ABSTRACT
The F box protein Skp2 is oncogenic. Skp2 and Skp2B, an isoform of Skp2 are overexpressed in breast cancer. However, little is known regarding the mechanism by which Skp2B promotes the occurrence and development of breast cancer. Here, we determined the expression and clinical outcomes of Skp2 in breast cancer samples and cell lines using breast cancer database, and investigated the role of Skp2 and Skp2B in breast cancer cell growth, apoptosis and cell cycle arrest. We obtained Skp2 is significantly overexpressed in breast cancer samples and cell lines, and high Skp2 expression positively correlated with poor prognosis of breast cancer. Both Skp2 and Skp2B could promote breast cancer cell proliferation, inhibit cell apoptosis, change the cell cycle distribution and induce the increased S phase cells and therefore induce cell proliferation in breast cancer cells. Moreover, the 2 isoforms could both suppress PIG3 expression via independent pathways in the breast cancer cells. Skp2 suppressed p53 and inhibited PIG3-induced apoptosis, while Skp2B attenuated the function of PIG3 by inhibiting PHB. Our results indicate that Skp2 and Skp2B induce breast cancer cell development and progression, making Skp2 and Skp2B potential molecular targets for breast cancer therapy.
KEYWORDS: breast cancer, PHB, PIG3, prognosis, proliferation, p53, Skp2, Skp2B
Introduction
F-box proteins function as the substrate recognition subunits of specific ubiquitin ligase complexes. 1 S phase kinase-associated protein 2 (Skp2), an F box protein, has been found to involve in the ubiquitin-dependent degradation of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1. 2,3 However, loss of p27Kip1 or low expression of p27Kip1 is a frequent event in several cancer types and is associated with poor prognosis. 4-7 The loss of p27Kip1 expression is often associated with increased degradation due to the overexpression of Skp2. 8 Under physiological conditions, the expression of Skp2 is maximal at the S, G2 phases and could primarily mediate p27 degradation at the G2, but not G0/1 phases. 9-11 As such, Skp2 was found to be indispensable for progression into mitosis in cell cycles. 10,12 It has been reported that Skp2 counteracts the transactivation function of p53 and suppresses apoptosis mediated by DNA damage or p53 stabilization. 13 Skp2 forms a complex with p300, and antagonizes the interaction between p300 and p53. 13 Skp2 amplification and overexpression can attenuate p53 activation and trigger uncontrolled cell proliferation, the combination of which promotes tumorigenesis. 13 Therefore, Skp2 is oncogenic and could act as a diagnostic marker and a potential molecular target for cancer therapy. 2,14
Skp2 contains 3 important domains: the F-box motif, which is situated at the N-terminus of the protein, the second consists of 10 consecutive leucine-rich repeats (LRR) which located near the C-terminus, and the C-terminus, as it folds back near the F-box and stabilizes the interaction with Skp1. 15 Skp2B is an isoform of Skp2 that differs at the C-terminus domain, that exon 10 is absent from Skp2B and replaced by exon 11. 16 As a consequence, Skp2 and Skp2B have distinct substrate specificities, and Skp2B does not affect the expression of p27. 15,16 However, Skp2B was also reported to attenuate the activity of p53 by inhibiting prohibitin (PHB), which is a new substrate of Skp2B and isolated in a 2-hybrid screen with the C-terminus domain unique to Skp2B. 17 However, it was reported that Skp2B does not show a significant effect on Skp2 substrates, and both Skp2 and Skp2B are overexpressed in breast cancer. 15 Little is known regarding the mechanism by which Skp2B promotes the occurrence and development of breast cancer. In the present study, we determined the expression and clinical outcomes of Skp2 in breast cancer samples and cell lines using breast cancer database, and investigated the role of Skp2 and Skp2B in breast cancer cell growth, apoptosis and cell cycle arrest. The p53-inducible gene 3 (PIG3), is a down-stream target of p53 involved in p53-initiated apoptosis. Our previous study shows that BRCA1 could regulate PIG3-mediated apoptosis in a p53-dependent manner. 18 In the present study, we also investigated whether Skp2 and Skp2B could regulate the expression of PIG3 in a p53-dependent manner.
Results
Skp2 is over-expressed in breast cancer samples and cell lines
We compared the expression levels of Skp2 across 41 analyses using Oncomine (www.oncomine.org) from original published data. We observed that Skp2 is significantly overexpressed in breast cancer samples in comparison with normal breast samples (Fig. 1A, P = 0.000486). We next interrogated the expression levels of Skp2 in various subtypes of breast cancer samples. As shown in Figure 1B, the expression levels of Skp2 were associated with the level of malignancy of breast cancer. The expression levels of Skp2 were in particular higher among tumors of the basal subtype when compared with other subtypes (Fig. 1B, P < 0.00001). The expression levels of Skp2 were higher among ER-negative breast cancer when compared with ER-positive breast cancer (Fig. 1B, P < 0.00001). Moreover, the over-expressed Skp2 was associated with higher tumor grade (Fig. 1B, P = 0.00085). To examine whether the above finding in breast cancer clinical samples can be similarly found in breast cancer cell lines in vitro, we obtained the Skp2 expression in the 55 breast cancer cell lines including Basal A, Basal B, and Luminal breast cancer cell lines (Fig. 1C). We observed a high level of Skp2 expression in Basal subtypes breast cancer cell lines when compared with Luminal subtypes breast cancer cell lines (Fig. 1D, P = 0.01422). Moreover, a significantly elevated Skp2 expression was found between triple negative breast cancer cell lines and HER2-enriched subtypes or HR-positive subtypes breast cancer cell lines (Fig. 1D, P = 0.00115). Taken together, these data indicated that Skp2 is overexpressed in breast cancer samples and cell lines.
Figure 1.
Skp2 is upregulated in breast cancer samples and cell lines. (A) Comparison of Skp2 expression across 41 analyses using Oncomine (www.oncomine.org) from original published data. (B) The expression levels of Skp2 across different subtypes of breast cancer samples. (C) Expression profiles of Skp2 mRNAs in 51 breast cancer cell lines. (D) The expression levels of Skp2 across different subtypes of the 51 breast cancer cell lines.
Skp2 expression is associated with poor prognosis of breast cancer
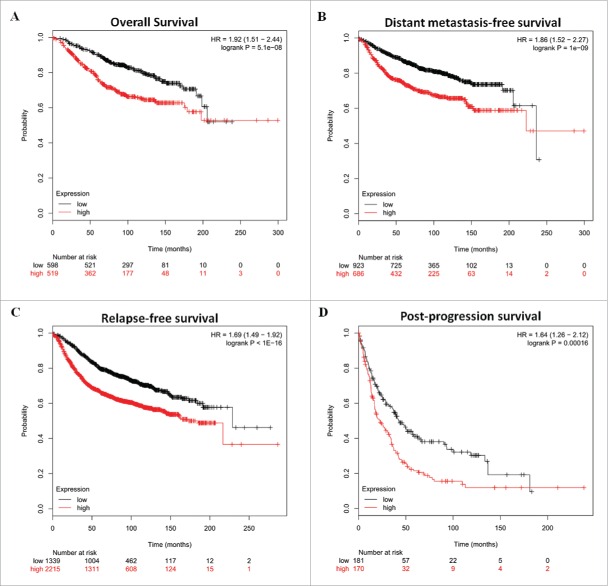
To investigate the role of Skp2 in the breast cancer clinical outcomes, we performed meta-analyses using Kaplan–Meier plotter online breast cancer survival analysis. As shown in Figure 2, high Skp2 expression positively correlated with reduced overall survival (Fig. 2A, HR = 1.92, 95%CI=1.51–2.44, P < 0.0001), distant metastasis-free survival (Fig. 2B, HR = 1.86, 95%CI=1.52–2.27, P < 0.0001), relapse-free survival (Fig. 2C, HR = 1.69, 95%CI=1.49–1.92, P < 0.0001) and post-progression survival (Fig. 2D, HR = 1.64, 95%CI=1.26–2.12, P = 0.00016), suggesting a prognostic value of Skp2 in breast cancers. Taken together, these findings suggest a potential role of Skp2 in breast tumorigenesis.
Figure 2.
Skp2 expression is associated with poor prognosis of breast cancer. Kaplan–Meier analyses of overall survival (A), distant metastasis-free survival (B), relapse-free survival (C), and post-progression survival (D) of breast cancer patients. P-values were calculated with log-rank (Mantel–Cox) test. Patients were stratified into ‘low’ and ‘high’ Skp2 expression based on auto select best cutoff.
Skp2 and Skp2B promote breast cancer cell proliferation and inhibit cell apoptosis
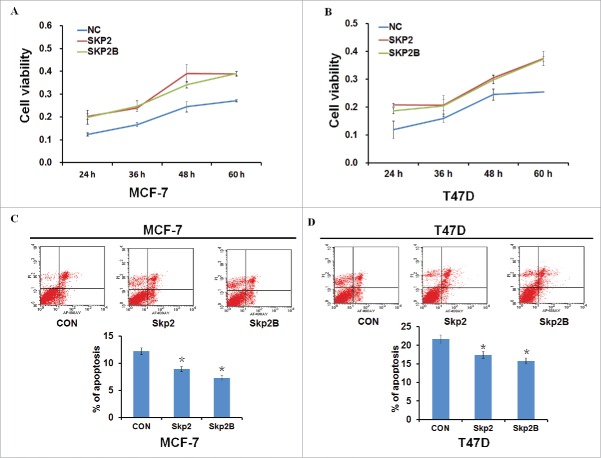
Skp2B, an isoform of Skp2 that differs at the C-terminus domain has also been found over-expressed in breast cancer. 15 To investigate the role of Skp2 and Skp2B in the breast cancer cell growth, plasmids encoding Skp2 or Skp2B were transfected into breast cancer cell lines MCF-7 and T47D, and cells were collected at different time points (24, 36, 48, or 60 h) to determine cell survival rates. As shown in Figures 3A–B, acceleration of cell proliferation occurred in a time-dependent manner following overexpression of both Skp2 and Skp2B. Furthermore, cell apoptosis was decreased in both MCF-7 and T47D cells following overexpression of either Skp2 or Skp2B (Figs. 3C–D). Collectively, these data indicated that Skp2 and Skp2B promote breast cancer cell proliferation and inhibit cell apoptosis.
Figure 3.
Skp2 and Skp2B promote breast cancer cell proliferation and inhibit cell apoptosis. (A, B) Cell viability was measured by MTT assay 24, 36, 48, or 60 h following transfection of plasmids encoding Skp2 or Skp2B into MCF-7 and T47D cell lines. (C, D) Apoptosis was detected by flow cytometry analysis following transfection of plasmids encoding Skp2 or Skp2B into MCF-7 and T47D cell lines. Data in are the mean of 3 independent experiments. *P < 0.05, as compared with untreated cells.
Skp2 and Skp2B change the cell cycle distribution
It has been reported that Skp2 mediates the ubiquitin degradation of several cyclin-dependent kinase (CDK) inhibitors, such as p21cip1, p27kip1, and p57kip2. 19-21 Moreover, Skp2 expression is low in the G0/G1 phase, but increases in the S phase. Overexpressed Skp2 in the G0/G1 phase could lead to an out-of-control G1/S phase, disruption of cell proliferation, and differentiation. 19,20,22 Next we sought to determine the effect of Skp2 and Skp2B on breast cancer cell cycle progression by flow cytometry. When MCF-7 cells were transfected with Skp2 or Skp2B vectors for 48 h, cells in G1 phase were decreased and S phase cells were increased (Fig. 4A). Transfection of T47D cells with Skp2B vectors could also increase S phase cells, while treatment of T47D cells with Skp2 vectors did not induce discernible changes in cell cycle distribution, which might be caused by the dose of Skp2 vectors (Fig. 4B). Taken together, these results showed that Skp2 and Skp2B could change the cell cycle distribution and induce the increased S phase cells and therefore induce cell proliferation in breast cancer cells.
Figure 4.
Skp2 and Skp2B led to breast cancer cell cycle arrest. The MCF-7 (A) and T47D (B) cells were transfected Skp2 or Skp2B vector or control vector for 48 h, and then cell cycle distribution was determined by flow cytometry analysis using PI staining. Data were represented as the average of 3 independent experiments. *P < 0.05, as compared with control vector transfected cells.
Skp2 and Skp2B regulate different downstream genes
To determine the effects of Skp2 and Skp2B on the expression of related downstream proteins, we transfected MCF-7 and T47D cells with a Skp2- or Skp2B- expressing plasmid for 48 h, and examined the related downstream proteins by western blotting (Figs. 5A–B). Decreased expression of prohibitin (PHB) was detected in Skp2B-transfected cells, though not in Skp2-transfected cells. In contrast, Skp2 suppressed levels of p27 protein. We also detected the effects of Skp2 and Skp2B on the expression of the p53-inducible gene 3 (PIG3), a downstream target gene of p53, which we have reported to be regulated by BRCA1 in a p53-dependent manner. 18 We found that overexpression of either Skp2 or Skp2B decreased both p53 and PIG3 protein levels. This suggests that Skp2 and Skp2B can both suppress p53-mediated PIG3 expression. Since Skp2 serves as an essential downstream effector of the AR, we found that both Skp2 and Skp2B transfection elicited only marginal changes in AR protein expression. 23 To further investigate the effects of Skp2 and Skp2B on PIG3 expression in the absence of p53, we detected the levels of related proteins in HCT116p53+/+ and HCT116p53−/− cells overexpressing Skp2 or Skp2B (Figs. 5C–D). HCT116p53−/− cells treated with Skp2B exhibited decreased PIG3, while no discernible changes were detected in Skp2-transfected HCT116p53−/− cells (Fig. 5D). Taken together, these results suggest that Skp2 suppresses p53-mediated PIG3 expression, while Skp2B suppresses PHB-mediated PIG3 expression.
Figure 5.

Skp2 and Skp2B regulate different downstream genes. p53, AR, PHB, PIG3, and p27 protein levels were determined by protein gel blotting and analyzed by grayscale software following transfection of Skp2 or Skp2B vector or control vector into MCF-7 (A), T47D (B), HCT116p53+/+ (C), and HCT116p53−/− (D) cells. Data are the mean of 3 independent experiments. *P < 0.05, as compared with control vector transfected cells.
Discussion
Skp2 and its isoform Skp2B have been reported to be overexpressed in a variety of cancer types where they contribute to malignant progression. 8,15 High levels of Skp2 protein are associated with ER negativity, low p27 expression, high proliferation rate, and poor survival. 24 Here, we determined the expression and clinical outcomes of Skp2 in breast cancer samples and cell lines using breast cancer database. We obtained Skp2 is significantly over-expressed in breast cancer samples and cell lines, while the overexpressed Skp2 was associated with higher tumor grade. Sun et al. also reported that Skp2 expression is significantly higher in patients with invasive breast cancer than in normal breasts. 25 Our data also showed that high Skp2 expression positively correlated with reduced overall survival (OS), distant metastasis-free survival (DFS), relapse-free survival (RFS) and post-progression survival (PPS), suggesting a prognostic value of Skp2 in breast cancers. We also investigated the role of Skp2 and Skp2B in breast cancer cell growth, apoptosis and cell cycle arrest. We report that both Skp2 and Skp2B could promote breast cancer cell proliferation, inhibit cell apoptosis, change the cell cycle distribution and induce the increased S phase cells and therefore induce cell proliferation in breast cancer cells. P53-inducible gene 3 (PIG3), a gene identified in an analysis of genes induced by p53 before the onset of apoptosis, has a potential role in reactive oxygen species (ROS) production and induces breast cancer cell apoptosis. 18,26, 27 It was reported that simultaneous phosphorylation of p53 at serine 15 and 20 increases expression of PIG3 and induces apoptosis. 28 However, acetylation of p53 at 320 and 373 lysine residues could also induces PIG3 expression. 29 That was because PIG3 have a p53-response element and p53 could increase transcriptional activation of PIG3 by this p53-binding site, (TGYCC)15, leading to an enhanced apoptotic response. 26 Our previous study has identified direct interactions of PHB with the (TGYCC)15 motif using ligand-chromatography combined with liquid chromatography-tandem mass spectrometry analyses. 30 In the present study, our data suggested that Skp2 suppresses p53-mediated PIG3 expression, while Skp2B suppresses PHB-mediated PIG3 expression.
It has been reported that Skp2 mediates the ubiquitin degradation of several cyclin-dependent kinase (CDK) inhibitors, such as p21cip1, p27kip1, and p57kip2. 19-21, 31 The expression of Skp2 is low in the G0/G1 phase, but increases in the S phase. Over-expressed Skp2 in the G0/G1 phase could lead to an out-of-control G1/S phase, disruption of cell proliferation, and differentiation. 19,20 Our data showed that both Skp2 and Skp2B could induce the increased S phase cells and therefore induce cell proliferation in breast cancer cells. Skp2 also could target several oncoproteins for ubiquitination and subsequent degradation, such as c-Myc. 32-34 However, it was reported that ubiquitination of c-Myc by Skp2 could enhance c-Myc-induced G1/S transition and c-Myc transactivation activity. 35,36 This is in agreement with our results.
Skp2 and Skp2B have both been reported to alter the activity of p53. 13,17 Skp2 can suppress p53-dependent apoptosis and Skp2B impairs p53 function via inhibition of prohibitin. 13,17 In the present study, we found the 2 isoforms could both suppress PIG3 expression via independent pathways in the breast cancer cells. Skp2 suppressed p53 and inhibited PIG3-induced apoptosis, while Skp2B attenuated the function of PIG3 by inhibiting PHB. This study is the first to demonstrate that enforced expression of Skp2 and Skp2B results in a decreased level of PIG3 through 2 different pathways. Emerging evidence has shown that androgen and the androgen receptor (AR) are involved in the regulation of Skp2. Recent studies have also shown that the AR is a robust upstream regulator of Skp2 through blocking its D-box-dependent degradation in prostate cancer cells. 23 Since Skp2 serves as an essential downstream effector of the AR, we found that both Skp2 and Skp2B transfection elicited only marginal changes in AR protein expression.
In conclusion, our results determined that Skp2 is significantly overexpressed in both breast cancer samples and cell lines, and Skp2 expression is associated with poor prognosis of breast cancer. Most importantly, both Skp2 and Skp2B could promote breast cancer cell proliferation, inhibit cell apoptosis, change the cell cycle distribution and induce the increased S phase cells and therefore induce cell proliferation in breast cancer cells. The two isoforms could both suppress PIG3 expression via independent pathways in the breast cancer cells. Skp2 suppressed p53 and inhibited PIG3-induced apoptosis, while Skp2B attenuated the function of PIG3 by inhibiting PHB. Our results indicate that Skp2 and Skp2B induce breast cancer cell development and progression, making Skp2 and Skp2B potential molecular targets for breast cancer therapy.
Materials and methods
Breast cancer molecular subtype and survival analysis
Cancer subtype-specific Skp2 gene expression analyses contained 55 breast cancer cell lines including Basal A, Basal B, and Luminal breast cancer cell lines, and an 1881-sample breast cancer data set consists of 1225 ER+ tumors, 395 ER- tumors were performed on data generated by the GOBO database 37 (http://co.bmc.lu.se/gobo/). The Skp2 gene expression analysis was performed on Oncomine (www.oncomine.org) data sets across 41 analyses after patient stratification based on the normal breast tissues versus breast cancer tissues from original published data. Kaplan–Meier survival analyses for disease outcomes were performed using the online database 38 (www.kmplot.com) and the percentiles of the patients between the upper and lower quartiles were auto-selected based on the computed best performing thresholds as cutoffs.
Cell lines and cell culture
Breast cancer cell lines MCF-7, and T47D were purchased in 2014 to 2015 from the Chinese Academy of Science Committee Type Culture Collection Cell Bank (Shanghai, China). Authenticity of these cell lines was done by Chinese Academy of Science Committee Type Culture Collection Cell Bank before purchase by STR DNA typing methodology. No authentication of these cell lines were done by the authors. Cells were cultured in RPMI 1640 or DMEM medium (GIBCO, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere containing 5% CO2. The HCT116 human colon cancer cell lines (p53+/+ and p53−/−) were generously provided by Dr. Bert Vogelstein (Johns Hopkins University). 30 Cells were grown in McCoy's 5A medium supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids and transient transfection
Skp2 and Skp2B plasmids were generously provided by Doris Germain. 17 Cells (1 × 106 cells/well) were plated in 6-well plates 24 h prior to transfection. Plasmids were then transfected into cells using TurboFect Transfection Reagent (Thermo Scientific) according to the manufacturer's protocol. Following incubation at 37°C for 24 h, cells were collected and lysed to verify the expression of related proteins by protein gel blot analysis.
MTT assay
A total of 1 × 104 cells per well were seeded into 96-well plates and transfected with required plasmids for 48 h. Following addition of 20 µL of 0.5 mg/mL MTT solution (Sigma) to each well, the medium was replaced with 200 µL DMSO after 4 h and vortexed for 10 min. Absorbance was measured at 490 nm with a microplate reader (BIO-RAD, USA) to determine the relative numbers of viable cells. Assays were performed independently 3 times.
Cell cycle analysis
Cells were treated with plasmids for 48 h, then harvested by trypsinization (no EDTA) and washed with phosphate-buffered saline (PBS). Analysis of the cell cycle was performed as previously described. 18,39,40 Each sample was tested in triplicate and untreated cells were used as controls.
Cell apoptosis analysis
Cells were treated with plasmids for 48 h, then harvested by trypsinization (no EDTA) and washed with phosphate-buffered saline (PBS). Analysis of the cell apoptosis was performed as previously described. 18,39,40 Each sample was tested in triplicate and untreated cells were used as controls.
Western blotting
Total protein was extracted using RIPA buffer supplemented with protease and phosphatase inhibitors, with protein concentrations determined using a BCA kit (Thermo Scientific). Approximately 20 µg protein was loaded per lane and separated on a sodium dodecylsulfate-polyacrylamide gel and blotted onto nitrocellulose. Blots were blocked with 5% dry milk in tris-buffered saline/0.1% tween-20 and incubated overnight with a diluted solution of primary antibody at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:5000) for 2 h. Antibodies used for western blot were: rabbit anti-Skp2/B antibody (ab19877, ab68455), rabbit anti-p53 antibody (ab32049), rabbit anti-PIG3 antibody (ab64798, sc-30068), mouse anti-p27 antibody (ab54563), and rabbit anti-PHB antibody (Ls-B7282), and rabbit anti-AR antibody (ab74272). Bands were normalized to GAPDH expression, which was used as an internal loading control. Results from at least 2 separate experiments were analyzed.
Statistical analysis
SPSS Statistics 19.0 (SPSS Inc.) was used for statistical analysis. Data were analyzed using one-way ANOVA or a Student's t-test. Data are presented as means ± SD of 3 independent experiments. The log-rank test was used to assess statistical significance of Kaplan-Meier plots. A P value of < 0.05 was considered statistically significant. *P < 0.05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the Research Innovation Program for College Graduates of Jiangsu Province (Grant No. KYLX15_0048), National Natural Science Foundation of China (No.81470357), a Foundation for Clinical Medicine Science and Technology Special Project of the Jiangsu Province, China (No. BL2014071).
References
- [1].Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013; 14:369-81; PMID:23657496; http://dx.doi.org/ 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chan CH, Morrow JK, Zhang S, Lin HK. Skp2: a dream target in the coming age of cancer therapy. Cell cycle 2014; 13:679-80; PMID:24526126; http://dx.doi.org/ 10.4161/cc.27853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1999; 1:193-9; PMID:10559916; http://dx.doi.org/ 10.1038/12013 [DOI] [PubMed] [Google Scholar]
- [4].Cariou S, Catzavelos C, Slingerland JM. Prognostic implications of expression of the cell cycle inhibitor protein p27Kip1. Breast Cancer Res Treatment 1998; 52:29-41; PMID:10066070; http://dx.doi.org/ 10.1023/A:1006154900130 [DOI] [PubMed] [Google Scholar]
- [5].Philipp-Staheli J, Payne SR, Kemp CJ. p27(Kip1): regulation and function of a haploinsufficient tumor suppressor and its misregulation in cancer. Exp Cell Res 2001; 264:148-68; PMID:11237531; http://dx.doi.org/ 10.1006/excr.2000.5143 [DOI] [PubMed] [Google Scholar]
- [6].Alkarain A, Jordan R, Slingerland J. p27 deregulation in breast cancer: prognostic significance and implications for therapy. J Mammary Gland Biol Neoplasia 2004; 9:67-80; PMID:15082919; http://dx.doi.org/ 10.1023/B:JOMG.0000023589.00994.5e [DOI] [PubMed] [Google Scholar]
- [7].Hershko DD, Shapira M. Prognostic role of p27Kip1 deregulation in colorectal cancer. Cancer 2006; 107:668-75; PMID:16826582; http://dx.doi.org/ 10.1002/cncr.22073 [DOI] [PubMed] [Google Scholar]
- [8].Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci U S A 2001; 98:5043-8; PMID:11309491; http://dx.doi.org/ 10.1073/pnas.081474898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, et al.. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 2000; 19:2069-81; PMID:10790373; http://dx.doi.org/ 10.1093/emboj/19.9.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell 2004; 6:661-72; PMID:15130491; http://dx.doi.org/ 10.1016/S1534-5807(04)00131-5 [DOI] [PubMed] [Google Scholar]
- [11].Hara T, Kamura T, Nakayama K, Oshikawa K, Hatakeyama S, Nakayama K. Degradation of p27(Kip1) at the G(0)-G(1) transition mediated by a Skp2-independent ubiquitination pathway. J Biol Chem 2001; 276:48937-43; PMID:11682478; http://dx.doi.org/ 10.1074/jbc.M107274200 [DOI] [PubMed] [Google Scholar]
- [12].Pagano M. Control of DNA synthesis and mitosis by the Skp2-p27-Cdk1/2 axis. Molecular cell 2004; 14:414-6; PMID:15149588; http://dx.doi.org/ 10.1016/S1097-2765(04)00268-0 [DOI] [PubMed] [Google Scholar]
- [13].Kitagawa M, Lee SH, McCormick F. Skp2 suppresses p53-dependent apoptosis by inhibiting p300. Molecular cell 2008; 29:217-31; PMID:18243116; http://dx.doi.org/ 10.1016/j.molcel.2007.11.036 [DOI] [PubMed] [Google Scholar]
- [14].Wu J, Huang YF, Zhou XK, Zhang W, Lian YF, Lv XB, Gao XR, Lin HK, Zeng YX, Huang JQ. Skp2 is required for Aurora B activation in cell mitosis and spindle checkpoint. Cell cycle 2015; 14:3877-84; PMID:26697838; http://dx.doi.org/ 10.1080/15384101.2015.1120916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Radke S, Pirkmaier A, Germain D. Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene 2005; 24:3448-58; PMID:15782142; http://dx.doi.org/ 10.1038/sj.onc.1208328 [DOI] [PubMed] [Google Scholar]
- [16].Ganiatsas S, Dow R, Thompson A, Schulman B, Germain D. A splice variant of Skp2 is retained in the cytoplasm and fails to direct cyclin D1 ubiquitination in the uterine cancer cell line SK-UT. Oncogene 2001; 20:3641-50; PMID:11439327; http://dx.doi.org/ 10.1038/sj.onc.1204501 [DOI] [PubMed] [Google Scholar]
- [17].Chander H, Halpern M, Resnick-Silverman L, Manfredi JJ, Germain D. Skp2B attenuates p53 function by inhibiting prohibitin. EMBO Rep 2010; 11:220-5; PMID:20134482; http://dx.doi.org/ 10.1038/embor.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang W, Luo J, Chen F, Yang F, Song W, Zhu A, Guan X. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget 2015; 6:7608-18; PMID:25797244; http://dx.doi.org/ 10.18632/oncotarget.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2). Curr Biol 2001; 11:263-7; PMID:11250155; http://dx.doi.org/ 10.1016/S0960-9822(01)00065-3 [DOI] [PubMed] [Google Scholar]
- [20].Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Molecular cell 2005; 20:9-19; PMID:16209941; http://dx.doi.org/ 10.1016/j.molcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- [21].Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U S A 2003; 100:10231-6; PMID:12925736; http://dx.doi.org/ 10.1073/pnas.1831009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Parameswaran B, Chiang HC, Lu Y, Coates J, Deng CX, Baer R, Lin HK, Li R, Paull TT, Hu Y. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell cycle 2015; 14:437-48; PMID:25659039; http://dx.doi.org/ 10.4161/15384101.2014.972901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang H, Sun D, Ji P, Mohler J, Zhu L. An AR-Skp2 pathway for proliferation of androgen-dependent prostate-cancer cells. J Cell Sci 2008; 121:2578-87; PMID:18628304; http://dx.doi.org/ 10.1242/jcs.030742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Investig 2002; 110:633-41; PMID:12208864; http://dx.doi.org/ 10.1172/JCI0215795 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [25].Sun YJ, Wang XK, Li BJ. S-phase kinase-associated protein 2 expression interference inhibits breast cancer cell proliferation. Genet Mol Res 2015; 14:9244-52; PMID:26345857; http://dx.doi.org/ 10.4238/2015.August.10.4 [DOI] [PubMed] [Google Scholar]
- [26].Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature 1997; 389:300-5; PMID:9305847; http://dx.doi.org/ 10.1038/38525 [DOI] [PubMed] [Google Scholar]
- [27].Lee JH, Kang Y, Khare V, Jin ZY, Kang MY, Yoon Y, Hyun JW, Chung MH, Cho SI, Jun JY, et al.. The p53-inducible gene 3 (PIG3) contributes to early cellular response to DNA damage. Oncogene 2010; 29:1431-50; PMID:20023697; http://dx.doi.org/ 10.1038/onc.2009.438 [DOI] [PubMed] [Google Scholar]
- [28].Amano T, Nakamizo A, Mishra SK, Gumin J, Shinojima N, Sawaya R, Lang FF. Simultaneous phosphorylation of p53 at serine 15 and 20 induces apoptosis in human glioma cells by increasing expression of pro-apoptotic genes. Journal of neuro-oncology 2009; 92:357-71; PMID:19357962; http://dx.doi.org/ 10.1007/s11060-009-9844-1 [DOI] [PubMed] [Google Scholar]
- [29].Terui T, Murakami K, Takimoto R, Takahashi M, Takada K, Murakami T, Minami S, Matsunaga T, Takayama T, Kato J, et al.. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer Res 2003; 63:8948-54; PMID:14695212 [PubMed] [Google Scholar]
- [30].Guan X, Liu Z, Wang L, Johnson DG, Wei Q. Identification of prohibitin and prohibiton as novel factors binding to the p53 induced gene 3 (PIG3) promoter (TGYCC) motif. Biochem Biophys Res Commun 2014; 443:1239-44; PMID:24388982; http://dx.doi.org/ 10.1016/j.bbrc.2013.12.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bencivenga D, Tramontano A, Borgia A, Negri A, Caldarelli I, Oliva A, Perrotta S, Della Ragione F, Borriello A. P27Kip1 serine 10 phosphorylation determines its metabolism and interaction with cyclin-dependent kinases. Cell cycle 2014; 13:3768-82; PMID:25483085; http://dx.doi.org/ 10.4161/15384101.2014.965999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jamal A, Swarnalatha M, Sultana S, Joshi P, Panda SK, Kumar V. The G1 phase E3 ubiquitin ligase TRUSS that gets deregulated in human cancers is a novel substrate of the S-phase E3 ubiquitin ligase Skp2. Cell cycle 2015; 14:2688-700; PMID:26038816; http://dx.doi.org/ 10.1080/15384101.2015.1056946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006; 6:369-81; PMID:16633365; http://dx.doi.org/ 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- [34].Sun XX, Sears RC, Dai MS. Deubiquitinating c-Myc: USP36 steps up in the nucleolus. Cell cycle 2015; 14:3786-93; PMID:26697836; http://dx.doi.org/ 10.1080/15384101.2015.1093713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell 2003; 11:1177-88; PMID:12769843; http://dx.doi.org/ 10.1016/S1097-2765(03)00173-4 [DOI] [PubMed] [Google Scholar]
- [36].von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al.. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Molecular cell 2003; 11:1189-200; PMID:12769844; http://dx.doi.org/ 10.1016/S1097-2765(03)00193-X [DOI] [PubMed] [Google Scholar]
- [37].Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One 2011; 6:e17911; PMID:21445301; http://dx.doi.org/ 10.1371/journal.pone.0017911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013; 8:e82241; PMID:24367507; http://dx.doi.org/ 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lai J, Nie W, Zhang W, Wang Y, Xie R, Wang Y, Gu J, Xu J, Song W, Yang F, et al.. Transcriptional regulation of the p73 gene by Nrf-2 and promoter CpG methylation in human breast cancer. Oncotarget 2014; 5:6909-22; PMID:25071010; http://dx.doi.org/ 10.18632/oncotarget.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nie W, Huang W, Zhang W, Xu J, Song W, Wang Y, Zhu A, Luo J, Huang G, Wang Y, et al.. TXNIP interaction with the Her-1/2 pathway contributes to overall survival in breast cancer. Oncotarget 2015; 6:3003-12; PMID:25605021; http://dx.doi.org/ 10.18632/oncotarget.3096 [DOI] [PMC free article] [PubMed] [Google Scholar]