SUMMARY
Cereblon (CRBN), a substrate receptor for the cullin–RING ubiquitin ligase 4 (CRL4) complex, is a direct protein target for thalidomide teratogenicity and antitumor activity of immunomodulatory drugs (IMiDs). Here we report that glutamine synthetase (GS) is an endogenous substrate of CRL4CRBN. Upon exposing cells to high glutamine concentration, GS is acetylated at lysines 11 and 14, yielding a degron that is necessary and sufficient for binding and ubiquitylation by CRL4CRBN and degradation by the proteasome. Binding of acetylated degron peptides to CRBN depends on an intact thalidomide-binding pocket but is not competitive with IMiDs. These findings reveal a feedback loop involving CRL4CRBN that adjusts GS protein levels in response to glutamine and uncover a new function for lysine acetylation.
Graphical abstract
eTOC blurb
Nguyen et al. demonstrate that glutamine induces acetylation of GS at lysines 11 and 14 to create an acetylated degron that binds CRL4CRBN, resulting in ubiquitylation and degradation of GS.
Introduction
Cereblon (CRBN) is a putative substrate receptor for a cullin–RING ubiquitin ligase 4 (CRL4) complex (Angers et al., 2006). Human CRBN was discovered as a gene that when mutated results in mild mental retardation (Higgins et al., 2004). Subsequent analysis of Crbn−/− knockout mice revealed that they are less prone to developing insulin resistance, fatty liver and visceral fat accumulation when fed a high-fat diet (Lee et al., 2013). To date, only four putative targets of CRBN have been identified: large conductance Ca2+-activated K+ channels (Jo et al., 2005), voltage-gated chloride channel (Hohberger and Enz, 2009), the developmental regulator MEIS2 (Fischer et al., 2014), and adenosine monophosphate-activated protein kinase (AMPK) (Lee et al., 2011b; Lee et al., 2014; Lee et al., 2013). AMPK activity is elevated in Crbn−/− mice, which could contribute to the resistance of these animals to developing metabolic syndrome on a high-fat diet. However, the mechanism by which CRBN recognizes AMPK or its other natural substrates remains unknown.
CRBN has achieved notoriety as the target that accounts for the teratogenic effects of thalidomide (Ito et al., 2010), which caused over 10,000 birth defects in humans before it was withdrawn from the market in 1962 (Lenz et al., 1962; Mcbride, 1961). Subsequently, CRBN was implicated as the target that accounts for the therapeutic activity of thalidomide and the related ‘immunomodulatory’ (IMiD) compounds lenalidomide and pomalidomide in multiple myeloma (Lopez-Girona et al., 2012; Zhu et al., 2011). CRBN must be present for myeloma cells to respond to IMiDs, suggesting that formation of an IMiD–CRBN complex underlies the therapeutic response. Recent studies have revealed that binding of IMiDs to CRBN promotes recruitment of neosubstrates, including Ikaros (IKZF1), Aiolos (IKZF3) (Kronke et al., 2014; Lu et al., 2014) and casein kinase 1A1 (CK1α) (Kronke et al., 2015) to CRL4CRBN, leading to their increased ubiquitylation and proteasome-dependent degradation. In contrast, IMiDs block endogenous CRBN substrate MEIS2 from binding to CRL4CRBN, resulting in inhibition of its ubiquitylation and degradation (Fischer et al., 2014). Because of the paucity of known CRL4CRBN ubiquitylation substrates, it remains unclear whether the therapeutic action of IMiDs might be modulated by general stabilization of CRBN’s natural substrates.
Glutamine synthetase (GS) plays a central role in metabolism, as glutamine is the key metabolite connecting carbon and nitrogen metabolism through the citric acid cycle. GS has different functions in different tissues. For example in liver it detoxifies ammonia, in brain it protects neurons against excitotoxicity by converting glutamate into glutamine, and in kidney it contributes to pH regulation (Taylor and Curthoys, 2004). Moreover, glutamine has a critical role in regulating mTOR signaling, translation, and autophagy to coordinate cell growth and proliferation (Nicklin et al., 2009). Mutations and deregulation of GS have been linked to human diseases, including congenital glutamine deficiency, Alzheimer’s disease, and cancers (Bott et al., 2015; Christa et al., 1994; Gunnersen and Haley, 1992; Haberle et al., 2005; Kung et al., 2011; Tardito et al., 2015). In keeping with its central role in carbon and nitrogen metabolism, prokaryotic GS is allosterically regulated by multiple end products of glutamine metabolism working in concert with cycles of reversible adenylylation/deadenylylation (Krajewski et al., 2008; Stadtman, 2001). Eukaryotic GS, by contrast, is not adenylylated and its regulation is poorly understood. It has been shown that GS is subject to feedback control by glutamine, which promotes its post-translational modification and degradation (Arad et al., 1976; Crook and Tomkins, 1978). However, the molecular basis for this regulation remains elusive.
Results
GS is an endogenous substrate of CRL4CRBN
To search for candidate CRBN substrates, including those that might be modulated by IMiDs, we used stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative mass spectrometry (Lee et al., 2011a) (Figure S1A). Comparison of heavy:light ratios of peptides indicated that CRBN, subunits of CRL4 (CUL4, DDB1, RBX1), and subunits of the CRL regulator CSN were recovered in equal amounts from DMSO or thalidomide-treated 293T cells stably expressing CRBN tagged with a Flag epitope at its N-terminus (FlagCRBN; all tagged proteins are indicated by a superscripted tag either before or after the name to indicate tagging at the N- or C-terminus) (Figure 1A). A number of putative substrates behaved like MEIS2 (Fischer et al., 2014) in that they were recovered in lesser amounts from cells treated with thalidomide (Table S1). Notably, glutamine synthetase (GS) behaved like Ikaros, Aiolos, and CK1α in that it was recovered in greater amounts (Figure 1A). Essentially identical results were obtained in a label-swap experiment (data not shown). To validate the mass spectrometry data, we immunoprecipitated FlagCRBN and immunoblotted for CRL4CRBN subunits and GS. Recovery of GS but not CRL subunits was stimulated by thalidomide (Figure S1B) and its analog lenalidomide (Len)(Figure 1B). A similar result was obtained when we evaluated the interaction of endogenous GS with endogenous CRBN (Figure 1C). An important distinction between GS and CRBN’s neosubstrates including IKZF1, IKZF3 and CK1α (Kronke et al., 2015; Kronke et al., 2014; Lu et al., 2014), is that we observed significant GS association with CRBN in the absence of IMiDs (Figures 1B, 1C, and S1B). Thus, we did not pursue further the relationship between GS and IMiDs.
Figure 1. GS is an endogenous substrate of CRL4CRBN.
(A) Identification of GS as a CRBN-interacting protein. 293T cells stably expressing FlagCRBN and grown in either ‘heavy’ or ‘light’ SILAC medium were treated with DMSO (light) or 50 µM thalidomide (heavy) for 4 h prior to lysis and immunoprecipitation (IP) with anti-Flag followed by mass spectrometry. The heavy:light ratios for GS and subunits of CRL4 and CSN are shown. The asterisk indicates a ratio that differs significantly from 1 (p-value 1×10−19). The data are an average of two experiments. Error bars indicate ± SD.
(B) GS binds CRBN. 293T cells stably expressing empty vector or wild-type FlagCRBN were treated with or without lenalidomide (10 µM) for 3 h. Protein extracts were immunoprecipitated with Flag antibody followed by Western blot analysis with the indicated antibodies. The ratio of GS bound to CRBN normalized to input GS is shown.
(C) Endogenous CRBN and GS interact. MM.1S cells were supplemented with DMSO or 1 µM lenalidomide 2 h prior to lysis and IP with mouse IgG control or CRBN antibodies. IP and input samples were fractionated by SDS-PAGE and immunoblotted with the indicated antibodies. Quantification was as described in (B).
(D–E) CRBN promotes GS ubiquitylation in cells (D) and in vitro (E). (D) 293T cells were transiently transfected with plasmids expressing GSFlag and HAubiquitin (HAUb). After 30h, cells were treated with 10 µM MG132 for 4 h, followed by cell lysis and Flag IP under denaturing conditions. The input and bound fractions were evaluated by immunoblotting with HA and Flag antibodies. Ubiquitin conjugates in the input are shown in Figure S1C. (E) 293T cells stably expressing FlagCRBN were treated with proteasome inhibitor (1 µM bortezomib) for 6 h. After IP with Flag antibody, in vitro ubiquitylation of endogenous, co-precipitated GS was carried out for 1 h at 30°C in the presence or absence of E1+E2 and HAUb. Where indicated, methylated ubiquitin (Me-Ub) or recombinant (r) CUL4A-RBX1 was added. Reactions were analyzed by SDS-PAGE and immunoblotting with GS antibody. (Ub)n indicates polyubiquitylation. S.E., L.E.: short and long exposures.
The apparent constitutive association of GS with CRBN suggested that GS might be a natural substrate for CRL4CRBN, albeit one that behaves markedly differently from MEIS2. To pursue this further, we sought to test whether ubiquitylation of GS was dependent on CRL4CRBN. In co-transfection assays, we observed incorporation of HAubiquitin into FlagGS (Figure 1D). Significantly, ubiquitin-modified FlagGS accumulated in cells in which the proteasome was inhibited with MG132, but was almost entirely absent upon depletion of endogenous CRBN (depletion was confirmed by immunoblot; Figure S1D). In addition, FlagCRBN promoted the in vitro ubiquitylation of co-precipitated endogenous GS when supplemented with E1, E2, ubiquitin, and ATP (Figure 1E, lane 6). GS polyubiquitylation was markedly enhanced by the addition of recombinant CUL4A-RBX1 purified from insect cells (Figure 1E, lane 3), whereas it was inhibited by addition of methylated ubiquitin. Collectively, these results argue that GS is an endogenous ubiquitylation substrate of CRL4CRBN.
CRL4CRBN directly controls the glutamine-induced degradation of GS
Glutamine regulates GS by altering the rate of degradation of the enzyme (Arad et al., 1976; Crook and Tomkins, 1978). Consistent with these reports, we observed that glutamine downregulated GS protein levels upon addition to glutamine-starved Hep3B cells (Figure 2A) as well as to multiple lung, breast, and glioblastoma cancer cell lines (Figure S1E). This effect was intermediate at the normal serum glutamine concentration (0.5 mM) and was saturated at 2 mM glutamine (Figure S1F), as reported previously (Crook and Tomkins, 1978). The glutamine-induced downregulation of GS in Hep3B cells was blocked by the addition of the proteasome inhibitor bortezomib or the NEDD8-activating enzyme inhibitor MLN4924 (Figure 2B), which inactivates Cullin-RING E3 ubiquitin ligase activity (Soucy et al., 2009). MLN4924 also inhibited glutamine-induced GS degradation in myeloma, breast, and lung cancer cell lines (Figure S2A–C). Most importantly, the glutamine-induced downregulation of GS in Hep3B cells was blunted upon disruption of CRBN loci by CRISPR/Cas9 (Figure 2C) or depletion of CRBN by shRNA knockdown (Figure S2D). Similar results were observed upon shRNA knockdown of CRBN in myeloma and lung cancer cells (Figure S2E & F). Consistent with a role for CRBN in GS degradation, the steady-state level of GS was elevated in CRBN-depleted cells (Figure S2G). For the Hep3B, myeloma, and lung cancer cell lines we confirmed that CRBN-dependent effects on GS downregulation were not due to changes in its mRNA level (Figure S2I–K). Two general trends in the data from different cell types are worth noting. First, glutamine does not induce complete degradation of GS; depending upon the cell line, the reduction ranged from 50–80%. Second, there remains a modest glutamine-stimulated loss of GS in cell lines depleted of CRBN by either shRNA or CRISPR/Cas9. It remains unclear if this is due to residual CRBN or the operation of an unknown secondary pathway. Nevertheless, it is clear that CRBN promotes rapid, glutamine-dependent downregulation of GS by a post-transcriptional mechanism that requires Nedd8 conjugation and proteasome activity.
Figure 2. CRBN is required for glutamine-induced degradation of GS.
(A) Glutamine regulates GS protein abundance. Hep3B cells were maintained in DMEM 10% FCS without glutamine for 48 h. The cells were then treated with glutamine (4 mM) for the indicated times. Equal amounts of protein extracts were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. GAPDH served as a loading control.
(B) Glutamine-induced GS degradation is blocked by the proteasome inhibitor bortezomib or the Nedd8-activating enzyme inhibitor MLN4924. Hep3B cells were starved of glutamine for 36 h, and then pretreated with or without bortezomib (200 nM) or MLN4924 (2 µM) for 30 min, followed by 4 mM glutamine treatment for 7 h. Cell lysates were analyzed by SDS-PAGE and immunoblotting with antibodies against GS, CRBN, and GAPDH. The relative ratio of GS:GAPDH, normalized to lane 1, is shown.
(C) Glutamine-induced GS degradation is promoted by CRBN. Wild-type (WT) and CRISPR/Cas9-derived CRBN-knockout (KO) Hep3B cells were starved of glutamine for 36 h, followed by addition of 4 mM glutamine for 0, 12 and 16 h. The relative ratio of GS:GAPDH protein level, normalized to that at 0-time, is shown. Note that this experiment was done with a pool of KO cells (i.e. non-clonal) and there appears to be a small amount of residual CRBN in the population.
(D–F) GS protein levels are elevated in the kidneys, skeletal muscles and lungs of Crbn−/− mice. Left panels: Tissue extracts prepared from total kidneys and skeletal muscles of wild-type (WT) and Crbn−/− (KO) mice were analyzed by SDS-PAGE and Western blotting, using GS, CRBN and GAPDH antibodies. n = 3–4 mice per group. Right panels: densitometric quantification of relative band intensities. Error bars represent the SEM.
(G) Crbn−/− mice exhibit an increased glutamine/glutamate ratio in serum. Glutamine and glutamate levels in serum of wild-type (WT) and homozygous mutant Crbn−/− mice (KO) were quantified by mass spectrometry. Glutamine/glutamate ratio was calculated and represented as mean ± SD; n = 6 mice per group (P = 0.02615 by t-test).
We next sought additional evidence for a physiological role of CRBN in regulating the abundance of GS in vivo. In mice, GS is highly expressed in the brain, liver, kidney and skeletal muscle (BioGPS.org; table S2). We therefore used immunoblotting to examine GS abundance in brain, liver, kidney, skeletal muscle and lung tissues of wild-type and Crbn−/−mice that were starved for 24 hours and re-fed for 4 hours to mimic glutamine starvation/re-feeding in cultured cells. As shown in Figures 2D–F, GS was elevated in the kidney, skeletal muscle and lung of Crbn−/− mice, but not in the brain and liver (data not shown). Skeletal muscle and lung GS plays a significant role in regulating the concentration of plasma glutamine (Hensley et al., 2013). Consistent with the accumulation of GS in these tissues, Crbn−/− mice exhibited an increased glutamine to glutamate ratio in serum (Figure 2G and table S3). Taken together, our data suggest that endogenous GS protein levels are negatively regulated by glutamine through a feedback loop involving CRL4CRBN.
The N-terminal extension of GS is required for its CRBN-dependent ubiquitylation
To identify the sequence in GS recognized by CRBN, we generated a series of deletion mutants and found that the N-terminal 24–amino acids were required for its constitutive interaction with CRBN (Figures 3A and 3B) and degradation (Figure 3C) in glutamine-supplemented medium. Intriguingly, this N-terminal segment is absent from bacterial GS enzymes but is conserved in GS throughout the chordate lineage (Figure S3). By analogy to other CRL enzyme–substrate interactions, we speculated that GS degradation might be controlled by a glutamine-dependent post-translational modification of its N-terminal segment. To test this idea, we mutated all Ser, Thr, and Tyr residues in the N-terminal 24 amino acids of GS to Ala, either individually or in combination. Notably, all mutants retained some binding to CRBN (Figure S4A), suggesting that phosphorylation does not play a major role. We next investigated whether a KxxK motif (lysines 11 and 14) that is highly conserved in chordates (Figure S3) was required for GS binding to CRBN. We analyzed double arginine substitutions (RR; keeps the positive charges) and double alanine substitutions (AA; neutralizes the positive charges) at K11 and K14 and found that the binding to CRBN, ubiquitylation, and degradation of GS were enhanced by the AA mutations but diminished or unaffected by the RR mutations (Figures 3D–F). To further investigate the N-terminal degron, we generated chimeric fusion proteins consisting of GS amino acids 1–25 fused to Myc-tagged GFP (Figure 4A). The N-terminal region of GS was sufficient to confer binding to and ubiquitylation by CRBN, as well as degradation (Figures 4B–D). However, all of these activities of the N-terminal degron were blocked by the RR substitutions. Together, these findings suggest that modification of lysine(s) in the N-terminal extension of GS was critical for its binding to and ubiquitylation by CRBN.
Figure 3. The N-terminal extension of GS and its KxxK motif promote binding to CRBN, ubiquitylation, and degradation.
(A–B) The N-terminal extension of GS is required to bind CRBN. (A) Schematic diagram of full-length (FL) human GS protein structure and deletion constructs used in (B). The GS degron (amino acids 1–24) recognized by CRBN is highlighted. (B) 293T cells stably expressing FlagCRBN were transfected with the indicated plasmids. After 36 h, cells were treated with 10 µM MG132 for 4 h. Cellular extracts were immunoprecipitated with Flag antibody, fractionated by SDS-PAGE and immunoblotted with Myc and Flag antibodies. *, indicates a non-specific band. A band ∼25 kDa represents IgG light chains (IgG-LC).
(C) The N-terminal extension of GS is required for degradation. 293T cells stably expressing Flag-HA-tagged GS (FHGS) or GS with deletion of the N-terminal 24 amino acids (d1FHGS) were cultured in complete DMEM with 2 mM glutamine, and treated with cycloheximide (CHX; 100 µg/ml) for 0, 2, 4, and 6 h. Cell lysates were analyzed by SDS-PAGE and immunoblotting with Flag and GAPDH antibodies. The relative ratio of GS:GAPDH, normalized to that of zero-time, is shown.
(D) The N-terminal KxxK motif modulates binding of GS to CRBN. 293T cells stably expressing FlagCRBN were transfected with empty vector (EV) or plasmids encoding the indicated GS mutants. After 36 h, cells were treated with 10 µM MG132 for 4 h. Cell extracts were immunoprecipitated with Flag antibody and the precipitated and input fractions were analyzed by SDS-PAGE and immunoblotting with DDB1, Myc, and Flag antibodies. WT: wild type. RR: K11R, K14R. AA: K11A, K14A.
(E) The N-terminal KxxK motif modulates ubiquitylation of GS. 293T cells were transfected with plasmids encoding HAUb and the indicated Myc-tagged GS mutants. After 24 h, the cells were treated with 10 µM MG132 for 4 h, followed by cell lysis, denaturation of the lysate proteins, and IP with anti-Myc. The input lysates and bound fractions were evaluated by SDS-PAGE and immunoblotting with HA and Myc antibodies.
(F) The N-terminal KxxK motif modulates degradation of GS. 293T cells were transfected with plasmids encoding wild type GSMyc or the RR and AA mutants. After 24–30 h, the cells growing in medium containing 2 mM glutamine were treated with 100 µg/ml cycloheximide (CHX). At the indicated times following addition of CHX, cells were harvested, and their content of GS and GAPDH was evaluated by immunoblotting. GSRR: K11R, K14R GS. GSAA: K11A, K14A GS.
Figure 4. The N-terminal extension of GS comprises a sufficient, KxxK-dependent ubiquitylation and degradation signal.
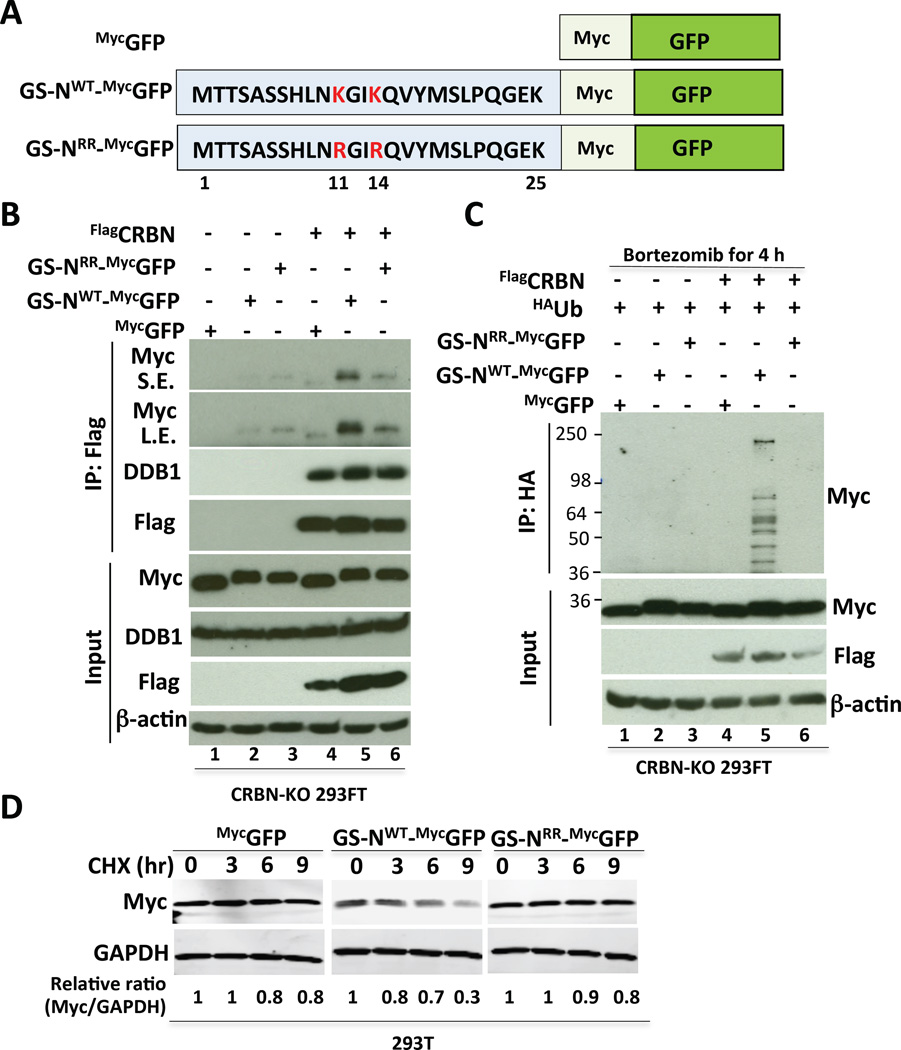
(A) Schematic of GS Degron-GFP fusion proteins. Wild type (GS-NWT) or mutant (GS-NRR) versions of the N-terminal extension (amino acids 1–25) of GS were fused to Myc-tagged GFP. RR refers to the double mutant in which K11 and K14 were changed to R.
(B) The N-terminal extension of GS is sufficient to bind CRBN in a manner that depends on an intact KxxK motif. CRBN-KO 293FT cells stably expressing MycGFP, GS-NWT_MycGFP, or GS-NRR_MycGFP fusion proteins were transfected with empty plasmid (lanes 1–3) or plasmid expressing FlagCRBN (lanes 4–6). After 36 h, cell extracts were immunoprecipitated with Flag antibody, fractionated by SDS-PAGE and immunoblotted with the indicated antibodies.
(C) The N-terminal extension of GS is sufficient to confer CRBN- and KxxK-dependent ubiquitylation. CRBN-KO 293FT cells stably expressing MycGFP, GS-NWT_MycGFP, and GS-NRR_MycGFP fusion proteins were transfected with plasmid expressing HAUb (lanes 1–6) and empty plasmid (lanes 1–3) or plasmid expressing FlagCRBN (lanes 4–6). After 48 h, cells were treated with bortezomib (1 µM) for 4 h prior to lysis and IP with HA antibody. Immunoprecipitates and input samples were fractionated by SDS-PAGE and immunoblotted with the indicated antibodies. The anti-HA blots are in Figure S4B.
(D) The N-terminal region of wild type GS is sufficient to confer degradation. 293T cells, stably expressing MycGFP, GS-NWT_MycGFP, and GS-NRR_MycGFP fusion proteins, grown in 2 mM glutamine were treated with 100 µg/ml cycloheximide (CHX) for the indicated times. Extracts were evaluated by SDS-PAGE and immunoblotting with Myc and GAPDH antibodies. The relative ratio of test protein:GAPDH, normalized to that of 0-time, is shown.
Glutamine-dependent acetylation of lysines 11 and 14 by CBP/p300 regulates GS degradation
Lysine residues can be targeted by multiple modifications, such as ubiquitylation, SUMOylation, methylation, and acetylation. Our observation that K->R substitutions at K11 and K14 in GS disrupted its binding to CRBN, ubiquitylation and degradation, whereas neutralizing K->A substitutions had the opposite effect, implicated a potential role for acetylation in neutralizing positive charges that impede interaction with CRBN. Acetylation was of particular interest, because proteomic studies uncovered modification of K14 on human, mouse and rat GS (Chen et al., 2012; Lundby et al., 2012; Svinkina et al., 2015; Weinert et al., 2013), and more generally revealed that reversible lysine acetylation plays an important role in regulating metabolic enzymes (Wang et al., 2010; Zhao et al., 2010). Moreover, a KxxK motif is known to be a potential target of the closely-related acetyltransferases CBP and p300 (Thompson et al., 2001). Consistent with GS being a potential CBP/p300 substrate, GSMyc co-immunoprecipitated with both p300HA and CBPHA (Figure S4C). Moreover, recombinant p300-HAT domain acetylated recombinant GS6xHis in vitro (Figure S4D). Furthermore, in 293FT CRBN knockout cells (CRBN-KO 293FT) (Lu et al., 2014), GSFlag acetylation was significantly enhanced upon coexpression with p300HA or treatment with histone deacetylase inhibitors (HDACi) (Figure 5A). To map the acetylation sites of GS, which has 19 lysines, GSFlag was immuno-purified from CRBN-KO 293FT cells and analyzed by mass spectrometry analysis. Two lysine residues, K11 and K14, were found to be independently acetylated based on the conclusive mass spectra (Figure S5A and S5B, and Table S4). However, we were unable to detect a tryptic peptide in which both residues were acetylated (see legend of Figure S5 for a detailed discussion). We also obtained strong evidence for acetylation of K25, K189 and K291, and modest evidence for K241 and K268 (Table S4). Given that our mutagenesis studies pointed to a critical role for lysine 11 and/or lysine 14 in mediating GS ubiquitylation and degradation via CRL4CRBN, we evaluated point mutants to determine whether these residues contributed to the signal observed in an anti-acetyllysine immunoblot of immunoprecipitated GS. Wild-type GSMyc, but not the corresponding AA or RR mutants, yielded a robust signal (Figure 5B and Figure S4E). We conclude that GS can be acetylated on K11 and K14 in cells.
Figure 5. p300-mediated acetylation promotes the degradation of GS.
(A) p300 promotes GS acetylation in cells. CRBN-KO 293FT cells were transfected with GSFlag and HA-tagged p300 (p300HA) plasmids. After 36 h, the cells were treated with or without HDAC inhibitors (1 µM TSA and 10 mM NAM) for 12 h. Cell lysates were immunoprecipitated with anti-Flag and precipitated and input fractions were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Ac-Lys refers to antibody that recognizes acetylated lysine.
(B) Lysines 11 and/or 14 are acetylation sites. Lysates from CRBN-KO 293FT cells transfected with plasmids expressing Myc-tagged wild type or RR (K11R/K14R) mutant GS were immunoprecipitated with anti-Myc, eluted with Myc peptide, and then analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. S.E., L.E.: short and long exposures.
(C) Glutamine induces p300-mediated acetylation of GS. CRBN-KO 293FT cells were transiently transfected with plasmids expressing wild type or RR mutant GSFlag. After 24 h, cells were starved of glutamine for 24 h, then pre-treated with or without 10 µM p300/CBP inhibitor C646 in fetal bovine serum-free DMEM medium for 2 h, followed by treatment with 4 mM glutamine for 2 h. The cell lysates were immunoprecipitated with anti-Flag, and then analyzed by SDS-PAGE and immunoblotting (IB) with the indicated antibodies. The relative ratio of acetylated GSFlag to total GSFlag protein (Ac-Lys/Flag ratio), normalized to that of untreated cells, is shown.
(D) HDAC inhibitors enhance glutamine-induced GS degradation. Hep3B cells were starved of glutamine for 24 h, and then supplemented (or not) with 4 mM glutamine for 12 h in the presence or absence of HDAC inhibitors SAHA (1 µM) and NAM (10 mM). Equal amounts of cell extracts were analyzed by SDS-PAGE and immunoblotting with antibodies against GS, CRBN, and GAPDH. The relative ratio of GS:GAPDH, normalized to that of untreated cells, is shown. S.E., L.E.: short and long exposures.
(E) Inhibition of the acetyltransferases p300 and CBP by C646 counteracts HDAC inhibitor-induced GS degradation. 293T cells were starved of glutamine for 24 h and then pretreated (or not) with 10 µM C646 in fetal bovine serum-free medium for 1 h. Afterwards, cells were treated with 4 mM glutamine in the presence or absence of HDAC inhibitors (1 µM TSA and 10 mM NAM) for 4 h. Equal amounts of cell extracts were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. The relative ratio of GS:GAPDH, normalized to that of untreated cells, is shown.
(F) HDAC inhibitor-induced GS degradation requires CRBN. Hep3B cells stably expressing control shRNA or different CRBN shRNAs were starved of glutamine for 48 h. Starved cells were mock-treated or supplemented with 4 mM glutamine and 1 µM TSA plus 10 mM NAM, as indicated, for 7 h. Cell lysates were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. The relative ratio of GS:GAPDH, normalized to that of untreated cells, is shown. S.E., L.E.: short and long exposures.
(G) CRBN interacts with acetylated endogenous GS. Cell extracts were prepared from 293T cells stably expressing FlagCRBN or empty vector. Immunoprecipitation (1st IP) was performed with anti-Flag antibody. One twenty-fifth of the unbound fractions was precipitated with anti-GS antibody (2nd IP; it was previously determined that using 25-fold less material in the 2nd IP would yield an equivalent amount of GS as the 1st IP). The precipitated fractions from 1st IP and 2nd IP were analyzed by SDS-PAGE and immunoblotting (IB) with indicated antibodies.
Covalent modifications that target CRL substrates for ubiquitylation are often tightly regulated. We therefore sought to address if K11 and/or K14 were acetylated in a glutamine and 300/CBP-dependent manner. To address these questions, CRBN-KO 293FT cells expressing wild type or RR-GSFlag and starved for glutamine were re-fed or mock treated, lysates of these cells were immunoprecipitated with anti-Flag, and the precipitates were blotted with acetyllysine antibody. Aetylation of GSFlag was markedly stimulated by glutamine but this effect was largely blocked by the RR mutations and the CBP/p300 inhibitor C646 (Bowers et al., 2010) (Figure 5C). A similar result was obtained for endogenous GS (Figure S4F), but the stimulatory effect of glutamine was less prominent.
If glutamine-stimulated acetylation of K11 and K14 on GS triggers its ubiquitylation and degradation, we reasoned that histone deacetylase inhibitors (HDACi) might enhance glutamine-induced downregulation of endogenous GS. Indeed, this was observed to be the case (Figure 5D; mRNA analysis in Figure S6A). The destabilizing effect of glutamine and HDACi was blunted by simultaneous treatment with C646 or a distinct p300/CBP inhibitor, garcinol (Figures 5E and S6B). HDACi-induced degradation of endogenous GS was mediated by CRBN because it was significantly attenuated by genetic (Figure 5F) or chemical (Figure S6C & D) reduction of CRL4CRBN activity.
The hypothesis that emerged from these studies is that high glutamine triggers acetylation of GS lysines 11 and/or 14, which then mediates binding to CRBN. To test this, we took advantage of our observation that acetylation at K11 and/or K14 made a dominant contribution to binding of the anti-acetyllysine antibody to GS. Lysates of 293T cells that stably expressed FlagCRBN were immunoprecipitated with anti-Flag and anti-GS, and the amount of each immunoprecipitate analyzed by blotting was adjusted such that the amount of GS was similar. As shown in Figure 5G, the GS bound to CRBN was enriched for acetylation compared to the unbound GS. We conclude that CRBN exhibits a strong preference for binding acetylated GS.
The N-terminal extension of GS comprises an acetyllysine degron that binds the C-terminal domain of CRBN
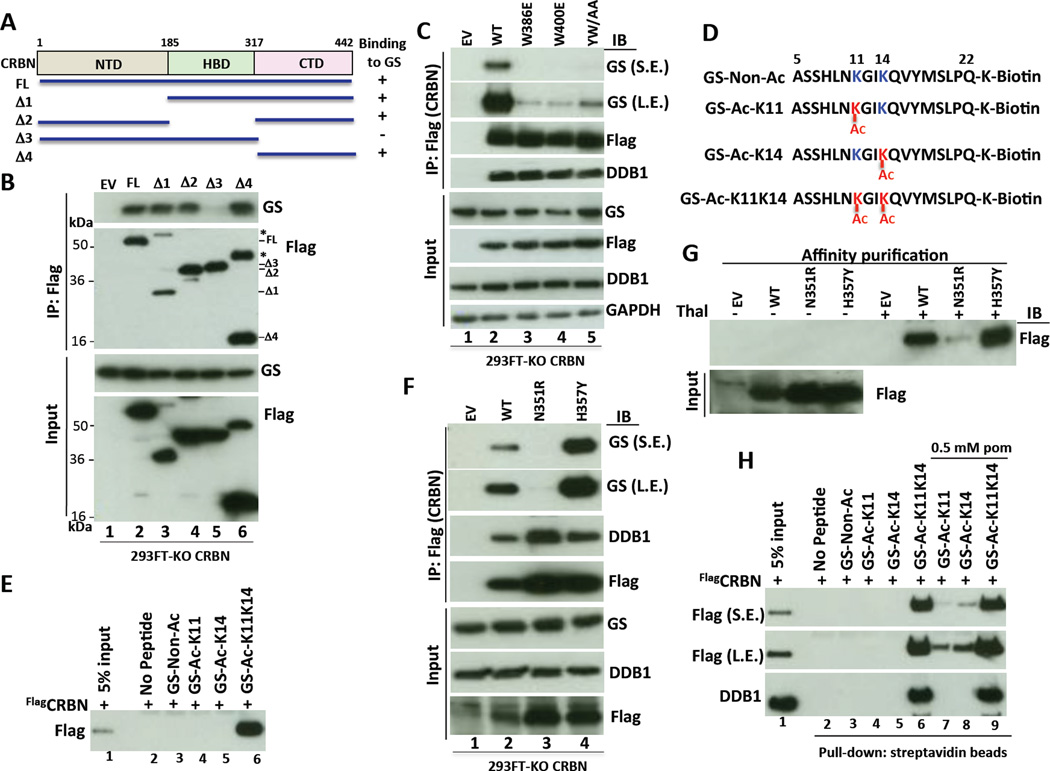
We next mapped the GS-binding region on CRBN by deletion analysis and found that the carboxy-terminal domain (CTD) was necessary and sufficient for binding endogenous GS (Figures 6A and B). The CTD harbors the IMiD binding pocket formed by Trp380, Trp386 and Trp400, with a phenylalanine residue at the base (Phe402) (Chamberlain et al., 2014; Fischer et al., 2014). These residues form a small hydrophobic pocket (tri-Trp pocket), which shows 100% conservation in CRBN orthologs across animal and plant kingdoms (Chamberlain et al., 2014; Fischer et al., 2014) (Figure S7A). We therefore speculated that the tri-Trp pocket might be important for GS binding. FlagCRBN-W386E and FlagCRBN-W400E, and the double point mutant FlagCRBN-Y384A/W386A (FlagCRBN-YW/AA, which is defective in IMiD binding (Ito et al., 2010; Lopez-Girona et al., 2012)) failed to bind endogenous GS (Figure 6C).
Figure 6. The N-terminal extension of GS comprises an acetylation-dependent degron for CRL4CRBN.
(A–B) GS binds the C-terminal domain of CRBN. (A) Schematic diagram of the structure of full-length (FL) human CRBN and the deletion constructs used in panel B. CRBN consists of the amino-terminal domain (NTD), the helical bundle domain (HBD) involved in DDB1 binding and the carboxy-terminal domain (CTD). (B) Cell extracts from CRBN-KO 293FT cells stably expressing full length FlagCRBN or deletion mutants were immunoprecipitated with Flag antibody and analyzed by SDS-PAGE and immunoblotting with GS and Flag antibodies. *, indicates uncleaved FlagCRBN-T2A–GFP forms, which were visible for all constructs on the uncropped film.
(C) Integrity of the ‘tri-Trp’ cavity in the CTD of CRBN is required for binding GS. Cellular extracts prepared from CRBN-KO 293FT cells stably expressing wild type (WT) FlagCRBN or the indicated mutants were subjected to IP with Flag antibody followed by SDS-PAGE and immunoblotting the precipitated and input fractions with the indicated antibodies. YW/AA corresponds to Y384A/W386A mutant. S.E., L.E.: short and long exposures.
(D) Design of GS N-terminal peptides. Where indicated, the K11 and K14 residues are acetylated.
(E) CRBN binds specifically to a GS N-terminal peptide acetylated on K11 and K14. Pull-down assays were performed using purified recombinant human FlagCRBN and immobilized non-acetylated or acetylated K11, K14, or K11K14 GS peptides (panel D) as indicated, and analyzed by SDS-PAGE and immunoblotting with anti-Flag.
(F–G) CRBN-N351R mutant does not bind to endogenous GS and thalidomide. (F) Cellular extracts prepared from CRBN-KO 293FT cells stably expressing wild type (WT) FlagCRBN or the indicated mutants were subjected to IP with Flag antibody followed by SDS-PAGE and immunoblotting the bound and input fractions with the indicated antibodies. (G) Thalidomide (Thal)-binding CRBN proteins were purified from CRBN-KO 293FT cells stably expressing empty vector or FlagCRBN (wild type or mutant) by using thalidomide-immobilized (+) or control (–) beads, and analyzed by SDS-PAGE and immunoblotting with Flag antibody. S.E., L.E.: short and long exposures.
(H) IMiDs do not compete out binding of GS to CRBN. Pull-down assays were performed in the presence or absence of pomalidomide (pom) as indicated, using FlagCRBN purified from CRBN-KO 293FT cells stably expressing FlagCRBN, and non-acetylated or acetylated biotin-GS peptides immobilized on streptavidin resin. The input and bound fractions were analyzed by immunoblotting with Flag and DDB1 antibodies. S.E., L.E.: short and long exposures.
To define in greater detail the molecular basis for GS–CRBN interaction, we generated biotinylated synthetic peptides (amino acids 5–22 of GS) that were unmodified, mono-acetylated on either K11 or K14, or di-acetylated (Figure 6D). The peptide acetylated at both K11 and K14, but not non-acetylated or mono-acetylated peptides, efficiently pulled-down recombinant FlagCRBN (Figure 6E). We further examined the effects of each acetylation site mutation of GSMyc on CRBN binding and ubiquitylation in vivo. Consistent with the peptide binding analysis, single substitutions at either K11 or K14 with alanine (A) resulted in significantly less binding and ubiquitylation, compared with double AA mutations at both K11 and K14 (Figure S6E, F). These results indicate that acetylation of GS on both lysine residues 11 and 14 contributes to CRBN binding, ubiquitylation and degradation.
To gain further insight into the mechanism by which CRBN binds the di-acetylated degron of GS, we made asparagine 351 to arginine (N351R) and histidine 357 to tyrosine (H357Y) (Figure S7A) point mutants based on examination of the crystal structure. We reasoned that the substitution of N351R might block degron binding by occluding the IMiD pocket whereas H357Y might enhance degron binding by decreasing the cationic charge surrounding the IMiD pocket. Remarkably, production and testing of the indicated FlagCRBN mutants confirmed these predictions (Figures 6F – G), suggesting that the GS-peptide binding site overlaps or is adjacent to the IMiD pocket. Enhanced binding of the H357Y mutant to the mono-acetylated K11 peptide but not the mono-acetylated K14 peptide (Figure S6G) further implicates electrostatics as contributing to recognition of the acetylated GS peptides.
Recent studies have reported that IMiD binding to CRBN promotes recruitment of neosubstrates (Kronke et al., 2014; Lu et al., 2014)(Kronke et al., 2015), whereas IMiDs block endogenous CRBN substrate MEIS2 from binding to CRL4CRBN (Fischer et al., 2014). We therefore examined whether IMiD binding to CRBN influences the interaction between the acetyl-degron and CRBN in vitro. Consistent with our initial findings in 293T and MM.1S cells (Figures 1B, 1C, and S1B), pomalidomide did not block FlagCRBN binding to di-acetylated peptide, but promoted the interaction between FlagCRBN and peptide mono-acetylated at K11 or K14 (Figure 6H).
Discussion
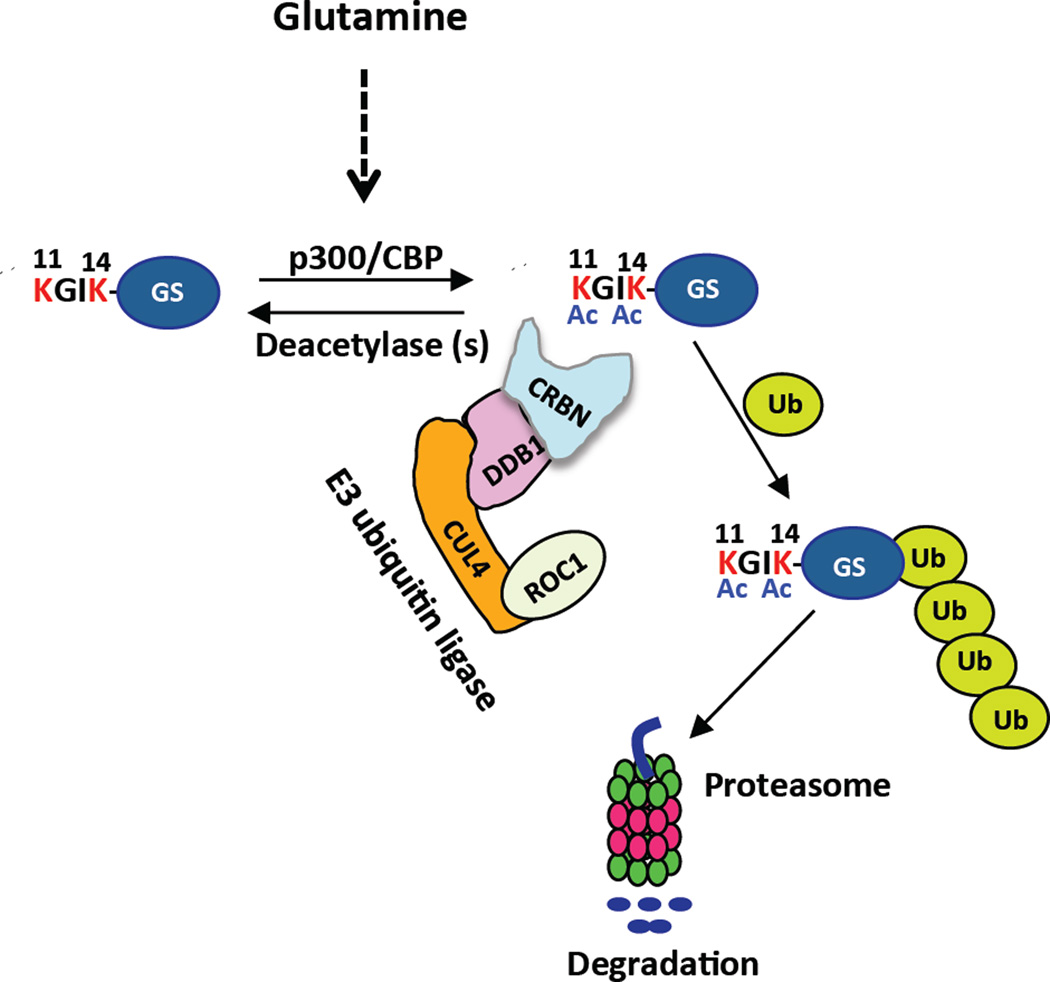
Almost sixty years ago, it was reported that mammalian GS is inactivated by extracellular glutamine (Demars, 1958; Paul and Fottrell, 1963). Subsequent work done prior to the discovery of the ubiquitin system suggested that glutamine stimulates the modification and degradation of GS enzyme through an unknown mechanism (Arad et al., 1976; Crook and Tomkins, 1978). Based on the findings reported here, we propose a model for regulation of glutamine-induced degradation of GS by CRL4CRBN (Figure 7). After exposure of cells to high glutamine, p300/CPB proteins acetylate GS at lysines 11 and 14 to create a degron that binds CRBN. Acetylated GS bound to CRBN is ubiquitylated and subsequently is degraded by the proteasome. Although we provide strong evidence to support these conclusions, we note that it has been difficult to identify by mass spectrometry acetylation of lysines 11 and 14 on GS bound to CRBN. This is probably due to low sequence coverage obtained for endogenous GS bound to CRBN. However, we have used antibodies against acetyl-lysine to show that CRBN enriches for acetylated forms of GS.
Figure 7. Proposed model for regulation of glutamine-induced degradation of GS by CRL4CRBN.
After exposure of cells to high glutamine, the N-terminal peptide of GS becomes exposed and p300/CPB acetylates it at lysines 11 and 14 to create a degron that binds CRBN, resulting in ubiquitylation and degradation of GS. For the sake of simplicity, other amino acids in the N-terminal extension of GS are omitted.
In mammals, GS is a homodecamer composed of two pentameric rings (Krajewski et al., 2008). The pentamer interface contains a network of electrostatic interactions involving both K11 and K14. In particular the ε-amino group of Lysine 11 is normally engaged in a salt bridge with aspartate 174 in the neighboring subunit (Figure S7B, C). We propose that high glutamine concentrations cause a conformational change in GS that disrupts this ionic interface and exposes the N-terminal extension such that lysines 11 and 14 can be bound by p300/CBP and acetylated. We currently do not understand how cells sense extracellular glutamine levels to induce K11 and K14 acetylation. Glutamine could potentially be sensed by mTOR (Nicklin et al., 2009) or perhaps directly by GS. Another question of interest is whether high glutamine results in processive or distributive acetylation of subunits in the decamer. Presumably, acetylation of a single subunit would suffice to target the decamer to CRBN. We do not know if all subunits are ubiquitinated and degraded in concert or the decamer is disassembled to enable degradation of only acetylated subunits. We note that a prior study reported that GS degradation in Schwann cells is mediated by ubiquitin ligase ZNRF1 (Saitoh and Araki, 2010), but regulation by glutamine levels was not investigated. ZNFR1 is expressed primarily in the nervous system (Araki et al., 2001), suggesting that it is unlikely to play a ‘pan-organismal’ role. ZNFR1 may contribute to residual GS degradation that we observed in CRBN-depleted cells.
Reversible lysine acetylation influences diverse biological processes. Recent proteomic analyses suggest that it regulates many metabolic enzymes (Choudhary et al., 2009; Wang et al., 2010; Zhao et al., 2010). It has been thought that lysine acetylation controls protein–protein interactions that regulate protein activity via acetyl-lysine-binding domains (Choudhary et al., 2014; Mujtaba et al., 2007). In some cases, lysine acetylation has been shown to compete with other post-translational modifications (e.g. ubiquitylation or SUMOylation), thereby controlling protein stability or transcriptional activity (Gronroos et al., 2002; Ito et al., 2002; Van Nguyen et al., 2012). Recent studies suggest that lysine acetylation can regulate the steady-state levels of metabolic enzymes by promoting their degradation through the ubiquitin-proteasome system or chaperone-mediated autophagy (Jiang et al., 2011; Lv et al., 2011). Collectively, these prior findings together with those reported here may have important implications for interpreting the clinical action of deacetylase inhibitors that alter the acetylome, since they may activate degradation of multiple proteins via CRBN or other pathways.
CRBN and its ‘tri-Trp’ pocket are evolutionarily conserved in plants and animals. This aromatic pocket binds thalidomide, lenalidomide, and pomalidomide, and is reminiscent of pockets found in proteins containing bromodomains (Dhalluin et al., 1999), plant homeodomain (PHD) fingers (Wysocka et al., 2006), chromodomains, Tudor domains, and malignant brain tumor (MBT) repeats, all of which have been implicated in binding to acetylated or methylated lysine (Taverna et al., 2007). Consistent with an important role for the tri-Trp pocket in recognition of natural substrates, mutations of residues that form the pocket eliminated binding of GS and its acetylated degron peptide, and also disrupt binding of the endogenous substrate MEIS2 (Fischer et al., 2014). However, IMiDs did not compete out binding of the acetylated GS degron peptide to CRBN. In fact, IMiDs actually enhanced binding of monoacetylated GS peptides. Moreover, mutation of the critical lysines 11 and 14 of GS to alanine enhanced binding and ubiquitylation of GS by CRBN. Together, these results suggest that acetylation neutralizes the positive charges on K11 and K14, which otherwise interfere with binding to CRBN. Our data suggest that the acetylated peptide binds adjacent to the IMiD pocket by a novel mechanism, the description of which awaits a crystal structure of the GS degron peptide bound to CRBN.
A role for CRBN in regulation of AMPK and fat accumulation has been demonstrated in a Crbn−/− mouse model (Lee et al., 2013). Our work establishes an unexpected molecular link between CRBN, acetylation, and metabolic control. We suggest that acetylation-dependent ubiquitylation by CRL4CRBN may be a general feature of metabolic regulation.
Experimental Procedures
In vitro ubiquitylation assay
The assays were performed as described (Duan et al., 2012; Kleiger et al., 2009). Briefly, 293T cells stably expressing FlagCRBN or empty vector were treated with Bortezomib (1 µM) for 6 h. Then, the cells were harvested and lysed in IP buffer and immunoprecipitated with Flag M2 agarose beads for 2–4 h at 4°C. After washing five times with IP lysis buffer and two times with ubiquitylation buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 0.2 mM CaCl2, 1 mM DTT, and 100 nM MG132), the beads were incubated at 300C for 1 h in 30 µl of ubiquitylation buffer containing E1 (0.5 µM), UbcH5a (0.5 µM), UbcH3 (1.67 µM), recombinant RBX1-CUL4A (250 nM), ubiquitin (60 µM), and ATP (4 mM). Where indicated, methylated ubiquitin (Me-Ub) was also added. Reactions were stopped by adding SDS sample buffer, separated by SDS-PAGE, transferred to a PVDF membrane, and subjected to immunoblot analysis.
Peptide pull-down assay
The assay was performed as described (Wysocka et al., 2005). FlagCRBN protein was immunoprecipitated from 293T cells stably expressing FlagCRBN using Flag antibody-conjugated agarose beads, and then eluted with Flag peptides. Biotinylated GS peptides were synthesized (Biomatik) and 5 µg was incubated with 20 µl of Dynabeads M-280 streptavidin (Life Technologies) in PBS for 1–2 h at room temperature. After washing three times with PBS-T (PBS containing 0.1% Tween-20), the beads were mixed with purified FlagCRBN in binding buffer (10 mM Tris [pH 7.6], 150 mM NaCl, 0.5% NP-40) containing a protease inhibitor cocktail, 1 mM DTT and 0.1% BSA for 2–4 h at 4°C. After binding, the beads were washed extensively in binding buffer containing 300 mM NaCl (stringent washing). The bound proteins were eluted in 2x SDS loading buffer, and analyzed by immunoblot.
Supplementary Material
Highlights.
-
➢
GS is an endogenous substrate of CRL4CRBN.
-
➢
CRL4CRBN directly mediates the glutamine-induced degradation of GS.
-
➢
Glutamine-stimulated acetylation of lysines 11 and 14 regulates GS degradation.
-
➢
The thalidomide-binding domain of CRBN binds to an acetyllysine degron of GS.
Acknowledgments
We thank W. Kaelin (Dana Farber Cancer Institute, Boston) for CRBN-KO 293FT cells. We also thank the Deshaies lab for helpful discussions, particularly E. Blythe, R. Mosadeghi, W. den Besten, A. Moradian (Proteome Exploration Laboratory/PEL) for their assistance and R. Verma for insightful advice. J.E.L. was supported by a grant from National Research Council of Science and Technology (DRC-14-2-KRISS). The PEL is funded by the Gordon and Betty Moore Foundation (Grant GBMF775) and the Beckman Institute. T.V.N is supported by the Vietnam Education Foundation, a Brian D. Novis Research Award from the International Myeloma Foundation, and the Leukemia & Lymphoma Society. R.J.D. is an Investigator of the Howard Hughes Medical Institute and this work was supported in part by HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
T.V.N. and R.J.D.: study conception and design, data analysis, drafting of manuscript with editorial assistance from other authors. T.V.N.: all cellular and molecular experiments. J. E.L.: mass spectrometry experiments in Figures 1A and S5, table S1. M.S. and S.H.: mass spectrometry data analysis. M.S.: protein sequence analysis. J.M.R. and S.G.L.: analysis of the mass spectrometry data in Figure S5. J.H. and B.K.: computational predictions of effects of mutations in the GS-binding site of CRBN and analysis of human GS structure in Figure S7B, C. J.H.Y. and J.S.J.: quantification of glutamine and glutamate in serum and analysis of data in Figure 2G and table S3. S.J.Y., S.J. J. and C.S.P.: mouse studies in Figure 2D–F. H.H.: provided essential reagents.
References
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Arad G, Freikopf A, Kulka RG. Glutamine-stimulated modification and degradation of glutamine synthetase in hepatoma tissue culture cells. Cell. 1976;8:95–101. doi: 10.1016/0092-8674(76)90190-2. [DOI] [PubMed] [Google Scholar]
- Araki T, Nagarajan R, Milbrandt J. Identification of genes induced in peripheral nerve after injury. Expression profiling and novel gene discovery. The Journal of biological chemistry. 2001;276:34131–34141. doi: 10.1074/jbc.M104271200. [DOI] [PubMed] [Google Scholar]
- Bott AJ, Peng IC, Fan Y, Faubert B, Zhao L, Li J, Neidler S, Sun Y, Jaber N, Krokowski D, et al. Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation. Cell Metab. 2015;22:1068–1077. doi: 10.1016/j.cmet.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chemistry & biology. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain PP, Lopez-Girona A, Miller K, Carmel G, Pagarigan B, Chie-Leon B, Rychak E, Corral LG, Ren YJ, Wang M, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nature structural & molecular biology. 2014;21:803–809. doi: 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Molecular & cellular proteomics : MCP. 2012;11:1048–1062. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Christa L, Simon MT, Flinois JP, Gebhardt R, Brechot C, Lasserre C. Overexpression of glutamine synthetase in human primary liver cancer. Gastroenterology. 1994;106:1312–1320. doi: 10.1016/0016-5085(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Crook RB, Tomkins GM. Effect of glutamine on the degradation of glutamine synthetase in hepatoma tissue-culture cells. The Biochemical journal. 1978;176:47–52. doi: 10.1042/bj1760047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demars R. The inhibition by glutamine of glutamyl transferase formation in cultures of human cells. Biochim Biophys Acta. 1958;27:435–436. doi: 10.1016/0006-3002(58)90367-6. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer ES, Bohm K, Lydeard JR, Yang H, Stadler MB, Cavadini S, Nagel J, Serluca F, Acker V, Lingaraju GM, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- Gunnersen D, Haley B. Detection of glutamine synthetase in the cerebrospinal fluid of Alzheimer diseased patients: a potential diagnostic biochemical marker. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11949–11953. doi: 10.1073/pnas.89.24.11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle J, Gorg B, Rutsch F, Schmidt E, Toutain A, Benoist JF, Gelot A, Suc AL, Hohne W, Schliess F, et al. Congenital glutamine deficiency with glutamine synthetase mutations. The New England journal of medicine. 2005;353:1926–1933. doi: 10.1056/NEJMoa050456. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology. 2004;63:1927–1931. doi: 10.1212/01.wnl.0000146196.01316.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohberger B, Enz R. Cereblon is expressed in the retina and binds to voltage-gated chloride channels. FEBS Lett. 2009;583:633–637. doi: 10.1016/j.febslet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Lee KH, Song S, Jung YK, Park CS. Identification and functional characterization of cereblon as a binding protein for large-conductance calcium-activated potassium channel in rat brain. J Neurochem. 2005;94:1212–1224. doi: 10.1111/j.1471-4159.2005.03344.x. [DOI] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol. 2008;375:217–228. doi: 10.1016/j.jmb.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Kronke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, Chamberlain PP, Mani DR, Man HW, Gandhi AK, et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS genetics. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Sweredoski MJ, Graham RL, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Molecular & cellular proteomics : MCP. 2011a;10 doi: 10.1074/mcp.M110.006460. M110 006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Jo S, Kim H, Lee J, Park CS. Functional modulation of AMP-activated protein kinase by cereblon. Biochim Biophys Acta. 2011b;1813:448–455. doi: 10.1016/j.bbamcr.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Lee KM, Yang SJ, Choi JH, Park CS. Functional effects of a pathogenic mutation in Cereblon (CRBN) on the regulation of protein synthesis via the AMPK-mTOR cascade. The Journal of biological chemistry. 2014;289:23343–23352. doi: 10.1074/jbc.M113.523423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Yang SJ, Kim YD, Choi YD, Nam JH, Choi CS, Choi HS, Park CS. Disruption of the cereblon gene enhances hepatic AMPK activity and prevents high-fat diet-induced obesity and insulin resistance in mice. Diabetes. 2013;62:1855–1864. doi: 10.2337/db12-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz W, Pfeiffer RA, Kosenow W, Hayman DJ. THALIDOMIDE AND CONGENITAL ABNORMALITIES. The Lancet. 1962;279:45–46. [Google Scholar]
- Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, Karasawa S, Carmel G, Jackson P, Abbasian M, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG., Jr The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcbride WG. Thalidomide and Congenital Abnormalities. Lancet. 1961;2:1358. [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J, Fottrell PF. Mechanism of D-glutamyltransferase repression in mammalian cells. Biochim Biophys Acta. 1963;67:334–336. doi: 10.1016/0006-3002(63)91836-5. [DOI] [PubMed] [Google Scholar]
- Saitoh F, Araki T. Proteasomal degradation of glutamine synthetase regulates schwann cell differentiation. J Neurosci. 2010;30:1204–1212. doi: 10.1523/JNEUROSCI.3591-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. The story of glutamine synthetase regulation. The Journal of biological chemistry. 2001;276:44357–44364. doi: 10.1074/jbc.R100055200. [DOI] [PubMed] [Google Scholar]
- Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, Jaffe JD, Kuhn E, Udeshi ND, Carr SA. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Molecular & cellular proteomics : MCP. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PO, Weinstock A, Wagner A, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nature cell biology. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L, Curthoys NP. Glutamine metabolism: Role in acid-base balance*. Biochem Mol Biol Educ. 2004;32:291–304. doi: 10.1002/bmb.2004.494032050388. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. The Journal of biological chemistry. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- Van Nguyen T, Angkasekwinai P, Dou H, Lin F-M, Lu L-S, Cheng J, Chin YE, Dong C, Yeh ET. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Molecular cell. 2012;45:210–221. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Braggio E, Shi CX, Bruins LA, Schmidt JE, Van Wier S, Chang XB, Bjorklund CC, Fonseca R, Bergsagel PL, et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.