Cellular signaling heavily relies on small molecule messengers, and many of these messengers are involved in multiple pathways. When analyzing signaling networks, it is typical procedure to establish whether a given small molecule is involved in the network. The task becomes very difficult if the messenger is involved in several points of the network or the regulation of more than one physiological response. Put differently, we usually have no clear idea which molecular mechanisms are used by cells to compute signaling outcome if multiple options are conceivable. A number of potential mechanisms for multifunctionality come to mind, for instance rapid metabolic conversion of signaling molecules with metabolites triggering a diverse set of targets. Another possibility has been widely underappreciated, namely the ability of cells to confine small molecule signaling events to particular compartments and organelles and thus use the same messenger for different tasks at different sites of the cell. Testing this hypothesis requires the experimental capacity to alter messenger levels with subcellular precision.
To date, only a limited repertoire of techniques for the manipulation of cells with subcellular resolution is available. Typically, locally restricted manipulations require the use of micromanipulators or other physically invasive techniques. Easier to apply are photoactivatable (‘caged’) molecules that are activated by light.1 The theoretical precision limit of photoactivation is defined by the diffraction limit of light, as is the case in microscopy. However in practice, even when illuminating through the objective of a confocal microscope, stray light often suffices to release active molecules outside the focal beam thus compromising the available spatial resolution.
In order to address this issue, we therefore thought of an alternative way to restrict the photorelease of the caged molecule within a given cell. Instead of trying to selectively photoactivate a fraction of an evenly distributed caged molecule at the desired area, we aimed at pre-localizing the caged molecule at its subcellular site of action. To achieve this, we used a highly negatively charged photoactivatable group (Fig. 1) to release hydrophobic signaling molecules in a spatially restricted fashion exclusively at the plasma membrane. Cellular effects induced by signaling molecules released at the plasma membrane were compared to photorelease of the same molecules at internal cell membranes by using non-charged caged derivatives. We chose arachidonic acid (AA) as our first target as it is a textbook example of a multifunctional signaling lipid. AA is involved in the regulation of various physiological functions of many cell types including apoptosis, insulin release and regulation of synaptic plasticity.2-3 Its metabolites include a large number of biologically important metabolites such as prostaglandins, leukotrienes as well as endocannabinoids.
Figure 1.
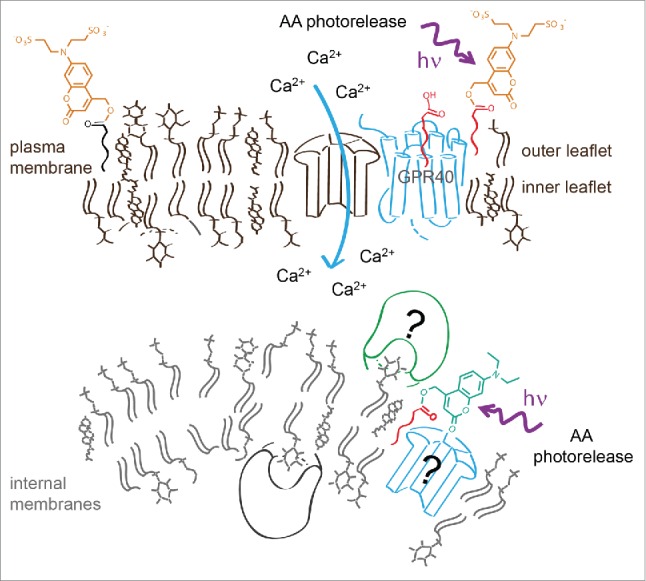
Controlled AA photorelease at different cellular membranes determined by using sulfonated and neutral caging groups. The negatively charged group prevents passage of the caged lipid over the plasma membrane, thus restricting its distribution to the outer membrane leaflet. Photorelease of AA at the plasma membrane or at internal membranes leads to significantly different modulation of intracellular calcium dynamics.
We aimed at analyzing the effects of spatially controlled AA release in pancreatic β-cells. AA is present in β-cells in fairly high concentrations (50–75 µM) and is known to modulate calcium signaling and thus insulin release.1 Indeed, factors that induce release of free AA in β-cells increase intracellular calcium levels and promote insulin secretion. On the other hand, overexpression of cytosolic phospholipase A2 in β-cells as well as their exposure to free fatty acid was reported to reduce the glucose-stimulated insulin secretion.4 We hypothesized that these findings actually point toward multiple signaling roles for AA in the regulation of insulin release. Therefore we tested if the precise cellular localization of the initial concentration increase is one of the major factors in determining the signaling outcome. We synthesized and used caged AA derivatives bearing either the novel sulfonated or a neutral caging group.5 We found that release of AA at the plasma membrane or at internal membranes leads to a significantly different modulation of intracellular calcium dynamics. Uncaging of AA at the plasma membrane induces calcium oscillations in non-oscillating cells and increases the duration of calcium transients in oscillating cells, leading to overall higher calcium levels. Release of AA at internal membranes results in transiently or permanently diminished calcium oscillations and lower average calcium levels. By applying spatially restricted photoactivation of AA we were thus able to establish that this signaling molecule is utilized in a dual role in β-cells and has directly opposing effects depending on cellular release sites. It should be mentioned that AA increases at the plasma membrane most likely triggered the fatty acid sensitive GRP40 receptors. How AA released in internal membrane is able to stop oscillations, however, remains a matter of future research.
In the future, spatially restricted photoactivation will help the analysis of small molecule-driven signaling events with much improved precision, particularly when it comes to lipid signaling. However, the extraordinary complexity inherent in lipid signaling events calls for a multidimensional approach. An optimized approach for identifying of lipid-binding proteins was very recently described by the Cravatt6 and our own group.7 Photocrosslinking was demonstrated to be useful for identification of proteins binding AA derivatives. Unfortunately, AA escaped analysis likely due its fast metabolism.6 Combining lipid-protein cross-linking with local uncaging may help to overcome this limitation and permit the determination of AA binding proteins responsible for modulating calcium signaling in the future thus significantly enhancing our understanding of the regulation of insulin secretion.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Hoglinger D, et al.. Biochim Biophys Acta 2014; 1841:1085-1096; PMID:24713581; http://dx.doi.org/ 10.1016/j.bbalip.2014.03.012 [DOI] [PubMed] [Google Scholar]
- [2].Brash AR. J Clin Invest 2001; 107:1339-1345; PMID:11390413; http://dx.doi.org/ 10.1172/JCI13210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Carta M, et al.. Neuron 2014; 81:787-799; PMID:24486086; http://dx.doi.org/ 10.1016/j.neuron.2013.12.028 [DOI] [PubMed] [Google Scholar]
- [4].Milne HM, et al.. Diabetes 2005; 54:116-124; PMID:15616018; http://dx.doi.org/ 10.2337/diabetes.54.1.116 [DOI] [PubMed] [Google Scholar]
- [5].Nadler A, et al.. Nat Commun 2015; 6:10056; PMID:26686736; http://dx.doi.org/ 10.1038/ncomms10056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Niphakis MJ, et al.. Cell 2015; 161:1668-1680; PMID:26091042; http://dx.doi.org/ 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haberkant P, et al.. ACS Chem Biol 2016; 11:222-230; PMID:26555438; http://dx.doi.org/ 10.1021/acschembio.5b00810 [DOI] [PubMed] [Google Scholar]


