Abstract
Telomeric DNA of Trypanosomatids possesses a modified thymine base, called base J, that is synthesized in a two-step process; the base is hydroxylated by a thymidine hydroxylase forming hydroxymethyluracil (hmU) and a glucose moiety is then attached by the J-associated glucosyltransferase (JGT). To examine the importance of JGT in modifiying specific thymine in DNA, we used a Leishmania episome system to demonstrate that the telomeric repeat (GGGTTA) stimulates J synthesis in vivo while mutant telomeric sequences (GGGTTT, GGGATT, and GGGAAA) do not. Utilizing an in vitro GT assay we find that JGT can glycosylate hmU within any sequence with no significant change in Km or kcat, even mutant telomeric sequences that are unable to be J-modified in vivo. The data suggests that JGT possesses no DNA sequence specificity in vitro, lending support to the hypothesis that the specificity of base J synthesis is not at the level of the JGT reaction.
Keywords: Glucosyltransferase, base J, Trypanosomatid, epigenetic, RNA Polymerase II transcription
Graphical abstract
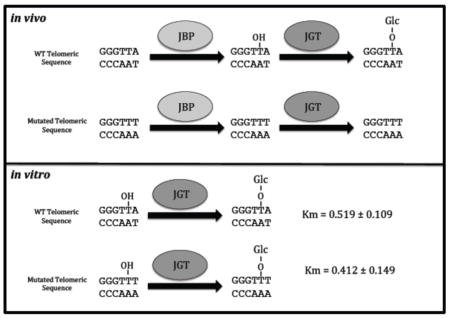
in vivo only particular DNA sequences can become J modified. The specificity of J modification is not due to the JGT.
Trypanosomatids, including the human pathogens Trypanosoma brucei, Trypanosoma cruzi, and Leishmania, possess a unique DNA modification within their genomes known as base J [1, 2]. Base J (β-D-glucopyranosyloxymethyluracil) is a hyper-modified thymine residue predominately present in repetitive sequences, such as telomeric repeats [2, 3]. While the function of base J in telomeric repeats is unknown, base J is also found at chromosome-internal regions at regions flanking polycistronic transcription units called divergent strand switch regions (dSSRs) and convergent strand switch regions (cSSRs), which are sites of RNA polymerase II (Pol II) transcription initiation and termination, respectively [4]. The loss of base J from these chromosome-internal sites led to alterations in transcription initiation and termination, and corresponding changes in gene expression [5–7]. While it is clear that base J represents a novel epigenetic mark involved in regulating Pol II transcription and gene expression, little is understood about what regulates the specific localization of base J in the genome.
Base J is synthesized in a two-step process. First, thymine residues in the context of DNA are hydroxylated by a thymidine hydroxylase (TH) forming 5-hydroxymethyluracil (hmU). A glucose moiety is then attached to hmU by a glucosyltransferase (JGT), to form base J. Two TH enzymes, JBP1 and JBP2, have been identified in trypanosomatids [8–11]. While both JBP1 and JBP2 stimulate de novo thymidine hydroxylation in vivo, the ability of JBP1 to bind J-DNA is thought to play a unique role in J propagation/maintenance [10, 12–14]. The simultaneous deletion of both JBP1 and JBP2 from T. brucei yields a cell line that is unable to synthesize base J, unless cells are fed hmU [9]. Studies such as these unambiguously identified JBP1/2 as the thymidine hydroxylases catalyzing the first step of base J synthesis as well as confirm that this step is independent from the subsequent step of glucose conjugation during base J synthesis. The glucosyltransferase involved in the second step of base J synthesis, base J associated GT (JGT), has recently been identified [15, 16]. Using recombinant protein we demonstrated that JGT utilizes UDP-glucose to transfer glucose to dsDNA substrates containing hmU [15]. In vivo, deletion of both JGT alleles in T. brucei results in complete loss of base J synthesis in the genome [15, 16]. These studies further confirm the two-step mechanism of J synthesis and indicate JGT is the only glucosyltransferase involved, catalyzing the final step of base J synthesis.
Despite the recent genome-wide data sets of DNA Jaylation patterns, and elucidation of the J-biosynthetic pathway, the rules that govern the establishment of DNA J patterns in trypanosomatids remain undefined. It is unclear what determines the specific localization of base J synthesis into specific sequences in the genome. While no consensus sequence or motif is evident from the genome-wide J analysis thus far, it is clear that, for at least the telomeric repeats, there is a sequence specificity component where in the top strand (GGGTTA), only the second T is modified [17, 18].
To gain systematic insight into the constraints that define endogenous Jaylation patterns, we integrated different DNA elements into episomes in L. major. To examine the ability of specific sequences to stimulate de novo J synthesis, DNA fragments were cloned into a PSP72 vector containing a hygromycin resistance gene and then transfected into Leishmania major and grown as episomes (Figure 1a). The episomes were then purified from L. major, digested with EcoRI and HindIII, and J synthesis assayed by anti-J IP-qPCR. This set up allows us to control for chromosomal environment and potential indirect effects such as those mediated by transcription. Furthermore, this approach allows us to measure the contributions of DNA sequence in establishing Jaylation patterns. To determine if the episome system would mimic the specificity of J synthesis in vivo, sequences representing J positive and J negative regions of the L. major genome were cloned into the PSP72 vector and assayed for base J synthesis after growth in L. major. Initially, this included J positive and J negative SSRs. Approximately 1kb genomic fragments representing a cSSR that normally contains base J (cSSR+) and a cSSR that lacks base J (cSSR-) were cloned into the PSP72 vector [6]. A similar approach was employed for dSSRs that do and do not contain J in vivo. In both cases, only the sequences that contain J in vivo are able to stimulate de novo J synthesis when cloned in the episome (Figure 1b and c). Thus, even when present outside of their normal chromatin context, i.e. within an episome instead of within the genome, DNA sequence specificity of J synthesis is maintained. This was also seen using a similar approach in L. tarentolae [17].
Fig. 1. Specific DNA sequences promote base J synthesis in vivo.
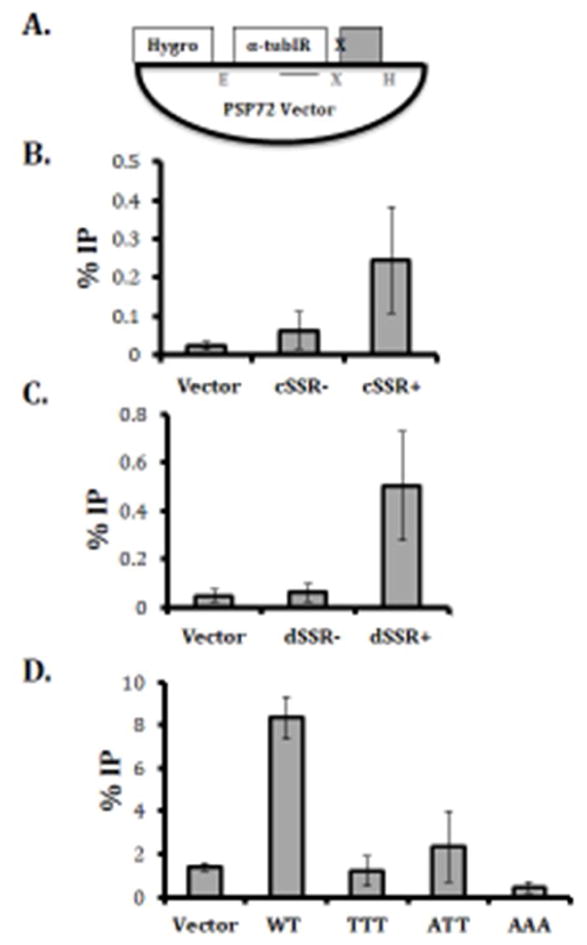
(A) Schematic of the plasmids containing strand switch regions and telomeric repeats. Fragments, indicated by the grey box, corresponding to ~1kb regions from SSRs and telomeric repeats were cloned into the XhoI (X) and HindIII (H) restriction site of the PSP72 Vector before transfection into wild type L. major. The hygromycin resistance gene (Hyg) proves a selectable marker after transfection. Plasmids were digested with EcoRI (E) and HindIII (H) and J levels stimulated by the cloned DNA fragment was determined by anti-J IP-qPCR as described in supplementary materials and methods. Solid line below tub IR indicates region amplified in qPCR. (B–D) The percent IP of J-containing DNA from an empty PSP72 vector, and PSP72 vector containing a SSR fragment and telomeric repeats. %IP was calculated relative to input DNA. (B) Convergent SSR that lacks base J (cSSR−) or contains base J (cSSR+) in the normal genomic context. (C) Divergent SSR that lacks base J (dSSR−) or contains base J (dSSR+) in the normal genomic context. (D) % J DNA IP resulting from 6 copies of the wild type GGGTTA telomeric sequence; GGGTTT (TTT), GGGATT (ATT), GGGAAA (AAA). Experiments were performed in triplicate and error bars are representative of standard error.
To further address the DNA sequence specificity, we assayed the telomeric repeat sequence in the episome system. A plasmid was generated that contains six copies of the WT telomeric repeat sequence (GGGTTA) and transfected into L. major. As expected, WT telomeric repeat sequence (GGGTTA) was able to stimulate J synthesis (Figure 1d). However, mutated telomeric sequences (GGGTTT, GGGATT, or GGGAAA) were unable to support J synthesis (Figure 1d). These data demonstrate that some aspect of J synthesis is DNA sequence specific, at least within telomeric DNA.
Taken together, these findings indicate that DNA Jaylation is largely regulated by cis-acting sequences and is thus genetically encoded. A functional role for interplay between DNA methylation and chromatin has been well established, making it possible that chromatin could be a determining factor in establishing global J patterns in trypanosomes. While these findings do not exclude the possibility that chromatin structure or other associated proteins are crucial in mediating local DNA Jaylation, they do show that local DNA sequence is a primary determinant of target specification for DNA Jaylation in trypanosomatids.
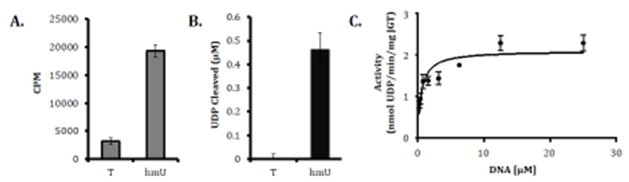
The current hypothesis is that the key regulatory step of J synthesis is the first step catalyzed by JBP1 and JBP2. Bypassing this first step, via feeding cells hmU, leads to J synthesis in regions of the genome that do not normally contain base J [9, 19]. This suggests that, regardless of where hmU is present, JGT will convert it to base J in a promiscuous (non-sequence-specific) manner. Thus, it follows that the specificity of base J localization is due to the JBP enzymes generating hmU at only specific sites throughout the genome; however, no direct evidence has confirmed this hypothesis. To address this, we tested synthetic telomeric DNA substrates in an in vitro GT assay to determine whether JGT can explain the sequence specificity of J synthesis in vivo. We took advantage of active recombinant JGT and an in vitro homogeneous bioluminescent UDP detection assay (UDP-Glo) that can detect the activity of glycosyltransferases that utilize UDP-sugars and release UDP as a product (see Supplemental Data 1 for details on methods). Using this assay we show that JGT is specific for hmU-containing dsDNA substrates and has no activity with unmodified dsDNA substrates (Figure 2b), consistent with assays directly measuring the transfer of glucose to DNA (Figure 2a), validating our use of UDP-Glo for further in vitro GT experiments.
Fig. 2. JGT is specific for hmU containing DNA but does not possess sequence specificity.
(A) Radiolabeled in vitro glucosyltransferase reaction. Recombinant JGT and UDP-[3H] glucose was incubated with 36nt long dsDNA substrates listed in supplementary material and methods containing one hmU modification (hmU) or unmodified dsDNA (T). CPM (counts per minute), indicative of the transfer of glucose to DNA, were read for each sample. Experiment was performed in triplicate and error bars are representative of standard error. (B) UDP-Glo in vitro glucosyltransferase reaction. Recombinant JGT was incubated with 36nt long dsDNA substrates listed in Table 1 containing one hmU modification (hmU) or unmodified dsDNA (T). The amount of UDP Cleaved, indicative of the transfer of glucose to DNA, was estimated from a standard curve of UDP for each sample. Experiment was performed in triplicate and error bars are representative of standard error. (C) Representative substrate–velocity curve of recombinant JGT. Recombinant JGT activity with the 15nt long hmU-containing ds DNA substrate (WT substrate) listed in Table I. Glucosylation reactions were conducted with hmU DNA substrate concentrations of 0, 0.195, 0.391, 0.781, 1.563, 3.125, 6.25, 12.5, and 25 μM and fixed enzyme and UDP-glucose concentrations of 18 μM and 1 mM, respectively. Kinetic experiments were performed in triplicate and error bars are representative of standard error. UDP cleaved is plotted vs substrate concentration, and nonlinear regression was performed to determine kinetic parameters.
To address sequence specificity, recombinant JGT was incubated with dsDNA substrates that correspond with the WT telomeric sequence (GGGTTA) as well as mutated sequences (GGGTTT and GGGATT) with an hmU positioned on the second T in the G-rich strand (WT2, TTT, ATT; Table 1). Michaelis-Menten curves were generated for each substrate (Figure 2b and data not shown) and hmU DNA kinetic parameters were determined (Table 1). We found no significant difference in either affinity (Km), p value > 0.05 or turnover (kcat), p value > 0.01 of the JGT when incubated with WT or mutated telomeric sequences. We also find no significant difference in affinity or turnover for WT telomeric sequences with hmU modification at different positions, including the first T on the G-strand that is not modified in vivo (WT1), or for hmU within a random (non-telomeric) DNA sequence (Random; Table 1). These data indicate that JGT is DNA sequence non-specific and thus, the enzyme presumably does not contribute to the telomeric DNA sequence specificity of J synthesis we characterized in vivo.
Table 1.
Oligonucleotides used as substrates for glucosyltransferase assay*
| Substrate | Sequence | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|---|
| WT2 | 5′-TTAGGGTTAGGGTTA-3′ 3′-AATCCCAATCCCAAT-5′ |
0.485 ± 0.022 | 0.519 ± 0.109 | 0.933 |
| WT1 | 5′-TTAGGGTTAGGGTTA-3′ 3′-AATCCCAATCCCAAT-5′ |
0.451 ± 0.025 | 0.523 ± 0.137 | 0.862 |
| WT3 | 5′-TTAGGGTTAGGGTTA-3′ 3′-AATCCCAATCCCAAT-5′ |
0.675 ± 0.037 | 0.519 ± 0.134 | 1.302 |
| TTT | 5′-TTTGGGTTTGGGTTT-3′ 3′-AAACCCAAACCCAAA-5′ |
0.703 ± 0.052 | 0.412 ± 0.149 | 1.705 |
| ATT | 5′-ATTGGGATTGGGATT-3′ 3′-TAACCCTAACCCTAA-5′ |
0.566 ± 0.034 | 0.267 ± 0.089 | 2.116 |
| Random | 5′-GTACGAGTCGAGTCA-3′ 3′-CATGCTCAGCTCAGT-5′ |
0.580 ± 0.021 | 0.313 ± 0.059 | 1.854 |
Bold T indicates the position of an hmU modification.
Glucosylation reactions were conducted with hmU DNA substrate concentration of 0, 0.195, 0.391, 0.781, 1.563, 3.125, 6.25, 12.5, and 25 μM and fixed enzyme and UDP-glucose concentrations of 18 μM and 1mM, respectively. UDP cleaved is plotted vs substrate concentration and nonlinear regression was performed to determine kinetic parameters.
These studies pave the way for the elucidation of the underlying molecular mechanisms involved in regulating J synthesis in vivo. While the results described here strongly support the importance of the first step of J synthesis, further work is required to fully understand the formation of hmU on specific sequences by the JBPs. Similar analysis of JBP catalysis, as we performed here for JGT, will help to shed light on primary DNA sequence requirements for hmU formation. In vitro analysis of the sequence specificity of the JBPs has, however, proven to be quite difficult. Currently, we are only able to express recombinant protein for JBP1. All attempts to express JBP2, the key de novo J synthesis enzyme, have failed. While we have had success demonstrating dioxygenase activity of JBP1 in vitro, the reaction is extremely inefficient [20]. Until we have a clear and robust assay for both hydroxylases, we are unable to fully characterize the contribution of this first step in determining the sequence specificity of J synthesis.
Supplementary Material
Highlights.
Only specific DNA sequences can promote base J synthesis in vivo.
Base J associated glucosyltransferase does not possess DNA sequence specificity.
Specific localization of base J throughout the genome is likely not due to the glucosyltransferase catalyzing the second step of base J synthesis.
Acknowledgments
We thank Dr. Lance Wells and Rob Bridger for performing MS analyses of the purified recombinant JGT preparation. Work performed in the Wells laboratory was supported by a P41 grant from NIGMS/NIH (P41GM103490). We would also like to thank Stephanie Halmo for help with kinetic assays and analyses, and Jessica Lopes da Rosa-Spiegler, David Reynolds, and Rudo Kieft for critical reading of the manuscript. This work was supported by National Institute of Health grant [RO1AI109108].
Abbreviations
- Base J
beta-D-glucopyranosyloxymethyluracil
- TH
thymidine hydroxylase
- JGT
base J-associated glucosyl transferase
- JBP
base J-binding protein
- UDP-Glc
uridine diphosphoglucose
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gommers-Ampt J, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. Beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 2.Van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc Natl Acad Sci. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Leeuwen F, De Kort M, van der Marel GA, Van Boom JH, Borst P. The modified DNA base beta-D-glycosyl-hydroxymethyluracil confers resistance to micrococcal nuclease and is incompletely recovered by 32P-postlabeling. Anal Biochem. 1998;258:223–229. doi: 10.1006/abio.1998.2587. [DOI] [PubMed] [Google Scholar]
- 4.Cliffe LJ, Siegel TN, Marshall M, Cross GA, Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekanayake D, Sabatini R. Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi. Modulation of nucleosome abundance, histone modification, and polymerase occupancy by O-linked thymine DNA glucosylation. Eukaryot Cell. 2011;10:1465–1472. doi: 10.1128/EC.05185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Luenen H, Farris C, Jan S, Genest PA, Tripathi P, Velds A, Kerkhoven R, Nieuwland M, Haydock A, Ramasamy G, Vainio S, Heidebrecht T, Perrakis A, Pagie L, Van Steensel B, Myler P, Borst P. Glucosylated Hydroxymethyluracil, DNA Base J, Prevents Transcriptional Readthrough in Leishmania. Cell. 2013;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds D, Cliffe L, Förstner K, Hon C, Siegel N, Sabatini R. Regulation of transcription termination by glucosylated hydroxymethyluracil, base J, in Leishmania major and Trypanosoma brucei. Nucleic Acids Res. 2014;42:9717–9729. doi: 10.1093/nar/gku714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, van Luenen HG, Rigden DJ, Sabatini R, Borst P. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Vainio S, Genest PA, ter Riet B, van Luenen H, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base. J Mol Biochem Parasitol. 2009;164:157–161. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Toaldo CB, Kieft R, Dirks-Mulder A, Sabatini R, van Luenen HG, Borst P. A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1. Mol Biochem Parasitol. 2005;143:111–115. doi: 10.1016/j.molbiopara.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross M, Kieft R, Sabatini R, Dirks-Mulder A, Chaves I, Borst P. J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol Microbiol. 2002;46:37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 14.Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO. 1999;18:6573– 6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullard W, Lopes da Rosa-Spiegler J, Liu S, Wang Y, Sabatini R. Identification of the glucosyltransferase that converts hydroxymethyluracil to base J in the trypanosomatid genome. J Biol Chem. 2012;289:20273–20282. doi: 10.1074/jbc.M114.579821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekara A, Merritta C, Baugha L, Stuarta K, Myler P. Tb927.10.6900 encodes the glucosyltransferase involved in synthesis of base J in Trypanosoma brucei. Molecular and Biochemical Parasitology. 2014;196:9–11. doi: 10.1016/j.molbiopara.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genest P, Baugh L, Taipale A, Zhao W, Jan S, van Luenen H, Korlach J, Clark T, Luong K, Boitano M, Turner S, Myler P, Borst P. Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing. Nucleic Acids Res. 2015;43:2102–2215. doi: 10.1093/nar/gkv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, Van der Marel GA, Van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Leeuwen F, Kieft R, Cross M, Borst P. Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol Cell Biol. 1998;18:5643–5651. doi: 10.1128/mcb.18.10.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cliffe L, Hirsch G, Wang J, Ekanayake D, Bullard W, Hu M, Wang Y, Sabatini R. JBP1 and JBP2 Proteins are Fe2+/2-Oxoglutarate-dependent dioxygenases regulating hydroxylation of thymidine residues in trypanosome DNA. J Biol Chem. 2012;287:19886–19895. doi: 10.1074/jbc.M112.341974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.