. Upregulation of inter-α-trypsin inhibitor heavy chain 4 and leucine-rich α-2-glycoprotiein was reported using either lectin enrichment or hydrazide chemistry-based enrichment.
Li et al. [11] employed hydrazide chemistry-based enrichment to study glycoproteins of different types of lung cancer tissues collected from bronchoalveolar lavage (BAL) fluids. They monitored eight glycoproteins showing elevation higher than twofold in cancer BAL compared with benign BAL, such as neutrophil elastase, integrin α-M, cullin-4B, napsin A, lysosome-associated membrane protein 2, cathepsin D, bactericidal/permeability-increasing fold containing family B member 2 and neutrophil gelatinase-associated lipocalin. Using an ELISA assay, napsin A was proposed as a good potential glycoprotein biomarker with the sensitivity of 84.21% and the specificity of 66.67%. As compared with the study by Zeng et al. [10], seven glycoproteins, including cathepsin D, complement factor I, kininogen-1, kallistatin, leucine-rich α-2-glycoprotiein, multimerin-1 and plasma protease C1 inhibitor, were also seen as lung cancer-related glycoproteins. This suggests that some of tumor-specific glycoproteins could be detected not only in lung parenchyma or fluids, but also in blood serum.
Haptoglobin is considered as a distinct lung cancer-related glycoprotein. It is an acute phase glycoprotein, which is produced and secreted by the liver. A substantial elevation of core-fucosylated glycopeptides was seen in lung cancer. This was supported by Tsai et al. [12] and Wang et al. [13] using different MS methods. Tsai et al. [12] employed MS methods for permethylated glycans released from purified haptoglobin. On the other hand, Wang et al. [13] have applied label-free MS/MS glycoproteomic methods. Glycopeptides were characterized by CID-ETD MS/MS. Both studies suggested that highly branched core-fucosylated glycan structures could be useful biomarkers for lung cancer. These results were confirmed by two different laboratories employing different methods, thus suggesting the validity of employing glycoproteins as effective cancer biomarkers.
Breast cancer
Breast cancer is the prevalent cancer among women accounting for 29% (232,340) of all new cancer cases among women in the USA [7]. Invasive breast cancer is likely to metastasis to other organs in the body. The majority of deaths from breast cancer are due to metastasis of breast cancer. Seventy percent of breast cancer cases metastasizes to bones. Bone metastasis can be successfully managed. Brain metastasis accounts for 10% of breast cancer metastasis. However, many of therapies for brain metastasis fail to penetrate the blood–brain barrier, causing tumor reoccurrence in the CNS. Therefore, a deeper understanding of proteins/glycoproteins at the earliest stage of breast cancer is expected to predict disease development and progression as well as to facilitate development of effective therapies.
Yang et al. [14] employed lectin enrichment technique to evaluate changes in glycoproteins in blood serum of patients diagnosed with ductal carcinoma in situ (DCIS) and invasive breast cancer (IBC). Multilectin affinity enrichment was applied using ConA, AIL and WGA to capture Man, Gal and GlcNAc/sialic acid residues, respectively. As a result, apolipoprotein C-III, ceruloplasmin, prothrombin, α-1-antichymotrypsin and pregnancy zone protein were detected with upregulation in DCIS compared with disease free subjects. They reported that fibrinogen β-chain and neuropilin-1 were observed with downregulation in DCIS. The comparable trends were detected in IBC, especially with a notable increase for pregnancy zone protein. Fibrinogen β-chain and neuropilin-1 were not detected in IBC. According to the ontology-based associations, lipid transport and metabolism, cell ion homeostatis and protease inhibitors were increased. The increase in lipid transport and specific metabolism is related to tumor growth and metastasis, while the increase in protease inhibitors is associated with the protection of tumor from invading leukocytes.
For better understanding of breast cancer metastasis, membrane glycoproteins are a promising target. Wang et al. [15] employed lectin enrichment to identify differentially expressed glycoproteins from two breast cell lines: a normal premalignant phenotype (MCF10AT1) and a malignant metastatic phenotype (MCF10CA1a). Sixteen glycoproteins were upregulated, while 11 membrane glycoproteins were downregulated in MCF10CA1a cell line relative to MCF10AT1. Most significant changes were observed in the case of γ-Glutamyl hydrolase (25-times upregulation), CD44 (2.5-times upregulation), galectin-3-binding protein (only detected in MCF10CA1a) and syndecan-1 (18.5 downregulation). Syndecan-1 is a four-member cell membrane proteoglycan that consists of the major heparan sulfate on the surface. It acts as coreceptor, especially for G protein-coupled receptor. It was previously reported that the loss of syndecan-1 occurs in association with aggressive phenotypes of cancers. In a similar way, the decreased expression of syndecan-1 was seen involving metastatic MCF10CA1a cell line.
Whelan et al. [16] reported the study of N-linked membrane glycoproteomics from three breast cancer cell lines using hydrazide chemistry-based enrichment. The cell lines were MCF-7, hormone receptor positive and HER2 negative; MDA-MB-453, hormone receptor negative and HER2 positive; and MDA-MB-468, hormone receptor (estrogen receptor and progesterone receptor) and HER 2 negative (triple negative). Twenty seven N-linked glycosylation sites from 25 glycoproteins were detected with only three glycosylation sites common in three cell lines: galectin-3 binding protein, oxygen-regulated protein and lysosome-associated membrane protein 1. Eight glycosylation sites were only observed in MCF-7, such as breast cancer 1, early onset isoform 1 (BRCA1) and thrombospondin 1. BRCA1 is a well known breast cancer biomarker suppressing uncontrolled tumor cell proliferation, while thrombospondin-1 is an angiogenesis inhibitor protein involving vascularization in breast cancer. On the other hand, ten glycosylation sites from eight glycoproteins were only detected in MDA-MB-453 cell lines, such as clusterin and prosaposin, while five glycosylation sites from four glycoproteins were observed in MDA-MB-468, such as CD44 antigen and human EGFR. Interestingly, the glycosylation site from EGFR was characterized which was not detectable in crude membrane fractions. This glycosylation site locates in the domain III of EGFR, in which contributes to ligand activation of the receptor. Thus, the conformation of EGFR can change due to the glycosylation, influencing the receptor activity [16].
Prostate cancer
Prostate cancer is the most common cancer among men in the USA [7]. Prostate-specific antigen is a US FDA approved diagnostic biomarker of prostate cancer. However, it is not a sensitive test since it cannot clearly differentiate between benign prostate hypertrophy and prostate cancer.
Chen et al. [17] reported new potential glycoprotein biomarkers associated with aggressive prostate cancer using hydrazide chemistry-based enrichment. Upregulation of cartilage oligomeric matrix protein and periostin while downregulation of membrane primary amine oxidase (VAP-1) was observed in aggressive prostate cancer. The performance of these potential biomarkers was validated by ELISA assays. Considering that VAP-1 regulates cell growth and differentiation, the decreased expression of this glycoprotein may be cytotoxic for cancer cells as killer cells. Also, VAP-1 is known to participate in the mediation of binding tumor-infiltrating lymphocytes to carcinomas. This suggested that the decreased expression of VAP-1 was associated with the tumor aggressiveness.
Yang et al. [18] have reported the successful isolation of cell-surface sialylated glycoproteins in prostate cancer cell lines (PC3-N2 and PC3-ML2) using click chemistry-based enrichment. N2 cell line is nonmetastatic, while ML2 is metastatic. Thirty six different cell-surface glycoproteins were observed in nonmetastatic N2 cell line. Eight of glycoproteins were involved in regulation of apoptosis pathway and cell adhesion. On the other hand, out of 44 different cell-surface glycoproteins in metastatic ML2 cell line, 11 glycoproteins were associated with cell movement, migration and cell invasion. Interestingly, six glycoproteins are involved in cell invasion, including basigin, peristin, glucose-6-phosphate isomerase, calreticulin, leucin-rich repeat-containing protein 15 and myristoylated alanine-rich protein kinase C substrate. Respective of nonmetastatic and metastatic phenotypes, those abovementioned cell-surface glycoproteins could be used as biomarkers.
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the fifth most common cancer in the world [19]. Currently, α-fetoprotein is widely used as a biomarker for HCC. However, the sensitivity and the specificity of α-fetoprotein are not highly favorable. Also, HCC metastasis is not successfully treated by therapies. Hence, it becomes crucial to develop reliable biomarkers for HCC diagnosis and metastasis prediction. The recent study by Qin et al. [20] suggested the possible use of new glycoprotein biomarkers, including α-1-antitrypsin, apolipoprotein A-I and fibrotectin 1 isoform 2. Those glycoproteins were enriched using WGA lectin. The expression of α-1-antitrypsin was elevated by 4.8-times in metastatic HCC, while that of apolipoprotein A-I and fibronectin 1 isoform 2 was decreased by 0.38- and 0.18-times, respectively.
On the other hand, hydrazide chemistry-based enrichment suggested the different glycoprotein biomarkers. Ishihara et al. [21] have reported eight differentially expressed glycoproteins as potential biomarkers for HCC. The elevated glycoproteins were afamin, ceruloplasmin, α-1-antichymotrypsin and multimerin-1, while the decreased glycoproteins were prothrombin, attractin, apolipoprotein B-100 and complement factor H. The abundances of α-1-antichymotrypsin showed elevated levels between disease free subjects and different stages of HCC. The abundance of this protein was almost four-times higher in stage IV relative to the matching control.
Chen et al. [22] demonstrated the usefulness of hydrazide chemistry-based enrichment in studying aberration of glycoproteins in cancer. In total, 38 glycoproteins were observed with substantial differentiation among HCC and disease-free subjects. Using ELISA assay, galectin-3 binding protein, insulin-like growth factor binding protein 3 (IGFBP-3) and thrombospondin-1 were further validated with p < 0.0085. The abundance of galectin-3 binding protein was increased in HCC by almost 1.31-times. On the other hand, the levels of IGFBP-3 and thrombos-pondin-1 were substantially decreased by 0.258- and 0.67-times, respectively.
Li et al. [23] have assessed N-glycosylations changes associated with two HCC cell lines with low- (MHCC97L) and high- (HCCLM3) metastatic potential. Two enrichment techniques were applied, namely hydrazide chemistry and HILIC. Prior to biomarker evaluation, they combined the results since the identified N-glycosylations sites by the two enrichment techniques were complementary. Out of 1213 unique N-glycosylation sites from 611 glycoproteins, 22 N-glycosylation sites from 20 glycoproteins were determined to be significantly altered. Fibronectin 1 (FN1), α-fetoprotein, zinc-binding α-2-glycoprotein 1, selenoprotein P, hepatocyte growth factor-like protein, dick-kopf-related protein 1 and kinnogen1 are glycoproteins localized in the extracellular space and are implicated in cell invasion or migration properties. They all exhibited upregulations in HCCLM3 cell lines.
It is not surprising that many of the identified glycoproteins from the plasma are also observed in the HCC cell lines, since the plasma proteins are secreted by the liver including ceruloplasmin, vitro-nectin, IGFBP, complement C proteins, hepatocyte growth factor-like protein, etc. However, several glycoproteins were observed with different abundances between the plasma and the cell lines. Also, different glycoproteins were determined to be statistically significant. Qin et al. [20] also investigated nonmetastatic and metastatic HCC, but the results were dissimilar. For example, they reported that FN1 isoform-2 was downregulated 0.18-times in metastatic HCC serum samples compared with nonmetastatic HCC serum samples. However, Li et al. [23] reported that FN1 is upregulated by 2.004–64.92-times (or only detected) in HCCLM3 cell line which has high-metastatic potential. Considering that FN1 isoform-2 is a truncated FN1 protein with matching 97% sequence, the alteration of the abundance between the cell lines and the plasma is surprising.
Esophageal adenocarcinoma
Esophageal adenocarcinoma (EAC) is one of the subtypes of esophageal cancer, the other being esophageal squamous cell carcinoma (ESCC). The incidence of EAC is increasing at a rate that surpasses that of other cancers over the past 25 years [19]. In the many cases, EAC is diagnosed at a late stage, thus leading to a 5-year survival rate of less than 20%. Moreover, 5-year survival drops to less than 4% with distant metastasis to other organs, which is the third lowest rate after liver/intrahepatic bile duct and pancreatic cancers. Like other cancers, EAC is believed to develop in a stepwise manner, and it is thought that its precursor lesion is high-grade dysplasia of metaplastic esophageal epithelium.
Assessing the glycopeptides/glycoprotein biomarkers associated with EAC and other esophageal diseases including high-grade dysplasia and Barrett disease was recently discussed in two publications [24,25]. Song et al. [24] used two enrichment techniques, LAC and hydrazide chemistry (HC), in conjunction with two tandem MS approaches, namely label-free LC-ESI-MS/MS and multiple reaction monitoring (MRM) LC-MS/MS, to assess glycopeptides/glycoproteins changes associated with esophagus diseases. MRM is widely used to quantify peptides or glycopeptides [26]. To improve the detection of glycopeptides, oxonium ions of glycopeptides are used as transitions, including m/z values at 138 (HexNAc–2H2O–CH2O), 274 (NeuAc–H2O), 366 (HexNAc+Hex) and 657 (HexNAc+Hex+NeuAc) as previously described [26]. Evaluations of glycopeptides/glycoprotein biomarkers using two enrichment techniques were deemed complementary. Spectral counting quantitation from LC-ESI-MS/MS analysis suggested that seven LAC-enriched glycoproteins and 11 HC-enriched glycoproteins are significantly different among disease free and disease groups, including GUGU-β form, ceruloplasmin, complement C3 or leucine rich α-2-glycoprotein. MRM quantitation suggested that 13 LAC-enriched glycopeptides and 10 HC-enriched glycosylation sites are potential biomarkers distinguishing disease free and disease groups. For example, by LAC enrichment, two fucosylated and sialylated glycopeptides attached to N453 site of hemopexin were significantly upregulated in EAC cohort. Hemopexin is an acute phase glycoprotein, which is induced after inflammation. It is mainly expressed/secreted by the liver. Fucosylated form of this glycoprotein has been shown to be significantly abundant in hepatocellular carcinoma [27]. By HC enrichment, the abundance of both glycosylation sites at N138 and N397 of ceruloplasmin showed a decrease in disease groups and these changes were statistically validated. However, this is not true if the microheterogeneity evolves. This is well described in the publication by Anoop et al. [25]. They developed a novel statistical method to efficiently quantify glycopeptides regarding the changes of site-specific glycosylation and overall protein glycosylation. They also used computational tools to identify glycopeptides, namely GlycoMapSera and GlycoFragWork [28]. Different glycoforms associated with ceruloplasmin showed the different abundances among disease free and disease groups. The abundance of disialylated biantennary glycopeptide attached to N138 glycosylation site decreases, while that of monofucosylated trisialylated triantennary glycopeptide on the same glycosylation site increases as the disease develops from Barrett’s disease to esophageal adenocarcinoma. The complex microheterogeneity thus requires in-depth investigations.
Conclusion & future perspective
Most of the FDA approved biomarkers for different types of cancer are glycoproteins, including prostate-specific antigen for prostate cancer, α-fetoprotein for liver cancer and MUC-1 for breast cancer. Moreover, most blood serum proteins (>75%) possess N-glycosylation motifs, thus suggesting that this many blood serum proteins are glycosylated. Glycoproteins are low-abundant proteins; therefore, effective analysis of glycoproteins requires the use of an enrichment method.
Glycoprotein enrichment can be attained through LAC, immunoaffinity, hydrazide chemistry, click chemistry and HILIC. Glycosylation site microheterogeneity information is lost through hydrazide chemistry-based enrichment. HILIC enrichment is achieved on glycopeptides, permitting efficient removal of peptides. More confident assignments of glycopeptides are thus effectuated. Assessing the changes associated with several glycoproteins in different types of cancer, including breast, prostate, lung, liver and esophagus, have been achieved using any of the abovementioned enrichment techniques.
Evaluation of potential biomarkers in glycoproteomics are attained regarding the levels of entire glycoproteins, an occupancy of site glycosylations, or an extent of glycans linked to different glycosylation sites. Validation of the changes associated with glycoproteins being demonstrated through enrichment techniques for cancer samples has not been conducted in clinical studies encompassing hundreds of samples. Accordingly, the development of glycoprotein enrichment methods capable of high throughput is what needed next for clinical validation of cancer biomarkers.
Executive summary.
Most of the US FDA approved biomarkers for different types of cancer are glycoproteins, including prostate-specific antigen for prostate cancer, α-fetoprotein for liver cancer and MUC-1 for breast cancer.
Most blood serum proteins (>75%) possess N-glycosylation motifs, thus suggesting that this many blood serum proteins are glycosylated.
Most glycoproteins are low-abundant proteins; therefore, effective analysis of glycoproteins requires the use of an enrichment method.
Glycoprotein enrichment can be attained through lectin affinity, immunoaffinity, hydrazide chemistry, click chemistry and hydrophilic interaction liquid chromatographic enrichment.
Glycosylation site microheterogeneity information is lost through hydrazide chemistry-based enrichment.
Hydrophilic interaction liquid chromatographic enrichment is achieved on glycopeptides, permitting efficient removal of peptides. More confident assignments of glycopeptides are thus effectuated.
Assessing the changes associated with several glycoproteins in different types of cancer, including breast, prostate, lung, liver and esophagus, has been achieved using any of the abovementioned enrichment techniques.
Evaluation of potential biomarkers in glycoproteomics are attained regarding the levels of entire glycoproteins, an occupancy of site glycosylations, or an extent of glycans linked to different glycosylation sites.
Validation of the changes associated with glycoproteins demonstrated through enrichment techniques for cancer samples has not been conducted in clinical studies encompassing hundreds of samples.
Footnotes
Financial & competing interests disclosure
This work is partially supported by a grant from the NIH (1R01GM112490-01) and the Cancer Prevention and Research Institute of Texas (CPRIT). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mechref Y, Hu Y, Garcia A, Hussein A. Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis. 2012;33(12):1755–1767. doi: 10.1002/elps.201100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW, Copeland RJ. Glycomics hits the big time. Cell. 2010;143(5):672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madera M, Mann B, Mechref Y, Novotny MV. Efficacy of glycoprotein enrichment by microscale lectin affinity chromatography. J Sep Sci. 2008;31(14):2722–2732. doi: 10.1002/jssc.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vector Laboratories. www.vectorlabs.com.
- 6.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 7.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 8.Heo SH, Lee SJ, Ryoo HM, Park JY, Cho JY. Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics. 2007;7(23):4292–4302. doi: 10.1002/pmic.200700433. [DOI] [PubMed] [Google Scholar]
- 9.Hongsachart P, Huang-Liu R, Sinchaikul S, et al. Glycoproteomic analysis of WGA-bound glycoprotein biomarkers in sera from patients with lung adenocarcinoma. Electrophoresis. 2009;30(7):1206–1220. doi: 10.1002/elps.200800405. [DOI] [PubMed] [Google Scholar]
- 10.Zeng X, Hood BL, Sun M, et al. Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J Proteome Res. 2010;9(12):6440–6449. doi: 10.1021/pr100696n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li QK, Shah P, Li Y, et al. Glycoproteomic analysis of bronchoalveolar lavage (BAL) fluid identifies tumor-associated glycoproteins from lung adenocarcinoma. J Proteome Res. 2013;12(8):3689–3696. doi: 10.1021/pr400274w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai HY, Boonyapranai K, Sriyam S, et al. Glycoproteomics analysis to identify a glycoform on haptoglobin associated with lung cancer. Proteomics. 2011;11(11):2162–2170. doi: 10.1002/pmic.201000319. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Hincapie M, Rejtar T, Karger BL. Ultrasensitive characterization of site-specific glycosylation of affinity-purified haptoglobin from lung cancer patient plasma using 10 mum i. d porous layer open tubular liquid chromatography-linear ion trap collision-induced dissociation/electron transfer dissociation mass spectrometry. Anal Chem. 2011;83(6):2029–2037. doi: 10.1021/ac102825g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52(10):1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Ao X, Vuong H, et al. Membrane glycoproteins associated with breast tumor cell progression identified by a lectin affinity approach. J Proteome Res. 2008;7(10):4313–4325. doi: 10.1021/pr8002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelan SA, Lu M, He J, et al. Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J Proteome Res. 2009;8(8):4151–4160. doi: 10.1021/pr900322g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Xi J, Tian Y, Bova GS, Zhang H. Identification, prioritization, and evaluation of glycoproteins for aggressive prostate cancer using quantitative glycoproteomics and antibody-based assays on tissue specimens. Proteomics. 2013;13(15):2268–2277. doi: 10.1002/pmic.201200541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Nyalwidhe JO, Guo S, Drake RR, Semmes OJ. Targeted identification of metastasis-associated cell-surface sialoglycoproteins in prostate cancer. Mol Cell Proteomics. 2011;10(6) doi: 10.1074/mcp.M110.007294. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 20.Qin X, Chen Q, Sun C, et al. High-throughput screening of tumor metastatic-related differential glycoprotein in hepatocellular carcinoma by iTRAQ combines lectin-related techniques. Med Oncol. 2013;30(1):420. doi: 10.1007/s12032-012-0420-8. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara T, Fukuda I, Morita A, et al. Development of quantitative plasma N-glycoproteomics using label-free 2-D LC-MALDI MS and its applicability for biomarker discovery in hepatocellular carcinoma. J Proteomics. 2011;74(10):2159–2168. doi: 10.1016/j.jprot.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Chen R, Tan Y, Wang M, et al. Development of glycoprotein capture-based label-free method for the high-throughput screening of differential glycoproteins in hepatocellular carcinoma. Mol Cell Proteomics. 2011;10(7) doi: 10.1074/mcp.M110.006445. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Jiang J, Zhao X, et al. N-glycoproteome analysis of the secretome of human metastatic hepatocellular carcinoma cell lines combining hydrazide chemistry, HILIC enrichment and mass spectrometry. PLoS ONE. 2013;8(12):e81921. doi: 10.1371/journal.pone.0081921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song E, Zhu R, Hammoud ZT, Mechref Y. LC–MS/MS quantitation of esophagus disease blood serum glycoproteins by enrichment with hydrazide chemistry and lectin affinity chromatography. J Proteome Res. 2014;13(11):4808–4820. doi: 10.1021/pr500570m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayampurath A, Song E, Mathur A, et al. Label-free glycopeptide quantification for biomarker discovery in human sera. J Proteome Res. 2014;13(11):4821–4832. doi: 10.1021/pr500242m. [DOI] [PubMed] [Google Scholar]
- 26.Song E, Pyreddy S, Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2012;26(17):1941–1954. doi: 10.1002/rcm.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debruyne EN, Vanderschaeghe D, Van Vlierberghe H, Vanhecke A, Callewaert N, Delanghe JR. Diagnostic value of the hemopexin N-glycan profile in hepatocellular carcinoma patients. Clin Chem. 2010;56(5):823–831. doi: 10.1373/clinchem.2009.139295. [DOI] [PubMed] [Google Scholar]
- 28.Mayampurath A, Yu CY, Song E, Balan J, Mechref Y, Tang H. Computational framework for identification of intact glycopeptides in complex samples. Anal Chem. 2014;86(1):453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]