Abstract
Background
Cell damage, tissue and vascular injury are associated with the exposure and release of intracellular components such as RNA, which promote inflammatory reactions and thrombosis. Based on the counteracting anti‐inflammatory and cardioprotective functions of ribonuclease A (RNase A) in this context, its role in an experimental model of heart transplantation in rats was studied.
Methods and Results
Inbred BN/OrlRj rat cardiac allografts were heterotopically transplanted into inbred LEW/OrlRj rats. Recipients were intravenously treated every other day with saline or bovine pancreatic RNase A (50 μg/kg). Toxic side effects were not found (macroscopically and histologically). Heart tissue flow cytometry and quantitative morphological analyses of explanted hearts at postoperative day 1 or postoperative day 4 showed reduced leukocyte infiltration, edema, and thrombus formation in RNase A‐treated rats. In allogeneic mixed lymphocyte reactions, RNase A decreased the proliferation of effector T cells. RNase A treatment of rats resulted in prolonged median graft survival up to 10.5 days (interquartile range 1.8) compared to 6.5 days (interquartile range 1.0) in saline treatment (P=0.001). Treatment of rats with a new generated (recombinant) human pancreatic RNase 1 prolonged median graft survival similarly, unlike treatment with (recombinant) inactive human RNase 1 (each 50 μg/kg IV every other day, 11.0 days, interquartile range 0.3, versus 8.0 days, interquartile range 0.5, P=0.007).
Conclusions
Upon heart transplantation, RNase administration appears to present a promising and safe drug to counteract ischemia/reperfusion injury and graft rejection. Furthermore, RNase treatment may be considered in situations of critical reperfusion after percutaneous coronary interventions or in cardiac surgery using the heart–lung machine.
Keywords: edema, extracellular RNA, ischemia/reperfusion injury, ribonuclease, transplantation
Subject Categories: Transplantation
Introduction
Following heart transplantation, acute graft rejection is often accompanied by edema, hemorrhage and vasculitis, polymorphic inflammatory leukocyte infiltration, as well as the damage and necrosis of cardiomyocytes.1, 2 Tissue injury leads to the release of intracellular components such as ribonucleic acids (RNA), now referred to as extracellular RNA (eRNA).3 The appearance of eRNA in connection with cardiovascular diseases and inflammation has recently been demonstrated to involve (1) leukocyte recruitment by eRNA‐induced proinflammatory cytokines,4 (2) eRNA‐promoted vascular permeability and edema formation,5 as well as (3) thrombus formation and vessel occlusion due to eRNA‐mediated activation of the contact phase of intrinsic blood coagulation6 (Figure 1). All these mechanism may contribute to the damaging nature of eRNA in pathophysiological situations of atherosclerosis or cardiac ischemia/reperfusion (I/R) injury, as shown by our recent work.7, 8
Figure 1.
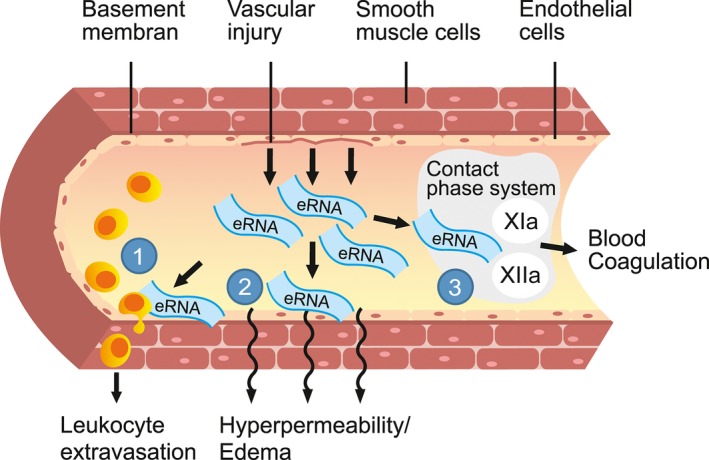
Multiple functions of extracellular ribonucleic acids (eRNA). Vascular injury leads to release of intracellular components such as RNA, referred to as eRNA.3 Extracellular RNA (1) promotes leukocyte recruitment as it mobilizes proinflammatory cytokines, (2) causes edema as it increases the permeability of blood vessels, and (3) serves as procoagulant cofactor by activating the contact phase system of intrinsic blood coagulation resulting in enhanced thrombus formation and vessel occlusion. All these mechanisms are supposed to be related to acute rejection reaction after organ transplantation. Illustration by Nina Bantschow.
The thermostable bovine pancreatic ribonuclease A (further called RNase A, 124 amino acids) is highly efficient in cleaving and destroying RNA.9 The well‐studied enzyme was first isolated and crystallized in the 1930/1940s,10, 11, 12 and sequenced and synthesized since the 1960s.13, 14 The human equivalent (human pancreatic ribonuclease, further called RNase 1, 128 amino acids) is distinguishable from bovine RNase A in its amino acid sequence,15 but is identical in its specific RNase cleaving activity.16
Human endothelial cells selectively express RNase 1; thus, the eRNA/RNase 1 system provides a new level of regulation of vascular homeostasis,17, 18 exemplified by the vessel‐protective functions of RNase administration in the following preclinical disease models: (1) antithrombotic function in arterial thrombosis in mice6; (2) prevention of edema and vascular occlusion in the superior sagittal sinus in a stroke model in rats5, 19, 20; and (3) cardioprotection in myocardial I/R injury in mice and in the isolated I/R Langendorff‐perfused rat heart.8
Based on these assumptions, it appears plausible to test the influence of RNase administration under conditions of acute organ rejection following heart transplantation.
Materials and Methods
Animals
For heart transplantation, BN/OrlRj rats served as donors and LEW/OrlRj rats as recipients (both from Janvier Labs, n=95 were included; exclusions due to technical failure). All animals were inbred males with an age of 8 to 10 weeks (200–300 g) and housed in the animal care facility of the Walter Brendel Centre, Munich. The protocol was approved by the national animal welfare authority (Administration of Upper Bavaria) and the animals received humane care in accordance with the Directive 2010/63/EU.21
Heterotopic Abdominal Heart Transplantation
The anesthetized (intramuscular anesthesia: fentanyl 0.005 mg/kg, medetomidine 0.15 mg/kg, midazolame 2.0 mg/kg; subcutaneous antagonists: naloxone 0.12 mg/kg, atipamezole 0.75 mg/kg, flumazenil 0.2 mg/kg; additional isoflurane 2% O2 via mask) donor rats (n=40) were systemically heparinized with 500 IE intravenously (IV). After perfusion with a cardioplegic solution (Crustadiol, Bretschneider, Köhler Chemie, Bensheim, Germany) the cardiac grafts were explanted, weighed, and transplanted into the abdomen of the anesthetized recipient rats (n=40) using common microvascular techniques for aorto‐aortic and pulmonary artery to inferior vena cava anastomoses, respectively (Figure 2).22 Postoperative subcutaneous analgesia consisted of metamizole (200 mg/kg) and buprenorphine (0.05 mg/kg).
Figure 2.
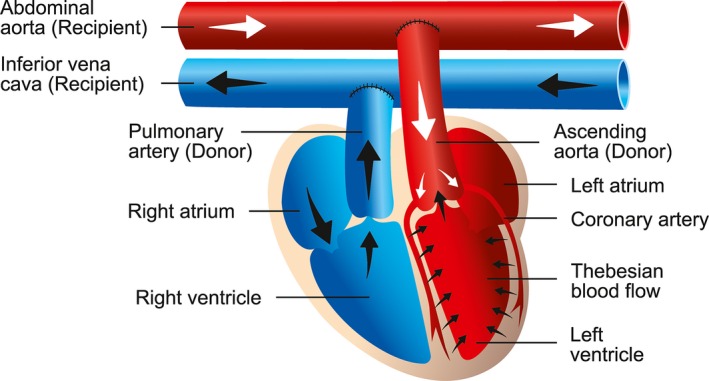
Method of heterotopic abdominal heart transplantation. The microsurgical technique consists of end‐to‐side anastomoses of the donor's aorta and the pulmonary artery trunk to the recipient's abdominal vessels, respectively. The heart is perfused via the coronary arteries. The coronary veins are joined together to form the coronary sinus, where the blood is delivered to the right atrium. The blood flow continues through the right ventricle and the pulmonary artery to the recipient's abdominal vena cava. Illustration by Nina Bantschow.
Study Medication and Treatment Groups
Three different RNases were used in the transplantation studies: (1) bovine pancreatic RNase A (Fermentas, Thermo Fisher Scientific, Waltham, MA), (2) enzymatically active and (3) inactive recombinant human pancreatic RNase 1 (Fischer/Griemert/Preissner, Giessen, manufactured by ProteoGenix, Schiltigheim, France). Following previous protocols,23, 24 RNase dosages of 50 μg/kg (dissolved in 1 mL saline 0.9%) were given 30 minutes before transplantations and then every other day (Figure 3). Animals of the controls were treated with 1 mL saline 0.9%.
Figure 3.
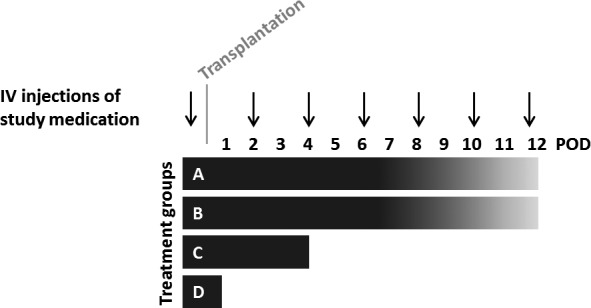
Study design and treatment groups. Study medications (RNase A, active and inactive RNase 1, and saline 0.9%) were intravenously injected 30 minutes before transplantations and then every other day. There were 4 different strategies: grafts of (A) RNase A or saline‐treated animals (n=6 in each group), and (B) active and inactive RNase 1–treated animals (n=4 in each group) were examined daily and harvested at the time point of total graft dysfunction (verified by echocardiography); (C) 4 days (POD 4) and (D) 1 day (POD1) after transplantation, the beating grafts of RNase A and saline‐treated animals were harvested (n=5 in each group, FACS and histological analyses were performed). FACS indicates fluorescence‐activated cell sorting; IV, intravenous; POD, postoperative day.
To evaluate the impact of RNase A on graft survival, six randomized allocated recipients were treated and sacrificed at the point of total graft rejection. In 2 additional treatment groups, the beating hearts were harvested either on the first postoperative day (POD 1, n=5) or on POD 4 (n=5), to investigate leukocyte infiltration in heart tissue (flow cytometry) and to analyze edema development and rejection reaction (histological analyses); at the early time point POD 1, our main focus was on the extent of I/R injury, whereas at POD 4 the ongoing cellular rejection was quantified. All RNase A–treated animals, and at the various time points, were compared with equal numbers of transplanted controls.
In order to prove that the ribonucleolytic activity of the RNase was responsible for the observed effects (inactive bovine RNase A is not available), active and inactive recombinant human RNase 1 were generated, studied, and compared (n=4 in each group).
Generation of Recombinant Ribonucleolytic Active and Inactive RNase 1
Total RNA was isolated from human cerebral microvascular endothelial cells (HCMEC‐D3), kindly supplied by P.O. Couraud, Paris25 to S. Fischer, B. Griemert, and K.T. Preissner, Giessen using the purification kit from Sigma (Heidelberg, Germany) followed by cDNA synthesis using the high‐capacity cDNA reverse transcription kit from Applied Biosystems (Darmstadt, Germany). To amplify the coding region of human RNase 1 by polymerase chain reaction (PCR), the following primers, containing specific restriction sites for EcoRI and XhoI (GenBank, accession no. NM_198232.2), were used: RNasepETfwd AGGAATTCATGGCTCTGGAGAAGTCTCTTCTG and RNasepETrev ATCTCGAGTGGTAGAGTCCTCCACAGAAGCATC and the Phusion High Fidelity DNA polymerase (Life Technologies, Darmstadt, Germany). Reaction tubes for PCR contained 10 mmol/L dNTPs, 10 mmol/L of each primer, 3% (v/v) dimethylsulfoxide, template, and 0.5 units of polymerase. Annealing temperature was 60°C and number of cycles was 30. The PCR product was purified using standard protocols (Qiagen, Venlo, Netherlands) and the GoTaq Green Kit (Promega, Mannheim, Germany) was used to produce polyA overhangs at the 3′‐end. Ten microliters of purified PCR product were transferred into the same volume of 2xGoTaq Green Polymerase Mix and incubated for 30 minutes at 72°C. The PCR product was subcloned into the pDRIVE vector using the PCR cloning kit from Qiagen. The ligated plasmid with inserted RNase 1 sequence was amplified by transformation into competent DH5α cells. The plasmid was isolated using standard protocols (Qiagen). Inserted RNase 1 cDNA was excised and cloned into the pET32a vector (Novagen, Merck, Darmstadt, Germany), whereby both inserted RNase 1 cDNA and vector were pretreated with suitable restriction enzymes (XhoI and EcoRI; Thermo Fisher Scientific, Waltham, MA). The ligated plasmid with inserted RNase 1 sequence was transformed into competent DH5α cells. Plasmid DNA was purified using standard protocols (Qiagen) and analyzed by digestion with restriction enzymes followed by gel electrophoresis and by sequencing the product.
Site‐directed mutagenesis was used to include point mutations in position 40, which will change CAT (histidin) to GTT (Valin). PCR was performed using 2 primers (each 10 mmol/L), complementary to each other, containing the mutant sequence (MutAkz1 CCAGCGGCAGCTTATGGACTCAGACAG and MutAkz2 CTGTCTGAGTCCATAACCTGCCGCTGG), dNTPs (10 mmol/L), 3% v/v dimethylsulfoxide, 250 ng of plasmid DNA (pET2a vector containing RNase 1 cDNA), 0.5 units of Phusion High Fidelity DNA polymerase (Life Technologies). Annealing temperature was 55°C for 10 minutes and PCR was performed with 20 cycles. PCR reaction was cooled down to room temperature and 10 units of DpnI was added and reaction mixture (20 μL) was incubated for 1 hour at 37°C. Five microliters of this PCR reaction tube was used for the transformation of competent JM109 cells. Plasmid DNA was purified (Qiagen, Hildesheim, Germany) and sequenced using pETfwd‐primers.
RNase 1 wild‐type (WT) and RNase 1 H40V cDNAs were used for cloning and for the expression of recombinant proteins (manufactured by Proteogenix, Schiltigheim, France). Final samples of RNase 1 WT (human active) and RNase 1 H40V (human inactive) were quality controlled by SDS‐PAGE. RNase activity was determined as described with minor modifications.18, 26 Activity of mutated protein was reduced to 3.6% compared to the activity of WT protein.
Graft Examination
The primary end point of the study was the graft survival, which was assessed by daily examination of the graft function by palpation and transabdominal echocardiography under isoflurane. The graft function was graduated in good contraction (+++), mild (++) or severe dysfunction (+), and no contraction (−), which led to termination of each experiment.27
Blood Parameters
Blood parameters (creatinine, urea, GPT/ALT, troponin I, hemoglobin, and leukocytes) were measured using routine hospital laboratory.
T Cell Proliferation Assay and Mixed Lymphocyte Reaction
Following magnetic bead isolation, CD4+ T cells were obtained using the human naive CD4+ T cell isolation Kit II (Miltenyi, Bergisch Gladbach, Germany). The analysis of effector T (Teff)‐cell proliferation was performed using a carboxyfluorescein succinimidyl ester dilution assay (5 mmol/L, Invitrogen, Carlsbad, CA). The proliferation of naive (resting) Teff cells were assessed and compared to the proliferation of polyclonal (anti‐CD3/anti‐CD28) activated Teff cells to verify their capacity of proliferation (maximum stimulus).
The role of RNase A in allogeneic stimulated Teff cells was evaluated using mixed lymphocyte reaction with unmatched and unrelated donor peripheral blood mononuclear cells as described before.28
Teff cell proliferation in mixed lymphocyte reaction was compared both in absence and as well as in presence of RNase A (10 μg RNase A). Analysis was performed in duplicates/triplicates by flow cytometry (fluorescence‐activated cell sorting, FACS Calibur, BD Bioscience, San Jose, CA) of carboxyfluorescein succinimidyl ester dilution of proliferating cells using the software FlowJo v10 (FlowJo, Tree Star Ashland, OR).28
Analyses of Explanted Grafts
At the termination of the transplantation experiments, the grafts were explanted, weighed, and then analyzed via pathohistology and quantitative morphological analyses (see Histology, Immunohistochemistry, and Quantitative Morphological Analyses section). The density of rat heart tissue was determined in 6 cases (1.052±0.062 g/cm3), using the fluid displacement method, described before.29 Briefly, the heart was cut into equidistant parallel slices of ≈3 mm thickness, orthogonal to the longitudinal axis of the heart. The slices were placed on the same section surfaces, and every second slab was systematically randomly allocated to heart tissue FACS, while qualitative and quantitative histological and immunohistochemical analyses were performed on the remaining slices (see Histology, Immunohistochemistry, and Quantitative Morphological Analyses section).30
Heart Tissue Flow Cytometry
Collagenase II (Biochrome, Berlin, Germany) digested heart tissue was stained using fluorochrome‐conjugated antibodies specific to rat CD45 (APC/Cy7; Biolegend, San Diego, CA), Ly6G (fluorescein isothiocyanate, Abcam, Cambridge, UK), or corresponding isotypes. Leukocyte subgroups were identified by FACS (Galios Flow Cytometer; Beckman Coulter, Pasadena, CA), using Kaluza Analysis Software (Beckman Coulter, Figure 4).
Figure 4.
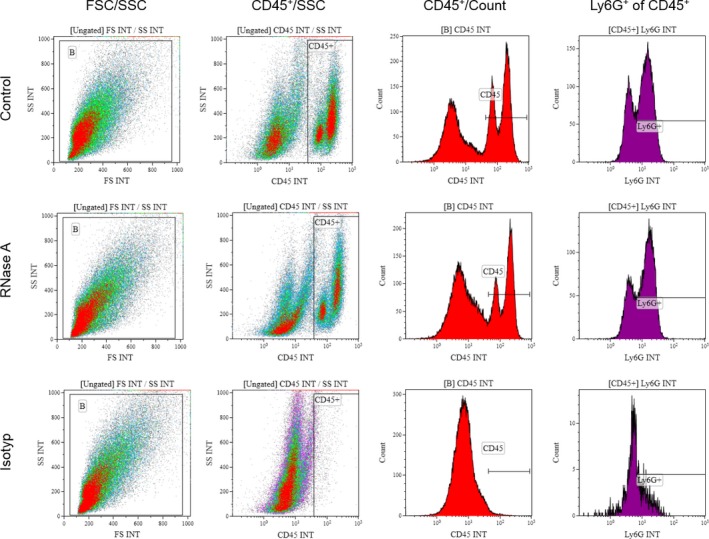
Gating strategy in heart tissue flow cytometry analysis. Cardiac graft tissue was stained for CD45 and Ly6G to quantify infiltration of leukocytes (CD45+ cells) or neutrophils (Ly6G+ cells), respectively.
Histology, Immunohistochemistry, and Quantitative Morphological Analyses
One of the heart tissue slices allocated to qualitative and quantitative histological and immunohistochemical analyses of the POD 1 group was immediately frozen for cryosectioning technique (Tissue Tek; Sakura, Torrance, CA) and stored at −80°C, while the other slices were immersion fixed in neutrally buffered (pH 7.6) 4% formaldehyde solution overnight at room temperature, followed by embedding in paraffin while maintaining their section surface orientation. Paraffin sections (≈3 μm thickness) were stained with hematoxylin and eosin, according to standard protocols. For detection of extravascular leukocyte subgroups in the myocardium, immunohistological staining was performed, using specific antibodies directed against macrophages (CD68; Acris, Herford, Germany, and Dako, Eching, Germany), T cells (CD3; Dako and Vector Laboratories, Burlingame, CA), and B cells (CD20; Santa Cruz, Dallas, TX, and Dako). Positive immunoreactivity was detected using the Vectastain ABC‐Kit Elite and diaminobenzidine (Dako) as chromophore. Quantitative morphological analyses were performed using stereological methods. The volume densities (stated in %) of the myocardium (myocardiocytes and interstitial tissue, excluding myocardial vessels) in the heart (VV(myocardium/heart)), of edema (optically empty extracellular space within the myocardium) in the myocardium (VV(edema/myocardium)), and of macrophages, T cells, and B cells in the myocardium (VV(CD68+/myocardium), VV(CD3+/myocardium), VV(CD20+/myocardium)) were calculated from their respective area densities, determined at ×200 magnification in systematically randomly sampled test‐fields per case, referring to 10±0.3% of the total tissue section area, using an automated stereology system (VIS‐Visiopharm Integrator System® Version 3.4.1.0 with newCAST® software; Visiopharm A/S, Hørsholm, Denmark).31 The sectional area densities of heart tissue, myocardium, macrophages, CD3‐positive T cells, and CD20‐positive B cells were determined by point counting. Per case, 800±16 points were counted for determination of VV(myocardium/heart), 5000±100 points for VV(edema/myocardium), 115 200±2304 points for VV(CD3+/myocardium), and 156 800±3136 points for each VV(CD68+/myocardium) and VV(CD20+/myocardium).31 The absolute volumes (stated in mm3) of the heart, the myocardium, and of edema, macrophages, T and B‐cells in the myocardium (V(heart), V(myocardium, heart), V(edema, myocardium), V(CD68+, myocardium), V(CD3+, myocardium), and V(CD20+, myocardium)) were calculated from the volume densities of the respective parameters and the absolute volume of the corresponding reference compartments (heart tissue or myocardium). The total volume of the heart was calculated from the weight and the density of the heart tissue. Cryosections were stained for leukocytes (CD45; Abcam Cambridge, UK; dky‐anti mouse‐Cy3; Jackson Immuno Research, West Grove, PA) and counterstained against α‐actinin (ACTN4‐Alexa488; Abcam, Cambridge, UK) or specifically for neutrophils (Ly6G, Abcam) and counterstained with phalloidin‐tetramethylrhodamine B isothiocyanate (Sigma‐Aldrich, St. Louis, MO).
Analysis of Toxic Side Effects
An assessment of toxic side effects was performed in 9 LEW/OrlRj rats by using the study dose of RNase A (50 μg/kg IV, dissolved in 1 mL saline), as well as a 20‐fold higher dose of RNase A (1000 μg/kg IV, dissolved in 1 mL saline), and for control 0.9% saline (1 mL IV), each administered every other day for 28 days (n=3 in each group). After 4 weeks the rats were sacrificed, blood samples were collected, followed by pathological examination according to standardized organ sampling and trimming procedures in rats of the Registry of Industrial Toxicology Animal‐data (RITA).32, 33, 34, 35 All organ samples were paraffin embedded and stained with hematoxylin and eosin; corresponding samples from livers were stained with periodic acid–Schiff according to standard protocols.
Statistical Analyses
Study design and calculated group size are based on the primary end point, the graft survival of bovine RNase A–treated animals compared to the control group (saline treated). Thus, graft survival in all RNase groups (bovine and human RNases) was analyzed using Kaplan–Meier estimator, and differences were assessed by log‐rank test. The graft survival time was specified as median days and interquartile range (IQR). Secondary end points were presented using the magnitude of effects (ie, effect size statistics) and their 95% CI. Standardized mean differences were compared between the groups (d statistics). According to Cohen (adapted for biological sciences, Nakagowa and Cuthill),36, 37 the dCohen results were classified in small (≥0.2 to <0.5), medium (≥0.5 to <0.8), and large (≥0.8) effects. Numerical data in d statistics are presented as means± SD. Analysis of the effect of RNase A on Teff cell proliferation was carried out by combining data of 2 independent experiments (n=6). For statistical analyses and diagrams, SPSS Statistics 22 (IBM, Armonk, NY), Excel 2010 (Microsoft, Redmond, WA), and Prism 6.00 for Windows and MacOSX (GraphPad Software, La Jolla, CA) were utilized.
Results
RNase A Treatment Induces No Toxic Side Effects
In accordance with the RITA,32, 33, 34, 35 treatment of healthy rats with saline or RNase A (even in the 20‐fold higher standard dose) showed neither macroscopic nor histological alterations, based on analyses of tissues and organs (including integumentary system, digestive system, respiratory system, male genital system, endocrine system, urinary system, nervous system, musculoskeletal system, cardiovascular system, and lymphoreticular system). Within the 4 weeks of treatment with saline or RNase A, all animals increased weight without any differences (by 13.8±1.9% in control group, 14.2±0.5% in RNase A–treated animals with study dose, and 13.5±3.6% in overdosed RNase A–treated animals, small effects according to Cohen's effect size statistics). After this period of time, the levels of blood parameters of animals treated with RNase A were comparable with those that received saline treatment (Table 1).
Table 1.
Blood Parameters of Nontransplanted Lewis (LEW/OrlRj) Rats After 28 Days of Intravenous Treatment With Saline or RNase A
| Blood Parameters | Treatment Groups | ||
|---|---|---|---|
| Control (Saline) | RNase A (50 μg/kg) | RNase A (1000 μg/kg) | |
| Creatinine, mg/dL | 0.7±0.2 | 0.6±0.0 | 0.6±0.1 |
| Urea, mg/dL | 54±3 | 41±2 | 47±3 |
| GPT/ALT, U/L | 64±4 | 44±6 | 54±3 |
| Leukocytes, K/μL | 9.8±1.8 | 9.2±0.9 | 10±0.9 |
| Hemoglobin, g/dL | 14.7±1.1 | 16.3±0.2 | 15.4±0.8 |
All parameters were in normal range compared to blood parameters of 10‐week‐old Lewis (LEW/OrlRj) rats (normal values provided by Janvier Labs [http://www.janvier-labs.com/tl_files/_media/images/FICHE_RESEARCH_MODEL_LEWIS.pdf, Technical Sheet 2013‐06‐ENG‐RM‐08, accessed: January 17, 2016]). Data represent mean±SD; n=3 in each group; RNase A, ribonuclease A; GPT/ALT, glutamate‐pyruvate transaminase or alanine transaminase, respectively.
Impact of RNase A Treatment on Leukocyte Activation and Recruitment
Following the described procedures of heterotopic heart transplantation, quantitative morphological analyses and FACS analyses, particularly regarding cell infiltration, were performed in beating grafts explanted at POD 1 and POD 4 (Table 2 and Figure 5, including representative immunofluorescence pictures).
Table 2.
Impact of RNase A Treatment on Inflammatory Cell Recruitment Into Graft Tissue
| POD 1 | POD 4 | |||||
|---|---|---|---|---|---|---|
| Control | RNase A | dCohen (95% CI) | Control | RNase A | dCohen (95% CI) | |
| Flow cytometry (% of total cells) | ||||||
| CD45+ | 44.9±5.0 | 35.1±9.2 | 1.33 (0.14/2.55) | 63.9±6.4 | 63.2±11.9 | 0.07 (1.17/1.31) |
| CD45+Ly6G+ | 32.3±5.0 | 24.2±6.5 | 1.38 (0.11/2.60) | 29.7±6.3 | 33.2±12.8 | 0.34 (0.94/1.56) |
| Quantitative morphological data: volume densities of inflammatory cells in the myocardium (%) | ||||||
| VV(CD68+/myocardium) | 0.47±0.05 | 0.35±0.10 | 1.53 (0.00/2.76) | 4.98±0.48 | 5.45±0.71 | 0.77 (0.58/1.97) |
| VV(CD3+/myocardium) | 1.91±0.26 | 1.12±0.15 | 3.78 (1.47/5.35) | 4.12±0.26 | 1.95±0.14 | 10.45 (5.12/13.75) |
| VV(CD20+/myocardium) | 0.34±0.05 | 0.25±0.03 | 2.34 (0.57/3.66) | 1.87±0.30 | 1.09±0.20 | 3.07 (1.04/4.51) |
| Quantitative morphological data: total volumes of inflammatory cells in the myocardium (mm³) | ||||||
| V(CD68+, myocardium) | 3.9±0.7 | 2.6±0.8 | 1.77 (0.17/3.02) | 46.2±8.5 | 41.8±6.1 | 0.53 (0.78/1.73) |
| V(CD3+, myocardium) | 15.8±2.9 | 8.3±1.4 | 3.33 (1.2/4.82) | 38.4±3.6 | 14.9±2.3 | 7.83 (3.73/10.4) |
| V(CD20+, myocardium) | 2.8±0.5 | 1.9±0.3 | 2.34 (0.62/3.76) | 17.1±1.9 | 8.1±1.7 | 4.43 (1.85/6.15) |
Following tissue digestion of explanted hearts and flow cytometry, the degree of infiltration of leukocytes (CD45+ cells) and neutrophils (CD45+Ly6G+ cells) in grafts derived from controls and the RNase A treatment group was determined as indicated. Volume densities of macrophages (VV(CD68+/myocardium)), T cells (VV(CD3+/myocardium)), and B cells (VV(CD20+/myocardium)) in the myocardium, and total volumes of macrophages (V(CD68+, myocardium)), T cells (V(CD3+, myocardium)), and B cells (V(CD20+, myocardium)) in the myocardium of explanted beating grafts at POD 1 and POD 4. Data represent means±SD; n=5 in each group; POD, postoperative day; RNase A, ribonuclease A; dCohen, effect size according to Cohen,36, 37 classified in small (≥0.2 to <0.5), medium (≥0.5 to <0.8), and large (≥0.8) effects; CD, cluster of differentiation.
Figure 5.

Infiltration of inflammatory and immune cells in grafted tissue. A, The distribution of leukocytes (CD45+ cells, red) in grafted heart tissue (green: α‐actin) at POD 1 in the control and RNase A treatment groups is depicted by representatives pictures; immunofluorescence was carried out with frozen heart tissue. B, Likewise, the distribution of neutrophils (Ly6G+ cells, green) at POD 1 for the control and RNase A groups is shown (red: Phalloidin counterstain for actin). C, The distribution of T cells (CD3+ cells, brown, IHC staining) in paraffin sections of explanted hearts at POD 4 is indicated for the control and RNase A treatment group in representatives pictures. Left ventricle, Scale bars = 100 μm. IHC indicates immunohistochemestry; POD, postoperative day.
After transplantation, the total number of leukocytes (CD45+) detected in the grafts by FACS were lower in the RNase A treatment group at POD 1 but no differences toward the control group were noted at POD 4. Posttransplantation on POD 1, higher numbers of neutrophils (CD45+Ly6G+) were counted in the grafts of saline‐treated rats, whereas on POD 4 neutrophils showed a higher number in the grafts of RNase A–treated rats. Saline‐treated animals, which received no additional immunosuppressive drugs, showed a diffuse infiltration of macrophages (CD68+), T cells (CD3+), and B cells (CD20+) on POD 1 and POD 4. In the RNase A treatment group, reduced infiltration of macrophages and T and B cells into the graft was seen at POD 1 and especially at POD 4.
Influence of RNase A Treatment on Proliferation of T Effector Cells
Unstimulated Teff cells exhibited a fraction of 2.2±1.2% proliferating cells, as determined by FACS. This value increased to 77.6±6.0% following exposure toward anti‐CD3/anti‐CD28 polyclonal stimulus (large effect according to Cohen's effect size statistics, dCohen=17.43, 95% CI 9.55/22.61, Figure 6A).
Figure 6.
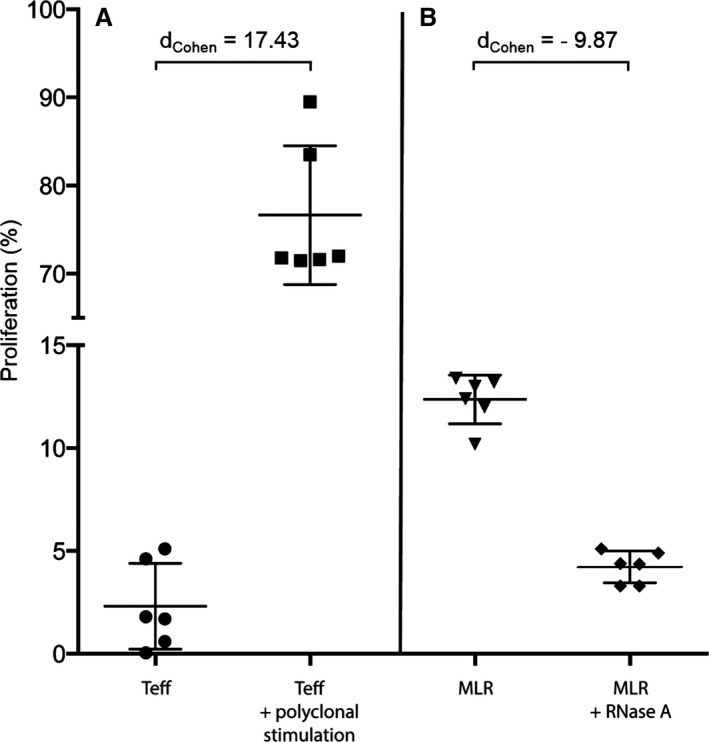
Influence of RNase A on mixed lymphocyte reaction. A, CD4+ Teff cells were isolated from naive human PBMCs and exposed to buffer only or towards polyclonal antiCD3/antiCD28 antibodies; proliferation of resting Teff cells was carried out by a CFSE dilution assay. B, The proliferation of allogeneic stimulated Teff cells was analyzed in MLR (co‐culture of isolated Teff cells with donor PBMCs in a ratio of 1:4) in the absence or presence of RNase A. Data represent the analysis of 2 independent experiments (n=6) and are shown as means±SD. CFSE indicates carboxyfluorescein succinimidyl ester; MLR, mixed lymphocyte reaction; PBMC, peripheral blood mononuclear cells; Teff, effector T cell.
Regarding the results of the mixed lymphocyte reaction in allogeneic co‐culture assays with unmatched and unrelated donor peripheral blood mononuclear cells, Teff cells showed a proliferative capacity of up to 12.0±1.0%, whereas in the presence of RNase A (10 μg/well) this value was decreased to 4.2±0.5% of all analyzed Teff cells (large effect, dCohen=9.87, 95% CI 5.29/12.92, Figure 6B).
Reduction of Edema Formation by RNase A Treatment
The mean pre‐implantation weight of the donor grafts in the graft survival groups was 0.94±0.03 g. After being rejected, the weight of the grafts in the saline treatment group showed an increase to 2.06±0.52 g, as opposed to a much smaller increase in donor graft weight in the RNase A treatment group (1.28±0.27 g, large effect, dCohen=1.88, 95% CI 0.41/3.07; Figure 7A). Quantitative morphological analyses evidenced less edema formation in RNase A–treated grafts as compared to the saline‐treated grafts, both at POD 1 and at POD 4 (Table 3 and Figure 7B).
Figure 7.
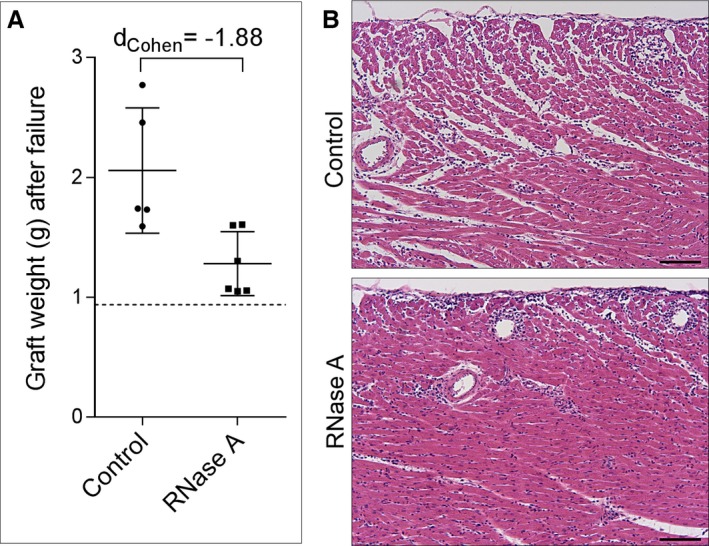
Graft weight upon complete dysfunction and explantation. A, The weight of grafted hearts was determined in the control and RNase A group as indicated, compared to the average weight of hearts prior to implantation (dashed line). B, Paraffin sections (left ventricle, hematoxylin and eosin staining) of explanted hearts at POD 4 show leukocyte infiltration and increased intercellular edema formation in the control group as compared to the RNase A group; scale bars 100 μm. POD indicates postoperative day.
Table 3.
Impact of RNase A Treatment on the Extent of Inflammatory Edema in POD 1 and POD 4 Beating Grafts
| POD 1 | POD 4 | |||||
|---|---|---|---|---|---|---|
| Control | RNase A | dCohen (95% CI) | Control | RNase A | dCohen (95% CI) | |
| Volume densities (%) | ||||||
| Vv(myocardium/heart) | 92.9±1.1 | 91.7±1.2 | 1.49 (0.02/2.27) | 90.7±3.2 | 87.4±2.2 | 1.2 (0.24/2.41) |
| Vv(edema/myocardium) | 5.8±0.8 | 4.0±0.4 | 2.94 (0.95/4.35) | 14.7±1.6 | 6.8±0.9 | 6.18 (2.84/8.33) |
| Absolute volumes (mm³) | ||||||
| V(heart) | 887.7±83 | 799.3±46.9 | 1.31 (0.16/2.53) | 1009.2±97.3 | 876.4±57.6 | 1.65 (0.09/2.98) |
| V(myocardium, heart) | 824.2±68.8 | 732.9±37.6 | 1.65 (0.09/2.89) | 916.2±103.7 | 765.2±43.0 | 1.89 (0.26/3.15) |
| V(edema, myocardium) | 47.8±9.4 | 29.1±2.0 | 2.74 (0.83/4.12) | 135.4±20.3 | 51.7±8.1 | 5.36 (2.38/7.3) |
Volume densities of the myocardium in the heart (VV(myocardium/heart)) and of edema in the myocardium (VV(edema/myocardium)). Total volumes of the heart (V(heart)), of the myocardium (V(myocardium, heart)), and of myocardial edema (V(edema, myocardium)). Data represent means±SD; n=5 in each group; POD, postoperative day; RNase A, ribonuclease A; dCohen, effect size according to Cohen,36, 37 classified in small (≥0.2 to <0.5), medium (≥0.5 to <0.8), and large (≥0.8) effects.
Prevention of Left Ventricular Thrombus Formation by RNase A Treatment
Rejected cardiac grafts of the saline as well as inactive RNase 1 treatment groups showed thrombus formation within the left ventricle as described in other studies using the same heterotopic abdominal heart transplantation technique.38 Grafts in the RNase A and active RNase 1 treatment groups maintained an unaltered left ventricle lumen even after loss of graft function as verified by transabdominal echocardiography (Figure 8).
Figure 8.
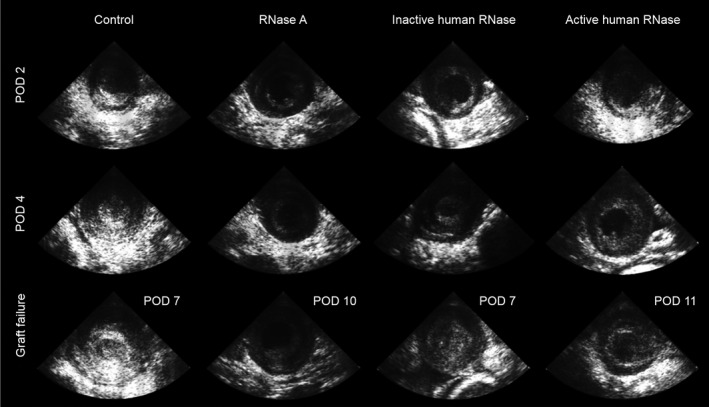
Thrombus formation in abdominal grafts as analyzed by echocardiography. At POD 2, POD 4, and the (variable) day of complete graft failure, echocardiography of left ventricles (LV) was performed on recipients of all treatment groups (bovine RNase A, inactive human RNase 1, active human RNase 1) and compared to control group (penetration depth 2 cm, 88 Hz). Note that rejected grafts presented LV thrombus formation in control and inactive human RNase 1 groups but not in bovine RNase A and active human RNase 1 treated hearts. POD indicates postoperative day.
Prolongation of Graft Survival by RNase A Treatment
As a primary biomarker for cardiac injury,39 serum troponin I was measured in transplanted animals. In comparison to the control group, troponin I levels were clearly lower in the RNase A treatment group, both at POD 1 (27.23±11.87 μg/L versus 5.32±1.35 μg/L, large effect according to Cohen's effect size statistics, dCohen=2.43, 95% CI 0.51/3.81) and POD 4 (4.07±1.21 μg/L versus 0.78±0.56 μg/L, large effect, dCohen=3.49, 95% CI 0.54/5.03). In addition to the decreased cardiac graft injury indicated by low troponin I levels, a tendency towards a lower cellular International Society for Heart and Lung Transplantation rejection grade was found in RNase A treatment at POD 4 (Table 4).
Table 4.
Grading of Acute Cellular Rejection
| Treatment Groups | Grade 0R (No Rejection) | Grade 1R (Mild Rejection) | Grade 2R (Moderate Rejection) | Grade 3R (Severe Rejection) |
|---|---|---|---|---|
| Control (n) | 0 | 1 | 3 | 1 |
| RNase A (n) | 0 | 2 | 3 | 0 |
At postoperative day 4, explanted beating grafts were graded based on a standardized grading system for the pathologic diagnosis of acute cellular rejection in cardiac biopsies recommended by the International Society for Heart and Lung Transplantation.2 n=5 in each treatment group. RNase A indicates ribonuclease A.
Treatment with RNase A improved cardiac median graft survival to 10.5 days (IQR 1.8) compared to 6.5 days (IQR 1.0) in saline‐treated animals (P=0.001, Figure 9A).
Figure 9.
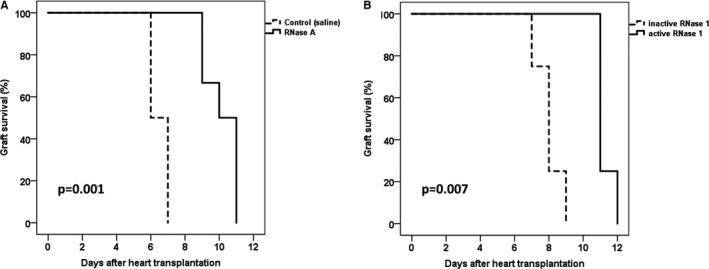
Kaplan–Meier estimation of graft survival. In both comparisons, the active enzyme (RNase A and RNase 1) was superior to the control (A) and accordingly inactive RNase 1 (B).
While treatment with enzymatically inactive RNase 1 revealed a median graft survival time similar to that of the saline control group (8.0 days, IQR 0.5), a prolonged graft survival was observed for the treatment group with active RNase 1 (11.0 days, IQR 0.3, P=0.007, Figure 9B). These results indicate that the efficacy of cardioprotection depends on the ribonucleolytic activity of RNase 1.
Discussion
The use of RNase as a potential drug in the immediate transplantation surrounding is based on its reported potent tissue‐ and cardioprotective functions. RNase A is an endogenous natural enzyme with the highest structural stability among all mammalian proteins, and has been demonstrated in several experimental in vivo studies of our group to counteract the damaging effect of eRNA in cardiovascular diseases. Here, the administration of even high doses of RNase A in rats for 28 days did not induce any destructive alterations in tissues and organs, as evidenced by macroscopic and histopathologic analyses.
Moreover, extracellular RNase appears to be a safe natural drug, since it acts in a nontoxic manner due to the ubiquitous presence of a highly reactive RNase inhibitor,40, 41 which prevents the destruction of intracellular RNAs upon exposure of RNase in all cell types. Overall, administration of both active pancreatic bovine RNase A as well as recombinant human RNase 1 resulted in a prolongation of graft survival in the described heart transplantation model, whereas enzymatically inactive RNase 1 and buffer alone were similarly ineffective. These data support the hypothesis that RNase 1 serves as a possible interventive regimen upon graft survival in heterotopic heart transplantation by degrading and removing detrimental eRNA.
Acute cellular rejection of a transplanted organ is primarily a T cell‐mediated event with infiltration of lymphocytes and monocytes/macrophages and resultant myocytolysis.42 In a previous study of our group, eRNA was demonstrated to induce leukocyte adhesion, which was mediated by activation of the vascular endothelial growth factor/vascular endothelial growth factor–receptor‐2 system and dependent on intercellular adhesion molecule‐1 and the β2‐integrin Mac‐1. Moreover, eRNA showed pro‐inflammatory properties by inducing tumor necrosis factor α release from monocytic cells.4 In the present study, RNase A–treated grafts showed a reduction of mostly T cells but also of B cells, indicating that RNase treatment resulted in eRNA degradation and hence reduced immune cell recruitment.
Yet, these connections still remain to be established in detail for the situation of heart transplantation. Another important factor in transplantation is I/R injury, which is associated with infiltration of neutrophils and macrophages.2 Macrophages have been shown to contribute to I/R injury in solid organ transplantation by generating inflammatory mediators.43 In the RNase A treatment group of the present study, reduced numbers of neutrophils and macrophages were detected in graft tissue in the early posttransplantation period as compared to the control group, reflecting decreased signs of I/R injury. Moreover, substantially decreased troponin I levels presenting the primary biomarker of cardiac damage39 were found in the RNase A treatment group as well. These promising results are reminiscent of the outcome of a recent study of cardiac I/R injury in rodents, where the molecular interplay between eRNA and tumor necrosis factor α was shown to be responsible to a large extent for myocardial damage and infarct size and where RNase A administration resulted in a significant reduction of this pathology.8
In case of graft rejection after transplantation, I/R injury accounts for accelerating transplant vasculopathy,44 which is associated with endothelial dysfunction45 that appears to be related to alloimmune responses such as episodes of acute and chronic allograft rejection and to nonalloimmunologic factors such as organ preservation, I/R injury, inflammation, and infections as well as to metabolic risk factors such as hyperlipidemia, diabetes, and hypertension.46 There are several mechanistic overlaps in the development of transplant vasculopathy and atherosclerosis.47 RNase was discussed to serve as a new treatment option against atherosclerosis.7 However, it remains to be investigated whether RNase administration could serve as a treatment option to prevent long‐term transplant vasculopathy in a model of chronic rejection.
Acute or chronic graft failure is based on combined processes, such as inflammation, thrombus formation, endothelium damage, and edema formation, which are associated with the appearance of eRNA, as demonstrated in several former studies. There, RNase application has been shown to remove the exposed eRNA that serves as cofactor for coagulation proteins and cytokines or permeability factors such as vascular endothelial growth factor A.5, 6, 23 In the present study of acute graft rejection, a similar mechanism is proposed, whereby treatment with RNase A resulted in diminished edema and thrombus formation related to increased graft survival. In fact, expression of vascular endothelial growth factor has been related to endothelial cell activation and phenotypic changes found in the microcirculation of cardiac allografts,48 thereby supporting our contention. Although ventricular thrombosis plays no role in clinical heart transplantation, in the presented animal model of unloaded left ventricle, thrombosis development is a common event.38 It is worth mentioning that all animals treated with active RNase exhibited no thrombosis. This supports the hypothesis that RNase counteracts the eRNA‐mediated activation of the contact phase of intrinsic blood coagulation.6
Finally, RNase may promote an immunosuppressive function, as indicated by its suppressive capacity on proliferating effector T cells verified by mixed lymphocyte reaction. Interestingly, it has been described already in the 1960s that intraperitoneal injection of an α2‐protein fraction from serum prolongs the survival of rat skin homografts by inhibition of antibody production to antigens.49 It was postulated that the respective fraction that contained RNase50 temporarily prevented antibody synthesis.51 Since after allogeneic transplantation, cellular rejection mechanisms are most important, humoral rejection mechanisms were not our major concern. Further studies dealing with antibody‐mediated rejection, eg, with sensitized animals,52 might help to clarify the contribution of eRNA and the possibility of RNase treatment in this respect.
However, the current experimental model provides new insights into the versatile role of ribonuclease as a potential drug to improve graft survival and that allows deciphering of the underlying mechanisms in more detail.
Conclusions
Our findings present evidence that RNase treatment provides a beneficial adjuvant therapy in heart transplantation, although it is difficult in the present model to differentiate between the detrimental impact of I/R injury versus rejection reaction. Mechanistically, we propose that enzymatically active RNase improves the survival of cardiac grafts by counteracting the adverse activities of eRNA and by acting as a yet to be defined mild immunosuppressive agent. Thus, RNase treatment should be considered as a universal potent adjuvant in situations of critical reperfusion such as percutaneous coronary intervention, cardiac surgery, or heart transplantation.
Sources of Funding
This work was in part supported by the Collaborative Research Center (SFB‐TR) 127, the Excellence Cluster Cardio‐Pulmonary System (ECCPS) of the Justus‐Liebig‐University, Giessen (Germany), and the individual grant FI‐543/2‐2, all funded by the German Research Foundation (Deutsche Forschungsgemeinschaft, Bonn, Germany), as well as by the von‐Behring‐Röntgen Foundation (Marburg, Germany) and the LOEWE‐Priority Program “Medical RNomics” (Wiesbaden, Germany).
Disclosures
None.
Acknowledgments
We thank Brita Zugenmaier for proofreading and the colleagues of the Walter Brendel Centre of Experimental Medicine (Ludwig‐Maximilians‐Universität München, Germany), particularly Franz Singer for his technical support concerning surgical equipment, and Mehdi Shakarami, head of animal facility, and his staff for the competency in animal housing. In particular, we are grateful to Sven Reese (Institute of Anatomy, Histology and Embryology, Ludwig‐Maximilians‐Universität München, Germany) for statistical advice and introduction to the method of effect size statistics according to Cohen.
(J Am Heart Assoc. 2016;5:e003429 doi: 10.1161/JAHA.116.003429)
Parts of this study were presented at the American Society of Transplantation (AST) Cutting Edge of Transplantation meeting in Chandler, Arizona, February 5–7, 2015.
References
- 1. Billingham ME, Cary NR, Hammond ME, Kemnitz J, Marboe C, McCallister HA, Snovar DC, Winters GL, Zerbe A. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Heart Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant. 1990;9:587–593. [PubMed] [Google Scholar]
- 2. Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, Andersen CB, Angelini A, Berry GJ, Burke MM, Demetris AJ, Hammond E, Itescu S, Marboe CC, McManus B, Reed EF, Reinsmoen NL, Rodriguez ER, Rose AG, Rose M, Suciu‐Focia N, Zeevi A, Billingham ME. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24:1710–1720. [DOI] [PubMed] [Google Scholar]
- 3. Fischer S, Preissner KT. Extracellular nucleic acids as novel alarm signals in the vascular system. Mediators of defence and disease. Hamostaseologie. 2013;33:37–42. [DOI] [PubMed] [Google Scholar]
- 4. Fischer S, Grantzow T, Pagel JI, Tschernatsch M, Sperandio M, Preissner KT, Deindl E. Extracellular RNA promotes leukocyte recruitment in the vascular system by mobilising proinflammatory cytokines. Thromb Haemost. 2012;108:730–741. [DOI] [PubMed] [Google Scholar]
- 5. Fischer S, Gerriets T, Wessels C, Walberer M, Kostin S, Stolz E, Zheleva K, Hocke A, Hippenstiel S, Preissner KT. Extracellular RNA mediates endothelial‐cell permeability via vascular endothelial growth factor. Blood. 2007;110:2457–2465. [DOI] [PubMed] [Google Scholar]
- 6. Kannemeier C, Shibamiya A, Nakazawa F, Trusheim H, Ruppert C, Markart P, Song Y, Tzima E, Kennerknecht E, Niepmann M, von Bruehl ML, Sedding D, Massberg S, Gunther A, Engelmann B, Preissner KT. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc Natl Acad Sci USA. 2007;104:6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simsekyilmaz S, Cabrera‐Fuentes HA, Meiler S, Kostin S, Baumer Y, Liehn EA, Weber C, Boisvert WA, Preissner KT, Zernecke A. Role of extracellular RNA in atherosclerotic plaque formation in mice. Circulation. 2014;129:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabrera‐Fuentes HA, Ruiz‐Meana M, Simsekyilmaz S, Kostin S, Inserte J, Saffarzadeh M, Galuska SP, Vijayan V, Barba I, Barreto G, Fischer S, Lochnit G, Ilinskaya ON, Baumgart‐Vogt E, Boning A, Lecour S, Hausenloy DJ, Liehn EA, Garcia‐Dorado D, Schluter KD, Preissner KT. RNase1 prevents the damaging interplay between extracellular RNA and tumour necrosis factor‐alpha in cardiac ischaemia/reperfusion injury. Thromb Haemost. 2014;112:1110–1119. [DOI] [PubMed] [Google Scholar]
- 9. Schein CH. From housekeeper to microsurgeon: the diagnostic and therapeutic potential of ribonucleases. Nat Biotechnol. 1997;15:529–536. [DOI] [PubMed] [Google Scholar]
- 10. Dubos RJ. The decomposition of yeast nucleic acid by a heat resistant enzyme. Science. 1937;85:549–550. [DOI] [PubMed] [Google Scholar]
- 11. Kunitz M. Isolation from beef pancreas of a crystalline protein possessing ribonuclease activity. Science. 1939;90:112–113. [DOI] [PubMed] [Google Scholar]
- 12. Kunitz M. Crystalline ribonuclease. J Gen Physiol. 1940;24:15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smyth DG, Stein WH, Moore S. The sequence of amino acid residues in bovine pancreatic ribonuclease: revisions and confirmations. J Biol Chem. 1963;238:227–234. [PubMed] [Google Scholar]
- 14. Gutte B, Merrifield RB. The total synthesis of an enzyme with ribonuclease A activity. J Am Chem Soc. 1969;91:501–502. [DOI] [PubMed] [Google Scholar]
- 15. Beintema JJ, Wietzes P, Weickmann JL, Glitz DG. The amino acid sequence of human pancreatic ribonuclease. Anal Biochem. 1984;136:48–64. [DOI] [PubMed] [Google Scholar]
- 16. Weickmann JL, Elson M, Glitz DG. Purification and characterization of human pancreatic ribonuclease. Biochemistry. 1981;20:1272–1278. [DOI] [PubMed] [Google Scholar]
- 17. Landre JB, Hewett PW, Olivot JM, Friedl P, Ko Y, Sachinidis A, Moenner M. Human endothelial cells selectively express large amounts of pancreatic‐type ribonuclease (RNase 1). J Cell Biochem. 2002;86:540–552. [DOI] [PubMed] [Google Scholar]
- 18. Fischer S, Nishio M, Dadkhahi S, Gansler J, Saffarzadeh M, Shibamiyama A, Kral N, Baal N, Koyama T, Deindl E, Preissner KT. Expression and localisation of vascular ribonucleases in endothelial cells. Thromb Haemost. 2011;105:345–355. [DOI] [PubMed] [Google Scholar]
- 19. Gerriets T, Stolz E, Walberer M, Muller C, Kluge A, Bachmann A, Fisher M, Kaps M, Bachmann G. Noninvasive quantification of brain edema and the space‐occupying effect in rat stroke models using magnetic resonance imaging. Stroke. 2004;35:566–571. [DOI] [PubMed] [Google Scholar]
- 20. Rottger C, Bachmann G, Gerriets T, Kaps M, Kuchelmeister K, Schachenmayr W, Walberer M, Wessels T, Stolz E. A new model of reversible sinus sagittalis superior thrombosis in the rat: magnetic resonance imaging changes. Neurosurgery. 2005;57:573–580; discussion 573‐580 [DOI] [PubMed] [Google Scholar]
- 21. Directive 2010/63/EU of the European parliament and of the council on the protection of animals used for scientific purposes. Official Journal of the European Union 2010. Available at: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32010L0063. Accessed March 4th, 2016.
- 22. Ono K, Lindsey ES. Improved technique of heart transplantation in rats. J Thorac Cardiovasc Surg. 1969;57:225–229. [PubMed] [Google Scholar]
- 23. Walberer M, Tschernatsch M, Fischer S, Ritschel N, Volk K, Friedrich C, Bachmann G, Mueller C, Kaps M, Nedelmann M, Blaes F, Preissner KT, Gerriets T. Rnase therapy assessed by magnetic resonance imaging reduces cerebral edema and infarction size in acute stroke. Curr Neurovasc Res. 2009;6:12–19. [DOI] [PubMed] [Google Scholar]
- 24. Fischer S, Gesierich S, Griemert B, Schanzer A, Acker T, Augustin HG, Olsson AK, Preissner KT. Extracellular RNA liberates tumor necrosis factor‐alpha to promote tumor cell trafficking and progression. Cancer Res. 2013;73:5080–5089. [DOI] [PubMed] [Google Scholar]
- 25. Weksler BB, Subileau EA, Perriere N, Charneau P, Holloway K, Leveque M, Tricoire‐Leignel H, Nicotra A, Bourdoulous S, Turowski P, Male DK, Roux F, Greenwood J, Romero IA, Couraud PO. Blood‐brain barrier‐specific properties of a human adult brain endothelial cell line. FASEB J. 2005;19:1872–1874. [DOI] [PubMed] [Google Scholar]
- 26. Sznajd J, Magdon M, Naskalski JW, Uracz R, Wojcikiewicz O. Serum ribonuclease activity in acute myocardial infarction. Cor Vasa. 1981;23:241–247. [PubMed] [Google Scholar]
- 27. Gordon CR, Matthews MS, Lefebvre DR, Strande LF, Marra SW, Guglielmi M, Hollenberg SM, Hewitt CW. A new modified technique for heterotopic femoral heart transplantation in rats. J Surg Res. 2007;139:157–163. [DOI] [PubMed] [Google Scholar]
- 28. Noyan F, Lee YS, Zimmermann K, Hardtke‐Wolenski M, Taubert R, Warnecke G, Knoefel AK, Schulde E, Olek S, Manns MP, Jaeckel E. Isolation of human antigen‐specific regulatory T cells with high suppressive function. Eur J Immunol. 2014;44:2592–2602. [DOI] [PubMed] [Google Scholar]
- 29. Scherle W. A simple method for volumetry of organs in quantitative stereology. Mikroskopie. 1970;26:57–60. [PubMed] [Google Scholar]
- 30. Howard CV, Reed MG. Unbiased Stereology. Coleraine, UK: QTP Publications; 2005. [Google Scholar]
- 31. Weibel ER. Stereological Methods, 1: Practical Methods for Biological Morphometry. London: Academic Press; 1979. [Google Scholar]
- 32. Bahnemann R, Jacobs M, Karbe E, Kaufmann W, Morawietz G, Nolte T, Rittinghausen S. RITA—registry of industrial toxicology animal‐data—guides for organ sampling and trimming procedures in rats. Exp Toxicol Pathol. 1995;47:247–266. [DOI] [PubMed] [Google Scholar]
- 33. Kittel B, Ruehl‐Fehlert C, Morawietz G, Klapwijk J, Elwell MR, Lenz B, O'Sullivan MG, Roth DR, Wadsworth PF; Group R, Group N . Revised guides for organ sampling and trimming in rats and mice—part 2. A joint publication of the RITA and NACAD groups. Exp Toxicol Pathol. 2004;55:413–431. [DOI] [PubMed] [Google Scholar]
- 34. Morawietz G, Ruehl‐Fehlert C, Kittel B, Bube A, Keane K, Halm S, Heuser A, Hellmann J; Group R, Group N . Revised guides for organ sampling and trimming in rats and mice—part 3. A joint publication of the RITA and NACAD groups. Exp Toxicol Pathol. 2004;55:433–449. [DOI] [PubMed] [Google Scholar]
- 35. Ruehl‐Fehlert C, Kittel B, Morawietz G, Deslex P, Keenan C, Mahrt CR, Nolte T, Robinson M, Stuart BP, Deschl U; Group R, Group N . Revised guides for organ sampling and trimming in rats and mice—part 1. Exp Toxicol Pathol. 2003;55:91–106. [PubMed] [Google Scholar]
- 36. Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed Hillsdale, NJ: Erlbaum; 1988. [Google Scholar]
- 37. Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol Rev Camb Philos Soc. 2007;82:591–605. [DOI] [PubMed] [Google Scholar]
- 38. Wen P, Wang X, Li T, Zhang B, Sun X, Qiu G, Fan J, Xing T, Luo Q, Tang H, Peng Z. Establishment of a novel volume‐loaded heterotopic heart transplantation model in rats. J Surg Res. 2013;183:435–441. [DOI] [PubMed] [Google Scholar]
- 39. Amsterdam EA, Wenger NK, Brindis RG, Casey DE Jr, Ganiats TG, Holmes DR Jr, Jaffe AS, Jneid H, Kelly RF, Kontos MC, Levine GN, Liebson PR, Mukherjee D, Peterson ED, Sabatine MS, Smalling RW, Zieman SJ; Members AATF . 2014 AHA/ACC guideline for the management of patients with non‐ST‐elevation acute coronary syndromes: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;130:2354–2394. [DOI] [PubMed] [Google Scholar]
- 40. Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007;368:434–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindenfeld J, Miller GG, Shakar SF, Zolty R, Lowes BD, Wolfel EE, Mestroni L, Page RL II, Kobashigawa J. Drug therapy in the heart transplant recipient: part I: cardiac rejection and immunosuppressive drugs. Circulation. 2004;110:3734–3740. [DOI] [PubMed] [Google Scholar]
- 43. Salehi S, Reed EF. The divergent roles of macrophages in solid organ transplantation. Curr Opin Organ Transplant. 2015;20:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goto R, Issa F, Heidt S, Taggart D, Wood KJ. Ischemia‐reperfusion injury accelerates human antibody‐mediated transplant vasculopathy. Transplantation. 2013;96:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Valantine HA. Cardiac allograft vasculopathy: central role of endothelial injury leading to transplant “atheroma”. Transplantation. 2003;76:891–899. [DOI] [PubMed] [Google Scholar]
- 46. Vassalli G, Gallino A, Weis M, von Scheidt W, Kappenberger L, von Segesser LK, Goy JJ; Working Group Microcirculation of the European Society of C . Alloimmunity and nonimmunologic risk factors in cardiac allograft vasculopathy. Eur Heart J. 2003;24:1180–1188. [DOI] [PubMed] [Google Scholar]
- 47. Guethoff S, Grinninger C, Kaczmarek I. Everolimus: sidekick against atherosclerosis? Atherosclerosis. 2013;231:27–28. [DOI] [PubMed] [Google Scholar]
- 48. Torry RJ, Labarrere CA, Torry DS, Holt VJ, Faulk WP. Vascular endothelial growth factor expression in transplanted human hearts. Transplantation. 1995;60:1451–1457. [DOI] [PubMed] [Google Scholar]
- 49. Mowbray JF. Effect of large doses of an alpha2‐glycoprotein fraction on the survival of rat skin homografts. Transplantation. 1963;1:15–20. [DOI] [PubMed] [Google Scholar]
- 50. Mowbray JF, Hargrave DC. Further studies on the preparation of the immunosuppressive alpha‐2 protein fraction from the serum and its assay in mice. Immunology. 1966;11:413–419. [PMC free article] [PubMed] [Google Scholar]
- 51. Mowbray JF, Scholand J. Inhibition of antibody production by ribonucleases. Immunology. 1966;11:421–426. [PMC free article] [PubMed] [Google Scholar]
- 52. Huang G, Wilson NA, Reese SR, Jacobson LM, Zhong W, Djamali A. Characterization of transfusion‐elicited acute antibody‐mediated rejection in a rat model of kidney transplantation. Am J Transplant. 2014;14:1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]


