Abstract
Recently, we identified an orphan Bombyx mori neuropeptide G protein-coupled receptor (BNGR)-A24 as an ion transport peptide-like (ITPL) receptor. BNGR-A24 belongs to the same clade as BNGR-A32 and -A33, which were recently identified as natalisin receptors. Since these three BNGRs share high similarities with known receptors for tachykinin-related peptides (TRPs), we examined whether these BNGRs can function as physiological receptors for five endogenous B. mori TRPs (TK-1–5). In a heterologous expression system, BNGR-A24 acted as a receptor for all five TRPs. In contrast, BNGR-A32 responded only to TK-5, and BNGR-A33 did not respond to any of the TRPs. These findings are consistent with recent studies on the ligand preferences for B. mori natalisins. Furthermore, we evaluated whether the binding of ITPL and TRPs to BNGR-A24 is competitive by using a Ca2+ imaging assay. Concomitant addition of a TRP receptor antagonist, spantide I, reduced the responses of BNGR-A24 not only to TK-4 but also to ITPL. The results of a binding assay using fluorescent-labeled BNGR-A24 and ligands demonstrated that the binding of ITPL to BNGR-A24 was inhibited by TK-4 as well as by spantide I, and vice versa. In addition, the ITPL-induced increase in cGMP levels of BNGR-A24-expressing BmN cells was suppressed by the addition of excess TK-4 or spantide I. The intracellular levels of cAMP and cGMP, as second messenger candidates of the TRP signaling, were not altered by the five TRPs, suggesting that these peptides act via different signaling pathways from cAMP and cGMP signaling at least in BmN cells. Taken together, the present findings suggest that ITPL and TRPs are endogenous orthosteric ligands of BNGR-A24 that may activate discrete signaling pathways. This receptor, which shares orthosteric ligands, may constitute an important model for studying ligand-biased signaling.
Introduction
Ion transport peptide (ITP) and its alternatively spliced variant, ITP-like (ITPL), are insect peptides that belong to the crustacean hyperglycemic hormone (CHH) superfamily [1–4]. These neuropeptides have been identified in many ecdysozoans, including insects, crustaceans, and nematodes. Members of this neuropeptide family are composed of 70–80 amino acid residues, showing structural similarity with three conserved intramolecular disulfide bonds. The insect members of this family, ITP and ITPL, were originally identified as regulators of ion and fluid transport across the locust ileum [5,6]. ITP and ITPL are differently expressed in various tissues, indicating they have extensive and discrete functions. In Drosophila, ITP functions in the regulation of the circadian rhythm [7–9] and has been suggested to be involved in ecdysis in the tobacco hornworm Manduca sexta [10]. ITPL plays a role in the ovarian maturation of the red flour beetle Tribolium castaneum [11]. However, the functions of ITP [12] and ITPL [13] in the silkworm Bombyx mori remain unclear.
Recently, by screening for orphan neuropeptide G protein-coupled receptors (GPCRs) in B. mori (BNGRs) [14], we successfully identified three BNGRs (BNGR-A2, -A24, and -A34) as functional receptors for endogenous ITP and ITPL [15]. The two ITP receptors, BNGR-A2 and -A34, and the ITPL receptor, BNGR-A24, belong to the class A (rhodopsin-like) GPCR family. While the two ITP receptors are newly deorphanized, BNGR-A24 appears to shares high similarity with tachykinin receptors.
Tachykinins and their structurally related peptides, comprise a large family of multifunctional brain-gut peptides found in both vertebrates and invertebrates [16–20]. One of the most well-known tachykinin family members is substance P. In invertebrates, tachykinin family members can be structurally divided into two types: invertebrate tachykinins (invTKs) containing a vertebrate-like C-terminal FXGLMa (‘a’ indicates C-terminal amidation); and tachykinin-related peptides (TRPs) with a C-terminal FXG/AXRa (Gly for the third residue appears more frequently) motif. TRPs are considered as functional counterparts of vertebrate tachykinins and are expressed in the nervous system, gastrointestinal tract, and many other tissues. An extensive range of biological activities, including modulation of muscle contraction, diuretic activity, neuromodulatory effects, and induction of adipokinetic hormone secretion, has been observed in insects using in vitro bioassays [19].
In B. mori, five endogenous TRPs are produced from a single precursor polypeptide encoded by the tachykinin gene [21]. Peptidomic analysis has demonstrated the presence of these TRPs in B. mori larvae, pupae, and adults [22]. To date, the physiological roles of TRPs have been demonstrated in the induction of pheromone biosynthesis [23] and the modulation of feeding behavior [24]. However, little is known regarding the biological functions of TRPs in B. mori.
Although several bioinformatic studies have identified the structure and presence of TRPs in various invertebrates, the functional characterization of TRP receptors has not been performed extensively thus far. In silico comparative analyses of GPCRs in different insect species have suggested that BNGR-A24, -A32, and -A33 are candidate TRP receptors in B. mori [14,15,25]. BNGR-A24 has been shown to respond to one TRP in D. melanogaster, DTK-6 [26] and BNGR-A32 and -A33 have been identified recently as receptors for endogenous natalisins, which function in reproduction and contain the TRP-like C-terminal motif (F/YXXXRa) [26]. To date, it remains unclear whether BNGR-A24, -A32, and -A33 function as receptors for the five endogenous TRPs in B. mori.
In the present study, we determined whether candidate BNGRs function as receptors for B. mori TRPs using a Ca2+ imaging technique. In addition, we evaluated the possibility of competitive binding in vivo, since BNGR-A24 functions as a physiological receptor for both TRP and ITPL.
Materials and Methods
Insects
Silkworm eggs of the B. mori racial hybrid, Kinshu × Showa, were purchased from Ueda Sanshu (Nagano, Japan) and used for all experiments. Larvae were reared in plastic containers at 26 ± 1°C with 70 ± 10% relative humidity under long-day lighting conditions (16 h light/ 8 h dark) and fed with SILKMATE 2S artificial diet (Nihon Nosan Kogyo, Yokohama, Japan).
Preparation of recombinant and synthetic peptides
Recombinant B. mori ITPL (rITPL) was prepared as previously reported [15]. rITPL was collected as inclusion bodies from an E. coli expression system and the inclusion body components were denatured and refolded. Purification of rITPL was performed by reversed-phase high performance liquid chromatography (HPLC) on a Shodex Asahipak ODP-50 column (10 mm inner diameter × 250 mm, Showa Denko, Tokyo, Japan), using a 30-min linear gradient of 20–50% acetonitrile, 0.05% trifluoroacetic acid, at a flow rate of 3 mL/min. Elution was monitored by absorbance at 280 nm. B. mori TRPs were chemically synthesized on an automated Apex 369 peptide synthesizer (AAPPTec, Louisville, KY, USA), using a standard Fmoc (N-[9-fluorenylmethoxycarbonyl]) solid-phase protocol and reagents purchased from Watanabe Chemical Industries (Hiroshima, Japan). Following deprotection and cleavage from the resin, the synthetic TRPs were purified by reversed-phase HPLC on a PEGASIL-300 ODS column (10 mm inner diameter × 250 mm, Senshu Kagaku, Tokyo, Japan), using a 30-min linear gradient of 10–40% acetonitrile, 0.05% trifluoroacetic acid, at a flow rate of 1 mL/min. Elution was monitored by absorbance at 225 nm. Synthetic spantide I was purchased from Peptide Institute (Osaka, Japan). Fluorescent-labeled peptides were prepared using rhodamine red (RR) succinimidyl ester (Life Technologies, Tokyo, Japan), according to manufacturer’s protocols, and were purified by reversed-phase HPLC under the same conditions as the TRPs.
Expression analysis by RT-PCR
Tissues were dissected in 0.9% NaCl solution from anesthetized B. mori larvae (three males and three females) at fifth instar day 2 that were fed ad libitum or starved for 24 h. Total RNA was extracted from the tissues using TRIzol Reagent (Life Technologies). cDNA synthesis was performed with SuperScript III (Life Technologies) and an oligo (dT)12-18 primer. Partial cDNA fragments of the target genes were amplified with Go Taq DNA polymerase (Promega, Tokyo, Japan) and the following primers: 5′-ATCAACGACGGCCAATACCC-3′ and 5′-TCCCGAAGAATCCCATCTGC-3′ for the tachykinin gene (tk, GenBank accession number NM_001130892); and 5′-GGTTCACTGTCGCGTTTCTA-3′ and 5′-TCTTTCCACGATCAGCTTCC-3′ for the ribosomal protein L32 gene (rpl32, GenBank accession number NM_001098282). The amplification conditions were as follows: 20 s (140 s for the first cycle) at 94°C, 30 s at 60°C, and 45 s at 72°C for 30 cycles (for tk) or 20 cycles (for rpl32). Densitometric analysis was performed for three independent experiments using the ImageJ software from the National Institutes of Health (http://imagej.nih.gov/ij/).
Ca2+ imaging assay of BNGRs
The assays were performed as previously reported [15]. Briefly, HEK293T cells seeded on a glass-based dish were co-transfected with pME18S plasmids carrying BNGRs and the promiscuous G protein alpha subunit, Gα15. Following 24 h incubation, the transfected cells were loaded with 500 μL of a Ca2+ indicator solution (2 μM Fluo-4 AM [Dojin Chemicals, Kumamoto, Japan] and 0.01% Pluronic-F 127 [Anaspec, San Jose, CA, USA] in Opti-MEM [Life Technologies]) and incubated at 37°C for 30 min in a dark room. The cells were washed twice with Ringer’s solution and then 800 μL Ringer’s solution was added to the cells. Within a given run time, 100 μL of the following compounds were added sequentially: Ringer’s solution (at 15 s, vehicle control), the peptide of interest or a mixture with its potential competitor (at 60 s), and 100 μM ATP (at 120 s, an experimental control). Fluo-4-derived fluorescence was chronologically monitored using a confocal laser-scanning microscope (FV-1000D; Olympus, Tokyo, Japan), with the preinstalled dye mode for Fluo-3. At least three independent experiments were performed. Dose-response curves were generated and EC50 values were calculated from the obtained fitting curves.
Competitive binding assay
Binding assays were performed as previously reported [15]. Briefly, CHO cells were transfected with 2 μg of pME18S plasmids harboring enhanced green fluorescent protein (EGFP)-fused BNGR cDNA. The binding assay was performed with cells 24 h post transfection. The cells were pre-incubated at 4°C for 30 min prior to the assay to minimize receptor internalization by ligand binding. Following the addition of 50 μL of non-labeled ligand as a potential competitor, 50 μL of RR-labeled ligand was added. Subsequent to incubation at 4°C for 60 min, the cells were washed twice with an ice-cold fixation buffer (4% formaldehyde in PBS) and then 500 μL of the fixation buffer was added. The fluorescence derived from EGFP and RR was observed using a confocal microscope (FV-1000D) with preinstalled dye modes for EGFP and DsRed, respectively. The fluorescent data obtained from six individual cells of at least two independent experiments were analyzed using Fluoview software (Olympus).
cAMP assay and competitive cGMP assay
BmN cells were plated in 12-well tissue culture plates (2 × 105 cells/well; Asahi glass, Yokohama, Japan) and maintained in normal growth medium (TC-100 [Applichem, Darmstadt, Germany], 10% FBS, and 50 μg/mL gentamycin). At 3 days post-seeding, the cells were pre-incubated in 800 μL of assay medium (TC-100 supplemented with 1 mM 1-methyl-3-isobutylxanthine [IBMX; Sigma-Aldrich, Tokyo, Japan]) and 1 mM phenylmethylsulfonyl fluoride for 10 min at 28°C. Following pre-incubation, 100 μL of a potential competitor and 100 μL of the peptide of interest were added sequentially to the medium and incubated at 28°C for 30 min. After incubation, the cells were washed twice with TC-100 and then immediately frozen in liquid nitrogen. Intracellular cAMP and cGMP were extracted by sonication with 300 μL 0.1 M HCl supplemented with 1 mM IBMX and then collected from the supernatant subsequent to centrifugation (4°C, 16,000 × g, 10 min). The extracted cAMP and cGMP were quantified in duplicate using a cAMP ELISA kit (Cayman Chemicals, Ann Arbor, MI, USA) and a cGMP EIA system (GE Healthcare, Hino, Japan), respectively. These experiments were repeated at least twice.
Results
Tissue distribution of the tachykinin gene
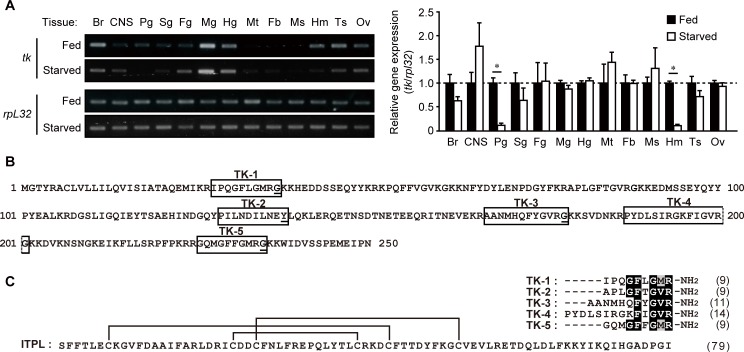
Previously, we showed that bngr-A24 is ubiquitously expressed in the tissues of fifth instar day 2 larvae and that gene expression of both bngr-A24 and itpl in the intestinal tissues is upregulated by 24 h starvation [15]. In order to determine in which tissues the tachykinin gene, tk, is expressed and whether tk expression is affected by feeding state, we analyzed the expression profile of tk by RT-PCR. In larvae fed ad libitum, tk expression was relatively higher in the midgut; moderate in the brain, hindgut, hemocytes, and reproductive tissues; and relatively lower in the central nervous system, prothoracic glands, silk gland, and foregut. Very low levels of PCR product were detected in the Malpighian tubules, fat body, or smooth muscle (Fig 1A, left panel, Fed). The tk expression profile in larvae following 24 h starvation was similar to that of larvae fed ad libitum (Fig 1A, left panel, Starved), except for a notable reduction of expression in the prothoracic glands and hemocytes (Fig 1A, right panel).
Fig 1. Endogenous TRPs in B. mori.
(A) Tissue distribution and expression profile of the tachykinin gene (tk) in B. mori larvae. Tissues from six individual fifth-instar day 2 larvae fed ad libitum or starved for 24 h were used for this semi-quantitative RT-PCR analysis. Ribosomal protein L32 gene (rpl32) was used as an experimental and expression control. The left panel is a representative image of the analyses. The right panel shows the densitometric analysis results from three independent experiments. The starved larvae data are depicted as the fold change over that in the same tissue of fed larvae (mean + S.D.). Asterisks indicate significant differences between the two groups for the same tissue (Fed vs. Starved; P < 0.05, unpaired t-test). Br, brain; CNS, central nervous system; Pg, prothoracic glands; Sg, silk gland; Fg, foregut; Mg, midgut; Hg, hindgut; Mt, Malpighian tubules; Fb, fat body; Ms, smooth muscle from the 8th body segment; Hm, hemocytes; Ts, testis; Ov, ovary. (B) Amino acid sequence of the polypeptide encoded by the tachykinin gene; this precursor is processed to produce five premature peptides (boxed), which are subsequently amidated with the C-terminal Gly residue serving as a donor (underlined) that requires maturation. (C) Sequence comparison of the five mature TRPs and ITPL present in B. mori. ITPL contains three intramolecular disulfide bonds (solid lines). The amino acid sequences of the five TRPs were aligned using ClustalW (http://clustalw.ddbj.nig.ac.jp); these peptides contain the consensus sequence (-Phe-X1-Gly-X2-Arg-NH2) of TRPs found in invertebrates. Numbers in parentheses indicate the number of amino acid residues.
Response of candidate BNGRs to B. mori TRPs
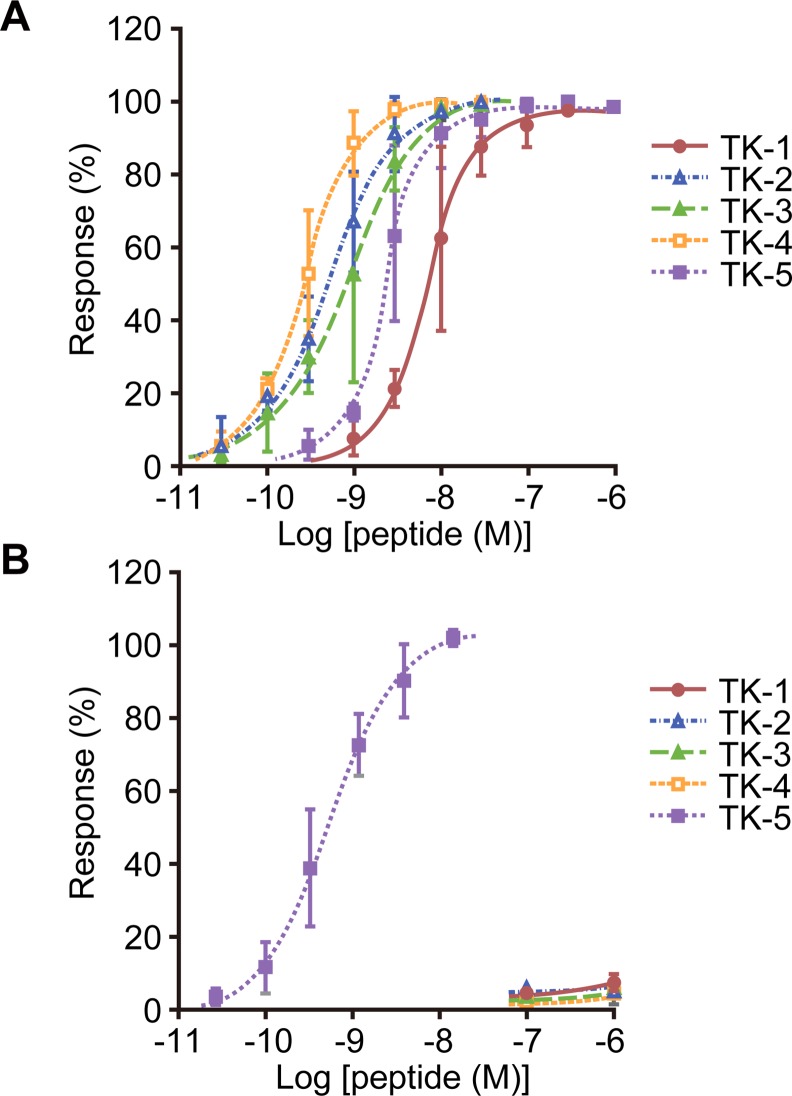
The B. mori tk gene encodes a 250-amino acid polypeptide (Fig 1B), which is a precursor of five TRPs, TK-1–5 (Fig 1C). In order to evaluate whether the three TRP receptor candidates, BNGR-A24, -A32, and -A33, function as receptors for B. mori TRPs, the response of these three BNGRs to chemically synthesized TRPs was examined using the Ca2+ imaging assay. When BNGR-A24 was heterologously expressed in HEK293T cells, it showed dose-dependent response to all five TRPs; the maximum response was observed against TK-4, with an EC50 = 0.27 nM (Fig 2A and Table 1). In addition, BNGR-A32 responded to TK-5 with an EC50 = 0.48 nM; however, almost no response was observed for TK-1–4 at concentrations as high as 1 μM (Fig 2B and Table 1). In contrast, BNGR-A33 did not exhibit an obvious response to a 1 μM concentration of any of the five TRPs. These results indicate that BNGR-A24 and -A32, but not BNGR-A33, function as receptors for endogenous TRPs in B. mori.
Fig 2. Responses of BNGR-A24 and BNGR-A32 to B. mori TRPs.
The response of HEK293T cells co-expressing BNGR-A24 (A) or BNGR-A32 (B) and promiscuous mouse Gα15 to the TRPs was monitored using the Ca2+ imaging technique. Dose-response curves are depicted as relative fluorescent intensity (mean ± S.D.; n = 3).
Table 1. EC50 of BNGR responses to B. mori TRPs.
| Receptor | Ligand | EC50 (nM)a |
|---|---|---|
| BNGR-A24 | TK-1 | 7.1 |
| TK-2 | 0.49 | |
| TK-3 | 0.79 | |
| TK-4 | 0.27 | |
| TK-5 | 2.3 | |
| BNGR-A32 | TK-5 | 0.48 |
aEC50 values were calculated using the Ca2+ imaging assays shown in Fig 2.
The EC50 values of BNGR-A32 to TK-1–4 and BNGR-A33 to TK-1–5 were not calculated because of their lower response levels.
Evaluation of the competitive binding of ITPL and TRPs to BNGR-A24
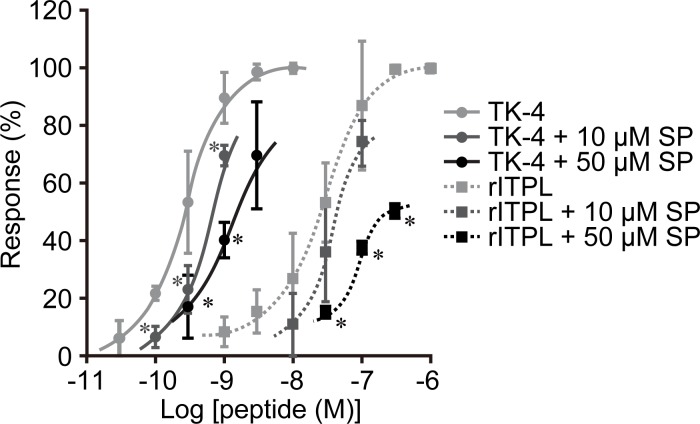
The results shown in Fig 2A and Table 1 and our previous report [15] indicate that BNGR-A24 is a functional receptor not only for ITPL but also for the five TRPs, suggesting the possibility of competitive binding of ITPL and the TRPs to BNGR-A24. To address this question, we examined whether the responses of BNGR-A24 to rITPL and the TRPs are affected by a potential competitor, spantide I, which is an antagonistic analog of substance P. Simultaneous treatment of BNGR-A24 with 10 μM or 50 μM of spantide I significantly decreased its response to TK-4 (Fig 3). The response of BNGR-A24 to TK-4 was successively decreased in a dose-dependent manner with increasing concentration of spantide I. These results indicate that spantide I functions as a TRP antagonist for BNGR-A24. Next, we examined the effect of simultaneous addition of spantide I and rITPL on ligand competition. The dose-response curve of 10–300 nM rITPL shifted in a similar manner to that observed for TK-4. Furthermore, when 50 μM of spantide I was added, the response of BNGR-A24 to rITPL was significantly lower than that without the competitor. These results indicate that ITPL binding to BNGR-A24 was inhibited by spantide I, a competitive antagonist of both tachykinins and TRPs.
Fig 3. Effect of spantide I on the response of BNGR-A24 to rITPL and TK-4.
Spantide I (SP), a potential competitor, was added together with rITPL or TK-4 and the response of BNGR-A24 was monitored using the Ca2+ imaging assay. Dose-response curves of the responses are presented as relative fluorescent intensity (mean ± S.D.; n = 3). The response curve of TK-4 with no competitor is the same as the one shown in Fig 2A. Asterisks indicate significant differences for treatment with the same ligand without spantide I (P < 0.05; Dunnett’s test).
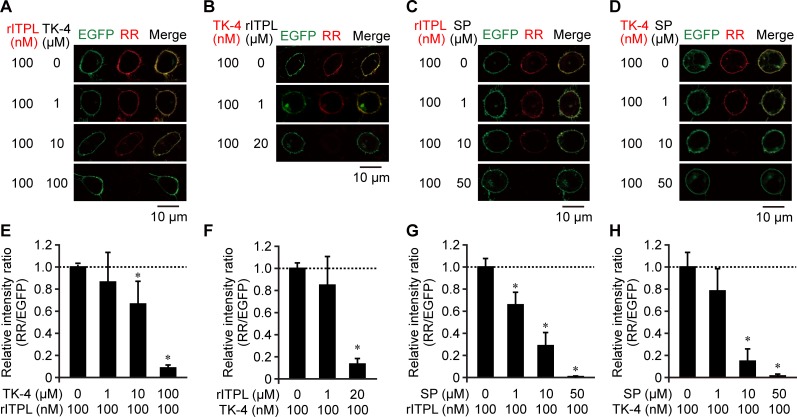
In order to directly demonstrate the competitive binding of ITPL and TRPs to BNGR-A24, we performed an in vitro binding assay using RR-labeled ligands and CHO cells expressing BNGR-A24. Following 30-min exposure of the cells to RR-labeled rITPL or TK-4, both red fluorescence (derived from the RR-labeled ligands) and green fluorescence (derived from EGFP C-terminally fused to BNGR-A24) were observed on the cell surface, indicating ligand-receptor binding (Fig 4). When 1–100 μM of unlabeled TK-4 was added prior to the addition of RR-labeled rITPL, the fluorescence of labeled rITPL on the cell surface gradually decreased with increasing TK-4 dose (Fig 4A and 4E). A similar result was obtained using RR-labeled TK-4 and unlabeled rITPL (Fig 4B and 4F). In this case, the addition of 20 μM rITPL reduced the ligand-derived signal by approximately 90% compared to that observed without the addition of unlabeled ligand. To confirm the results obtained from the Ca2+ imaging assay, we examined binding in the presence of spantide I, a competitor of rITPL and TK-4; our results show that the fluorescence intensity of the RR-labeled ligands decreased in response to increasing doses of spantide I (Fig 4C, 4D, 4G and 4H). Addition of 50 μM spantide I was sufficient for nearly complete quenching of the fluorescence derived from RR-labeled rITPL and TK-4. This competitive effect of spantide I against rITPL suggests that other TRPs in addition to TK-4 act as competitors against ITPL binding to BNGR-A24.
Fig 4. In vitro competitive binding assay of rITPL and TK-4 to BNGR-A24.
(A–D) Microscopic imaging. The binding of RR-labeled ligands (A and C, rITPL; B and D, TK-4) and CHO cells expressing EGFP-fused BNGR-A24 was examined in the presence of potential competitors (A, TK-4; B, rITPL; C and D, spantide I [SP]). EGFP and RR fluorescences were observed by confocal microscopy. Co-localization is presented as yellow in the merged images (Merge). Representative images of cells from at least two independent experiments are shown. (E–H) Relative fluorescent intensity obtained from experiment images (A)–(D), respectively. RR fluorescent intensity was normalized by EGFP intensity per image and is indicated as fold change over that without competitor (mean + S.D.; n = 6). Asterisks indicate significant differences compared to ‘no competitor’ data (P < 0.05, Dunnett’s test).
Effect of ITPL and TRP competition for BNGR-A24 on signal transduction
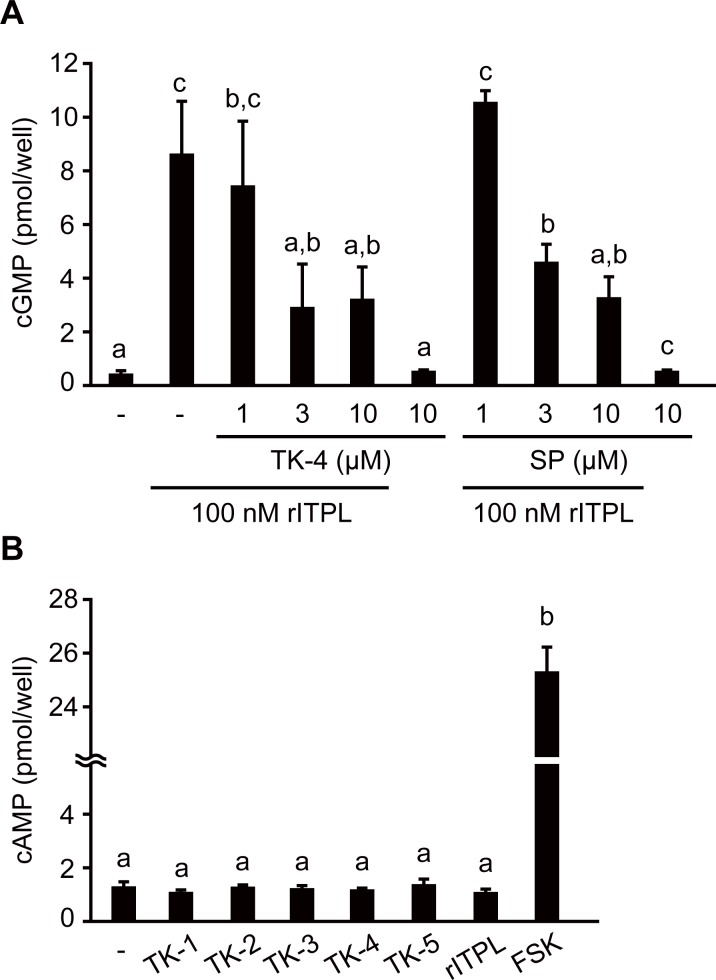
Previously, we showed that BNGR-A24 is expressed in B. mori ovary-derived BmN cells and that rITPL increased intracellular cGMP levels via BNGR-A24 [15]. The results of the current study show that 30-min exposure to 100 nM rITPL caused significant elevation of cGMP levels in the presence of the phosphodiesterase inhibitor IBMX (Fig 5A). In contrast, exposure to TK-4 at concentrations as high as 10 μM failed to increase cGMP levels (Fig 5A), suggesting that the effect of TK-4 on BNGR-A24 does not alter intracellular cGMP production in BmN cells. Since ITPL and TK-4 are competitive ligands of BNGR-A24, we hypothesized that TK-4 might inhibit the cGMP signal induced by ITPL via the binding of ITPL to BNGR-A24. To confirm this hypothesis, we added an excess of TK-4 to BmN cells exposed to rITPL and evaluated the stimulatory effect of ITPL on the cGMP signal. Addition of ≥3 μM TK-4 resulted in a significant decrease in rITPL-induced cGMP levels (Fig 5A), supporting our hypothesis. A similar result was obtained when spantide I was used as a potential competitor of ITPL. These findings suggest that the intracellular signaling pathways of TK-4 and ITPL via BNGR-A24 are different from each other, and that each of these ligands could inhibit the signaling cascade of the other through competitive binding to BNGR-A24. We also examined the effects of the peptides on intracellular levels of cAMP as a second messenger candidate responsible for the signaling pathways of TRPs and ITPL; 30-min exposure to 1 μM five TRPs or rITPL failed to stimulate cAMP production (Fig 5B). These results suggest that TRPs stimulate BNGR-A24 via different signaling pathway from cAMP and cGMP signaling at least on BmN cells.
Fig 5. Effects of TK-4, rITPL, and spantide I on cyclic nucleotide levels in BmN cells.
Intracellular cGMP (A) and cAMP (B) levels in BmN cells following 30-min incubation with the peptide and reagent of interest at the indicated concentrations were measured. (A) Effects of TK-4 and spantide I on the response of BmN cells to rITPL. The cGMP levels of the cells exposed to 100 nM rITPL with TK-4 or spantide I (SP) were determined. Responses to TK-4 alone, spantide I alone, rITPL alone, and vehicle treatment (-) were also examined. (B) The cAMP levels of the cells after incubation with 1 μM of five TRPs or rITPL. Responses to vehicle treatment (-) and 10 μM forskolin (FSK; positive control) were also examined. Data are presented as the mean + S.D. (n = 3 or 4). Different letters indicate significant differences (P < 0.05; Tukey’s HSD).
Discussion
Previously, we reported that BNGR-A24 is an ITPL receptor [15]. The present study showed that BNGR-A24, as well as BNGR-A32, are physiological receptors for endogenous TRPs. Our data also demonstrated the competitive binding of TRPs and ITPL to BNGR-A24, which may lead to ligand-specific signals.
BNGR-A24 as a TRP receptor
Our results clearly indicate that BNGR-A24 is a functional receptor of five endogenous TRPs in B. mori, as predicted based on a prior report showing that heterologous DTK-6 activates BNGR-A24 [26]. The binding of TK-4 to BNGR-A24 was inhibited by the addition of spantide I. This result is consistent with previous findings that spantide I and its derivatives, spantide II and III, exhibit antagonistic effects against TRP receptors in D. melanogaster (Drosophila tachykinin receptor [DTKR] and neurokinin receptor from Drosophila [NKD]) [27,28] and a TRP receptor in the stable fly Stomoxys calcitrans [29]. A more recent study has shown that BNGR-A24 expressed in HEK293 and Sf21 cells responds to five B. mori TRPs, as well as to an additional TRP-like peptide (KPQFFVGVKa) derived from the same tachykinin precursor [30]. The authors found that TK-5 generates a weaker response than the other TRPs in the Ca2+ imaging assay in HEK293 cells; this is inconsistent with our data demonstrating that BNGR-A24 responds similarly to all five B. mori TRPs with nanomolar-order EC50s. Our results showing that of the five TRPs examined, BNGR-A24 exhibits the weakest response to TK-1 (Fig 2A and Table 1), are similar to those of the assay in Sf21 cells [30]. These differences in response may be due to variations in the assay systems; in particular, our heterologous expression system utilized co-expression of a promiscuous mouse Gα15 subunit, which couples with most GPCRs, and its consequent signaling via the increase in intracellular Ca2+ levels [31,32]. In this context, our results suggest that HEK293 cells do not possess an appropriate G protein alpha subunit for BNGR-A24 and that Gα15 might compensate for the activity of BNGR-A24 via TK-5 stimulation.
Relationship between TRPs and natalisins in B. mori
Of the three putative TRP receptors in B. mori, BNGR-A24, an ortholog of DTKR [14,15,25], is functional as a receptor for all five TRPs. In contrast, the NKD orthologs, BNGR-A32 and BNGR-A33 [14,15,25], respond only to TK-5 or none of the TRPs, respectively. These results are consistent with previous reports of D. melanogaster TRPs and their receptors demonstrating inconsistencies between the response of DTKR to six endogenous TRPs [27] and the response of NKD to one TRP, DTK-6 [28]. A more recent study has shown that NKD-type GPCRs in various insect species, including BNGR-A32 and -A33, are characterized as natalisin receptors [26]. BNGR-A32 and -A33 have distinct preferences for ligands; the former receptor responds to natalisins with the C-terminal FXXXRa motif, and the latter is a receptor for YXXXRa-containing natalisins. Three natalisins (NTL-1, -3, and -5) acting on BNGR-A32 with EC50 = 15–160 nM, share C-terminal F(F/W)(G/A)XRa consensus sequences, which, except for the hydrophobic aromatic residue at the second position, are similar to those of TRPs. The second characteristic Phe residue is found in TK-5, but not in the other B. mori TRPs (Fig 1C). This may explain why BNGR-A32 responded only to TK-5 (Fig 2B). The natalisins of two other lepidopterans, Danaus plexippus (8/15) and M. sexta (12/15) also contain the F(F/W)(G/A)XRa motif [26]. Since this motif is not found within natalisins in other insect orders [26], it may be a characteristic of ligands specific to lepidopteran natalisin/TRP receptors. Taken together, these findings indicate that BNGR-A32 is shared by TRPs and natalisins containing the FFXXRa motif. Sharing of the same GPCR by neuropeptides of different families has been reported for RFXa-possessing peptides [33].
Competitive action of ITPL and TRPs via BNGR-A24
Our finding that endogenous ITPL and TRPs bind to the same receptor was unexpected, since these peptides belong to two distinct peptide families with dissimilar amino acid sequences and structural characteristics. B. mori TRPs are linear polypeptides composed of 9–14 amino acids with a C-terminal pentapeptide consensus motif (Fig 1C), whereas B. mori ITPL is a 79-amino acid neuropeptide folded by three intramolecular disulfide bonds (Fig 1C), which is the characteristic of CHH family peptides [1–4]. The C-terminal region seems to be important for BNGR-A24 recognition of ITPL; B. mori ITP, which contains the same 40-amino acid N-terminus as ITPL [34], exhibits much lower affinity to BNGR-A24 than ITPL [15]. However, unlike ITP and most CHH family peptides that are C-terminally amidated, the C-terminal sequence of ITPL is not subject to amidation and differs from the consensus C-terminal motif found in TRPs. Therefore, the binding mechanism of ITPL to BNGR-A24 is dissimilar to that of the TRPs. Since our results indicate competitive binding of ITPL and TRPs to BNGR-A24, the regions on the receptor responsible for interacting with these two ligands must be sufficiently proximal to affect the binding of each ligand.
The response of BNGR-A24 to TRPs was 3.7–96-fold higher than to ITPL, with EC50 = 26 nM [15]. Consistent with the signaling of other TRPs [20], it has shown that B. mori TRP signaling via BNGR-A24 is mediated by cAMP and Ca2+, concomitant with the recruitment of two G protein alpha subunits, Gs and Gq, in heterologous HEK293 and Sf21 cells [30]. However, the present study showed that none of endogenous B. mori TRPs induced intracellular cAMP production in BmN cells (Fig 5B), suggesting that different signaling pathways such as Ca+ signaling are responsible for TRPs function at least on BmN cells. We also demonstrated that ITPL failed to increase cAMP levels in BmN cells (Fig 5B); these results are inconsistent with a previous report showing that ITP plays a role in ion fluid modulation in the locust ileum via both cAMP and cGMP [35], although ITPL has not been examined. These results indicate that ITPL is not a partial agonist of TRPs, but rather an agonist acting through a distinct signaling pathway. Our data also showed that TRP failed to increase cGMP levels in BmN cells and that ITPL-activated cGMP production was inhibited by TK-4 and spantide I (Fig 5A). The presence of excess TK-4 inhibited the interaction of ITPL with BNGR-A24 and vice versa (Fig 4), indicating that these peptides act as ‘orthosteric ligands’ rather than ‘allosteric modulators’ [36,37].
As discussed above, our results suggest that TRP and ITPL signaling are independent; thus, these two families of neuropeptides may exert distinct functions via the same receptor. Such ‘ligand-biased signaling’ is an accepted concept, according to which multiple orthosteric ligands have the ability to bias their signaling between different G proteins and/or between G proteins and β-arrestins, contributing to stabilization of the receptor in a ligand-specific conformation [37]. Such a ligand signaling selectivity has been observed for the TRP receptor in S. calcitrans when the receptor was expressed in heterologous S2 cells; substitution of Ala by Gly within the C-terminal FXG/AXRa motif of an endogenous TRP resulted in full agonistic activity against Ca2+ signaling, whereas cAMP was not affected [38]. Another study has shown the significance of β-arrestin signaling in the anti-apoptotic effects exerted by substance P via the neurokinin 1 receptor [39], suggesting that tachykinin receptors may function via β-arrestin signaling in addition to G protein-mediated signaling. Although the functional consequences of β -arrestin-mediated signaling have yet to be determined, the vertebrate tachykinin receptors, Drosophila DTKR and NKD, have been shown to recruit β-arrestin 2 in a heterologous expression system following activation by an endogenous ligand [40]. Since it remains unclear whether the ITPL signaling effector couples directly with BNGR-A24 leading to activation of guanylyl cyclases, it is important to determine which signaling pathway is facilitated by ITPL following its binding to BNGR-A24.
The fact that ITPL and TRPs act competitively on the same receptor also points to a functional relationship. Previous studies have suggested feeding-stimulatory effects of B. mori TK-1 and TK-2 [24] and the participation of BNGR-A24 in feeding [30]; thus, TRP and ITPL signaling may coordinate to regulate feeding behavior via BNGR-A24. Our previous findings indicate that starvation may potentiate ITPL signaling by up-regulating both the ligand and its receptor in intestinal tissues [15]. The present finding that tk expression levels were unchanged in intestinal tissues following 24-h starvation (Fig 1A), suggests that locally expressed TRPs in these tissues may not be involved in regulation of the ITPL signaling, at least at the transcriptional level.
In conclusion, our present study demonstrates that endogenous TRPs and ITPL act competitively on the same receptor, BNGR-A24. In addition, BNGR-A32 functions as a physiological receptor for TK-5 and natalisins containing the C-terminal F(F/W)(G/A)XRa consensus sequence. These findings provide an important clue to understanding the biological functions of ITPL, TRPs, and natalisins. Our data suggest the ligand-biased signaling systems via BNGR-A24, although the downstream effectors of TRPs and ITPL remain to be elucidated. Since the occurrence of such ligand-biased signaling systems involving endogenous ligands of different peptide families is rare, these GPCRs constitute interesting models for understanding how complicated signaling systems contribute to coordinated regulation. Our present data also further our understanding of ligand-receptor co-evolution; tachykinin family peptides, including TRPs, are found in vertebrates and invertebrates, while CHH family peptides, including ITPL, are unique to ecdysozoan species. Since BNGR-A24 is the only TRP receptor-like GPCR identified as an ITPL receptor, the next crucial step would be to determine whether the competitive binding of TRPs and CHH family peptides is a common phenomenon in ecdysozoan species.
Acknowledgments
We thank Dr. Takeshi Kawai of the Department of Applied Biological Chemistry, the University of Tokyo, for technical support regarding chemical peptide synthesis.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the JSPS KAKENHI (Grant-in-Aid for JSPS Fellows [22-1424]) to CN-O (URL: https://www.jsps.go.jp/english/e-grants/) and MEXT KAKENHI (Grant-in-Aid for Scientific Research (C) [24580157]) to SN (URL: https://www.jsps.go.jp/english/e-grants/) and the NAITO Foundation to SN (URL: https://www.naito-f.or.jp/en/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Keller R. Crustacean neuropeptides: structures, functions and comparative aspects. Experientia. 1992;48: 439–448. [DOI] [PubMed] [Google Scholar]
- 2.Webster SG, Keller R, Dircksen H. The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen Comp Endocrinol. 2012;175: 217–233. 10.1016/j.ygcen.2011.11.035 [DOI] [PubMed] [Google Scholar]
- 3.Katayama H, Ohira T, Nagasawa H. Crustacean peptide hormones: Structure, gene expression and function. Aqua-BioScience Monogr. 2013;6: 49–90. [Google Scholar]
- 4.Chung JS, Zmora N, Katayama H, Tsutsui N. Crustacean hyperglycemic hormone (CHH) neuropeptides family: Functions, titer, and binding to target tissues. Gen Comp Endocrinol. 2010;166: 447–454. 10.1016/j.ygcen.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 5.Audsley N, McIntosh C, Phillips JE. Isolation of a neuropeptide from locust corpus cardiacum which influences ileal transport. J Exp Biol. 1992;173: 261–274. [DOI] [PubMed] [Google Scholar]
- 6.Meredith J, Ring M, Macins A, Marschall J, Cheng NN, Theilmann D, et al. Locust ion transport peptide (ITP): primary structure, cDNA and expression in a baculovirus system. J Exp Biol. 1996;199: 1053–1061. [DOI] [PubMed] [Google Scholar]
- 7.Damulewicz M, Pyza E. The clock input to the first optic neuropil of Drosophila melanogaster expressing neuronal circadian plasticity. PLoS One. 2011;6: e21258 10.1371/journal.pone.0021258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johard HAD, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, et al. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516: 59–73. 10.1002/cne.22099 [DOI] [PubMed] [Google Scholar]
- 9.Hermann C, Saccon R, Senthilan PR, Domnik L, Dircksen H, Yoshii T, et al. The circadian clock network in the brain of different Drosophila species. J Comp Neurol. 2013;521: 367–388. 10.1002/cne.23178 [DOI] [PubMed] [Google Scholar]
- 10.Drexler AL, Harris CC, dela Pena MG, Asuncion-Uchi M, Chung S, Webster S, et al. Molecular characterization and cell-specific expression of an ion transport peptide in the tobacco hornworm, Manduca sexta. Cell Tissue Res. 2007;329: 391–408. [DOI] [PubMed] [Google Scholar]
- 11.Begum K, Li B, Beeman RW, Park Y. Functions of ion transport peptide and ion transport peptide-like in the red flour beetle Tribolium castaneum. Insect Biochem Mol Biol. 2009;39: 717–725. 10.1016/j.ibmb.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 12.Endo H, Nagasawa H, Watanabe T. Isolation of a cDNA encoding a CHH-family peptide from the silkworm Bombyx mori. Insect Biochem Mol Biol. 2000;30: 355–361. [DOI] [PubMed] [Google Scholar]
- 13.Dai L, Zitnan D, Adams ME. Strategic expression of ion transport peptide gene products in central and peripheral neurons of insects. J Comp Neurol. 2007;500: 353–367. [DOI] [PubMed] [Google Scholar]
- 14.Yamanaka N, Yamamoto S, Zitnan D, Watanabe K, Kawada T, Satake H, et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One. 2008;3: e3048 10.1371/journal.pone.0003048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai C, Mabashi-Asazuma H, Nagasawa H, Nagata S. Identification and characterization of receptors for ion transport peptide (ITP) and ITP-like (ITPL) in the silkworm Bombyx mori. J Biol Chem. 2014;289: 32166–32177. 10.1074/jbc.M114.590646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinhoff MS, von Mentzer B, Geppetti P, Pothoulakis C, Bunnett NW. Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol Rev. 2014;94: 265–301. 10.1152/physrev.00031.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Loy T, Vandersmissen HP, Poels J, Van Hiel MB, Verlinden H, Vanden Broeck J. Tachykinin-related peptides and their receptors in invertebrates: a current view. Peptides. 2010;31: 520–524. 10.1016/j.peptides.2009.09.023 [DOI] [PubMed] [Google Scholar]
- 18.Satake H, Aoyama M, Sekiguchi T, Kawada T. Insight into molecular and functional diversity of tachykinins and their receptors. Protein Pept Lett. 2013;20: 615–627. [DOI] [PubMed] [Google Scholar]
- 19.Nässel DR. Tachykinin peptides In: Kastin A, editor. Handbook of biologically active peptides. 2nd ed. San Diego: Academic Press; 2013. pp. 315–320. [Google Scholar]
- 20.Vanden Broeck J, Torfs H, Poels J, Van Poyer W, Swinnen E, Ferket K, et al. Tachykinin-like peptides and their receptors. A review. Ann N Y Acad Sci. 1999;897: 374–387. [DOI] [PubMed] [Google Scholar]
- 21.Roller L, Yamanaka N, Watanabe K, Daubnerová I, Zitnan D, Kataoka H, et al. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38: 1147–1157. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Ning X, Zhang Y, Chen W, Zhao Z, Zhang Q. Peptidomic analysis of the brain and corpora cardiaca-corpora allata complex in the Bombyx mori. Int J Pept. 2012;2012: 640359 10.1155/2012/640359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fónagy A, Matsumoto S, Schoofs L, De Loof A, Mitsui T. In vivo and in vitro pheromonotropic activity of two locustatachykinin peptides in Bombyx mori. Biosci Biotechnol Biochem. 1992;56: 1692–1693. [DOI] [PubMed] [Google Scholar]
- 24.Nagata S, Morooka N, Matsumoto S, Kawai T, Nagasawa H. Effects of neuropeptides on feeding initiation in larvae of the silkworm, Bombyx mori. Gen Comp Endocrinol. 2011;172: 90–95. 10.1016/j.ygcen.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 25.Fan Y, Sun P, Wang Y, He X, Deng X, Chen X, et al. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2010;40: 581–591. 10.1016/j.ibmb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Lkhagva A, Daubnerova I, Chae H-S, Simo L, Jung S-H, et al. Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc Natl Acad Sci. 2013;110: E3526–3534. 10.1073/pnas.1310676110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poels J, Verlinden H, Fichna J, Van Loy T, Franssens V, Studzian K, et al. Functional comparison of two evolutionary conserved insect neurokinin-like receptors. Peptides. 2007;28: 103–108. [DOI] [PubMed] [Google Scholar]
- 28.Poels J, Birse RT, Nachman RJ, Fichna J, Janecka A, Vanden Broeck J, et al. Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides. 2009;30: 545–556. 10.1016/j.peptides.2008.10.012 [DOI] [PubMed] [Google Scholar]
- 29.Torfs H, Poels J, Detheux M, Dupriez V, Van Loy T, Vercammen L, et al. Recombinant aequorin as a reporter for receptor-mediated changes of intracellular Ca2+ -levels in Drosophila S2 cells. Invert Neurosci. 2002;4: 119–124. [DOI] [PubMed] [Google Scholar]
- 30.He X, Zang J, Li X, Shao J, Yang H, Yang J, et al. Activation of BNGR-A24 by direct interaction with tachykinin-related peptides from the silkworm Bombyx mori leads to the Gq- and Gs-coupled signaling cascades. Biochemistry. 2014;53: 6667–6678. 10.1021/bi5007207 [DOI] [PubMed] [Google Scholar]
- 31.Kostenis E, Waelbroeck M, Milligan G. Techniques: promiscuous Galpha proteins in basic research and drug discovery. Trends Pharmacol Sci. 2005;26: 595–602. [DOI] [PubMed] [Google Scholar]
- 32.Simon MI. G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J Biol Chem. 1995;270: 15175–15180. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H, Wei Z, Nachman RJ, Adams ME, Park Y. Functional phylogenetics reveals contributions of pleiotropic peptide action to ligand-receptor coevolution. Sci Rep. 2014;4: 6800 10.1038/srep06800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dircksen H. Insect ion transport peptides are derived from alternatively spliced genes and differentially expressed in the central and peripheral nervous system. J Exp Biol. 2009;212: 401–412. 10.1242/jeb.026112 [DOI] [PubMed] [Google Scholar]
- 35.Audsley N, Jensen D, Schooley DA. Signal transduction for Schistocerca gregaria ion transport peptide is mediated via both cyclic AMP and cyclic GMP. Peptides. 2013;41: 74–80. 10.1016/j.peptides.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 36.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9: 373–386. 10.1038/nrd3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khoury E, Clément S, Laporte SA. Allosteric and biased g protein-coupled receptor signaling regulation: potentials for new therapeutics. Front Endocrinol (Lausanne). 2014;5: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poels J, Van Loy T, Franssens V, Detheux M, Nachman RJ, Oonk HB, et al. Substitution of conserved glycine residue by alanine in natural and synthetic neuropeptide ligands causes partial agonism at the stomoxytachykinin receptor. J Neurochem. 2004;90: 472–478. [DOI] [PubMed] [Google Scholar]
- 39.DeFea KA, Vaughn ZD, O’Bryan EM, Nishijima D, Déry O, Bunnett NW. The proliferative and antiapoptotic effects of substance P are facilitated by formation of a beta -arrestin-dependent scaffolding complex. Proc Natl Acad Sci U S A. 2000;97: 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson EC, Bohn LM, Barak LS, Birse RT, Nässel DR, Caron MG, et al. Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-beta-arrestin2 interactions. J Biol Chem. 2003;278: 52172–52178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.