ABSTRACT
The PHD zinc finger protein Jade-1S is a component of the HBO1 histone acetyltransferase complex and binds chromatin in a cell cycle-dependent manner. Jade-1S also acts as an E3 ubiquitin ligase for the canonical Wnt effector protein β-catenin and is influenced by CK1α-mediated phosphorylation. To further elucidate the functional impact of this phosphorylation, we used a stable, low-level expression system to express either wild-type or mutant Jade-1S lacking the N-terminal CK1α phosphorylation motif. Interactome analyses revealed that the Jade-1S mutant unable to be phosphorylated by CK1α has an increased binding affinity to proteins involved in chromatin remodelling, histone deacetylation, transcriptional repression, and ribosome biogenesis. Interestingly, cells expressing the mutant displayed an elongated cell shape and a delay in cell cycle progression. Finally, phosphoproteomic analyses allowed identification of a Jade-1S site phosphorylated in the presence of CK1α but closely resembling a PLK1 phosphorylation motif. Our data suggest that Jade-1S phosphorylation at an N-terminal CK1α motif creates a PLK1 phospho-binding domain. We propose CK1α phosphorylation of Jade 1S to serve as a molecular switch, turning off chromatin remodelling functions of Jade-1S and allowing timely cell cycle progression. As Jade-1S protein expression in the kidney is altered upon renal injury, this could contribute to understanding mechanisms underlying epithelial injury repair.
KEYWORDS: Cell cycle, CK1α, Jade-1S, mitosis, NuRD complex, PLK1, proliferation, S-phase
Introduction
Cell cycle re-entry and subsequent mitosis of resident cells are critical events in renal tissue regeneration. Tubular epithelial cells of the human kidney possess an intrinsic capacity for self-renewal during homeostasis and after injury. A growing body of evidence supports a model whereby tubule repair is predominantly attributed to mature epithelial cells that dedifferentiate and re-enter the cell cycle after injury.1-6 The molecular mechanisms underlying this process require that polarized structures such as adherens junctions and primary cilia disassemble, while signaling cascades reflective of an embryonic, stem cell, or proliferative status are resumed. Evidence suggests transient activation of multiple signaling pathways such as transforming growth factor-β and canonical Wnt signaling to be required for repair, while prolonged activation contributes to disease states including fibrosis and cyst formation.7–9 It is therefore imperative to understand the molecular mechanisms that distinguish these cell fate decisions, in order to maximize repair capacity in disease and acute kidney injury.
The short isoform of the protein Gene for Apoptosis and Differentiated Epithelia (Jade-1S, also known as Plant Homeodomain Finger (PHF)-17) was identified as a binding partner of the von Hippel-Lindau tumor suppressor and is highly expressed in kidney epithelial cells.10 Jade1S was further characterized as a zinc-finger DNA binding protein with histone-acetyltransferase and transcription factor activity11 and subsequently shown to promote histone H4 acetylation by functioning as a member of the HBO-1 histone acetylation complex.12 The HBO-1/Jade-1 complex binds DNA in a cell-cycle-dependent manner13 and is required for H4 acetylation during epithelial cell regeneration14 as well as during DNA damage response signaling.15 Jade-1S can furthermore act as an E3-ubiquitin ligase for both nuclear and cytosolic β-catenin to negatively regulate canonical Wnt signaling.16 In addition to concentrating in the nucleus of interphase cells, we have identified a dynamic accumulation of Jade-1S at the primary cilium and centrosome,17 organelles which are inherently linked to the cell cycle and have been previously associated with inhibition of canonical Wnt signaling.8,18,19 Regulation of the protein abundance and phosphorylation status of Jade-1S is influenced by the ciliopathy gene product nephronophthisis-4 (NPHP4) and the canonical Wnt pathway kinase casein-kinase-1α (CK1α).17,20
Jade-1S function therefore centers on important events during injury repair: e.g. DNA damage, Wnt signaling regulation, DNA transcription and epigenetic modifications. Here, we show that loss of CK1α-mediated phosphorylation alters the interactome profile of Jade-1S with respect to protein complexes involved in epigenetic modification, and that this is accompanied by an impaired rate of cell proliferation due to a prolonged S-phase. Mass spectrometry analyses further revealed an intriguing phosphorylation motif linking Jade-1S to Polo-like kinase-1 (PLK1), a well-known mitotic kinase with important roles in S-phase, and we demonstrate that PLK1 and Jade-1S interact in a CK1α-dependent manner.
Results
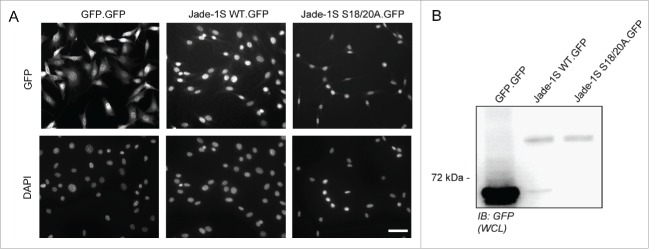
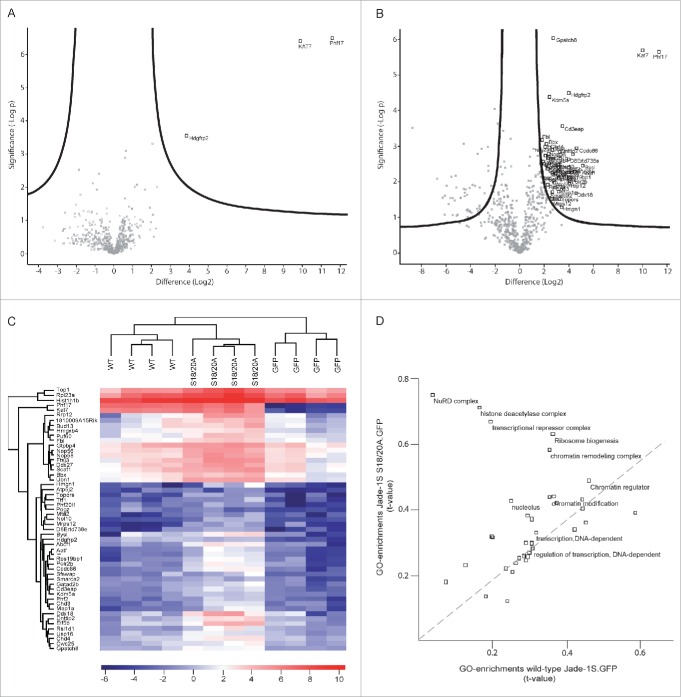
To assess how loss of Jade-1S N-terminal phosphorylation by CK1α could influence protein function in cycling cells, we sought to identify key associated protein complexes by generating polyclonal cell lines expressing low levels of wild-type or mutant Jade-1S and using a pulldown strategy for unbiased analyses of the dynamic interactome by mass spectrometry.21 We used a single-copy integration Flp-In strategy22 in NIH-3T3 cells to express C-terminal GFP-tagged wild-type Jade-1S (Jade-1S.GFP), CK1α phosphorylation motif mutant Jade-1S (Jade-1S S18/20A.GFP) or control GFP only (GFP.GFP) proteins in cycling cells. Cells with successful integration were selected using hygromycin B and maintained as polyclonal lines. All Jade-1S expressing cell lines demonstrated a weaker, predominantly nuclear GFP signal compared to the GFP.GFP cells (Fig. 1A). GFP-fusion proteins were expressed at predicted sizes as visualised by western blot with equal expression of Jade-1S wild-type and mutant species (Fig. 1B). We performed quantitative label-free proteomics with stringent statistical criteria to define the most highly abundant interactome members for each fusion protein. The CK1α phosphorylation motif mutant Jade-1S S18/20A.GFP pulldown substantially differed from wild-type Jade-1S.GFP expressing cells, whereby the Jade-1S S18/S20A.GFP mutant had an increased number of significant interacting partners (Figs. 2A, 2B; supplementary Tables 1–2). The efficiency of Jade-1S.GFP purification did not differ between cell lines (Figs. 2A, 2B). Hierarchical clustering analysis of protein intensities clearly distinguished both Jade-1S expressing lines from the control GFP.GFP line (Fig. 2C), and identified both Phf17 (Jade-1S) and Kat7 (HBO1) in both Jade-1S expressing lines. 2D-gene ontology (GO)-enrichment analysis demonstrated that Jade-1S S18/20A.GFP had increased affinity compared to wild-type Jade-1S.GFP for proteins related to nucleosome remodelling deacetylase (NuRD) complex function, histone deacetylation, transcriptional repression, ribosome biogenesis, and chromatin remodelling (Fig. 2D). Kinases were not detected in this analysis, likely due to the typically transient nature of endogenous kinase interaction. These data demonstrate that phosphorylation of Jade-1S at residues S18 and S20 dramatically alters Jade-1S protein-protein interactions (PPI) in cycling cells.
Figure 1.
Generation of Flp-In NIH-3T3 cells. A) Low-level stable expression of GFP, Jade-1S WT or Jade-1S S18/20A coding sequences was achieved using the Flp-In™-3T3 system with the C-terminally GFP tagged vector pgLAP5. All Flp-In cell lines were positive for GFP expression as assessed by immuno-fluorescence (top panel) after PFA-fixation. Exposure time for GFP.GFP cells was 50% of that required to visualize the Jade-1S.GFP-expressing lines. DAPI co-stain indicates nuclei (bottom panel). Scale bar = 50 µm. B) Confluent cell lines were prepared as whole cell lysates (WCL) and assessed by immunoblot for GFP expression. IB, immunoblot.
Figure 2.
Jade-1S interactome is altered by S18/20A mutation. Volcano plots showing significant interactors of wild-type Jade-1S.GFP (A) and Jade-1S S18/20A.GFP (B). The decadic logarithm of the p-value is plotted against the logarithmic fold change of the respective Jade-1S IP as compared to the control IP. Proteins determined to be interactors are labeled with the gene symbol. C) Hierarchical clustering of protein intensities (LFQ values) analyzing the effect of phospho-ablating mutations on the Jade interactors. Results from 4 independent experimental replicates cluster for wild-type Jade-1S.GFP (WT), Jade-1S S18/20A.GFP (S18/20A) and control GFP.GFP (GFP) expressing cell lines. HBO1 was a strong interacting partner for both Jade-1S expressing cell lines but not for the control GFP.GFP cell line. The profile for the Jade-1S S18/20A.GFP line showed an overall increase in interaction partners (–= uncharacterized protein C1orf131 homolog). D) 2D-GO-enrichment analysis showing significantly enriched GO terms. The scatter plot shows comparison between the significant GO-enrichments within the Jade-1S S18/20A.GFP interactome (Y-axis) and the wild-type Jade-1S.GFP (X-axis) interactome. The dashed line indicates GO terms enriched equally in both samples. The mutant Jade-1S S18/20A.GFP line showed increased interaction compared to the wild-type Jade-1S.GFP line for proteins related to the NuRD complex, histone deacetylation, transcriptional repression, ribosome biogenesis, and chromatin remodeling.
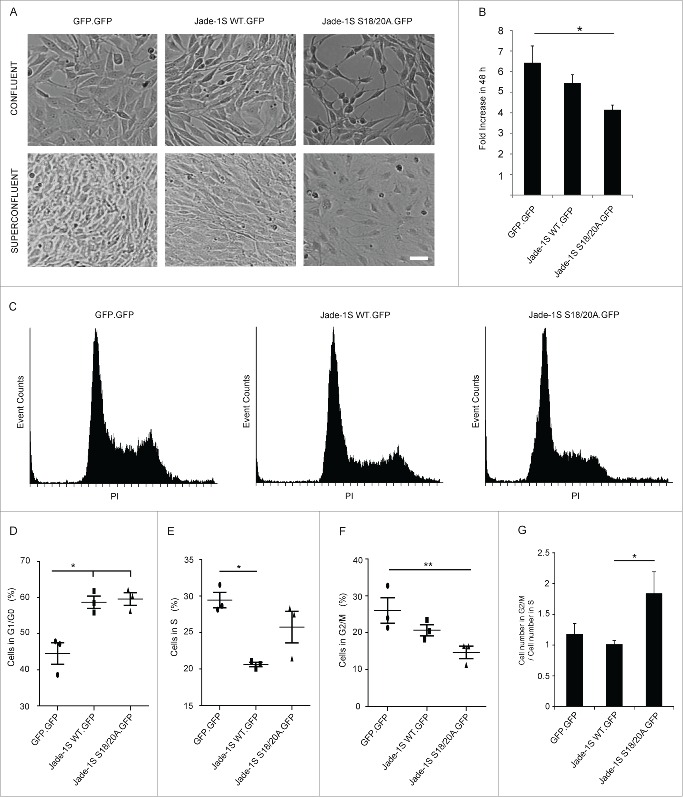
During these experiments, it was evident that the mutant Jade-1S S18/20A.GFP expressing cells displayed a prominent elongated cell morphology which was not present in the other polyclonal lines generated (Fig. 3A). This effect was most apparent as cells approached confluency, and was not prominent if cells were seeded densely enough to be confluent immediately after splitting. If cells were allowed to grow from sub- to super-confluency, the Jade-1S S18/20A.GFP-expressing cells developed extensive membrane networks but cells failed to reach the same density that was seen in both other polyclonal lines. We furthermore noted that both Jade-1S expressing cell lines proliferated more slowly than control cells. Analysis of exponentially growing cells using an automated counting system confirmed that within a 48 hour period Jade-1S S18/20A.GFP expressing cells proliferated significantly less than control GFP.GFP cells (Fig. 3B). Flow cytometry was used to determine whether the delayed proliferation could be attributed to an arrest in a specific cell cycle phase (Figs. 3C-F). Wild-type Jade-1S.GFP and Jade-1S S18/20A.GFP polyclonal cell lines had a significantly higher percentage of cells in G1/G0 than control GFP.GFP cells. Furthermore, compared to the control GFP.GFP cells, there were significantly fewer wild-type Jade-1S.GFP expressing cells in S-phase and significantly fewer Jade-1S S18/20A.GFP expressing cells in G2/M. The Jade-1S S18/20A.GFP expressing cells showed a 2:1 ratio for cells in S-phase compared to G2/M, whereas both other lines approached an expected 1:1 ratio (Fig. 3G). Together these data indicate that protein abundance and phosphorylation status of Jade-1S both affect cell cycle progression.
Figure 3.
Growth of Flp-In NIH-3T3 cells is altered by single-copy Jade-1S insertion. A) The mutant Jade-1S S18/20A.GFP cell line displayed an elongated cell shape as cells approached confluency. If allowed to grow until super-confluent, Jade-1S S18/20A.GFP cells displayed increased membrane spreading, and nuclei did not achieve close proximity as was seen in the other cells. Scale = 50 µm. B) Cells were seeded at 200 000 per 10 cm culture dish and counted 48h and 96h later. Fold change from 48h to 96h is shown. The Jade-1S S18/20A.GFP line proliferated significantly more slowly than the control GFP.GFP line (F(2,4) = 8.553, p = 0.0359; Tukey's post hoc, * = p <0.05). C) Representative histograms of propidium iodide (PtdIns) staining of each Flp-In cell line. Cells were seeded at 2 × 10e6 /10 cm dish and harvest after 24 hours for flow cytometry. D, E, F) Quantification of flow cytometry data where the first histogram peak corresponds to cells in G1/G0 phase (2n DNA) and the second peak to G2/M (4n DNA), with S phase corresponding to cells with an intermediary DNA status. The percentage of cells in cell cycle phases G1/G0 (D), S (E), and G2/M (F) was calculated for each Flp-In cell line using Cyflogic software. Results were analyzed using repeated measures ANOVA to maintain integrity of individual experimental repetitions. The wild-type Jade-1S.GFP and the mutant Jade-1S S18/20A.GFP cell lines had a significantly increased percentage of cells in G1/G0 (D) (F(2,4) = 13.77, p = 0.0161; Tukey's post hoc *, p < 0.05; **, p < 0.01). A significantly smaller percentage of wild-type Jade-1S expressing cells were in S-phase compared to the control GFP.GFP expressing cells (E) (F(2,4) = 11.15, p = 0.0231; Tukey's post hoc *, p<0.05). The mutant Jade-1S S18/20A.GFP cell line had a significantly lower percentage of cells in G2/M than the GFP.GFP control line (F) (F(2,4) = 16.97, p = 0.0111; Tukey's post hoc **, p < 0.01). G) The ratio of percentage cells in G2/M to S-phase was significantly increased in cells expressing Jade-1S S18/20A.GFP (F(2,4) = 8.497, p = 0.0363; Tukey's post hoc *, p < 0.05). Errors bars represent SEM.
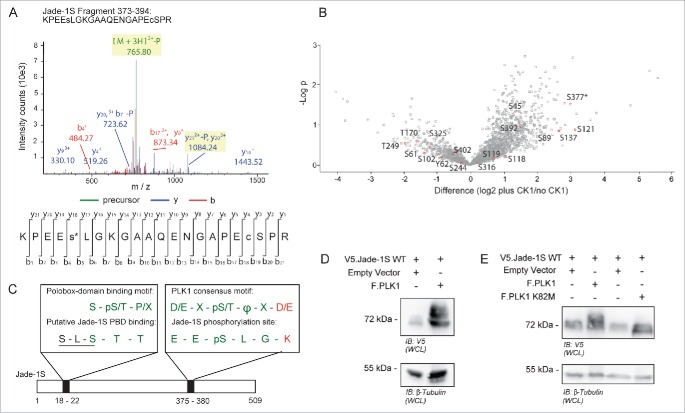
To further investigate a link between Jade-1S phosphorylation and cell cycle, we used mass spectrometry to analyze Jade-1S phosphorylated by CK1α and sought to identify sites matching known mitotic kinase motifs. We identified a novel phosphorylation site at Serine (S) 377 (Fig. 4A) which, with the exception of one amino acid residue (K380, highlighted in red, Fig. 4C), corresponded to a PLK1 consensus phosphorylation motif.23 Phosphorylation at S377 was one of the most prominent Jade-1S modifications induced by CK1α, as illustrated by the high fold change and significance obtained in a quantitative phosphoproteomic analysis comparing Jade1S phosphorylation with or without over-expressed CK1α (Fig. 4B). We have previously demonstrated that CK1α phosphorylates the N-terminus of Jade-1 at an unprimed, non-canonical S-L-S phosphorylation motif at amino acids 18-20.20 This motif overlaps with the minimal sequence requirements for a polo-box domain (PBD) phospho-binding motif24 (highlighted in green, Fig. 4C). Our identification of these residues as corresponding to PLK1 phosphorylation site and PBD-binding motif is supported by the Group-based Prediction System for Polo (GPS-Polo 1.0), a freely available online tool that predicts PBD-based phospho-binding and PLK1 phosphorylation sites.25 GPS-Polo 1.0 predicts residue T21 as one of 4 low-threshold predicted PLK phospho-binding motifs on Jade-1S, and residue S377 as one of 4 high-threshold predicted PLK1 phosphorylation sites. Using tagged proteins expressed transiently in human embryonic kidney (HEK) 293T cells, we verified that over-expression of Flag-tagged PLK1 (F.PLK1) leads to a shift in expression size of V5-tagged Jade-1S (V5.Jade-1S; Fig. 4D), which did not occur in the presence of a kinase-inactive version of F.PLK1 harbouring a K82M mutation in the ATP binding motif26 (Fig. 4E). Together, these data suggest that PLK1 phosphorylates Jade-1S in a CK1α-dependent manner.
Figure 4.
Identification of Jade-1S site S377 phosphorylated in the presence of over-expressed CK1α. A) MS2 spectrum demonstrating Serine 377 phosphorylation on the Jade-1S fragment 373-394. B) Quantitative phosphoproteomic analysis of Jade phosphorylation change in the absence and presence of CK1α in whole cells. The S377 site is marked by an asterisk. Only high-quality (class I) sites are shown, with all Jade-1S sites labeled accordingly. C) Schematic illustrating the amino acid sequences of the N-terminal CK1α phosphorylation site (S18/S20, underlined) corresponding to a putative Polobox-binding domain, and the C-terminal S377 motif corresponding to a PLK1 phosphorylation consensus motif. Green represents shared amino acid sequences while red represents those that differ (X = any amino acid; Φ = hydrophobic amino acid; p = phosphorylated). D and E) 293T cells were transiently transfected with constructs as indicated and processed as whole cell lysates 24 hours later. Expression of V5.Jade-1 was heavily modified by overexpressed F.PLK1 (D). This effect was not seen when V5.Jade-1 was over-expressed with a kinase-dead mutant of F.PLK1 (E). IB, immunoblot. WCL, whole cell lysate.
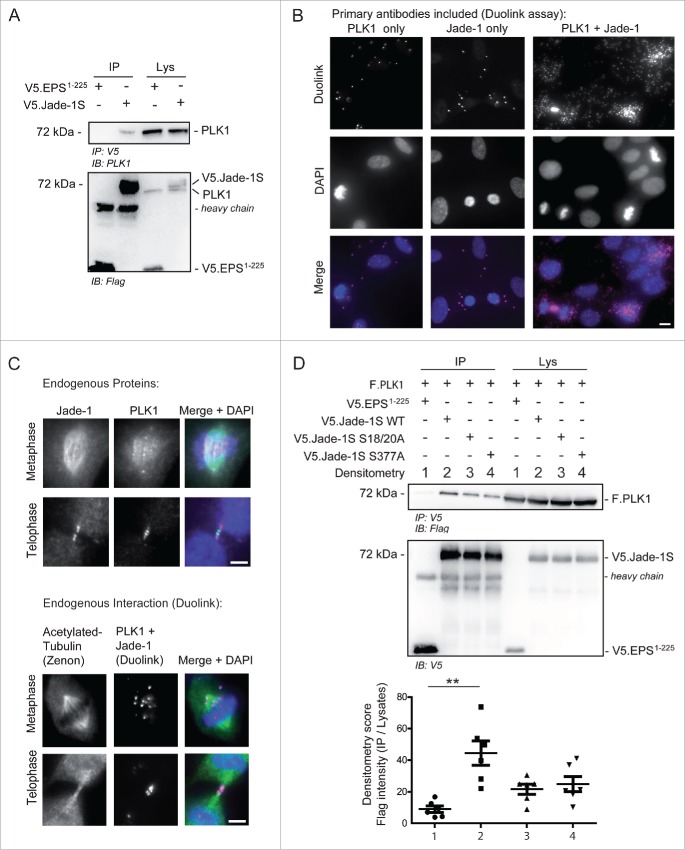
To confirm a relationship between Jade-1S and PLK1 in cycling cells, we first verified a PPI between V5.Jade-1S and endogenous PLK1 (Fig. 5A) by co-immunoprecipitation experiments. We furthermore confirmed endogenous PPI in telomerase immortalized human retinal pigment epithelium (RPE-1) cells by using a Duolink Proximity-Ligation-Assay (PLA), where proteins of interest are first labeled with appropriate primary antibodies, and subsequently with species-specific oligonucleotide-tagged probes. Fluorescence is only observed if 2 individually labeled proteins are physically close enough to allow the oligonucleotide-tag ligation and rolling-circle signal amplification.27 Using this strategy, we showed that signal amplification was present in interphase RPE-1 cells where both endogenous Jade-1S and PLK1 were labeled, and was highly augmented in dividing RPE-1 cells (Fig. 5B). To obtain a clearer view of Duolink fluorescence in mitotic cells, we optimised the staining protocol so that the overall signal was substantially reduced (see Methods), and co-labeled acetylated-tubulin using Zenon labeling.28 With this protocol we were able to clearly visualize PPI between Jade-1S and PLK1 at the metaphase spindle and the cytokinetic bridge; this interaction was corroborated by viewing Jade-1S and PLK1 co-localization at these regions by traditional immunofluorescence (Fig. 5C). We furthermore demonstrated that co-precipitation of PLK1 with Jade-1S was influenced by both the putative PLK1 phosphorylation and binding sites. When wild-type or mutant V5.Jade-1S and F.PLK1 were co-expressed in HEK 293T cells, both V5.Jade-1S mutants did not co-precipitate F.PLK1 as efficiently as wild-type V5.Jade-1S did (Fig. 5D), indicating that the interaction of Jade-1S and PLK1 is influenced by CK1α.
Figure 5.
Endogenous Jade-1 and PLK1 interaction is dependent on a CK1α phosphorylation motif. A) 293T cells were transiently transfected with V5.Jade-1S or a control protein (V5.EPS1-225), harvested after 24 hours and the V5 tag was immuno-precipitated (IP). Endogenous PLK1 co-precipitated with V5.Jade-1S but not the control protein. B) RPE-1 cells grown on coverslips were PFA-fixed at 80% confluency. Endogenous PLK1 (left panel), endogenous Jade-1 (middle panel), or both (right panel) were immuno-labeled with appropriate primary antibodies and incubated with PLUS / MINUS PLA probes. The Duolink signal (magenta) labels interacting proteins and was increased from background when both Jade-1 and PLK1 primary antibodies were included, and further increased in mitotic cells. Merge with DAPI to label nuclei, scale bar = 10 µm. C) Comparison of co-localization (top panel set) vs. interaction (bottom panel set) of endogenous Jade-1 and PLK1 in metaphase and telophase. RPE-1 cells were grown to 80% confluency on coverslips and PFA-fixed. Cells were immuno-labeled with both anti-Jade-1 and anti-PLK1 antibodies and visualised using species-specific fluorescent secondary antibodies (top panel set; Jade-1 = green; PLK1 = magenta) or a Duolink PLA reaction (bottom panel set, magenta), in which case tubulin was subsequently co-labeled using a commercially available Zenon labeling kit (green). Both endogenous Jade-1 and PLK1 were visualised at the metaphase spindle and cytokinetic bridge (top panel). Interaction between Jade-1 and PLK1 was seen at the metaphase spindle and cytokinetic bridge (bottom panel). To allow for more specific vizualization of protein interaction, all background signal was virtually eliminated (see Methods). Merge with DAPI, scale bar = 5µm. D) 293T cells were transiently transfected with constructs as indicated for 24 hours. Co-precipitation of F.PLK1 with mutant V5.Jade-1S constructs lacking either the N-terminal CK1α phosphorylation site (Jade-1S S18/20A) or lacking the S377 putative PLK1-phosphorylation site (Jade-1S S377A) was reduced compared to co-precipitation with wild-type V5.Jade-1S (Jade-1 WT). Densitometry analysis (bottom panel) showed that F.PLK1 co-precipitation was significantly elevated from that obtained using a control protein (V5.EPS1-225) only with Jade-1 WT and not either of the mutants (F(3,15) = 6.736, p = 0.0043; Tukey's post hoc **, p < 0.01) IB, immunoblot. Lys, IP lysates.
Discussion
Jade-1S has previously been suggested to function as an “epigenetic integrator at the crossroads of DNA replication, DNA repair, transcription, cell cycles, and development.”29 Our data demonstrates that the Jade-1S interactome was significantly altered if the CK1α phosphorylation motif was disrupted, whereby Jade-1S S18/20A.GFP had increased affinity for binding partners associated with all of these processes: enriched interacting partners included the nucleosome remodelling and deacetylation NuRD complex, histone deacetylation, chromatin remodelling, transcriptional repression, and ribosome biogenesis. Identified members of the NuRD complex include Chd3, Chd4, and MTA2 which are central to processes associated with stemness, aging and cancer,30 areas highly relevant to injury repair. The NuRD complex function is further associated with transcriptional repression,31,32 possibly countering the transcriptional activation properties of Jade-1S.11 HBO1/Kat7 was identified as a strong interacting partner of both wild-type and mutant Jade-1S. Jade-1S is a known component of the HBO1 histone acetylation complex and facilitates DNA synthesis which is required for S-phase progression;14 however, histone deacetylation is important for continued progression through G2 to mitosis.33 It is therefore interesting to note that the Jade-1S S18/20A.GFP expressing cells proliferated significantly more slowly than control GFP.GFP cells. The increased affinity of the CK1α phosphorylation motif mutant Jade-1S S18/20A.GFP to proteins involved with histone deacetylation and transcriptional repression could reflect an inability to “turn off” a normal function of wild-type Jade-1S, delaying cell cycle progression.
To further examine delayed proliferation, we investigated whether cycling cells showed altered accumulation in a specific cell cycle phase. Flow cytometry demonstrated that compared to the control GFP.GFP expressing cells, a smaller percentage of wild-type Jade-1S.GFP cells were in S-phase and a smaller percentage of mutant Jade-1S S18/20A.GFP expressing cells were in G2/M. Both of the Jade-1S expressing polyclonal lines had a higher percentage of cells in G0/G1 than control cells. It was recently shown that Jade-1S depletion facilitated the rate of progression through mitosis to G1 in synchronised cells, but that overexpression delayed cytokinesis.34 Our data supports a model whereby Jade-1S acts to restrain cell cycle rate, and suggests additional roles of Jade-1S throughout the cell cycle. Low-level over-expression of both Jade-1S versions enhanced the percentage of cells in G1/G0, and proliferation data supports a mild inhibition of cell cycle rate for these polyclonal lines; however, G2/M:S ratios suggest that every Jade1S.GFP expressing cell that enters S-phase progresses to G2/M, whereas only half of those expressing Jade-1S S18/20A.GFP do. This indicates that expression of this mutant acts in a dominant negative manner causing an additional impairment in S-phase due to a lack of phosphorylation by CK1α. Other reports also describe a growth disadvantage associated with Jade-1S,35,36 and show that endogenous Jade-1S protein is not well expressed in several highly proliferative cancer cell lines.36 Endogenous Jade-1S protein levels were also shown to decrease after injury and increase during repair;14 however, in this study the cell cycle marker Ki67 protein expression increased before Jade-1S and decreased immediately after Jade-1S protein levels peaked, leaving open the possibility that the increase in Jade-1S is causal for the Ki67/cell cycling decrease.
The specific accumulation of Jade-1S S18/20A.GFP cells in S-phase could be due to the increased affinity of the Jade-1S S18/20A protein to interacting partners important for chromatin remodelling. We observed Jade-1S S18/20A.GFP expressing cells to have an elongated cell morphology which has previously been associated with a depletion of PLK1 in hTERT-RPE1 cells due to disrupted S-phase progression.37 PLK1 is a well-known mitotic kinase with multiple functions including critical roles in centrosome maturation, microtubule-kinetochore attachment and metaphase spindle elongation, and cytokinesis,38–41 but which is also active during interphase.42 Our previous data has highlighted the importance of phosphorylation in modifying Jade-1S activity20 and we now identify PLK1 as an interacting partner of Jade-1S. The interaction between Jade-1S and PLK1 is dependent in part on an intact Jade-1S phosphorylation site identified by mass spectrometry, which is located at S377 and corresponds to a PLK1 phosphorylation motif, as well as a putative N-terminal PBD-recognition site, which overlaps with a previously identified unprimed CK1α phosphorylation motif at S18-S20.20 We previously noted that direct phosphorylation by CK1α does not account for a vastly modified Jade-1S N-terminus seen in the presence of overexpressed CK1α in intact cells. We now suggest that PLK1 could play an important role in Jade-1S phosphorylation, in a CK1α-dependent manner. A cooperative role for PLK1 and CK1α was recently described in cell cycle progression in lung cancer context.43 In the case of Jade-1S, phosphorylation by CK1α at S18 would create a classical primed CK1α motif predicted leading to further phosphorylation by CK1α at T21 and fulfilling the minimum requirements for PBD-based phospho-binding by PLK1, allowing subsequent phosphorylation by PLK1 at one or all of the 9 putative PLK1 phosphorylation motifs predicted by GPS-Polo 1.0 including the site S377. Using the Duolink PLA strategy we demonstrated in situ that the PLK1 and Jade-1S protein complex is present in interphase cells and upregulated in mitotic cells, predominantly at the metaphase spindle and the cytokinetic bridge, therefore also suggesting roles for this complex during mitosis. It should be noted that the polyclonal antibody used to visualize Jade-1S does not distinguish between the long and short isoforms; therefore, we also verified PPI with Jade-1S by expressing only the short isoform and demonstrating co-precipitation (Fig. 5A). Indeed, over-expressing kinase active F.PLK1 heavily modifies the appearance of V5.Jade-1S in western blot: multiple bands with higher molecular weight can be seen (Fig. 4D) compared to the previously reported modification by F.CK1α where phosphorylated V5.Jade-1S was visualised as a double-band.20 Of interest, activation of PLK1 prior to mitosis onset is achieved through phosphorylation by Aurora A,44,45 a kinase previously noted to indirectly lead to Jade-1S phosphorylation.13
In light of the interphase roles of PLK1,42 it is particularly interesting that upregulated interactors identified for the mutant Jade-1S S18/20A included TOPORS, which, like the Jade1S interacting partner Kat7/HBO1, is associated with recognition of genotoxic stress15,46 and regulated by PLK1.47,48 Given a requirement for PLK1 in cell cycle progression especially after DNA damage,49,50 and the links between tissue repair and Jade1S described above, a relationship between Jade-1S and PLK1 during the cell cycle is intriguing. Mechanisms underlying S-phase delay could involve impaired regulation of the Jade1/HBO1 histone acetylation complex, and may directly involve PLK1 as PLK1-mediated phosphorylation of HBO1 contributes to pre-replicative complex formation and DNA replication licensing.47
In summary, a mutant version of Jade-1S that cannot be phosphorylated by CK1α shows disrupted PLK1 interaction and increased complex formation with proteins involved in chromatin remodelling, delaying growth when stably expressed in cycling cells in part due to impaired S-phase progression. Our data are consistent with a model suggesting multiple Jade-1S roles throughout the cell cycle progression, as is the case for both PLK142 and CK1α.51–53 As DNA synthesis is upregulated during kidney injury repair,1 identification of proteins and their roles in this process will be a major contribution to understanding cell fate decisions to maximise healthy repair rather than a fibrotic or cystic response. We previously showed that NPHP4, which localizes to the primary cilium, a post-mitotic structure, stabilises de-phosphorylated nuclear Jade-1S.17 As Jade-1S does not associate with DNA during mitosis,13 and our data show that a low-level increase of nuclear Jade-1S influences G1/G0 accumulation, we suggest Jade-1S phosphorylation by CK1α and PLK1 to act as a molecular switch: necessary to remove Jade-1S from the nucleus prior to mitosis and influencing cell cycle exit after. Such a role for Jade-1S has implications for injury repair processes particularly relevant to understanding tubular epithelial repair in kidney disease.
Materials and methods
Plasmids & Antibodies - FLAG- or V5-tagged coding sequences were generated by PCR from the fetal human kidney cDNA library (Stratagene, La Jolla, CA, USA) and inserted into a modified pcDNA6 vector (Thermofisher, Carlsbad, CA, USA) using standard cloning techniques. To generate Flp-In cell lines, the Flp recombinase expressing plasmid pOG44 (Thermofisher, Carlsbad, CA, USA) was co-transfected with pgLAP5 (gift from Peter Jackson, Addgene plasmid # 19706) containing appropriate cDNA insert transferred via LR clonase reaction (Gateway Technology, Thermofisher, Carlsbad, CA, USA). Site-directed mutagenesis was achieved by PCR-amplification using the relevant wild-type plasmid as a template together with primers containing required alterations. The PCR product was incubated with 1 µl of Dpn1 (2 hours at 37°C) and heat inactivated (70°C for 10 minutes) to digest methylated template DNA. All plasmids were verified by automated DNA sequencing. Antibodies were obtained from Sigma-Aldrich (St. Louis, MO, USA; monoclonal mouse anti-FLAG (M2); monoclonal mouse anti-β-tubulin; monoclonal mouse anti-acetylated-tubulin clone 6-11B), Serotec (Puchheim, Germany; monoclonal mouse anti-V5), Proteintech (Manchester, UK; polyclonal rabbit anti-Jade-1), Abcam (Cambridge, UK; monoclonal mouse anti-PLK1) and Santa Cruz (Dallas, Texas, USA; monoclonal mouse anti-GFP).
Cell Culture – hTert RPE-1 cells (ATCC, Manassas, Virginia, USA) were cultured in DMEM-F12 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% Fetal Bovine Serum (FBS), 2 mM GlutaMAX™ (Thermofisher, Darmstadt, Germany) and 2.6 g/L sodium bicarbonate. Flp-In™-3T3 (Thermofisher, Carlsbad, CA, USA) and HEK 293T (gift from Gerd Walz, University of Freiburg) cell lines were cultured in DMEM with GlutaMAX™ (Thermofisher, Darmstadt, Germany) supplemented with 10% FBS. For transfection experiments, HEK 293T cells were seeded in 10 cm or 6-well dishes, grown to 60% confluency and transiently transfected with plasmid DNA using the calcium-phosphate method as described previously.54,55 To generate cell lines stably expressing the gene of interest, Flp-In™-3T3 cells were grown to 80% confluency in a 6-well plate and transfected with Lipofectamine 2000 (Thermofisher, Carlsbad, CA, USA) according to the manufacturer's instructions. Twenty-four hours post-transfection, medium including 160 µg/ml of hygromycin B (Invivogen, San Diego, CA, USA) was exchanged every 2 d until single colonies formed. All cell lines were passaged regularly before reaching confluency.
Immunoprecipitation (IP) and Whole Cell Lysates (WCL). Cells grown in 6-well dishes were harvested in 1 ml of cold phosphate-buffered saline (PBS), and the centrifuged pellet was boiled in Laemmli buffer at 95°C for 10 minutes to obtain a whole cell lysate. For co-IP experiments, cells grown in 10 cm dishes were harvested in cold PBS, centrifuged, and the cell pellet was used as previously described.17 Proteins were immunoprecipitated using or 1 µg of V5 antibody coupled to Protein G beads (GE Healthcare, Solingen, Germany) and analyzed by single or duplicate SDS-PAGE gels. Densitometry values were obtained using ImageJ (version 1.49d, National Institutes of Health, USA) and calculated as follows: Each protein band measured value was subtracted by measured local background. The calculated protein precipitate intensity was divided by the intensity of the respective protein lysate band on the same membrane to yield the percentage precipitated for each condition. Total values in each experimental replicate were set to 100% in order to control for exposure times between images.
Immunofluorescence (IF) – hTert RPE-1 cells grown on glass coverslips were fixed with 4% PFA and washed with Dulbecco's PBS (DPBS), permeabilized with 0.05% (v/v) Triton-X100 and blocked with 5% normal donkey serum. Primary antibodies were incubated sequentially for 1 hour each with anti-Jade1 used first. Secondary antibodies were incubated for 30 minutes. Coverslips were washed with DPBS between each step, and extensively before mounting onto glass slides using Prolong Gold antifade reagent with DAPI (Thermofisher, Carlsbad, CA, USA) mounting medium. In situ PPI was analyzed using the Duolink® In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's protocol except where noted in the Fig. legends, in which case the following amendments were made: coverslips were washed under gentle agitation for 5×5 minutes after primary antibody incubation, 4×5 minutes after ligation, and 4×2 minutes after amplification. The amount of PLA probe solution used was reduced from 8 µl each to 6 µl each. Anti-acetylated-tubulin co-staining was achieved using Zenon Alexa Fluor 488 Mouse IgG2b Labeling Kit (Thermofisher, Carlsbad, CA, USA) according to the manufacturer's instructions after the final washing step of the Duolink protocol.
Microscopy – All fluorescent images were acquired using an Axiovert 200 microscope (objectives: Plan Apochromat ×20/0.8, C-Apochromat ×63/1.22 W) equipped with an axiocam MRm and apotome system (Carl Zeiss MicroImaging, Jena, Germany). Images were captured using Axiovision 4.8 (Carl Zeiss MicroImaging, Jena, Germany). Bright-field images were taken of living cells in cell culture dishes using a JuLI Smart fluorescent cell analyzer microscope (Digital-Bio/NanoEn Tek, Seoul, Korea).
Proliferation Assay – Cells were seeded at 200,000 cells / 10 cm culture dish and trypsinised at 48 and 96 hours post-seeding. Trypsinised cells were resuspended in a total volume of 10 ml culture medium and a 50 µl sample was removed for cell counting using a CASY® Cell Counter and Analyzer System (Model TT, Roche Innovatis AG, Basel, Switzerland). After the sample for counting was removed, the resuspended cells were left in the same culture dish to continue to proliferate. Results were scored as the percentage increase from the 48 hour to 96 hour measurements.
Flow Cytometry – Flp-in NIH-3T3 cell lines were harvested in StemPro® Accutase® Cell Dissociation Reagent (Thermofisher, Darmstadt, Germany) and washed with 2 × 5 ml BSA (1% w/v). The cell pellet was resuspended in 100 µl sodium chloride (0.9% w/v) and added dropwise while vortexing to 5 ml ice cold methanol. The cell suspension was incubated for 45 minutes on ice and centrifuged at 240 × g for 5 minutes at 4°C. The cell pellet resuspended in 1 ml of a 2N hydrogen chloride solution containing 0.5% (v/v) Triton X-100 and incubated for 40 minutes at room temperature, after which time cells were pelleted at 100 × g for 5 minutes at 4°C and resuspended in 500 µl of propidium iodide (PI) solution (0.38 M sodium citrate, 0.75 mM PtdIns, 0.73 mM RNAse A, pH 7.0), incubated for 30 minutes at room temperate and washed twice with PBS. PI intensity was analyzed using BD FACSCanto I with FACS Diva software (BD Biosciences, San Jose, CA, USA). Percentage of cells with DNA amounts corresponding to specific cell cycle phases were analyzed using Cyflogic software (version 1.2.1, CyFlo Ltd, Turku, Finland).
Statistical Analyses – Differences were assessed by repeated measures ANOVA with Tukey's post-hoc analyses. Data for all graphs are presented as mean +/− standard error of the mean (s.e.m.).
Mass Spectrometry and Phosphoproteomic Analysis – Phosphoproteomic analysis of whole cell lysates was performed as previously described with modifications.56,57 Harvested HEK 293T cells were lysed in a buffer containing 8 M urea and 50 mM ammonium bicarbonate including 1x Pierce protease and phosphatase inhibitor cocktail (Thermo Scientific, Waltham, MA, USA). 400 µg of total protein (determined with a commercial BCA assay kit) was reduced using DTT and alkylated using Iodoacetamide as previously described and peptides were digested using trypsin at a w:w ratio of 1:50 at 37°C for 16 hours. After desalting procedure, phosphopeptides were enriched using Fe-NTA immobilised metal affinity chromatography columns (Thermo Scientific, Waltham, MA, USA). Samples were cleaned and desalted using stage tip.58 Cleaned peptides were analyzed using an LTQ Orbitrap Discovery mass spectrometer (Thermo Scientific, Waltham, MA, USA) coupled to a Proxeon EASY-nLC II nano-LC system (Proxeon/Thermo Scientific, Waltham, MA, USA). Peptides were separated on a 15 cm, 75 µm reverse phase column (Proxeon/Thermo Scientific, Waltham, MA, USA). Gradient elution was performed from 10 to 40% acetonitrile within 90 minutes at a flow rate of 250 nl/min. Intact peptides were detected at a resolution of 30,000 in the mass-to-charge (m/z) range 200–2000 (MS). Internal calibration was performed with m/z 445.120025 as a lock mass. A top5-MS2 scan method was used. Generated raw data were searched using the SEQUEST algorithm against a recent version of the Uniprot human database (released 2012-06-01). The protein and site FDR was adjusted to be lower than 0.01 with PhosphoRS score higher than 0.8.59 Analysis and visualization of spectra were performed using the ProteomeDiscoverer software 1.4 (Thermo Scientific, Waltham, MA, USA).
Interactome Analyses – For each experimental replicate, 5 × 15 cm dishes (80% confluency, containing ˜6 × 107 cells) were used. Cells were processed as described for IP (above) with an additional sonification step of prior to first centrifugation in lysis buffer at 30% power for 7 seconds (Digital Sonifier Model 450, Branson Ultrasonics, Danbury, CT, USA). After ultra-centrifugation, the supernatant was incubated at 4°C for 2 hours with µMACS Anti-GFP MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Beads were washed on µ columns (Miltenyi Biotec, Bergisch Gladbach, Germany) 3 times with Wash Buffer 1 (50 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, and 0.05% IGEPal-CA-630) and 5 times with Wash Buffer 2 (50 mM Tris pH 7.5 and 150 mM NaCl). This procedure was modified from the protocol by Hubner and Mann.60 Purified protein complexes were then eluted nonspecifically by direct in-column digestion with trypsin as previously described.21 Digestion was stopped the next day by adding 1% trifluoracetic acid. Label-free pull-downs were performed as 4 biological replicates. Clean-up of peptides was performed using C18 StageTips (Sp301; Thermo Scientific, Waltham, MA, USA).58 Peptides were separated using a 60 minutes nLC-MS/MS gradient on a 50 cm C18 column (Dr. Maisch HPLC GmbH, Ammerbuch, Germany) with a column oven. Gradient buffers were buffer A, 0.1% FA and Buffer B, 80% ACN and 0.1%FA. The buffer B concentration was raised to 38% during the gradient, followed by 5 minutes in 100% buffer B. Peptides were then sprayed into a Q Exactive Plus mass spectrometer (Thermo, Bremen, Germany). The setting for peptide fragmentation on a Q Exactive plus mass spectrometer were as follows: A MS1 scan (Resolution 70.000, Scan range 200-2000 m/z) was followed by 10 MS2 scans acquired in the orbitrap (Resolution 25.000,) using HCD fractionation. Dynamic exclusion was enabled (10s).
Bioinformatic Analyses – Analysis of raw files was performed using the MaxQuant suite algorithms with the LFQ option enabled (v. One.3.0.5 or 1.5.0.1).61 Raw file spectra were searched against the mouse uniprot reference database using the target-decoy strategy (reversed database). Mass accuracy was 20 ppm in the first search and 6 ppm in the second search with enabled deisotoping option. For fragment ions, mass tolerance was 0.5 Da. Fixed modifications were carbamidomethylation of cysteines (+57 Da). Variable modification was oxidation of methionine (+16 Da) or – in case of expected phosphopeptides – phosphorylation on S,T,Y. A number of 4 modifications was allowed. Protein, peptide, and site false discovery rate (FDR) were adjusted to <0.01. All proteins identified with one or more peptides were taken for label-free quantification (LFQ) analysis (Fast LFQ option enabled). LFQ intensities for respective protein groups or site intensities were uploaded in Perseus62 and analyzed as previously described.21 Briefly, raw LFQ intensities were logarithmised and normalized by subtraction of the mean. At least 2 LFQ values per protein group needed to be present for the analysis. Contaminants and proteins resolved by posttranslational modification-sites only were removed from the dataset. To replace nonquantified values with low intensities, data imputation was performed based on normal distribution of LFQ intensities (downshift = 1.8 SD, width = 0.3). Significant interactors were determined using a 2-sample analysis (t-test). Permutation-based FDR cutoff in Perseus was used to correct for multiple testing. 2D GO enrichment was performed as previously described (FDR cutoff = 0.05).62
Supplementary Material
Abbreviations
- PHD
Plant Homeodomain Finger
- Jade-1S
Gene for Apoptosis and Differentiated Epithelia, isoform1 short
- HBO1
Histone acetyltransferase bound to ORC
- CK1α
Casein-kinase-1α
- PLK1
Polo-like kinase-1
- NPHP4
Nephronophthisis-4
- GFP
Green Fluorescent Protein
- NuRD
Nucleosome remodelling deacetylase
- PPI
Protein-protein interaction
- PBD
Polo-box domain
- HEK
Human embryonic kidney
- PLA
Proximity-Ligation-Assay
- RPE
Retinal pigment epithelium.
Acknowledgments
We thank Stefanie Keller, Ruth Herzog, Gunter Rappl, and Astrid Wilbrandt-Hennes for technical assistance and members of the laboratories for helpful discussions.
Funding
This work was supported by Deutsche Forschungsgemeinschaft Grants SCHE1562-2 (to B. S.) and SFB832 (to B. S. and T. B.). L.B. was supported by the Köln Fortune Program / Faculty of Medicine (University of Cologne). M.M.R. was supported by a UoC Postdoc grant (University of Cologne) and by the Fritz-Scheler-Stipendium (KfH Stiftung für Präventivmedizin).
References
- [1].Bonventre JV. Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J Am Soc Nephrol JASN 2003; 14 Suppl 1:S55-61; PMID:12761240; http://dx.doi.org/ 10.1097/01.ASN.0000067652.51441.21 [DOI] [PubMed] [Google Scholar]
- [2].Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell 2008; 2:284-91; PMID:18371453; http://dx.doi.org/ 10.1016/j.stem.2008.01.014 [DOI] [PubMed] [Google Scholar]
- [3].Kramann R, Kusaba T, Humphreys BD. Who regenerates the kidney tubule? Nephrol Dial Transplant Off Publ Eur Dial Transpl Assoc - Eur Ren Assoc 2015; 30:903-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD. Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci U S A 2014; 111:1527-32; PMID:24127583; http://dx.doi.org/ 10.1073/pnas.1310653110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Smeets B, Boor P, Dijkman H, Sharma SV, Jirak P, Mooren F, Berger K, Bornemann J, Gelman IH, Floege J, et al.. Proximal tubular cells contain a phenotypically distinct, scattered cell population involved in tubular regeneration. J Pathol 2013; 229:645-59; PMID:23124355; http://dx.doi.org/ 10.1002/path.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vogetseder A, Palan T, Bacic D, Kaissling B, Le Hir M. Proximal tubular epithelial cells are generated by division of differentiated cells in the healthy kidney. Am J Physiol Cell Physiol 2007; 292:C807-13; PMID:16987990; http://dx.doi.org/ 10.1152/ajpcell.00301.2006 [DOI] [PubMed] [Google Scholar]
- [7].Ishibe S, Cantley LG. Epithelial-mesenchymal-epithelial cycling in kidney repair. Curr Opin Nephrol Hypertens 2008; 17:379-85; PMID:18660674; http://dx.doi.org/ 10.1097/MNH.0b013e3283046507 [DOI] [PubMed] [Google Scholar]
- [8].Lancaster MA, Gleeson JG. Cystic kidney disease: the role of Wnt signaling. Trends Mol Med 2010; 16:349-60; PMID:20576469; http://dx.doi.org/ 10.1016/j.molmed.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thomasova D, Anders H-J. Cell cycle control in the kidney. Nephrol Dial Transplant Off Publ Eur Dial Transpl Assoc - Eur Ren Assoc 2015; 30:1622-1630. [DOI] [PubMed] [Google Scholar]
- [10].Zhou MI, Wang H, Ross JJ, Kuzmin I, Xu C, Cohen HT. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J Biol Chem 2002; 277:39887-98; PMID:12169691; http://dx.doi.org/ 10.1074/jbc.M205040200 [DOI] [PubMed] [Google Scholar]
- [11].Panchenko MV, Zhou MI, Cohen HT. von Hippel-Lindau partner Jade-1 is a transcriptional co-activator associated with histone acetyltransferase activity. J Biol Chem 2004; 279:56032-41; PMID:15502158; http://dx.doi.org/ 10.1074/jbc.M410487200 [DOI] [PubMed] [Google Scholar]
- [12].Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Côté J, Panchenko MV. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem 2008; 283:28817-26; PMID:18684714; http://dx.doi.org/ 10.1074/jbc.M801407200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Siriwardana NS, Meyer R, Havasi A, Dominguez I, Panchenko MV. Cell cycle-dependent chromatin shuttling of HBO1-JADE1 histone acetyl transferase (HAT) complex. Cell Cycle Georget Tex 2014; 13:1885-901; http://dx.doi.org/ 10.4161/cc.28759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Havasi A, Haegele JA, Gall JM, Blackmon S, Ichimura T, Bonegio RG, Panchenko MV. Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am J Pathol 2013; 182:152-62; PMID:23159946; http://dx.doi.org/ 10.1016/j.ajpath.2012.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wan G, Hu X, Liu Y, Han C, Sood AK, Calin GA, Zhang X, Lu X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J 2013; 32:2833-47; PMID:24097061; http://dx.doi.org/ 10.1038/emboj.2013.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, et al.. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 2008; 10:1208-16; PMID:18806787; http://dx.doi.org/ 10.1038/ncb1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Borgal L, Habbig S, Hatzold J, Liebau MC, Dafinger C, Sacarea I, Hammerschmidt M, Benzing T, Schermer B. The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate β-catenin signaling. J Biol Chem 2012; 287:25370-80; PMID:22654112; http://dx.doi.org/ 10.1074/jbc.M112.385658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen M-H, Chuang P-T, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol 2008; 10:70-6; PMID:18084282; http://dx.doi.org/ 10.1038/ncb1670 [DOI] [PubMed] [Google Scholar]
- [19].Gerdes JM, Liu Y, Zaghloul NA, Leitch CC, Lawson SS, Kato M, Beachy PA, Beales PL, DeMartino GN, Fisher S, et al.. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet 2007; 39:1350-60; PMID:17906624; http://dx.doi.org/ 10.1038/ng.2007.12 [DOI] [PubMed] [Google Scholar]
- [20].Borgal L, Rinschen MM, Dafinger C, Hoff S, Reinert MJ, Lamkemeyer T, Lienkamp SS, Benzing T, Schermer B. Casein Kinase 1 Alpha Phosphorylates the Wnt-Regulator Jade-1 and Modulates its Activity. J Biol Chem 2014; 289(38):26344-56; PMID:25100726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kohli P, Bartram MP, Habbig S, Pahmeyer C, Lamkemeyer T, Benzing T, Schermer B, Rinschen MM. Label-free quantitative proteomic analysis of the YAP/TAZ interactome. Am J Physiol Cell Physiol 2014; 306:C805-18; PMID:24573087; http://dx.doi.org/ 10.1152/ajpcell.00339.2013 [DOI] [PubMed] [Google Scholar]
- [22].O'Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 1991; 251:1351-5; http://dx.doi.org/ 10.1126/science.1900642 [DOI] [PubMed] [Google Scholar]
- [23].Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J Biol Chem 2003; 278:25277-80; PMID:12738781; http://dx.doi.org/ 10.1074/jbc.C300126200 [DOI] [PubMed] [Google Scholar]
- [24].Elia AEH, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 2003; 299:1228-31; PMID:12595692; http://dx.doi.org/ 10.1126/science.1079079 [DOI] [PubMed] [Google Scholar]
- [25].Liu Z, Ren J, Cao J, He J, Yao X, Jin C, Xue Y. Systematic analysis of the Plk-mediated phosphoregulation in eukaryotes. Brief Bioinform 2013; 14:344-60; PMID:22851512; http://dx.doi.org/ 10.1093/bib/bbs041 [DOI] [PubMed] [Google Scholar]
- [26].Lee KS, Yuan YL, Kuriyama R, Erikson RL. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol 1995; 15:7143-51; PMID:8524282; http://dx.doi.org/ 10.1128/MCB.15.12.7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, et al.. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 2006; 3:995-1000; PMID:17072308; http://dx.doi.org/ 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- [28].Tang X, He J, Partin J, Vafai A. Comparative analysis of direct fluorescence, Zenon labeling, and quantum dot nanocrystal technology in immunofluorescence staining. J Immunoassay Immunochem 2010; 31:250-7; PMID:20623410; http://dx.doi.org/ 10.1080/107391-49.2010.488620 [DOI] [PubMed] [Google Scholar]
- [29].Calvi BR. HBO1:JADE1 at the cell cycle chromatin crossroads. Cell Cycle Georget Tex 2014; 13:2322; http://dx.doi.org/ 10.4161/cc.29832 [DOI] [PubMed] [Google Scholar]
- [30].Zhang Y. Biology of the Mi-2/NuRD Complex in SLAC (Stemness, Longevity/Ageing, and Cancer). Gene Regul Syst Biol 2011; 5:1-26; http://dx.doi.org/ 10.4137/GRSB.S6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 1998; 395:917-21; PMID:9804427; http://dx.doi.org/ 10.1038/27699 [DOI] [PubMed] [Google Scholar]
- [32].Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a Novel Complex with Both ATP-Dependent Chromatin-Remodeling and Histone Deacetylase Activities. Mol Cell 1998; 2:851-61; PMID:9885572; http://dx.doi.org/ 10.1016/S1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- [33].Mikhailov A, Shinohara M, Rieder CL. Topoisomerase II and histone deacetylase inhibitors delay the G2/M transition by triggering the p38 MAPK checkpoint pathway. J Cell Biol 2004; 166:517-26; PMID:15302851; http://dx.doi.org/ 10.1083/jcb.200405167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Siriwardana NS, Meyer RD, Panchenko MV. The novel function of JADE1S in cytokinesis of epithelial cells. Cell Cycle Georget Tex 2015; 14:2821-34; http://dx.doi.org/ 10.1080/15384101.2015.1068476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry A-J, Côté V, et al.. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 2009; 33:257-65; PMID:19187766; http://dx.doi.org/ 10.1016/j.molcel.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou MI, Foy RL, Chitalia VC, Zhao J, Panchenko MV, Wang H, Cohen HT. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc Natl Acad Sci U S A 2005; 102:11035-40; PMID:16046545; http://dx.doi.org/ 10.1073/pnas.0500757102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lei M, Erikson RL. Plk1 depletion in nontransformed diploid cells activates the DNA-damage checkpoint. Oncogene 2008; 27:3935-43; PMID:18297112; http://dx.doi.org/ 10.1038/onc.2008.36 [DOI] [PubMed] [Google Scholar]
- [38].Ebrahimi S, Fraval H, Murray M, Saint R, Gregory SL. Polo kinase interacts with RacGAP50C and is required to localize the cytokinesis initiation complex. J Biol Chem 2010; 285:28667-73; PMID:20628062; http://dx.doi.org/ 10.1074/jbc.M110.103887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol 1995; 129:1617-28; PMID:7790358; http://dx.doi.org/ 10.1083/jcb.129.6.1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Petronczki M, Lénárt P, Peters J-M. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell 2008; 14:646-59; PMID:18477449; http://dx.doi.org/ 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- [41].van Vugt MATM, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene 2005; 24:2844-59; PMID:15838519; http://dx.doi.org/ 10.1038/sj.onc.1208617 [DOI] [PubMed] [Google Scholar]
- [42].Takaki T, Trenz K, Costanzo V, Petronczki M. Polo-like kinase 1 reaches beyond mitosis–cytokinesis, DNA damage response, and development. Curr Opin Cell Biol 2008; 20:650-60; PMID:19000759; http://dx.doi.org/ 10.1016/j.ceb.2008.10.005 [DOI] [PubMed] [Google Scholar]
- [43].Bowman BM, Sebolt KA, Hoff BA, Boes JL, Daniels DL, Heist KA, Galbán CJ, Patel RM, Zhang J, Beer DG, et al.. Phosphorylation of FADD by the kinase CK1α promotes KRASG12D-induced lung cancer. Sci Signal 2015; 8:ra9; PMID:25628462; http://dx.doi.org/ 10.1126/scisignal.2005607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Macůrek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 2008; 455:119-23; PMID:18615013; http://dx.doi.org/ 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- [45].Seki A, Coppinger JA, Jang C-Y, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 2008; 320:1655-8; PMID:18566290; http://dx.doi.org/ 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Seong KM, Nam SY, Kim J-Y, Yang KH, An S, Jin Y-W, Kim CS. TOPORS modulates H2AX discriminating genotoxic stresses. J Biochem Mol Toxicol 2012; 26:429-38; PMID:22972498; http://dx.doi.org/ 10.1002/jbt.21438 [DOI] [PubMed] [Google Scholar]
- [47].Wu Z-Q, Liu X. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci U S A 2008; 105:1919-24; PMID:18250300; http://dx.doi.org/ 10.1073/pnas.0712063105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yang X, Li H, Deng A, Liu X. Plk1 phosphorylation of Topors is involved in its degradation. Mol Biol Rep 2010; 37:3023-8; PMID:19821153; http://dx.doi.org/ 10.1007/s11033-009-9871-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hyun S-Y, Hwang H-I, Hwan H-I, Jang Y-J. Polo-like kinase-1 in DNA damage response. BMB Rep 2014; 47:249-55; PMID:24667170; http://dx.doi.org/ 10.5483/BMBRep.2014.47.5.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].van Vugt MATM, Brás A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell 2004; 15:799-811; PMID:15350223; http://dx.doi.org/ 10.1016/j.molcel.2004.07.015 [DOI] [PubMed] [Google Scholar]
- [51].Brockman JL, Gross SD, Sussman MR, Anderson RA. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc Natl Acad Sci 1992; 89:9454-8; PMID:1409656; http://dx.doi.org/ 10.1073/pnas.89.20.9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Honaker Y, Piwnica-Worms H. Casein kinase 1 functions as both penultimate and ultimate kinase in regulating Cdc25A destruction. Oncogene 2010; 29:3324-34; PMID:20348946; http://dx.doi.org/ 10.1038/onc.2010.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nguyen HQ, Nye J, Buster DW, Klebba JE, Rogers GC, Bosco G. Drosophila casein kinase I alpha regulates homolog pairing and genome organization by modulating condensin II subunit Cap-H2 levels. PLoS Genet 2015; 11:e1005014; PMID:25723539; http://dx.doi.org/ 10.1371/journal.pgen.1005014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Benzing T, Gerke P, Höpker K, Hildebrandt F, Kim E, Walz G. Nephrocystin interacts with Pyk2, p130(Cas), and tensin and triggers phosphorylation of Pyk2. Proc Natl Acad Sci U S A 2001; 98:9784-9; PMID:11493697; http://dx.doi.org/ 10.1073/pnas.171269898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schermer B, Höpker K, Omran H, Ghenoiu C, Fliegauf M, Fekete A, Horvath J, Köttgen M, Hackl M, Zschiedrich S, et al.. Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J 2005; 24:4415-24; PMID:16308564; http://dx.doi.org/ 10.1038/sj.emboj.7600885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rinschen MM, Yu M-J, Wang G, Boja ES, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor–dependent signaling pathways in renal collecting duct cells. Proc Natl Acad Sci U S A 2010; 107:3882-7; PMID:20139300; http://dx.doi.org/ 10.1073/pnas.0910646107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rinschen MM, Wu X, König T, Pisitkun T, Hagmann H, Pahmeyer C, Lamkemeyer T, Kohli P, Schnell N, Schermer B, et al.. Phosphoproteomic Analysis Reveals Regulatory Mechanisms at the Kidney Filtration Barrier. J Am Soc Nephrol JASN 2014; 25(7):1509-22; PMID: 24511133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003; 75:663-70; PMID:12585499; http://dx.doi.org/ 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- [59].Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 2011; 10:5354-62; PMID:22073976; http://dx.doi.org/ 10.1021/pr200611n [DOI] [PubMed] [Google Scholar]
- [60].Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods San Diego Calif 2011; 53:453-9; http://dx.doi.org/ 10.1016/j.ymeth.2010.12.016 [DOI] [PubMed] [Google Scholar]
- [61].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367-72; PMID:19029910 [DOI] [PubMed] [Google Scholar]
- [62].Cox J, Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 2012; 13 Suppl 16:S12; http://dx.doi.org/ 10.1186/1471-2105-13-S16-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.