Abstract
Xanthomonas citri subsp. citri (Xcc) A strain causes citrus bacterial canker, a serious leaf, fruit and stem spotting disease of several Citrus species. X. alfalfae subsp. citrumelonis (Xac) is the cause of citrus bacterial spot, a minor disease of citrus nursery plants and X. campestris pv. campestris (Xc) is a systemic pathogen that causes black rot of cabbage. Xanthomonas spp. form biofilms in planta that facilitate the host infection process. Herein, the role of extracellular DNA (eDNA) was evaluated in the formation and stabilization of the biofilm matrix at different stages of biofilm development. Fluorescence and light microscopy, as well as DNAse treatments, were used to determine the presence of eDNA in biofilms and bacterial cultures. DNAse treatments of Xcc strains and Xac reduced biofilm formation at the initial stage of development, as well as disrupted preformed biofilm. By comparison, no significant effect of the DNAse was detected for biofilm formation by Xc. DNAse effects on biofilm formation or disruption varied among Xcc strains and Xanthomonas species which suggest different roles for eDNA. Variation in the structure of fibers containing eDNA in biofilms, bacterial cultures, and in twitching motility was also visualized by microscopy. The proposed roles for eDNA are as an adhesin in the early stages of biofilm formation, as an structural component of mature bacterial aggregates, and twitching motility structures.
Introduction
Biofilms adhere bacteria to surfaces within a tridimensional structure that protects the bacterium against antibiotics and abiotic stresses [1–4]. Most biofilms are composed of 10% bacteria and 90% extracellular polymeric matrix [5]. The extracellular matrix provides resistance to temperature, pH and other deleterious conditions and also confers mechanical stability. Biofilms are composed of water, proteins, exopolysaccharide, lipopolysaccharide, lipids, surfactants and extracellular DNA (eDNA) [5,6].
eDNA is an important component of the extracellular matrix in biofilms for several Gram-positive, Gram-negative bacteria [7–12] and archaea [13]. The presence of eDNA in the extracellular matrix was described in 1956 [14] and its importance in biofilm formation was first demonstrated for Pseudomonas aeruginosa in the early stages of bacterial adhesion [10]. Meanwhile, in clinical isolates of the same species, eDNA was found to be important for stability of the biofilm [11]. In P. aeruginosa biofilms, eDNA coats the surface like a net in which bacteria are attached, and may also form a mushroom-like cap over the outer surface of the aggregate [15]. Besides biofilm formation, eDNA contributes to bacterial resistance to antibiotics, e.g., a Bacillus cereus mutant, that did not produce eDNA, was more susceptible to actinomycin D than the wild type [9]. eDNA has also been demonstrated to act as nutrient source of phosphorus, nitrogen and carbon. DNAse in P. aeruginosa secreted via the Type II Secretion System is related with nutrient acquisition, biofilm dispersal and horizontal gene transfer [16–18]. At high concentrations, eDNA produces antimicrobial activity by chelating cations and destabilizing the bacterial outer membrane through lipopolysaccharide modification [19]. These findings identify eDNA as an important component of the extracellular bacterial matrix that plays a role in several processes related to bacterial colonization and virulence.
Xanthomonas citri subsp. citri (Xcc) (formerly X. axonopodis pv. citri) is the causal agent of Asiatic citrus bacterial canker (CBC), a serious disease of many citrus species [20,21]. X. alfalfae subsp. citrumelonis (Xac) is the cause of citrus bacterial spot (CBS), a minor foliar disease of young citrus plants in nurseries [22]. Xcc produces necrotic lesions on leaves, twigs and fruits that reduces fruit quality and marketability and restricts commercialization of plants and fruits in markets free of CBC [20]. Xcc A strain type is by far the most severe and widespread CBC pathogen and affects the widest range of Citrus species. Within Xcc A strains, two narrow host range variants have been described, Xcc A* and Aw from Southwest Asia and Florida, respectively, that cause CBC on Mexican lime (Citrus aurantifolia) [23–27]. X. campestris pv. campestris (Xc) is the causal agent of black rot, one of the most important diseases of crucifers worldwide. Black rot is a systemic vascular disease that causes symptoms including marginal leaf chlorosis, necrosis, darkening of leaf veins and vascular tissue within the stem [28,29].
Xcc and Xc produce biofilm to facilitate the infection process [3,30,31]. Several mechanisms in Xcc contribute to biofilm formation during the host-pathogen interaction: adhesins, the type III Secretion System, lipopolysaccharide, exopolysaccharide, type IV pili associated to twitching motility and chemotaxis [31–41]. Furthermore, biofilm formation in planta is related to the different host range of Xcc strains [42].
The roles that eDNA play in attachment and biofilm formation are not widely characterized for plant-bacterial interactions. Indeed, presence of eDNA in the extracellular matrix and in the different phases of biofilm formation have not been studied for Xanthomonas spp. Herein, we report on the contribution of eDNA to biofilm formation and the related twitching motility for several Xcc strains with different host range and ability to form biofilms, and compare these features with those of Xc, a systemic pathogen of cabbage, and Xac, a citrus pathogen able to infect and produce biofilm on citrus leaves [20,22,42]. In these studies, presence of eDNA was corroborated after DNase treatments and visualized by fluorescence staining of bacterial cultures, biofilms and bacteria undergoing twitching motility associated with bacterial aggregation.
Results
Detection of eDNA in Xanthomonas citri subsp. citri strains
Presence of eDNA during biofilm formation was confirmed by SYTO-9 staining of bacterial cultures at different stages (Figs 1 and 2, right side). Furthermore, eDNA was observed in bacterial cultures at the exponential growth phase (Fig 3), after plate growth (S1 Fig) and in twitching motility assays (Fig 4, right side). In addition, CV staining of the same cultures (Figs 1, 2 and 4) revealed similar extracellular structures to those observed with SYTO-9 and fluorescence microcopy. Microscopy observations of GFP tagged cells stained with propidium iodide (S1 Fig) indicated that fiber fluorescence is due to the presence of eDNA and not caused by bacterial cell aggregation. In all assays bacterial cells in a bacillary shape were distinguishable from the containing DNA fibers. These results confirm that most of the extracellular fibers have a high eDNA content.
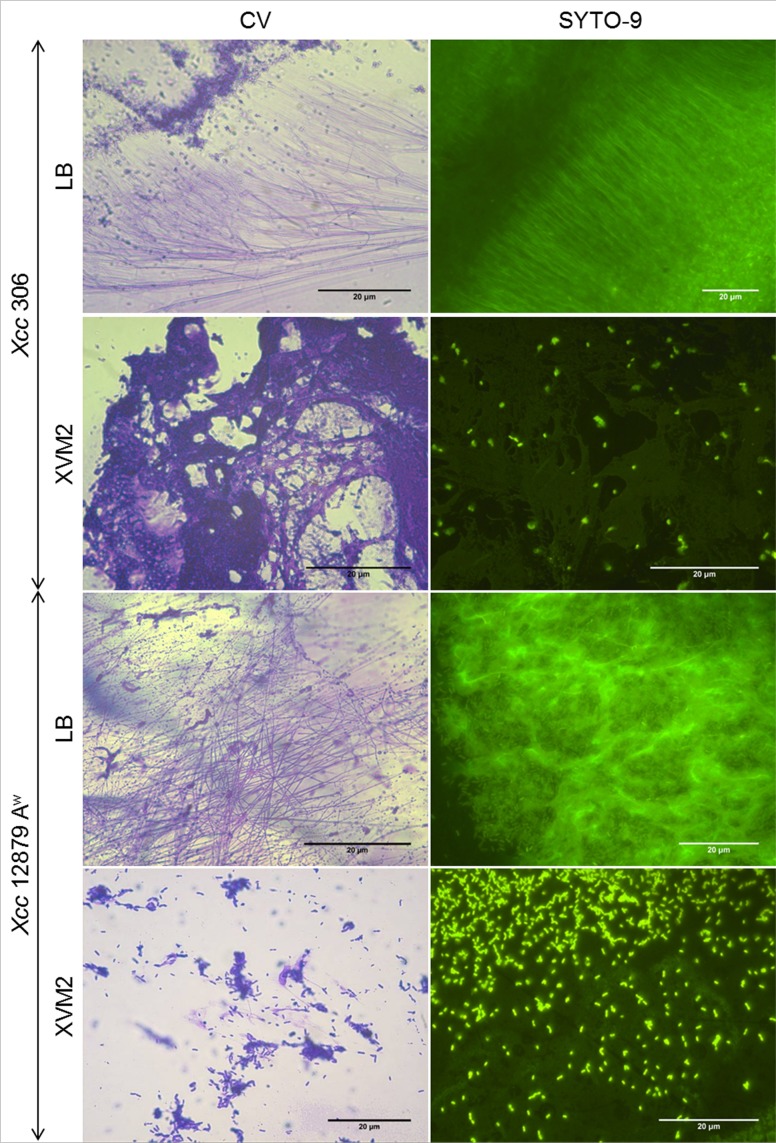
Fig 1. Presence of eDNA at the early stages of biofilm formation in Xcc.
Representative light (crystal violet, CV staining) and fluorescence (SYTO-9 staining) images of 72 h static cultures on LB or XVM2 media. Fibers were observed after both staining for strains Xcc 306 and Xcc 12879 Aw at the early stages of biofilm formation. Fibers interconnected cells at different stages of aggregation, from one to several cells are shown. In XVM2 medium, fibers were thicker and more uniform after staining with CV and SYTO-9. eDNA in XVM2 medium appeared to cover the surface like a sheet in contrast to individual fibers produced in LB medium.
Fig 2. Presence of eDNA in preformed biofilms of Xcc.
Representative light (CV staining) and fluorescence (SYTO-9 staining) images of mature biofilms on LB or XVM2 media. Both Xcc 306 and Xcc 12879 Aw strains were more aggregated in XVM2 than LB. A high level of aggregation for strain Xcc 12879 Aw in XVM2 made it difficult to observe eDNA fibers in aggregates. In XVM2, strain Xcc 306 was less aggregated and CV and SYTO-9 staining revealed eDNA surrounding the cells. In LB both staining revealed long fibers interconnecting aggregates.
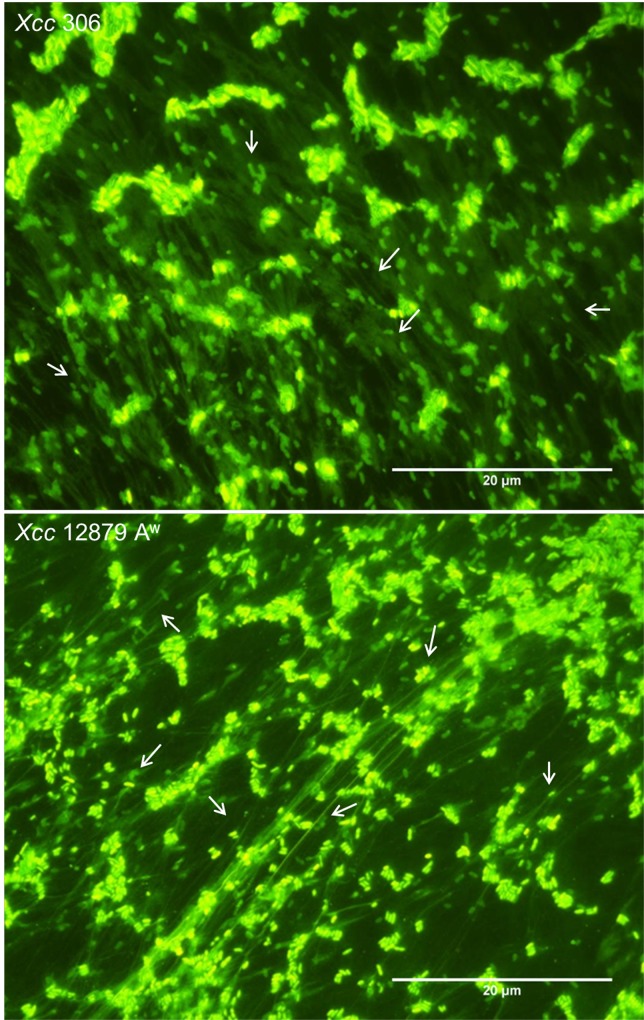
Fig 3. eDNA in exponential growth phase of Xcc.
Representative fluorescence microscopy images of Xcc 306 and Xcc 12879 Aw in LB broth at exponential growth phase stained with SYTO-9. A high level of eDNA fibers (marked with white arrows) were produced by both strains. Bacillary shape bacteria are distinguishable from the fibers that appeared to connect cells over a long distance. Xcc Aw 12879 fibers were well developed while strain Xcc 306 fibers were more diffuse.
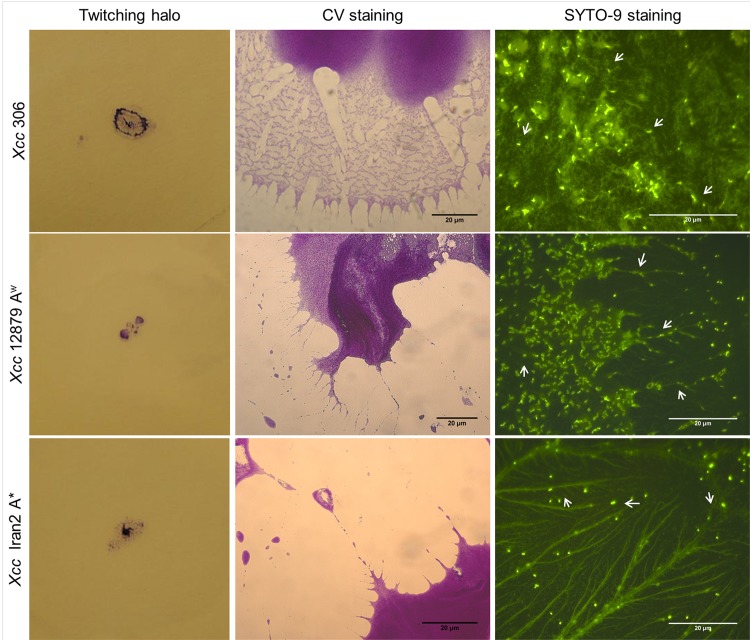
Fig 4. Presence of eDNA in Xcc twitching motility.
Twitching halos from Xcc 306, 12879 Aw and Iran2 A* and light and fluorescence microscopy of twitching halos stained with CV or SYTO-9. Twitching motility was observed between the plate surface and the medium. CV and SYTO-9 staining of strains revealed similar twitching structures. Fibers observed after CV staining appeared to have a high eDNA content. Twitching fibres were longer for Xcc 12879 Aw and Iran2 A* than for Xcc 306. Some fibers are marked with white arrows to highlight diference from bacillary shape bacteria associated.
Microscopic observations of biofilm development were in agreement with previously reported studies, since Xcc aggregation was higher in minimal than nutrient rich medium [31,42]. Less bacterial aggregation and biofilm formation for both wide and narrow host range strains of Xcc, Xcc 306 and Xcc 12879 Aw, were observed in LB medium than in XVM2 (Figs 1 and 2). The difference in biofilm formation between culture media was more marked for Xcc 12879 Aw than Xcc 306 [42].
eDNA appeared to be associated with long fibers of variable thickness and to an amorphous mass surrounding bacterial cells (Figs 1, 2, 3, 4 and S1 Fig). Fibers containing eDNA were observed in static cultures after 72 h (early stage of biofilm formation) as well as in mature biofilms (Figs 1 and 2). For strains Xcc 306 and Xcc 12879 Aw on LB and XVM2 media, the fibers appeared to interconnect bacterial cells within the aggregates. However fibers in LB medium were longer, thinner and less connective than in XVM2 medium. The short and branched filaments in XVM2 appeared to provide higher bacterial connectivity and aggregate stability. Differences in the development of eDNA fibers were also observed between Xcc 306 and Xcc 12879 Aw in exponential growth phase (Fig 3) and plate growth (S1 Fig) in LB medium. Similar results were observed in the twitching motility assay (Fig 4); fibers produced by strains Xcc 12879 Aw and Iran 2 A* were longer than the fibers produced by Xcc 306 that correlated to the different phenotype of the twitching colonies demonstrated for narrow and wide host range strains.
Importance of eDNA in Xanthomonas biofilm formation
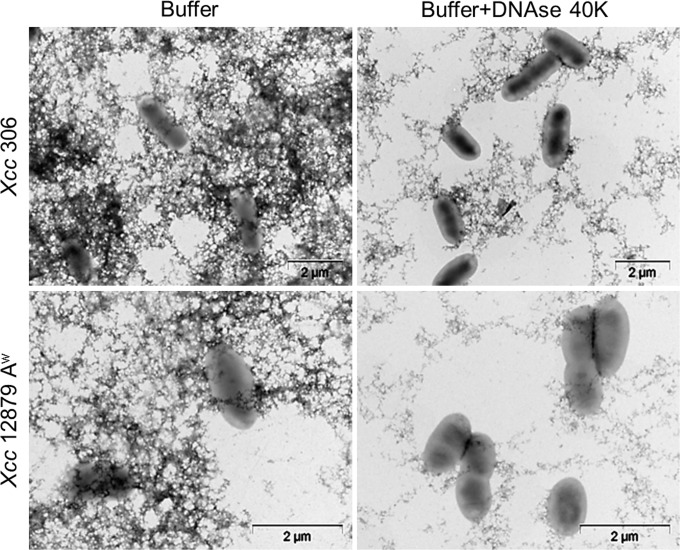
The role of eDNA in biofilm formation was estimated by measuring biofilm formation after DNAse I treatment at 0, 24, 48 or 72 hours post seeding (hps) by staining biofilms with crystal violet (CV) as described in Material and Methods. Transmission electron microscopy of Xcc cultures treated with DNAse showed reduction of extracellular fibers (Fig 5), confirming the DNAse activity and therefore the eDNA content. The bacterial population was not reduced by DNAse treatment (S2 Fig) and cell division in LB medium was shown under DNAse treatment (Fig 5).
Fig 5. Effect of DNAse in the extracellular matrix of Xcc.
Transmission Electron Microscopy of Xcc 306 and 12879 Aw strains in exponential growth phase, after DNAse and no-DNAse (Buffer) treatments. DNAse treated bacteria showed less extracellular structures than de buffer control. The remaining structures observed after treatment could be associated to extracellular proteins. Bacterial cell division is shown under DNAse treatment.
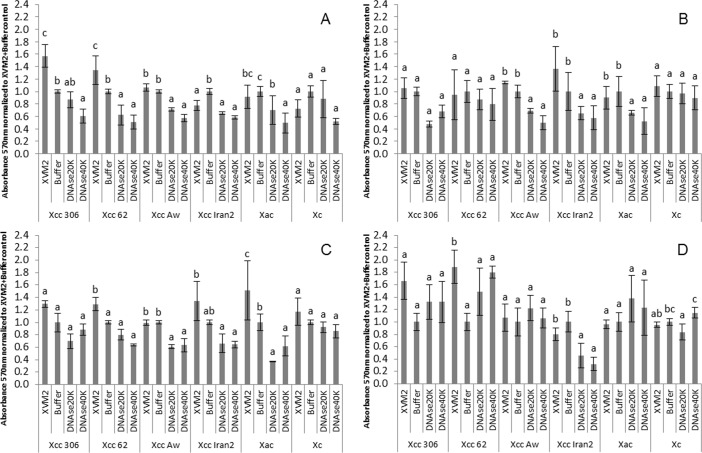
Biofilm formation was compared in XVM2 medium, XVM2 medium plus buffer, and XVM2 medium plus buffer and DNAse I at 20 or 40 Kunitz mL-1. In most cases, reduction of biofilm formation was dependent on the timing of DNAse I treatment, but was not dependent on DNAse I concentration (Fig 6).
Fig 6. Effect of DNAse I during biofilm formation in Xanthomonas.
Biofilm formation after treatment with DNAse I at different times after bacterial seeded with different Xanthomonas species and strains (see Table 1). A, DNAse added at 0 h post seeding (hps); B, DNAse added at 24 hps; C, DNAse added at 48 hps; and D, DNAse added at 72 hps. The absorbance values were normalized to the control XVM2 plus buffer in order to compare the response for different strains. Error bars represent the standard deviation. Graphs are a representative assay of at least three assays with three replicates per assay. Statistical analyses were performed using STATGRAPHICS Plus, version 5.1 (Copyright Manugistics Inc.).
Reduction of biofilm was significant (p<0.05) for citrus xanthomonads and DNAse I application time. In contrast, DNAse I did not produce a statistically significant reduction of biofilm formation by Xc 1609 strain (p>0.05). Citrus Xanthomonas strains varied widely in response to DNAse I (Fig 6). In wide host range strains Xcc 306 and Xcc 62 biofilm formation was statistically reduced (p<0.05) when DNAse I was added at 0 hps but no reduction was observed at longer exposure times (p>0.05). For narrow host range strains, Xcc 12879 Aw and Iran2 A*, and Xac F1 biofilm formation was reduced by DNAse I treatmentsat 0, 24 and 48 hps. Furthermore, biofilm produced by the Iran2 A* strain was reduced at 72 hps after DNAse I treatment.
Importance of eDNA in preformed biofilm
To evaluate the role of eDNA in mature biofilms, DNAse I treatments were performed for 1 h or overnight. Biofilm disruption was observed after both incubation periods but overnight treatment had greater effect (Fig 7). DNAse I treatment for 1 h did not affect strains Xcc 306 and Xc 1609, but significantly (p<0.05) ruptured biofilm for strains Xcc 62, 12789 Aw, Iran 2 A* and Xac F1 (Fig 7A). Overnight DNAse treatment of Xcc strains affetcted up to 80–90% of the biofilm and no differences were observed between wide and narrow host range Xcc strains (Fig 7B). Similar treatment did not significantly (p>0.05) disrupt biofilm of Xc 1609 compared to the buffer control. In summary, biofilm disruption by DNAse I was greater for Xcc strains than Xac F1 or Xc 1609.
Fig 7. Xanthomonas biofilm rupture by DNAse I.
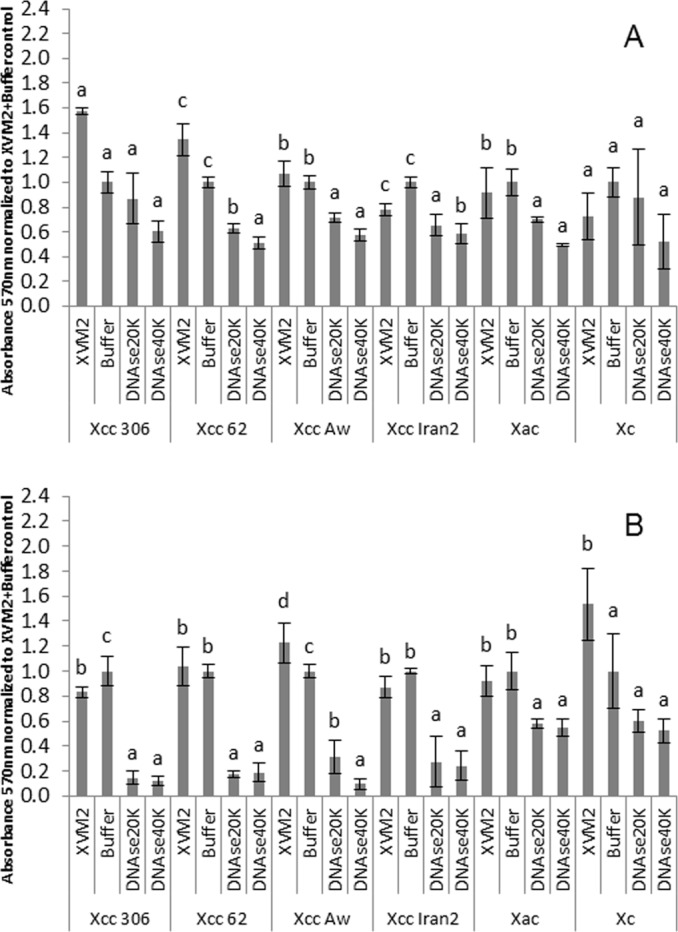
A, DNAse treatment of 1 h duration. B, DNAse treatment for an overnight period. The absorbance values were normalized to the control XVM2 plus buffer in order to compare the response for different strains. Error bars represents the standard deviation. Graphs are a representative assay of at least three assays with three replicates per assay
Discussion
Biofilm formation is an important virulence factor in Xcc [3,31]. Furthermore several differences in biofilm structures and biofilm formation have been observed between wide and narrow host range strains of Xcc which was related to their ability to infect the host [42]. The variation in function of eDNA in biofilm formation between wide and narrow Xcc strains could also be associated with the ability to form biofilms and their structure.
Extracellular DNA (eDNA) is an important component of the biofilm matrix in several bacteria, playing roles in adhesion, bacterial aggregate stability, nutrition, protection and gene exchange [9,10,16–19]. In this study, eDNA was detected either at different stages of the biofilm or in cultures and during twitching motility in citrus Xanthomonas strains with different host range. The presence of eDNA in biofilms for citrus Xanthomonas was determined and compared to Xc.
eDNA in Xcc has been observed in fibers interconnecting bacterial cells, possibly associated with other extracellular components including proteins and polysaccharides. Amorphous eDNA was also observed which is possibly eDNA that had been recently secreted, but not processed into fibers. Fibers were observed during biofilm formation in LB and XVM2 media, however those fibers formed in XVM2 appeared to be thicker and more uniform than those in LB medium, which is consistent with previous reports of higher biofilm formation in XVM2 medium. In previous reports [31,42] biofilm formation varied between Xcc 306 and 12879 Aw, especially in LB medium. In XVM2, a culture medium that mimics apoplastic conditions, several pathogenicity factors are expressed for Xcc strains [26,43] and among them, eDNA could be considered an important virulence component in the biofilm for plant infection.
In P. aeruginosa, eDNA is a factor associated with twitching motility [44]. Twitching is enabled through the extension and retraction of the type IV pilus [45] which binds to DNA [46,47]. Herein, SYTO-9 staining of twitching assay plates revealed the presence of fibers containing eDNA during this type of movement recently been described for Xcc [32,48]. Moreover, twitching structures varied among narrow and wide host range strains of Xcc as also observed for biofilm formation on plant surfaces [42]. Twitching fibers of narrow host range strains were longer than those of the wide host range strains which had more interconnecting fibers, this may be related with the variable twitching halo shape between wide and narrow host range Xcc strains. Interestingly, Xcc cells were always connected by fibers containing eDNA. In P. aeruginosa, eDNA along with surfactants, were secreted onto the culture plate surface as the bacterium was undergoing motility [44]. In our study, Xcc appeared to track the eDNA in fibers possibly through the type IV pilus, since no surfactant activity was detected for Xcc (results not shown). Our hypothesis is that eDNA is associated with fibers on the plant surface in the initial stage of the colonization process, that involves motility and bacterial aggregation. Bacteria twitch across the surface by binding the type IV pilus with eDNA fibers deposited on the surface.
Biofilms treatments with DNAse confirmed the role of eDNA in biofilm formation in citrus related Xanthomonas. In many cases, DNAse treatment reduced biofilm formation depending on the strain and the biofilm stage. DNAse did not cause mortality or reduce bacterial growth rate, but was limited to the effect on biofilm formation (S2 Fig). When DNAse treatment terminated, bacterial attachment to the surfaces and aggregation resumed thereafter.
eDNA has been described as an adhesin in B. cereus [9], Staphylococcus epidermis, S. aureus, S. mutants [8] and P. aeruginosa [10]. Herein, similar eDNA properties were demonstrated for citrus strains of Xanthomonas. eDNA appeared to be important for the early stages of attachment by wide host range Xcc strains, whereas narrow host range strains required eDNA for biofilm development over an extended period, concurring with their higher requirements for biofilm formation. In planta, at 24 hpi, wide host range Xcc strains produced more complex structures in biofilms than the narrow host range Xcc strains and Xac strain which aggregated by producing long fibers [42]. The less interconnected fibers of the narrow host range Xcc strains and Xac strain may have rendered them more susceptible to DNAse disruption of aggregated cells and altered their biofilm formation. Other extracellular structures such as fimbria or flagella could influence biofilm formation and those structures may possibly be associated with eDNA since most of the fibers observed with CV stain were also stained with SYTO-9.
The role of eDNA in the stability of biofilms is important for bacteria including E. coli, B. subtilis [49], S. mutants [50], some strains of P. aeruginosa [11] and, as this study reports, strains of Xcc and Xac. Mature biofilms of citrus strains, but not the crucifer strain Xc 1609, were disrupted when exposed to DNAse. This observation suggests that the systemic pathogen Xc has a low dependence on eDNA for biofilm formation, surface attachment and stability of the biofilm or that eDNA is associated with other structures, making them inaccessible to DNAse I. Our hypothesis is that Xc moves systemically and aggregation would not be occurring after the initial stage of the plant colonization [51]. Hence, the role of eDNA in the different bacteria, depends on the structure and function of biofilm in plant colonization process in the Xanthomonas strain-host interaction.
Biofilm has been described as major factor for Xcc virulence and survival in citrus [3,31]. Several factors influence biofilm for wide host range Xcc strains. Mutants in the type IV pili produced biofilm but less structured than that produced by the wild type strain [32]. Non-fimbrilar adhesins have been described in the initial attachment of Xcc to abiotic surfaces [33]. Our findings provide a clue to a plausible role of eDNA acting in early stages of the biofilm as an adhesin in conjunction probably with other adhesins. Many studies describe the disruption of eDNA in animal bacterial pathogens, for examples, use of recombinant DNAse as a treatment against P. aeruginosa for cystic fibrosis patients [52] and extracellular DNAse (NucB) from B. licheniformis to inhibit and break bacterial biofilms [53]. Although biofilm formation and motility are well-described virulence factors for many plant pathogenic bacteria, occurrence and function of eDNA in plant pathogens in these processes is less well known. The determination of roles of eDNA in plant pathogenesis may lead to a better understanding of the plant-pathogen interaction and perhaps the development of tactics to disrupt eDNA to effect bacterial disease control.
Material and Methods
Bacterial strains, culture media and growth conditions
Bacterial strains used in this study and their natural hosts are listed in Table 1. Two wide host range strains of Xcc A, and two narrow host range strains, A* and Aw, were evaluated along with X. alfalfae subsp. citrumelonis and X. campestris pv. campestris. Bacterial strains were routinely grown in Luria Bertani (LB) broth (10 g tryptone, 5 g yeast extract and 5 g sodium chloride per litre) or on LB plates (1.5% bacteriological agar) at 27°C for 48 h.
Table 1. Strains of Xanthomonas spp. used in the study.
| Strain | Taxon, or disease and CBC type | Natural Host |
|---|---|---|
| Xcc 306 | Xanthomonas citri subsp. citri, CBCa A | Citrus spp. |
| Xcc 62 | Xanthomonas citri subsp. citri, CBC A | Citrus spp. |
| Xcc Iran 2 | Xanthomonas citri subsp. citri, CBC A* | C. aurantifolia |
| Xcc 12879 Aw | Xanthomonas citri subsp. citri, CBC Aw | C. aurantifolia |
| Xac F1 | Xanthomonas alfalfae subsp. citrumelonis, CBSb | Citrus spp. |
| Xc 1609 | Xanthomonas campestris pv. campestris CBRc | Cabbage |
| 306 pUFZ75d | Xanthomonas citri subsp. citri, CBC A | |
| 12879 pUFZ75e | Xanthomonas citri subsp. citri, CBC Aw |
In addition and to discriminate bacterial cells from eDNA contained in fibres, GFP tagged strains of Xcc 306 and Xcc 12879 Aw were used [3,42]. GFP expressing strains were grown on LB broth or plates with the addition of kanamycin at 50 μl mL-1 (LB+K).
eDNA staining
To detect eDNA, samples were stained with SYTO-9 from the Live/Dead Bacterial viability kit (Molecular Probes Europe BV; Leiden, The Netherlands) and observed with fluorescence microscopy. SYTO-9 stains DNA and is also able to penetrate bacterial membranes [54,55]. Crystal Violet (CV) staining was also performed to visualize extracellular structures as previously described for flagella visualization of Xcc [56]. To determinate the presence of eDNA in biofilms, a Xcc 306 and Xcc 12879 Aw colony was taken and seeded into 5 mL LB medium for an overnight period, a 60 μL aliquot was then seeded in 30 mL of LB medium an incubated for an overnight period. Finally bacteria in exponential growth phase (0.5–0.6 ODs) were washed twice in 10mM MgCl2 and diluted to a concentration of 108 cfu mL-1 in 25 mL of XVM2 (20 mm NaCl, 10 mm (NH4)2SO4, 5 mm MgSO4, 1 mm CaCl2, 0.16 mm KH2PO4, 0.32 mm K2HPO4, 0.01 mm FeSO4, 10 mm fructose, 10 mm sucrose, 0.03% casamino acid) [43,57] or LB media, and incubated without shaking at 27°C in a 50 mL borosilicate flask. In order to detect eDNA at the early stages of biofilm, bacteria growing and deposited on bottom of the flask were collected at 72 hours post seeding (hps) and stained either with SYTO-9 for 20 min in the dark or with CV. For mature biofilm eDNA detection, after 72 hps of static growth in 50 mL borosilicate flask, the liquid medium was decanted and the flasks were incubated an additional 72 h in dried condition in order to simulate natural situation. To stain mature biofilm, bacteria were collected by adding 50 μL of sterile distilled water (SDW) and scraping them from the bottom of the flask followed by the staining procedures described above.
To detect eDNA in exponential growth phase bacteria, an aliquot of 50 μL was collected from LB liquid cultures of Xcc 306 and Xcc 12879 Aw and deposited onto a glass slide following the staining procedures described above.
To detect eDNA in plate growth Xcc 306 and Xcc 12879 Aw carrying the GFP plasmid pUFZ75 [3,42] were collected from LB+K, stained with propidium iodide from the Live/Dead Bacterial viability kit (Molecular Probes Europe BV; Leiden, The Netherlands), deposited on a glass slide and observed with fluorescence microscopy.
To evaluate twitching motility, bacteria in exponential growth phase were washed twice with 10 mM MgCl2 and suspended in 1.0 mL of MgCl2 in a 1.5 mL centrifuge tube. Bacteria were then centrifuged at 10,000 g for 10 min and the supernatant was discarded. Bacteria were inoculated with a toothpick in PYM agar medium (Peptone 0.5%, Yeast extract 0.3%, Malt extract 0.3% and bacteriological agar 1%) supplemented with 2% glucose. Plates were incubated at 27°C for 7 d. In order to estimate the twitching production and observe the twitching halo the medium was removed and plates were washed and stained with 0.3% of CV. To observe extracellular structures and eDNA the twitching colony was stained with SYTO-9 or CV. After 20 min of staining, plates were washed with SDW and observed under the light and fluorescence microscope.
Determination of eDNA in biofilms
To confirm the presence of eDNA in biofilm, DNAse (Deoxyribonuclease I from bovine pancreas (2000 units mg-1); Sigma Aldrich Inc., St. Louis, MO, USA) was added at different concentrations to static cultures of Xanthomonas strains at different times.
In order to visualize the effect of DNAse in the extracellular matrix, bacterial broth cultures were treated with DNAse at a 40 Kunitz mL-1 concentration and observed by transmission electron microscopy (TEM) after staining with 1% uranyl acetate.
To evaluate biofilm formation, bacterial cultures in exponential growth phase, performed as described above, were washed twice with 10 mM MgCl2 and suspended at 108 cfu mL-1 in XVM2 medium. XVM2 medium has been demonstrated to increase biofilm formation for Xcc strains when compared to other media such as LB [31,42]. Microtiter plate wells were filled with 200 μL of the bacterial culture and incubated without shaking for 72 h at 27°C. After removing the medium, bacteria were incubated for an additional 72 h in dried conditions, as described above for staining mature biofilms. Biofilm formation was measured after rinsing the plates with SDW and staining with 0.3% CV for 15 min. Excess stain was removed by rinsing the plates with SDW. Residual CV in each well was solubilized in 200μL of an acetone (20%), ethanol (80%) mixture and the absorption of the extract measured in a microtiter plate reader set at A570 nm wavelength. Absorption values for each strain and treatment were calculated as the mean of readings from three wells from at least three different assays. The means were compared by analysis of variance (ANOVA) and separated by Student-Newman-Keuls (SNK) multiple range test using Statgraphics Plus for Windows 4.1 (Statistical Graphics, Rockville, MD).
For the different treatments, DNAse I was suspended in 0.15 M of NaCl and then in DNAse I reaction buffer (10mM of MgCl2 and 50% of glycerol suspended in 10mM of Tris-HCl at pH 7.5) as previously described [7]. DNAse I was added at concentrations of 20 or 40 Kunitz mL-1 at 0, 24, 48 and 72 hps to evaluate eDNA effect on biofilm development or on preformed biofilm. In order to determine if variation in biofilm was affected by the reaction buffer, a control treatment with the buffer without DNAse I was included.
Conclusions
Presence of eDNA in Xcc with different host range is described as an important component of filaments connecting bacteria during bacterial growth, biofilm formation and twitching motility. In addition, fibers comprising eDNA were thicker and more connective during biofilm formation by different Xcc strains in certain culture media.
The role of eDNA in biofilm formation was determined for citrus and crucifer pathogenic xanthomonads. Whereas DNAse treatments did not produce a significant effect on biofilm of Xc 1609 strain, for citrus xanthomonads, both Xcc and Xac, biofilms were reduced after DNAse treatments performed at early stages of biofilm formation as well as in mature aggregates. Furthermore, DNAse effects differed between narrow and wide host range strains of Xcc in early stages of biofilm; the narrow host range strains were affected over a longer period of time. Once the biofilm was established no differences were shown between the two Xcc strain types. Those results demonstrate the role of eDNA for biofilm formation and maintenance for citrus xanthomonads strains, at the early stage of biofilms and as structural component.
The presence and potential importance of eDNA in biofilm formation by citrus xanthomonads opens the door for future studies of new control methods for citrus bacterial canker disease based on the disruption of biofilms or interference in their formation.
Supporting Information
Representative fluorescence microscopy images of Xcc 306 and Xcc 12879 Aw transformed with plasmid pUFZ75 [3,42] after 48 hpi plate growth in LB medium stained with propidium iodide. eDNA fibers are shown in red and in some areas marked with white arrows.
(TIF)
Bacterial population of xanthomonads strains was estimated in biofilm induction condition (XVM2 medium and static growth) after DNAse treatment in order to demonstrate that DNAse did not influence bacterial population and therefore biofilm formation. No differences were observed at 0, 24 or 72 hours for the treatments assayed, solely DNAse at 40 Kunitz mL-1 treatment showed higher population, however biofilm formation after this treatment showed the minor biofilm formation.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Citrus Research and Development Foundation project CRDF546 (JHG JC) and Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) project RTA2008-00048 (JC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Busscher HJ, van der Mei HC (2012) How do bacteria know they are on a surface and regulate their response to an adhering state? PLoS Pathog 8: e1002440 10.1371/journal.ppat.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova OE, Sauer K (2012) Sticky Situations: Key Components That Control Bacterial Surface Attachment. Journal of Bacteriology 194: 2413–2425. 10.1128/JB.00003-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cubero J, Gell I, Johnson EG, Redondo A, Graham JH (2011) Unstable green fluorescent protein for study of Xanthomonas citri subsp. citri survival on citrus. Plant Pathology 60: 977–985. [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322. [DOI] [PubMed] [Google Scholar]
- 5.Flemming H-C, Wingender J (2010) The biofilm matrix. Nat Rev Micro 8: 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 6.Strelkova EA, Pozdnyakova NV, Zhurina MV, Plakunov VK, Belyaev SS (2013) Role of the extracellular polymer matrix in resistance of bacterial biofilms to extreme environmental factors. Microbiology 82: 119–125. [DOI] [PubMed] [Google Scholar]
- 7.Conover MS, Mishra M, Deora R (2011) Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS One 6: e16861 10.1371/journal.pone.0016861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP (2010) Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol 76: 3405–3408. 10.1128/AEM.03119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilain S, Pretorius JM, Theron J, Brözel VS (2009) DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75: 2861–2868. 10.1128/AEM.01317-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS (2002) Extracellular DNA required for bacterial biofilm formation. Science 295: 1487 [DOI] [PubMed] [Google Scholar]
- 11.Nemoto K, Hirota K, Murakami K, Taniguti K, Murata H, Viducic D et al. (2003) Effect of varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy 49: 121–125. 10.1159/000070617 [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon W et al. (2008) Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiology 8: 173 10.1186/1471-2180-8-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chimileski S, Franklin MJ, Papke RT (2014) Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC biology 12: 65 10.1186/s12915-014-0065-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catlin WB (1956) Extracellular deoxyribonucleic acid of bacteria and a deoxyribonuclease inhibitor. Science 124: 441–442. [DOI] [PubMed] [Google Scholar]
- 15.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S et al. (2006) A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Molecular Microbiology 59: 1114–1128. [DOI] [PubMed] [Google Scholar]
- 16.Bakkali M (2013) Could DNA uptake be a side effect of bacterial adhesion and twitching motility? Archives of Microbiology 195: 279–289. 10.1007/s00203-013-0870-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakubovics NS, Shields RC, Rajarajan N, Burgess JG (2013) Life after death: the critical role of extracellular DNA in microbial biofilms. Lett Appl Microbiol 57: 467–475. 10.1111/lam.12134 [DOI] [PubMed] [Google Scholar]
- 18.Mulcahy H, Charron-Mazenod L, Lewenza S (2010) Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol 12: 1621–1629. 10.1111/j.1462-2920.2010.02208.x [DOI] [PubMed] [Google Scholar]
- 19.Mulcahy H, Charron-Mazenod L, Lewenza S (2008) Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4: e1000213 10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham JH, Gottwald TR, Cubero J, Achor DS (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol Plant Pathol 5: 1–15. MPP197 [pii]; 10.1046/j.1364-3703.2004.00197.x [DOI] [PubMed] [Google Scholar]
- 21.Schaad NW, Postnikova E, Lacy GH (2006) Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol 29: 690–695. 10.1016/j.syapm.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 22.Graham JH, Gottwald TR (1990) Variation in aggressiveness of Xanthomonas campestris pv. citrumelo associated with citrus bacterial spot in Florida citrus nurseries. Phytopathology 80: 190–196. 10.1094/Phyto-80-190 [DOI] [Google Scholar]
- 23.Cubero J, Graham JH (2002) Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Applied Environmental Microbiology 68: 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Stall RE, Jones JB, Cubero J, Gottwald TR, Graham JH et al. (2004) Detection and characterization of a new strain of citrus canker bacteria from Key/Mexican lime andalemow in South Florida. Plant Disease 88: 1179–1188. [DOI] [PubMed] [Google Scholar]
- 25.Jalan N, Kumar D, Yu F, Jones JB, Graham JH, Wang N (2013) Complete genome sequence of Xanthomonas citri subsp. citri Strain Aw 12879, a restricted-host-range citrus canker-causing bacterium. Genome Announcements 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jalan N, Kumar D, Andrade MO, Yu F, Jones JB, Graham JH et al. (2013) Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. Bmc Genomics 14: 551 10.1186/1471-2164-14-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernière C, Hartung JS, Pruvost OP, Civerolo EL, Álvarez AM, Maestri P et al. (1998) Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. European Journal of Plant Pathology 104: 477–487. [Google Scholar]
- 28.Hayward AC (1993) The hosts of Xanthomonas In: Swings JG, Civerolo EL, editors. Xanthomonas. Springer; Netherlands: pp. 1–119. [Google Scholar]
- 29.Vicente JG, Holub EB (2013) Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Molecular Plant Pathology 14: 2–18. 10.1111/j.1364-3703.2012.00833.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crossman L, Dow JM (2004) Biofilm formation and dispersal in Xanthomonas campestris. Microbes and Infection 6: 623–629. [DOI] [PubMed] [Google Scholar]
- 31.Rigano LA, Siciliano F, Enrique R, Sendin L, Filippone P, Torres PS et al. (2007) Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact 20: 1222–1230. 10.1094/MPMI-20-10-1222 [DOI] [PubMed] [Google Scholar]
- 32.Dunger G, Guzzo CR, Andrade MO, Jones JB, Farah CS (2014) Xanthomonas citri subsp. citri type IV pilus is required for twitching motility, biofilm development, and adherence. MPMI 27: 1132–1147. 10.1094/MPMI-06-14-0184-R [DOI] [PubMed] [Google Scholar]
- 33.Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J (2009) A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One 4: e4358 10.1371/journal.pone.0004358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Wang N (2011) The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Molecular Plant Pathology 12: 381–396. 10.1111/j.1364-3703.2010.00681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malamud F, Torres PS, Roxana R, Rigano LA, Enrique R, Bonomi H et al. (2011) Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 157: 819–829. mic.0.044255–0 [pii]; 10.1099/mic.0.044255-0 [DOI] [PubMed] [Google Scholar]
- 36.Malamud F, Homem RA, Conforte VP, Yaryura PM, Castagnaro AP Marano MR et al. (2013) Identification and characterization of biofilm formation-defective mutants of Xanthomonas citri subsp. citri. Microbiology 159: 1911–1919. 10.1099/mic.0.064709-0 [DOI] [PubMed] [Google Scholar]
- 37.Yaryura PM, Conforte VP, Malamud F, Roeschlin R, De Pino V, Castagnaro AP et al. (2015) XbmR, a new transcription factor involved in the regulation of chemotaxis, biofilm formation and virulence in Xanthomonas citri subsp. citri. Environ Microbiol n/a. [DOI] [PubMed] [Google Scholar]
- 38.Zimaro T, Thomas L, Marondedze C, Sgro G, Garofalo C, Ficarra F et al. (2014) The type III protein secretion system contributes to Xanthomonas citri subsp. citri biofilm formation. BMC Microbiology 14: 96 10.1186/1471-2180-14-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimaro T, Thomas L, Marondedze C, Garavaglia B, Gehring C, Ottado J et al. (2013) Insights into Xanthomonas axonopodis pv. citri biofilm through proteomics. BMC Microbiology 13: 186 10.1186/1471-2180-13-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreira L, Facincani A, Ferreira C, Ferreira R, Ferro M, Gozzo F et al. (2014) Chemotactic signal transduction and phosphate metabolism as adaptive strategies during citrus canker induction by Xanthomonas citri. Functional and Integrative Genomics 15: 197–210. 10.1007/s10142-014-0414-z [DOI] [PubMed] [Google Scholar]
- 41.Sgro GG, Ficarra FA, Dunger G, Scarpeci TE, Valle EM, Cortadi A et al. (2012) Contribution of a harpin protein from Xanthomonas axonopodis pv. citri to pathogen virulence. Molecular Plant Pathology 13: 1047–1059. 10.1111/j.1364-3703.2012.00814.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sena-Vélez M, Redondo C, Gell I, Ferragud E, Johnson E, Graham JH et al. (2015) Biofilm formation and motility of Xanthomonas strains with different citrus host range. Plant Pathology 64: 767–775. 10.1111/ppa.12311 [DOI] [Google Scholar]
- 43.Astua-Monge G, Freitas-Astua J, Bacocina G, Roncoletta J, Carvalho SA, Machado MA (2005) Expression profiling of virulence and pathogenicity genes of Xanthomonas axonopodis pv. citri. Journal of Bacteriology 187: 1201–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM et al. (2013) Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proceedings of the National Academy of Sciences 110: 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattick JS (2002) Type IV pili and twitching motility. Annu Rev Microbiol 56: 289–314. 10.1146/annurev.micro.56.012302.160938 [DOI] [PubMed] [Google Scholar]
- 46.Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M et al. (2013) Specific DNA recognition mediated by a type IV pilin. Proceedings of the National Academy of Sciences 110: 3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Schaik EJ, Giltner CL, Audette GF, Keizer DW, Bautista DL, Slupsky CM et al. (2005) DNA binding: a novel function of Pseudomonas aeruginosa type IV Pili. Journal of Bacteriology 187: 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sena-Vélez M, Ferragud E, Redondo C, Johnson EG, Graham JH, Girón JA et al. (2012) Comparative study of different host range strains of Xanthomonas citri subsp. citri: chemotaxis and biofilm formation. XII International Citrus Congress 197. [Google Scholar]
- 49.Shields RC, Mokhtar N, Ford M, Hall MJ, Burgess JG, ElBadawey MR et al. (2013) Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS One 8: e55339 10.1371/journal.pone.0055339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ et al. (2014) Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. Journal of Bacteriology 196: 2355–2366. 10.1128/JB.01493-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL (2003) Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A 100: 10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs HJ, Borowitz DS, Christiansen DH, Morris EM, Nash ML, Ramsey BW et al. (1994) Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. New England Journal of Medicine 331: 637–642. [DOI] [PubMed] [Google Scholar]
- 53.Nijland R, Hall MJ, Burgess JG (2010) Dispersal of Biofilms by Secreted, Matrix Degrading, Bacterial DNase. PLoS One 5: e15668 10.1371/journal.pone.0015668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Böckelmann U, Janke A, Kuhn R, Neu TR, Wecke J, Lawrence JR et al. (2006) Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiology Letters 262: 31–38. [DOI] [PubMed] [Google Scholar]
- 55.Lawrence JR, Swerhone GDW, Leppard GG, Araki T, Zhang X, West MM et al. (2003) Scanning Transmission X-Ray, Laser Scanning, and Transmission Electron Microscopy Mapping of the Exopolymeric Matrix of Microbial Biofilms. Appl Environ Microbiol 69: 5543–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraiselburd I, Alet AI, Tondo ML, Petrocelli S, Daurelio LD, Monzón J et al. (2012) A LOV protein modulates the physiological attributes of Xanthomonas axonopodis pv. citri relevant for host plant colonization. PLoS One 7: e38226 10.1371/journal.pone.0038226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wengelnik K, Marie C, Russel M, Bonas U (1996) Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction ofthe hypersensitive reaction. Journal of Bacteriology 178: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative fluorescence microscopy images of Xcc 306 and Xcc 12879 Aw transformed with plasmid pUFZ75 [3,42] after 48 hpi plate growth in LB medium stained with propidium iodide. eDNA fibers are shown in red and in some areas marked with white arrows.
(TIF)
Bacterial population of xanthomonads strains was estimated in biofilm induction condition (XVM2 medium and static growth) after DNAse treatment in order to demonstrate that DNAse did not influence bacterial population and therefore biofilm formation. No differences were observed at 0, 24 or 72 hours for the treatments assayed, solely DNAse at 40 Kunitz mL-1 treatment showed higher population, however biofilm formation after this treatment showed the minor biofilm formation.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.