Summary
Across eukaryotic species, mild mitochondrial stress can have beneficial effects on the lifespan of organisms. Mitochondrial dysfunction activates an unfolded protein response (UPRmt), a stress signaling mechanism designed to ensure mitochondrial homeostasis. Perturbation of mitochondria during larval development in C. elegans not only delays aging but also maintains UPRmt signaling, suggesting an epigenetic mechanism that modulates both longevity and mitochondrial proteostasis throughout life. We identify the conserved histone lysine demethylases jmjd-1.2/PHF8 and jmjd-3.1/JMJD3 as positive regulators of lifespan in response to mitochondrial dysfunction across species. Reduction-of-function of the demethylases potently suppresses longevity and UPRmt induction while gain-of-function is sufficient to extend lifespan in an UPRmt-dependent manner. A systems genetics approach in the BXD mouse reference population further indicates conserved roles of the mammalian orthologs in longevity and UPRmt signaling. These findings illustrate an evolutionary conserved epigenetic mechanism that determines the rate of aging downstream of mitochondrial perturbations.
Introduction
Aging is a deleterious and complex process characterized by the progressive decline of cellular and organismal homeostasis amid constantly increasing levels of entropy (Kirkwood, 2005) and represents the primary risk factor in major human pathologies including cancer, diabetes, cardiovascular disorders, and neurodegenerative diseases (Lopez-Otin et al., 2013). Aging, however, is not only a response to experiences incurred toward the end of an organism's life: it is shaped and determined by experiences that have accumulated from the earliest stages of development, and even during the generations that came before it. The organism's perpetuation of historic cues – how its patterns of gene expression change and become cemented in response to stresses that may have occurred long in the past or in parental populations – have proven to significantly affect long-term health and longevity, the mechanistic details of which are only beginning to emerge. It is thus that we now know, for example, that the transgenerational inheritance of chromatin marks can increase lifespan for multiple generations in C. elegans (Greer et al., 2010; Han and Brunet, 2012; Lopez-Otin et al., 2013; Rando and Chang, 2012) underscoring the importance of understanding not only the genetic but also the epigenetic contributions to the complex and disordered aging process.
One of the most dramatic examples in which early events have effects on longevity is found in the nematode C. elegans, in which mitochondrial stress during development can cause nearly a doubling of the animal's lifespan (Dillin et al., 2002a). The timing and degree of mitochondrial dysfunction is highly selective: it must occur during a specific L3/L4 larval transition in order to cause lifespan extension, a time during which a heavy amount of germline-specific mitochondrial biogenesis also occurs (Rea et al., 2007; Tsang and Lemire, 2002). In contrast, mitochondrial dysfunction that is too severe or which is implemented too early or late can have a negative effect on lifespan. In many cases, titrating a level of dysfunction is absolutely required in order to observe an extension of lifespan (Rea et al., 2007). Longevity caused by mitochondrial dysfunction also often fails to generate universal health benefits, as organisms may live longer but exhibit developmental delay and a drastic reduction in reproductive fitness (Dillin et al., 2002a; Lee et al., 2003). These effects are surprisingly conserved: in yeast, flies, and mice, mitochondrial dysfunction can delay the aging process (Copeland et al., 2009; Dell'agnello et al., 2007; Dillin et al., 2002a; Feng et al., 2001; Kirchman et al., 1999; Lee et al., 2003; Liu et al., 2005), but, when occurring later in life, has deleterious effects and is associated with age-onset neurodegenerative diseases, directly contributing to their pathologies (Schon and Przedborski, 2011).
Within mitochondria, intricate surveillance systems monitor the quality of existing and newly synthesized mitochondrial proteins to ensure mitochondrial homeostasis (Baker et al., 2011). The mitochondrial unfolded protein response (UPRmt) consists of a signaling cascade that results in upregulation of nuclear-encoded genes to alleviate the stress sensed in mitochondria. Perception of misfolding in mitochondria requires the protease ClpP, which generates a mitochondrial derived signal to activate downstream genes (Haynes et al., 2007). ClpP triggers the activation of the ubiquitin-like protein UBL-5, which acts as a coactivator of the transcription factor DVE-1. UBL-5 and DVE-1 respond to mitochondrial perturbation to increase expression of mitochondrial chaperones, including hsp-6 and hsp-60 (Benedetti et al., 2006). In parallel, the bZIP transcription factor ATFS-1 coordinates a wide cellular response to mitochondrial stress (Nargund et al., 2012, 2015).
Previously, several genes characterized for their role in the regulation of the UPRmt were identified as specific requirements for the long lifespan of animals with reduced mitochondrial function (Durieux et al., 2011; Houtkooper et al., 2013). The requirements for these genes in electron transport chain (ETC)-mediated longevity suggested that the function of the UPRmt might have a beneficial effect on the organism and was required to maintain the longer lifespans in the mitochondrial mutants. In this model, mitochondrial dysfunction early in development was capable of imposing a mild, hormetic stress that could remodel gene expression patterns, perpetuating a beneficial effect long into adulthood (Durieux et al., 2011). In keeping with this hypothesis, induction of the UPRmt during early larval stages persists long into adulthood, suggesting that the animal may possess an epigenetic mechanism to maintain the activation of stress responses and ensure increased resistance to future mitochondrial insults (Durieux et al., 2011). In yeast, mitochondrial stress generated by reactive oxygen species (ROS) causes an epigenetic remodeling that extends lifespan (Schroeder et al., 2013), and forced expression of UPRmt genes in Drosophila is sufficient to preserve mitochondrial function (Owusu-Ansah et al., 2013).
In this study we identify two conserved histone lysine demethylases as regulators of the ETC longevity pathway. Using RNAi-based screens in C. elegans, we isolated the conserved histone demethylases jmjd-1.2 and jmjd-3.1 as potent suppressors of longevity in response to mitochondrial perturbations. We demonstrate that both jmjd-1.2 and jmjd-3.1 are necessary and sufficient for activation of the UPRmt in C. elegans. Moreover, our experiments identify jmjd-1.2 and jmjd-3.1 as positive regulators of a longevity response that genetically requires UPRmt signaling. Using transcriptome analysis, we demonstrate that jmjd-1.2 and jmjd-3.1 coordinate the transcriptional response to mitochondrial stress. Furthermore, using a systems genetics approach, we find that the mammalian orthologs exhibit positive genetic correlations with UPRmt core components in the BXD mouse genetic reference population (Andreux et al., 2012; Wu et al., 2014). Together, these data reveal a conserved epigenetic mechanism that determines longevity and stress signaling in response to mitochondrial dysfunction.
Results
Mitochondrial ETC-Mediated Longevity Requires the Histone Lysine Demethylases jmjd-1.2 and jmjd-3.1
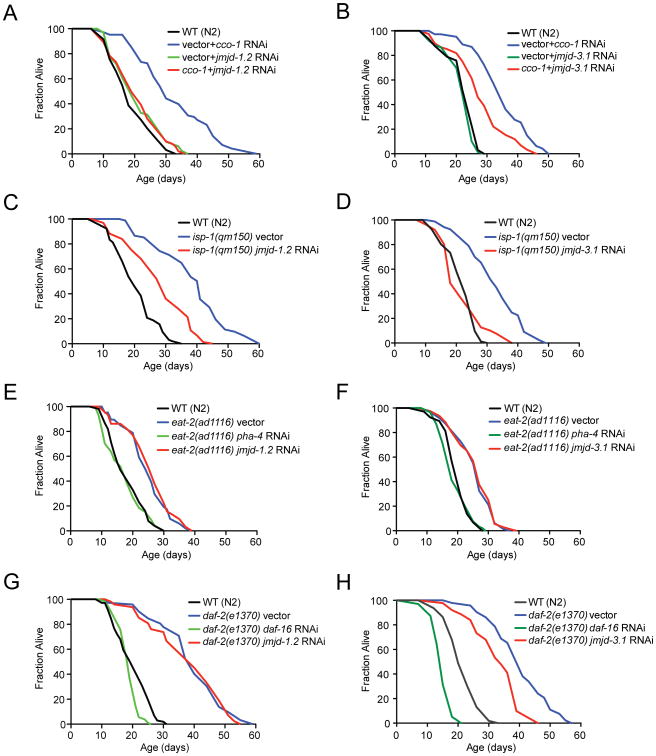
We performed an RNAi-based screen to identify genes specifically required for the ETC-mediated longevity in C. elegans. Through these analyses, we identified a putative histone lysine demethylase of the JumonjiC (JmjC)-domain-containing protein family, jmjd-1.2 (F29B9.2), (Kooistra and Helin, 2012), as a potent suppressor of the ETC longevity pathway (Figure 1A and Table S1). We hypothesized that other JmjC domain containing demethylases may have similar effects on the regulation of lifespan. From the JmjC orthologs found in C. elegans, we identified a second histone lysine demethylase, jmjd-3.1 (F18E9.5), that was also an effective suppressor of ETC-mediated longevity (Figure 1B and Table S1). In their roles as demethylases, jmjd-1.2 and jmjd-3.1 have both distinct and overlapping specificity for histone modifications, most notably against H3K27me2/3 (Agger et al., 2007; Kleine-Kohlbrecher et al., 2010). We found that both jmjd-1.2 and jmjd-3.1 were required for the extended lifespan caused by point mutation in the Rieske iron-sulfur protein (complex III), isp-1(qm150) (Feng et al., 2001) (Figures 1C, D). Downregulation of the closely related H3K27me2/3 demethylases jmjd-3.2, jmjd-3.3 and utx-1, however, did not significantly affect lifespan extension by cco-1 RNAi (Figure S1A), highlighting a specific role of jmjd-1.2 and jmjd-3.1 for ETC-mediated longevity.
Figure 1. Lifespan Extension by Mitochondrial ETC Perturbation Requires the Histone Demethylases jmjd-1.2 and jmjd-3.1.
(A) and (B) Knockdown of jmjd-1.2 (A) or jmjd-3.1 (B) suppresses cco-1-mediated lifespan extension.
(C) jmjd-1.2 is partially required for longevity of isp-1(qm150) mutant animals.
(D) jmjd-3.1 is required for longevity of isp-1(qm150) mutant animals.
(E) and (F) Dietary restriction-mediated longevity of eat-2(ad1116) animals is not affected by jmjd-1.2 (E) or jmjd-3.1 (F) knockdown.
(G) Longevity of daf-2(e1370) mutant animals is not affected by jmjd-1.2 knockdown.
(H) Longevity of daf-2(e1370) mutant animals partially depends on jmjd-3.1. See also Figure S1 and Table S1 for lifespan statistics.
Next, we assessed whether jmjd-1.2 and jmjd-3.1 were specific to lifespan extension in response to mitochondrial perturbation. The mitochondrial ETC longevity pathway acts in parallel to several other lifespan extending paradigms, including the insulin/IGF longevity response (Kenyon et al., 1993), dietary restriction (DR) (Lakowski and Hekimi, 1998), and germline-mediated longevity (Hsin and Kenyon, 1999). Neither jmjd-1.2 nor jmjd-3.1 downregulation altered longevity of dietary restricted eat-2(ad1116) animals (Figures 1E, F). We further found that jmjd-1.2 had no effect on the lifespan of animals in which stress responses of the ER or cytosol were constitutively activated (Baird et al., 2014; Taylor and Dillin, 2013) (Figures S1B-D) while transcription factors FoxO/daf-16 and FoxA/pha-4 successfully suppressed lifespan in our analyses (Figures 1E-H, Table S1). In contrast, while jmjd-1.2 RNAi did not affect the extended longevity seen in daf-2(e1370) mutant animals or germline deficient glp-1(e2141) animals (Figures 1G, S1B), we observed that the long lifespan daf-2(e1370) mutant worms depended on jmjd-3.1, albeit partially (Figure 1H). Recently jmjd-3.1 was also reported to be required for the lifespan extension of germline-deficient glp-1 animals (Labbadia and Morimoto, 2015). Collectively, these data indicate that while jmjd-3.1 is involved in several lifespan extension paradigms, both histone demethylases are required for ETC-mediated longevity.
Overlapping Temporal Requirements of JmjC Demethylase Activity and ETC-Mediated Longevity
One of the most remarkable features of ETC-mediated longevity in C. elegans lies in its precise temporal requirements: Reduction of ETC function during a narrow window of development is sufficient to extend lifespan throughout life (Dillin et al., 2002a; Rea et al., 2007). In contrast, loss of ETC function in adulthood is sufficient to reduce ATP production but can no longer affect lifespan. We reasoned that the activity of histone demethylases might be required during development and therefore performed RNAi shifting experiments in which we reduced ETC activity during development and restored them during adulthood by shifting young adults to Dicer (dcr-1) RNAi to inhibit the RNAi machinery (Dillin et al., 2002a, 2002b). We found that reduction of jmjd-1.2 during larval development was sufficient to block cco-1 mediated longevity (Figure S1E). jmjd-1.2 RNAi during adulthood, in contrast, reduced cco-1 lifespan partially (Figure S1F). jmjd-3.1 was partially required for cco-1 mediated longevity during larval development but had only a minor effect during adulthood (Figures S1G, H). These data suggest that both jmjd-1.2 and jmjd-3.1 are required for an initial response to ETC deficiency during development that is maintained into adulthood.
JMJD-1.2 and JMJD-3.1 Regulate the Mitochondrial Unfolded Protein Response (UPRmt)
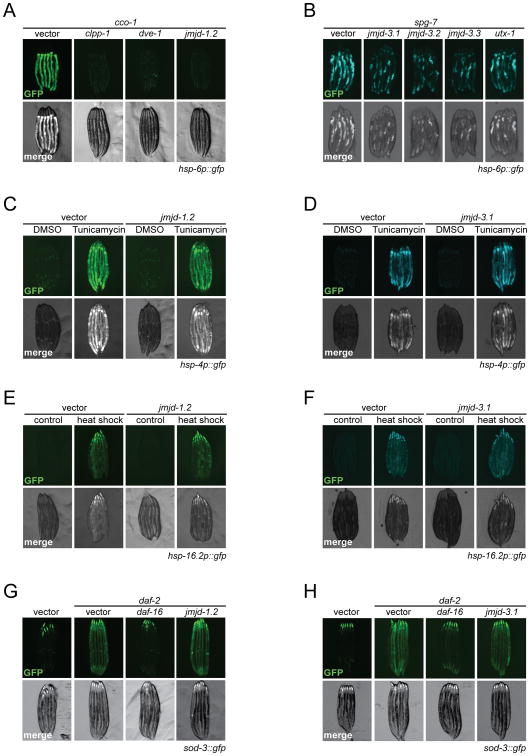
Previously, the UPRmt was found to be required for the extended longevity of mitochondrial mutants (Durieux et al., 2011). We therefore asked whether jmjd-1.2 and jmjd-3.1 had any effect on the induction of the UPRmt in C. elegans. In these analyses, cco-1 RNAi was used as a potent inducer of both hsp-6 and hsp-60 endogenous transcripts, robustly turning on the transcriptional hsp-6p∷gfp reporter (Yoneda et al., 2004). As reported, cco-1 mediated UPRmt activation required the ClpP protease clpp-1 and the homeodomain transcription factor dve-1 (Haynes et al., 2007) (Figures 2A and S2B). Similarly, RNAi of either jmjd-1.2 or jmjd-3.1 suppressed the hsp-6p∷gfp reporter activation elicited by cco-1 (Durieux et al., 2011) (Figures 2A and S2A), spg-7 (Yoneda et al., 2004) (Figures 2B and S2C) or mrps-5 (Houtkooper et al., 2013) knockdowns (Figure S2D).
Figure 2. jmjd-1.2 and jmjd-3.1 are Necessary and Specific for Induction of the UPRmt.
(A) Fluorescent micrographs of hsp-6p∷gfp UPRmt reporter animals treated with the indicated RNAi at day 1 of adulthood. Knockdown of jmjd-1.2 suppresses cco-1-mediated UPRmt induction in hsp-6p∷gfp reporter animals.
(B) Knockdown of jmjd-3 histone demethylase family members suppresses spg-7-mediated UPRmt induction.
(C) and (D) Fluorescent micrographs of hsp-4p∷gfp UPRer reporter animals. Induction of the UPRer response in hsp-4p∷gfp UPRer reporter animals by tunicamycin treatment is not affected by neither jmjd-1.2 (C) or jmjd-3.1 (D) RNAi.
(E) and (F) Fluorescent micrographs of hsp-16.2p∷gfp reporter animals. Induction of the heat shock response in hsp-16.2p∷gfp reporter animals occurs independently of jmjd-1.2 (E) or jmjd-3.1 (F) RNAi.
(G) and (H) Fluorescent micrographs of sod-3∷gfp reporter animals treated with the indicated RNAi. daf-16 RNAi was used as a positive control. See also Figures S2 and S3.
We tested additional JmjC-containing proteins for their effects on UPRmt following mitochondrial stress (Figure S2A). Interestingly, RNAi against the H3K27 demethylase family members jmjd-3.2, jmjd-3.3 and utx-1 also substantially decreased UPRmt activation, while other JmjC-domain containing demethylases had no significant effect on UPRmt activation (Figures 2B, S2A). As an alternative approach, we used the H3K27 demethylase inhibitor GSK-J4 (Heinemann et al., 2014) to examine potential activity towards UPRmt induction. GSK-J4 treatment of hsp-6p∷gfp reporter animals grown on mrps-5 RNAi suppressed UPRmt activation (Figure S2E) indicating that H3K27 demethylase activity was required for the perpetuation of mitochondrial stress signaling. As H3K27 methylation can be established by the Polycomb repressive complex 2 (PRC2) in C. elegans (Bender et al., 2004), we asked whether reduction of polycomb components is sufficient to trigger the UPRmt. However, knockdown of individual PRC2 components mes-2, mes-3 and mes-6 in C. elegans did not activate UPRmt signaling (Figure S2F).
We next examined a potential contribution of jmjd-1.2 and jmjd-3.1 on additional cellular stress responses including the unfolded protein response of the endoplasmic reticulum (UPRer) (Walter and Ron, 2011) and the cytosolic heat shock response (HSR) (Morimoto, 2011). RNAi against jmjd-1.2 did not affect induction of UPRer target genes, including the ER chaperone BiP (hsp-4), in response to UPRer stimulation with tunicamycin (Figures 2C, S3A). In contrast, jmjd-3.1 RNAi revealed minor but statistically significant effects on several UPRer target genes, which became evident only upon monitoring gene expression independent of the hsp-4p∷gfp reporter (Figure 2D, S3B), consistent with recent findings (Labbadia and Morimoto, 2015). Moreover, RNAi against jmjd-1.2 or jmjd-3.1 did not affect the heat shock-induced expression of the HSR chaperone hsp-16.2 (Figures 2E, F; S3C, D). Finally, jmjd-1.2 and jmjd-3.1 had no effect on the expression of the antioxidant enzyme sod-3, which is induced by reduced insulin/IGF-1 signaling (Figures 2G, H). Collectively, these data indicate that jmjd-1.2 specifically modulates the UPRmt, while jmjd-3.1 appears to be moderately involved in the transcriptional response to ER stress.
Overexpression of jmjd-1.2 or jmjd-3.1 is Sufficient for Lifespan Extension and UPRmt Induction
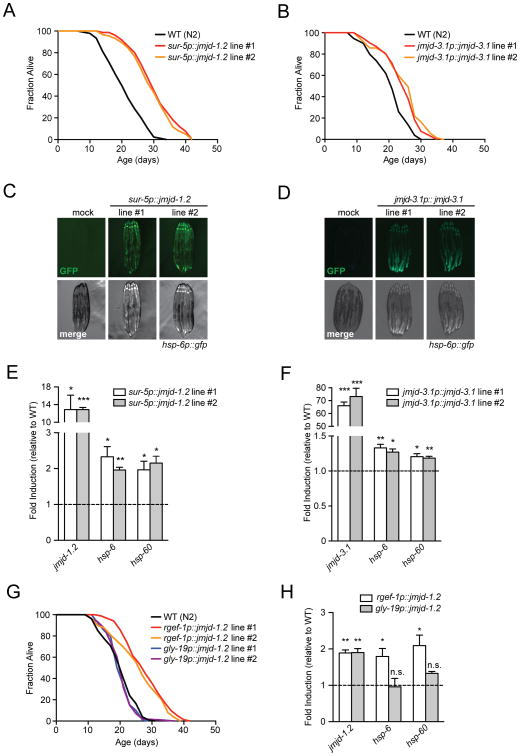
The results of the previous experiments indicated that both jmjd-1.2 and jmjd-3.1 were required for ETC-mediated longevity and UPRmt induction. We next tested their sufficiency to extend lifespan and increase mitochondrial stress signaling. We generated transgenic strains expressing jmjd-1.2 under control of the ubiquitous sur-5 promoter and monitored lifespan. For control, we generated an enzymatically inactive version of jmjd-1.2 that harbors a point mutation in the catalytic histidine of the JmjC domain (jmjd-1.2H508A). jmjd-1.2 overexpression significantly increased longevity when compared to wild type animals (Figure 3A), while the catalytically-inactive jmjd-1.2H508A had no effect (Figure S4A). We also generated strains overexpressing jmjd-3.1 under the control of its endogenous promoter (jmjd-3.1p∷jmjd-3.1) and found these animals to be long-lived (Figure 3B), in agreement with previous reports (Labbadia and Morimoto, 2015).
Figure 3. jmjd-1.2 and jmjd-3.1 Overexpression is Sufficient for Lifespan Extension and UPRmt Induction.
(A) and (B) Overexpression of jmjd-1.2 and jmjd-3.1 extends C. elegans lifespan. Lifespan analysis of two independent transgenic lines of sur-5p∷jmjd-1.2 (A) or jmjd-3.1p∷jmjd-3.1 (B) expressing animals compared to WT (N2) animals.
(C) and (D) Fluorescent micrographs of hsp-6p∷gfp UPRmt reporter animals expressing either sur-5p∷jmjd-1.2 (C) or jmjd-3.1p∷jmjd-3.1 (D) transgenes in two independent lines analyzed at day 1 of adulthood.
(E) and (F) Transcript levels in two independent lines of sur-5p∷jmjd-1.2 (E) or jmjd-3.1p∷jmjd-3.1 (F) expressing animals at day 1 of adulthood were measured by qRT-PCR. Results are shown relative to transcript levels in WT (N2) animals, with error bars indicating mean ± SEM. *** denotes p < 0.001, **p < 0.01, *p < 0.05.
(G) Neuron-specific overexpression of jmjd-1.2 is sufficient to extend lifespan. Lifespan analysis of two independent lines expressing either pan-neuronal (rgef-1p∷jmjd-1.2) or intestinal (gly-19p∷jmjd-1.2) transgenes compared to WT (N2) animals.
(H) Transcript levels in animals expressing either pan-neuronal (rgef-1p∷jmjd-1.2) or intestinal (gly-19p∷jmjd-1.2) transgenes at day 1 of adulthood were measured by qRT-PCR. Results are shown relative to transcript levels in WT (N2) animals, with error bars indicating mean ± SEM. ** denotes p < 0.01, *p < 0.05, n.s. = p > 0.05. See also Figure S4 and Table S1 for lifespan statistics.
Intriguingly, overexpression of either jmjd-1.2 or jmjd-3.1 was sufficient to induce the UPRmt (Figures 3C-F). To identify the tissues in which expression was required, we generated transgenic strains expressing jmjd-1.2 in either neurons or the intestine. Neuronal overexpression of jmjd-1.2 was sufficient for lifespan extension and UPRmt activation, while expression in the intestine did not (Figures 3G, H, S4B). Similarly, overexpression of the catalytically inactive jmjd-1.2H508A variant failed to activate the UPRmt (Figure S4B). Moreover, jmjd-1.2 overexpression had no effect on mRNAs levels of UPRer and HSR target genes indicating that jmjd-1.2 selectively modulates the UPRmt (Figures S4C, D). jmjd-3.1 overexpression, however, lead to a significant increase in expression of some UPRer target genes, but not of the HSR (Figures S4E, F).
The UPRmt Is a Genetic Requirement for JmjC demethylase-mediated Longevity
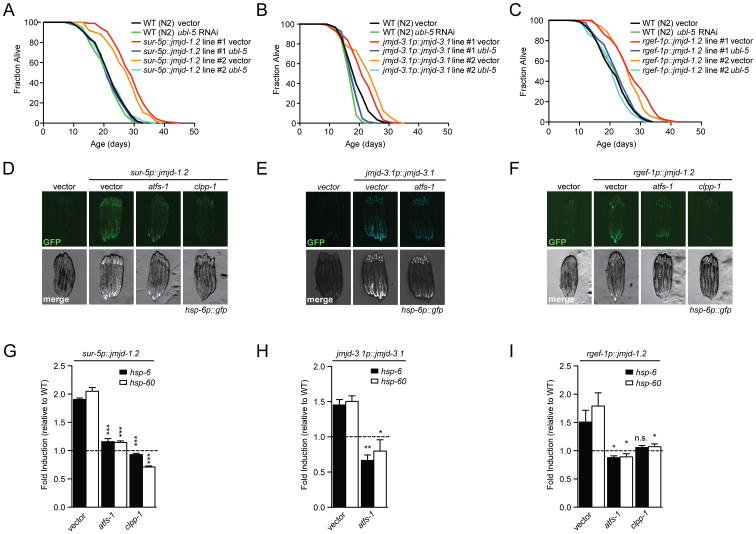
The ubiquitin-like protein UBL-5 is required for increased longevity of the ETC mutant isp-1(qm150) (Durieux et al., 2011) (Figure S5A). Therefore, we tested whether the long lifespan of jmjd-1.2 and jmjd-3.1 overexpressing animals depended upon ubl-5. Intriguingly, ubl-5 RNAi fully suppressed increased longevity of both jmjd-1.2 and jmjd-3.1, as well as neuron-specific jmjd-1.2 overexpressing animals (Figures 4A-C). Notably, ubl-5 was not required for increased longevity of animals ubiquitously expressing heat shock factor-1 (HSF-1) (Baird et al., 2014) (Figure S5B) and had no effect on wild type lifespan (Durieux et al., 2011). These data strongly suggest that the UPRmt is an essential and specific requirement in H3K27 demethylase gain-of-function models.
Figure 4. The UPRmt is a Genetic Requirement for the Pro-Longevity Response upon jmjd-1.2 and jmjd-3.1 Overexpression.
(A), (B) and (C) ubl-5 RNAi suppresses lifespan extension upon jmjd-1.2, jmjd-3.1 or neuronal jmjd-1.2 overexpression. Lifespan analysis of two independent lines of sur-5p∷jmjd-1.2 (A), jmjd-3.1p∷jmjd-3.1 (B), neuronal rgef-1p∷jmjd-1.2 (C) and WT (N2) animals grown on empty vector control or ubl-5 RNAi.
(D), (E) and (F) Fluorescent micrographs of hsp-6p∷gfp UPRmt reporter animals expressing the indicated transgenes treated with the indicated RNAi, at day 1 of adulthood.
(G), (H) and (I) Transcript levels of canonical UPRmt targets assessed by qRT-PCR in sur-5p∷jmjd-1.2 (G), jmjd-3.1p∷jmjd-3.1 (H) or neuronal rgef-1p∷jmjd-1.2 (I) transgenic animals at day 1 of adulthood treated with the indicated RNAi. Results are shown relative to transcript levels in WT (N2) animals grown on the indicated RNAi, with error bars indicating mean ± SEM. *** denotes p < 0.001, **p < 0.01, *p < 0.05, n.s. = p > 0.05. See also Figure S5 and Table S1 for lifespan statistics.
Since both jmjd-1.2 and jmjd-3.1 overexpression induced the UPRmt, we also tested the requirement of the UPRmt core components clpp-1 and atfs-1 for UPRmt activation in response to demethylase overexpression. RNAi against atfs-1 and clpp-1 in strains overexpressing jmjd-1.2 or jmjd-3.1 efficiently abrogated UPRmt signaling, suggesting that jmjd-1.2 and jmjd-3.1 function upstream of the core transcriptional UPRmt machinery in C. elegans (Figures 4D-I).
JMJD-1.2 and JMJD-3.1 Overexpression Recapitulates the Transcriptional Response to Mitochondrial Stress
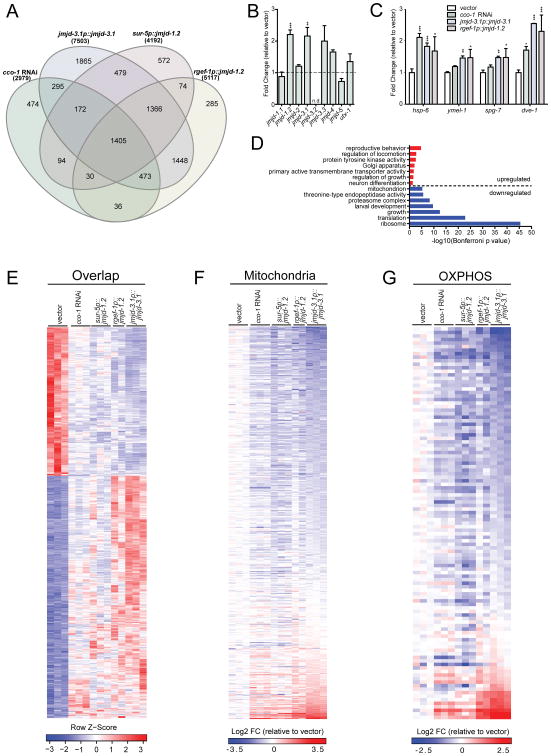
To understand the molecular mechanisms that underlie the extended longevity in response to mitochondrial ETC perturbation and histone demethylase overexpression on a whole genome level, we performed RNA-seq analysis of strains ubiquitously overexpressing jmjd-3.1 or jmjd-1.2, or neuronally-expressed jmjd-1.2, and compared them to cco-1 RNAi treated animals. Our analysis identified ∼3,000-7,500 differentially expressed genes (DEGs) in each condition (adjusted p-value < 0.05), with the greatest number of changes observed in the jmjd-3.1p∷jmjd-3.1 strain (Figure 5A). Strikingly, almost half the genes differentially expressed upon cco-1 RNAi treatment (1405/2979) were also significantly differentially expressed in all three demethylase overexpression strains, and 84% (2505/2979) were in common with at least one of the examined overexpression strains (Figure 5A). Among the overlapping DEGs, 99% (1385/1405) of gene expression changes go in the same direction in all 4 conditions (Figure 5E). These data indicate that an overwhelming majority of the gene expression changes induced by cco-1 RNAi are recapitulated by overexpression of jmjd-1.2 or jmjd-3.1. Furthermore, an examination of all nine JmjC-domain encoding genes revealed that jmjd-1.2 and jmjd-3.1 were specifically upregulated in response to cco-1 RNAi treatment (Figure 5B). These data place jmjd-1.2 and jmjd-3.1 downstream of mitochondrial stress and support a model in which a majority of the transcriptional response to mitochondrial stress is mediated by jmjd-1.2 and jmjd-3.1.
Figure 5. ETC Perturbation and JMJD Overexpression Share Common Lifespan Extension Mechanisms.
(A) Venn diagram of differentially expressed genes (DEGs) in cco-1 RNAi treated worms and transgenic overexpression lines jmjd-3.1p∷jmjd-3.1, sur-5p∷jmjd-1.2, rgef-1p∷jmjd-1.2, compared to wild type N2 worms on empty vector control, as measured by RNA-seq (Benjamini-Hochberg adjusted p value < 0.05). See Table S2 for complete list of DEGs.
(B) Transcriptional upregulation of jmjd-1.2 and jmjd-3.1 upon cco-1 RNAi. Gene expression analysis of all nine JmjC domain encoding genes in RNA-seq samples, expressed as fold change relative to wild type N2 on empty vector control. Results are expressed as mean ±SEM of normalized count values (n=3, Benjamini-Hochberg adjusted p values (padj) calculated by DESeq2, jmjd-1.2 padj=6.93E-09, jmjd-3.1 padj=0.002, jmjd-3.3 padj=0.452, jmjd-4 padj=0.085. n.d.= not detected, ** denotes p < 0.01, ***p < 0.001).
(C) UPRmt gene expression in RNA-seq samples, expressed as fold change relative to wild type N2 on empty vector control. Results are expressed as mean ±SEM of normalized count values. * denotes p < 0.05, **p < 0.01, ***p < 0.001).
(D) Representative top GO terms of upregulated and downregulated genes in the 1405 overlapping DEGs (Bonferroni adjusted p value<0.05). See also Table S2.
(E) Gene expression heatmap of 1405 overlapping DEGs in all 4 conditions described in(A). DESeq2-normalized count values were used for calculations. The indicated Row Z-scores reflect the number of standard deviations each replicate is apart from the mean gene expression value over all conditions. See also Table S2.
(F) Heatmap of 470 mitochondrial genes from GO cellular component category mitochondrion (GO:0005739). Fold change (FC) was calculated by comparing normalized count values of each condition to wild type N2 empty vector control, then transformed to log2 scale. See also Table S2.
(G) Heatmap of 111 OXPHOS genes from GO biological process category oxidative phosphorylation (GO:0006119) and manual annotation. See also Table S2.
The 1405 overlapping genes were subjected to Gene Ontology (GO) analysis using the DAVID database (Huang et al., 2009). Among the upregulated genes, signaling and cell communication related terms were enriched (Figure 5D). Importantly, larval development and growth, as well as multiple protein processing pathways, such as translation and proteasome were extensively downregulated, consistent with global remodeling of the proteome in response to stress (Figure 5D). Whereas the majority of mitochondrial genes were significantly reduced in all examined conditions (Figure 5F, G), many of the UPRmt components, such as the chaperone hsp-6, the proteases ymel-1 and spg-7 as well as the transcriptional regulator dve-1 were induced (Figure 5C). Remarkably, OXPHOS genes in jmjd-1.2 and jmjd-3.1 overexpression strains were downregulated to a similar extent as in cco-1 RNAi treated animals (Figure 5G). Surprisingly, there were only a few OXPHOS components that were significantly upregulated in all conditions, specifically cyc-1 from complex III and mtDNA encoded nduo-2, nduo-4 and nduo-5. Intriguingly, this finding is reminiscent of mito-nuclear protein imbalance, a phenomenon of contrasting expression between nuclear- and mtDNA encoded OXPHOS components which tightly couples UPRmt activation with longevity across organisms (Houtkooper et al., 2013).
Mammalian PHF8 and JMJD3 Correlate with Lifespan and UPRmt Activation
To assess the potential roles of jmjd-1.2 and jmjd-3.1 in regulating UPRmt and longevity in mammals, we investigated their murine homologs Phf8 and Jmjd3, respectively, in the GeneNetwork database (www.genenetwork.org), which contains a vast collection of clinical and molecular (transcript and protein expression) phenotypes from the BXD mouse genetic reference panel (GRP) (Andreux et al., 2012; Wu et al., 2014). Importantly, variations within these datasets reflect mild, natural variations in gene expression patterns found in isogenic populations and are not reliant upon more deleterious genetic manipulations of mitochondrial function.
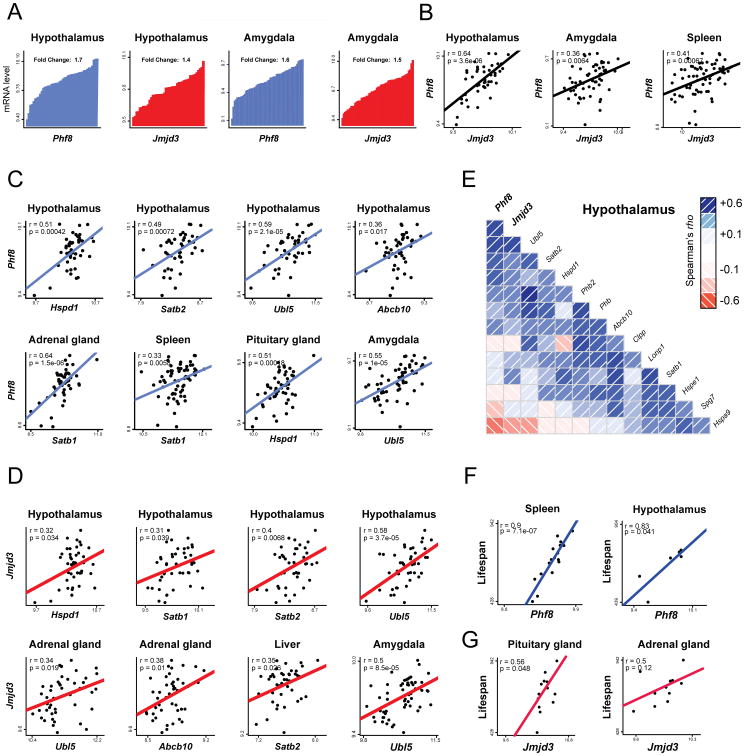
Both the transcripts for Phf8 and Jmjd3 showed high levels of variability in expression across the tissues and strains examined (Figure 6A). In tissue-specific datasets, natural variations in Phf8 expression in the hypothalamus, spleen, and amygdala positively correlated with Jmjd3 expression (Figure 6B), suggesting a correlative genetic interaction between the two enzymes. Importantly, expression levels of both histone demethylases were also positively correlated with UPRmt related genes, including Hspd1 (encoding the mitochondrial chaperone HSP60), Satb1, Satb2 (orthologs of the C. elegans UPRmt regulator dve-1), Abcb10 (ortholog of the mitochondrial peptide exporter haf-1) (Haynes et al., 2010) and Ubl5 (ortholog of ubl-5) across an array of tissues (Figures 6C, D). The strongest associations were observed in the hypothalamus, in which UPRmt related genes in addition to Phf8 and Jmjd3 formed a connected correlation network (Figure 6E). In accordance with the impact of jmjd-1.2 and jmjd-3.1 on lifespan regulation in C. elegans, we observed correlations between Phf8 and Jmjd3 expression and lifespan in the spleen and hypothalamus or pituitary and adrenal glands of mice, respectively (Figures 6F, G).
Figure 6. Positive Correlations between Phf8, Jmjd3, Lifespan and UPRmt transcripts in the BXD Mouse Genetic Reference Population.
(A) Variation of Phf8 and Jmjd3 mRNA levels in hypothalamus (n=44) and amygdala(n=56) across BXD mouse strains. Each bar represents mRNA levels from a pool of approximately five animals per strain.
(B) Positive correlations between Phf8 (y-axis) and Jmjd3 (x-axis) expression in hypothalamus (n=44), amygdala (n=56) and spleen (n=67).
(C) Positive correlations between Phf8 (y-axis) and selected UPRmt genes (x-axis) transcripts in various tissues (n=46 for adrenal gland, n=49 for pituitary gland).
(D) Positive correlations between Jmjd3 (y-axis) and selected UPRmt genes (x-axis) transcripts in various tissues (n=46 for liver).
(E) Spearman's correlation co-expression network for Phf8, Jmjd3 and UPRmt genes in hypothalamus. Blue correlations are positive, red correlations are negative – intensity of the colors corresponds to correlation coefficients.
(F) Pearson correlations of Lifespan versus Phf8 transcript levels in either spleen (left) or hypothalamus (right) of BXD mice.
(G) Pearson correlations of Lifespan versus Jmjd3 transcript levels in either pituitarygl and (left) or adrenal gland (right) of BXD mice.
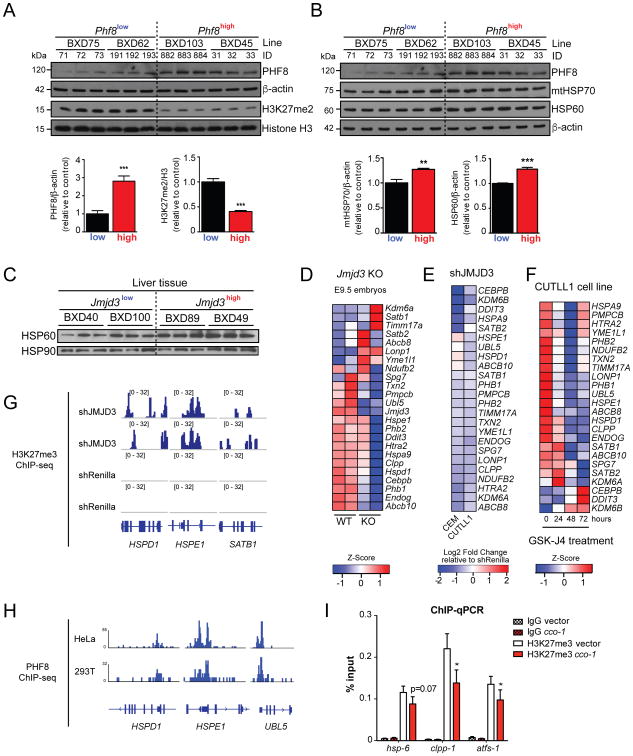
Across strains expressing variable levels of Phf8 mRNA, using immunoblotting, we found a correlative change of PHF8 protein, which was paralleled by a reduction of global H3K27me2 levels (Figure 7A). Increased PHF8 protein levels were also associated with higher abundance of UPRmt marker proteins, such as the chaperones mtHSP70 and HSP60 (Figure 7B), reminiscent of the transcript correlations (Figure 6C). Similarly, higher JMJD3 levels were associated with an increase of HSP60 protein (Figure 7C).
Figure 7. Conserved UPRmt Gene Regulation Mechanisms Through H3K27 Demethylases.
(A) and (B) Immunoblot analysis of tissue protein lysates using the indicated antibodies. Increased Phf8 expression correlates with reduced H3K27me2 (A) and higher levels of mitochondrial chaperones (B) in the hypothalamus of the indicated BXD mouse strains. β-actin and Histone H3 were used as loading controls (upper panels). Densitometric quantifications of immunoblot signals normalized to either β-actin or Histone H3 (lower panels). Data represent the mean+ SEM. *** denotes p < 0.001, **p < 0.01.
(C) Immunoblot analysis of BXD liver tissue protein lysates using the indicated antibodies. HSP90 was used as a loading control.
(D) Heatmap of selected UPRmt transcripts in WT and Jmjd3 KO mice embryos at E9.5 (GSE40332). Low expression is shown in blue, while high expression is in red.
(E) Heatmap of fold change in UPRmt transcripts upon shJMJD3 treatment relative to shRenilla in human T-cell lymphoblastic leukemia CEM and CUTLL1 cell lines (GSE56696).
(F) Heatmap of selected UPRmt transcripts in the human T-cell lymphoblastic leukemia CUTLL1 cell line upon treatment with the H3K27 demethylase inhibitor GSK-J4 (GSE56696).
(G) ChIP-Seq profiles of H3K27me3 enrichment at selected UPRmt genes in the CUTLL1 cell line upon shJMJD3 and shRenilla treatments (GSE56696).
(H) ChIP-Seq profiles of PHF8 binding at selected UPRmt gene promoters in HeLa and 293T cells (GSE20725).
(I) ChIP-qRT-PCR analysis of H3K27me3 enrichment at UPRmt genes at L3 stage of cco-1 RNAi treated worms compared to empty vector control. IgG antibody was used as a control. Results are expressed as percent of input, with error bars indicating mean ± SEM (n=7, * denotes p < 0.01).
We hypothesized that a relationship between the expression of markers of UPRmt, Phf8, and Jmjd3 also might be detectable in mammalian Jmjd3 loss-of-function experiments, which have been published previously (GEO datasets GSE40332, GSE56696) (Ntziachristos et al., 2014). Consistent with our prediction, in Jmjd3 knockout mouse embryos (GSE40332), the expression of various UPRmt related transcripts was decreased (Figure 7D). Likewise, knockdown of JMJD3 by shRNA in two human T-cell lymphoma cell lines (CUTLL1 and CEM) decreased the expression levels of multiple transcripts related to the UPRmt (Figure 7E). Similarly, treatment of CUTLL1 cell line with the H3K27 demethylase inhibitor GSK-J4 (GSE56696) (Ntziachristos et al., 2014) also decreased the levels of multiple UPRmt transcripts in a time-dependent fashion (Figure 7F). Overall, these data in both the murine GRP, mice with loss-of-function alleles, or cell lines with modified levels or activity of PHF8 and/or JMJD3 collectively suggest that expression levels of these demethylases positively correlate with UPRmt expression.
Finally, we examined ChIP-seq analysis of CUTLL1 cells treated with shJMJD3 (GSE56696) for evidence of direct JmjC regulation of chromatin at coding regions of UPRmt target genes. In these analyses, we found that H3K27me3 enrichment on HSPD1, HSPE1, and SATB1 genes was increased upon JMJD3 knockdown (Figure 7G), indicating that the repressive H3K27me3 mark may be actively removed from the coding regions of these genes by JMJD3, thereby allowing for their expression. In another independent ChIP-seq dataset (GSE20725) (Fortschegger et al., 2010), PHF8 was found to bind to coding regions of UPRmt genes HSPD1, HSPE1 and UBL5 in both HeLa and 293T cells (Figure 7H).
In addition to these in silico analyses, we examined H3K27me3 occupancy at coding regions of UPRmt regulators during mitochondrial stress in larval stage C. elegans by ChIP-qRT-PCR. We observed a substantial decrease in H3K27me3 abundance on hsp-6, clpp-1 and atfs-1 genes upon cco-1 RNAi during early larval development (Figure 7I). Taken together, these data suggest that PHF8 and JMJD3 may be conserved positive regulators of UPRmt and lifespan from C. elegans to mammals, through the modulation of the H3K27 methylation status at coding regions of key UPRmt genes.
Discussion
The distinct timing requirements of lifespan extension due to mild mitochondrial dysfunction during larval development led to the early proposal of an epigenetic mechanism that determines ETC-mediated longevity of C. elegans (Dillin et al., 2002a; Durieux et al., 2011). Remarkably, the discovery that the UPRmt is not only a genetic requirement but also shares these overlapping timing requirements with ETC-mediated longevity reinforces the idea that primary mitochondrial perturbations establish an epigenetic memory which sets the rate of aging of the entire organism and protects its mitochondrial proteome from future insults (Durieux et al., 2011). The identification of jmjd-1.2/PHF8 and jmjd-3.1/JMJD3 provides a molecular explanation for these observations and suggests the epigenetic regulation of transcriptional outputs during mitochondrial stress. We find jmjd-1.2 to be not only necessary and specific, but also sufficient for both induction of the UPRmt and extension of lifespan. jmjd-3.1 is also necessary and sufficient for UPRmt induction and lifespan extension, but has overlapping roles in ER stress regulation (Labbadia and Morimoto, 2015).
The two histone demethylases differ in their substrate specificity: While jmjd-3.1/JMJD3 is associated with the removal of H3K27me2/3 epigenetic marks (Agger et al., 2007), jmjd-1.2/PHF8 has activity towards a wider range of substrates, including H3K9me1/2, H3K27me2 and H4K20me1 (Feng et al., 2010; Fortschegger et al., 2010; Kleine-Kohlbrecher et al., 2010; Liu et al., 2010; Qi et al., 2010). Both genes are associated with the positive regulation of gene expression and the removal of repressive marks. This raises the intriguing possibility that both enzymes might function in a linear pathway to sequentially demethylate H3K27me3 and thereby activate gene expression in response to mitochondrial perturbations. This hypothesis is supported by the mitochondrial stress-induced reduction of H3K27me3 occupancy at UPRmt response genes during larval development in worms (Figure 7I). Our results are therefore consistent with the idea that removal of repressive marks might allow access of the core UPRmt transcriptional machinery to induce mitochondrial stress signaling (Haynes et al., 2010; Nargund et al., 2012).
How is mitochondrial stress sensed by jmjd-1.2/PHF8 and jmjd-3.1/JMJD3? An attractive idea is that both jmjd-1.2 and jmjd-3.1 themselves are targets of the transcriptional response to mitochondrial stress as indicated in our transcriptomics analyses (Figure 5B). Another possibility is that acute adaptations in mitochondrial metabolism may promote activity of JmjC demethylases. Since both JMJD3 and PHF8 belong to the family of 2-oxoglutarate dependent oxygenases, it appears likely that elevated levels of the TCA cycle intermediate alpha-ketoglutarate (α-KG) might contribute to their increased activity (Teperino et al., 2010). Intriguingly, the exogenous supplementation of α-KG has recently been found to extend lifespan of C. elegans (Chin et al., 2014). While RNAi knockdown experiments for both jmjd-1.2 and jmjd-3.1 revealed robust suppression of ETC-mediated longevity and UPRmt induction, we did not find strong effects on lifespan and hsp-6 transcriptional induction in the analysis of the respective mutant strains suggesting that an acute depletion of the enzymes during larval development rather than chronic deficiency is necessary to unmask their role for these phenotypes (Figure S6).
In this work, we identify neurons as a key tissue to promote longevity and UPRmt induction upon neuron-specific jmjd-1.2 overexpression. Neuronal jmjd-1.2 is not only sufficient to mediate a robust UPRmt induction but also extends C. elegans lifespan in an UPRmt-dependent manner. These findings were corroborated in mice and appear to be conserved in the BXD mouse reference population. Across various neuronal subregions, high levels of PHF8 and JMJD3 correlate with increased expression of mammalian UPRmt core components (CLPP, ABCB10, SATB1/2, UBL5) and downstream mitochondrial chaperones HSP60 (HSPD1) and mtHSP70 (HSPA9) illustrating that both demethylases might control an integrated transcriptional network promoting mammalian longevity.
We previously demonstrated that mito-nuclear protein imbalance induces a robust UPRmt and is linked to increased lifespan, both in nematodes and in BXD mice (Houtkooper et al., 2013). Interestingly, mito-nuclear imbalance might also play a role in longevity mediated by overexpression of jmjd-1.2 and jmjd-3.1, as suggested by our RNA-seq analysis (Figure 5G). As further proof of concept that similar stressors could activate the UPRmt across species, we recently found that expression of Cox5b and Spg7 correlate negatively with the UPRmt network, indicating that their low abundance likely triggers the UPRmt in mammals (Wu et al., 2014). These data indicate that the UPRmt pathway is active in vivo in mammals under physiological, non-stress conditions. Based on the observed positive correlations between PHF8, JMJD3 and UPRmt in multiple tissues, we suggest that the regulation of UPRmt by the H3K27 demethylases PHF8 and JMJD3 is also conserved in BXD mice under basal conditions. Of note, this association between histone demethylases and UPRmt seems in certain tissues linked to mouse lifespan regulation, although further mechanistic work is required to ascertain this relation. Our bioinformatics analysis and literature discussed above suggests that PHF8 and JMJD3 regulate UPRmt genes by removing repressive H3K27 methylation marks from their coding regions. Identifying the exact mechanism of regulation and whether there is a developmental aspect of the UPRmt pathway in mammals, as demonstrated in C. elegans (Durieux et al., 2011), remains an important direction for future work.
Collectively, our data corroborate the increasing body of literature in which epigenetic marks that control chromatin states, including histone methylation, represent a hallmark of aging (Greer et al., 2010; Han and Brunet, 2012; Lopez-Otin et al., 2013; Rando and Chang, 2012). Mitochondrial perturbations early in life have long-lasting effects on gene expression, and within this work we provide a mechanistic understanding of how this might be achieved. Our results thus reveal a conserved mode for the regulation of stress response and lifespan dependent on mitochondrial function.
Experimental Procedures
Additional details are provided in the Extended Experimental Procedures.
Lifespan Analysis
Lifespan experiments were conducted at 20°C as previously described (Durieux et al., 2011). Lifespans were performed on nematodes fed HT115 bacteria expressing the indicated RNAi, using the pre-fertile period of adulthood as day 0. Animals were transferred to fresh plates every other day until day 12. Prism 5 software was used for statistical analysis to determine significance calculated using the log-rank (Mantel-Cox) method. Lifespan experiments involving RNAi shifting to Dicer (dcr-1) RNAi were performed as described (Dillin et al., 2002a, 2002b). Briefly, for lifespans with selective RNAi only during development, animals were grown on cco-1+jmjd-1.2 or cco-1+jmjd-3.1 RNAi bacteria and transferred to dcr-1 dsRNA expressing bacteria at L4 stage. For lifespans with RNAi during adulthood, animals were grown on empty vector RNAi control bacteria and transferred to cco-1+jmjd-1.2 or cco-1+jmjd-3.1 RNAi bacteria at L4 stage.
Gene Expression Analysis
C. elegans were age synchronized by egg bleaching and cultivated on nematode growth (NGM) plates containing HT115 bacteria expressing the indicated RNAi constructs at 20°C and harvested at day 1 of adulthood. Animals were collected in M9 buffer, centrifuged at 1000 × g for 30 sec, resuspended in Trizol (Life Technologies) and snap frozen in liquid nitrogen. After several freeze-thaw cycles, total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. 1 μg of total RNA was subjected to cDNA synthesis using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative Real-Time PCR reactions were performed with the SYBR Select Master Mix (Applied Biosystems) in Optical 384-well MicroAmp plates (Applied Biosystems) using a QuantStudio 6 Flex (Applied Biosystems).
Fluorescence Microscopy
For fluorescence microscopy, animals were blindly chosen under the light microscope (at random) from a population, immobilized with 100 μg/ml levamisole (Sigma) and images were then captured using a Leica M250FA automated fluorescent stereo microscope equipped with a Hamamatsu ORCA-ER camera.
Immunoblot Analysis
Mouse tissue extracts were prepared in modified RIPA buffer using a hand-held homogenizer (UltraTurrax). Crude lysates were centrifuged at 10,000 × g at 4°C for 10 min and total protein amount was determined with the DC Protein Assay (BioRad). Supernatants were supplemented with 4x SDS sample buffer, boiled for 5 min at 95°C and resolved by standard SDS-PAGE. Proteins were transferred to PVDF membranes (Immobilon). Equal loading was assessed with anti-β-actin (Abcam) antibodies.
Supplementary Material
Acknowledgments
We thank Dr. Y. Tian for help with reporter crosses, Lawrence Joe for preparing the RNA-seq libraries and Drs. C. Riera, P. Douglas and N. Baird for scientific insight. We thank Franziska Lorbeer for help with data analysis and Ed Ralston for help with gamma irradiation. We are grateful to Drs. A. Salcini, A. Carrano, M. Hansen, R. O'Sullivan and C. Holmberg for reagents. Some of the nematode strains used in this work were provided by the Japanese National BioResource Project and the CGC, which is supported by the NIH-Officer of Research Infrastructure Programs (P40 OD010440). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. C.M. was supported by a postdoctoral fellowship from the Glenn Center for Aging Research at the Salk Institute for Biological Studies. S.D.J was supported by American Heart Association grant #15POST22510020. K.K.S. was supported by a grant from the Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by the Howard Hughes Medical Institute (HHMI), NIH (R01 ES021667) to A.D., the Glenn Foundation for Medical Research to C.M. and S.C.W., the EPFL, NIH (R01AG043930), SwissCancerLeague (KFS-3082-02-2013), Systems X (SySX.ch 2013/153) and the SNSF (31003A-140780) to JA.
Footnotes
Accession Numbers: RNA-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE78990.
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, six Supplemental Figures, four Supplemental Tables and can be found with this article online at.
Author Contributions: C.M., V.J., J.A. and A.D. conceived the study and wrote the manuscript with input from all co-authors. C.M. performed C. elegans RNAi screens, lifespan assays, RNA-seq sample preparations, fluorescence microscopy, qRT-PCR experiments, immunoblots related to jmjd-1.2/PHF8 and analyzed data. V.J. performed RNA-seq analysis, C. elegans lifespan assays, fluorescence microscopy, qRT-PCR experiments and immunoblots related to jmjd-3.1/JMJD3 and analyzed data. J.D. generated transgenic C. elegans lines by microinjection. O.M. performed some of the C. elegans experiments and generated jmjd-3.1 overexpressing C. elegans strains. S.D.J. performed qRT-PCR experiments and analyzed data. P.M.Q. performed mammalian microarray and ChIP-seq analyses. K.K.S. helped with the RNA-seq sample preparations and performed RNA-seq gene expression analysis. S.N.U. performed C. elegans crosses, backcrosses and strain integrations of jmjd-1.2 overexpressing lines. E.G.W. performed initial bioinformatic analyses on BXD mouse tissues. L.M. and C.M. identified jmjd-3.1 as a UPRmt regulator in a screening campaign. V.M. conducted the C. elegans lifespan suppressor screen. S.C.W. conceived the high-throughput C. elegans RNAi screens and wrote the manuscript. R.J.S. provided intellectual input and supported the early phase of the project.
J.A. is the Nestlé Chair in Energy Metabolism at the EPFL and a cofounder of Mitobridge, Inc. and declares no financial interest related to this work. A.D. is a cofounder of Proteostasis Therapeutics, Inc. and Mitobridge, Inc. and declares no financial interest related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Johan Auwerx, Email: admin.auwerx@epfl.ch.
Andrew Dillin, Email: dillin@berkeley.edu.
References
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, Schoonjans K, Williams RW, Auwerx J. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR, 3rd, Manning G, Dillin A. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 2014;346:360–363. doi: 10.1126/science.1253168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol CB. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, Diep S, Lomenick B, Meli VS, Monsalve GC, et al. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002a;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002b;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FMA, Timmers HTM, Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010;30:3286–3298. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Brunet A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012;22:42–49. doi: 10.1016/j.tcb.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, Labelle M, Gerlach LO, Birk P, Helin K. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514:E1–E2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. The heat shock response systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol Cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 Is a Cell-Nonautonomous Regulator of Stress Resistance and Longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochem Biophys Res Commun. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.