ABSTRACT
All organisms ensure once and only once replication during S phase through a process called replication licensing. Cdt1 is a key component and crucial loading factor of Mcm complex, which is a central component for the eukaryotic replicative helicase. In higher eukaryotes, timely inhibition of Cdt1 by Geminin is essential to prevent rereplication. Here, we address the mechanism of DNA licensing using purified Cdt1, Mcm and Geminin proteins in combination with replication in Xenopus egg extracts. We mutagenized the 223th arginine of mouse Cdt1 (mCdt1) to cysteine or serine (R-S or R-C, respectively) and 342nd and 346th arginines constituting an arginine finger-like structure to alanine (RR-AA). The RR-AA mutant of Cdt1 could not only rescue the DNA replication activity in Cdt1-depleted extracts but also its specific activity for DNA replication and licensing was significantly increased compared to the wild-type protein. In contrast, the R223 mutants were partially defective in rescue of DNA replication and licensing. Biochemical analyses of these mutant Cdt1 proteins indicated that the RR-AA mutation disabled its functional interaction with Geminin, while R223 mutations resulted in ablation in interaction with the Mcm2∼7 complex. Intriguingly, the R223 mutants are more susceptible to the phosphorylation-induced inactivation or chromatin dissociation. Our results show that conserved arginine residues play critical roles in interaction with Geminin and Mcm that are crucial for proper conformation of the complexes and its licensing activity.
KEYWORDS: Cdc2/Cyclin B, Cdc7 kinase, Cdk2/Cyclin A, Cdt1, cell cycle, DNA helicase, DNA replication, DNA-binding activity, licensing, Mcm
Introduction
During the course of DNA replication, multiprotein complexes, called pre-replicative complexes (pre-RCs), containing Orc1–6 (origin recognition complex 1–6), Cdc6, Cdt1 and Mcm2∼7 (minichromosome maintenance 2∼7), need to be sequentially assembled to establish ‘licensed’ origins.1,2 Cdt1 plays a critical role in the licensing process to load the hexameric Mcm2∼7 complex (an eukaryotic replicative helicase) onto chromatin during G1 phase. At the onset of S phase, Cdt1 is rendered inactive through ubiquitin-dependent proteolysis in a CDK-dependent and PCNA (proliferating cell nuclear antigen)-dependent manners3-7 and through the inhibitor Geminin.8 Geminin directly binds to Cdt1 and prevents the loading of the Mcm2∼7 complex onto DNA.8-12 This double regulation ensures that Cdt1 is not present during S phase, resulting in prevention of DNA re-replication.
To strictly enforce once per cell cycle replication, it is necessary to coordinate the temporal and spatial regulation of licensing. The level of Cdt1 protein oscillates during cell cycle (high in the G1 phase, low in S phase, and high again at the M-G1 transition).13 Geminin accumulates during S, G2, and M phases, and is destabilized during G1 phase of the cell cycle.8,9 It has been proposed that Cdt1 associates dynamically with chromatin throughout G1 phase and the licensing inhibitor Geminin is recruited simultaneously by Cdt1 onto chromatin, and that the stoichiometry of the Cdt1:Geminin complex could act as a molecular switch that determines when the origins of replication are on (licensing) or off (inhibition).14-17 In addition, crystallographic studies suggest the presence of 2 forms that are recruited to chromatin. One is a “permissive” 1:2 Cdt1-Geminin heterotrimer complex that is able to load Mcm; the other is an “inhibitory” 2:4 Cdt1-Geminin heterohexamer complex that is unable to engage Mcms, thus preventing new pre-RCs formation.18 Phosphorylation also regulates the function of Cdt1, since a mutant Cdt1 with a 6-amino-acid deletion, which was not efficiently phosphorylated during mitosis and was more strongly associated with chromatin, showed enhanced licensing activity.19 It is known that Cdk (cyclin-dependent kinasea) and Cdc7 kinase are involved in regulation of Cdt1.2,6,20,21
It was proposed that 2 Cdt1 molecules recruit double hexameric Mcm2∼7 complex loading during pre-RC formation.22 Previously, we reported that mouse Cdt1 forms a complex with Mcm4/6/7, Mcm2/3/4/5/6/7 and Mcm2/4/6/7 and that Cdt1 significantly stimulates the DNA binding and helicase activities of Mcm4/6/7, an active helicase complex.23 It was reported that C-terminal region of Cdt1 is required for interaction with Mcm.11 More recently, charge complementarity between the C-terminal conserved residues of Cdt1 and Mcm6 was shown to be important for the Cdt1-Mcm2∼7 interaction and licensing activity by mutagenesis analysis.23-26 However, the Cdt1-mediated Mcm2∼7 chromatin loading and licensing is more complex, and the central domain of Cdt1 was also reported to be required for interaction with Mcm2∼7 and licensing activity.17,27 Thus, it remains controversial how Cdt1 acts during Mcm loading.
Alignment of Cdt1 sequences revealed that Cdt1 R223 and arginine finger-like residues (R342, 346) are well conserved in human, mouse, Xenopus, and Drosophila. A mutant of Drosophila Cdt1 containing cystine substitution at this conserved R223 exhibits female-sterile phenotype, suggesting that it is functionally important.28 We mutagenized the corresponding arginine (R223) of mouse Cdt1 to cystine or serine and arginine finger-like residues (R342, 346) to alanine. We report here that mutations of these 3 arginine residues (R-S, R-C and RR-AA) profoundly affect DNA replication, pre-RC formation, interaction with Mcm and Geminin and phosphorylation, suggesting they are critical for licensing activity of Cdt1.
Results
Construction and purification of mutant Cdt1 proteins with amino acid substitutions at conserved arginine residues
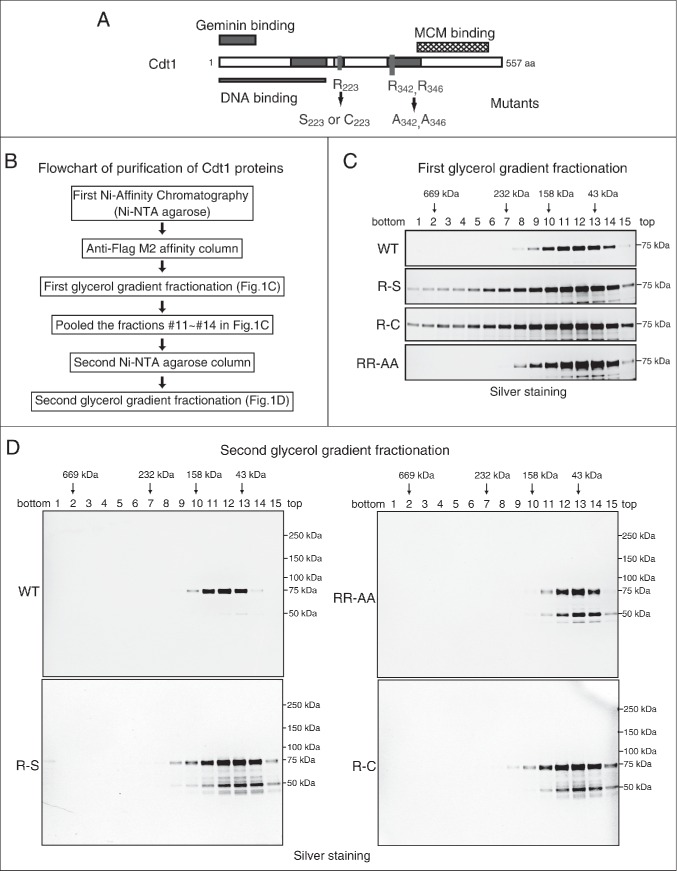
We previously reported that the mouse Cdt1 forms complexes with both Mcm2∼7 and hexameric Mcm4/6/7 through direct interaction with Mcm2 and Mcm6 subunit.23 We have aligned the sequences of Cdt1 from different species (human, mouse, Xenopus, and Drosophila) and have identified 3 conserved arginine residues (R223, R342 and R346) (Fig. S1). A Drosophila mutant carrying arginine to cysteine amino acid substitution at R223 exhibits female-sterile phenotype, suggesting functional importance of this residue.28 Furthermore, alignment with E. coli DnaA protein revealed similarity of the sequences around R342 and R346 of Cdt1 with those around Arg281 and Arg285 of DnaA (Fig. S1B). Both arginine residues are required for DnaA functions and Arg285 residue constitutes the arginine finger.29 Thus, we mutagenized R223 of mouse Cdt1 to cystine or serine and R342/R346 to alanine (Fig. 1A). We have realized that the plasmids expressing the wild-type and mutant Cdt1 inadvertently contained the following mutations; Ile24->Thr24 in wild-type and RR-AA, and Pro61-> Ser61 in R-S/R-C. These amino acid changes occur at non-conserved residues and it is more likely that they do not affect the functions of Cdt1, although we cannot completely rule out the possibility that they may affect the functions in some unknown way.
Figure 1.
Purification of various recombinant Cdt1 proteins. (A) Schematic representation of Cdt1 protein showing the Geminin- and Mcm-binding motifs, and the mutagenized arginines. The two regions of Cdt1 required for association with Geminin are indicated by gray boxes. The C-terminal region of Cdt1 required for interact with MCM is indicated by a cross-hatched box. (B) Diagram explaining the procedure of purification of Cdt1 protein. (C) Wild-type or various mutant forms of Cdt1 protein, expressed in insect cells, were purified by Ni-NTA and anti-Flag antibody beads, and then were fractionated on 15-35% glycerol gradient (38,000 rpm for 17 hr). (D) After first glycerol gradient fractionation, the fractions from 10 to 14 of mCdt1 were pooled and further purified with Ni-NTA column and glycerol gradient fractionation. Each fraction was subjected to SDS-PAGE (5–20% polyacrylamide), followed by staining with silver. Degradation products (˜50 kDa) were co-purified with the full-length proteins in case of the mutant proteins.
We expressed wild-type and mutant Cdt1 proteins using the baculo-virus expression system and purified them through the consecutive steps involving nickel agarose affinity chromatography, anti-Flag M2 antibody-agarose affinity chromatography, and glycerol gradient sedimentation (Fig. 1B). The purified Cdt1 proteins appear to be almost homogeneous in silver staining (Fig. 1C). However, in helicase assays, contaminated nucleases were detected which specifically degraded the single stranded DNA in the fractions near the bottom (Fig. S2C, lower). Further purification by the mono-Q column also could not remove the nuclease (Fig. S2A and S2B). The contaminating nuclease prohibited accurate measurement of the helicase activity (Fig. S2B and S2C, middle). Thus, Cdt1 proteins were further purified by applying the first glycerol gradient fractions (11 to 14) to the second Ni-sepharose column and glycerol gradient fractionation (Fig. 1D). As a result, we were able to obtain fractions virtually free from contaminating nucleases. All the mutants Cdt1 migrated at around 60 kDa (monomer position) in glycerol gradient as the wild-type protein, although the peak fraction of the mutants was smaller than the wild-type by one fraction. Thus, the results suggest that the mutation did not significantly affect the size or the shape of the protein.
Licensing and DNA replication activities of mutant Cdt1
In order to evaluate the licensing and DNA replication activities of Cdt1, the wild type and mutated Cdt1 proteins were assayed for their ability to restore the replication and Mcm-loading activities in Xenopus egg extract immunodepleted of endogenous Xenopus Cdt1 (xCdt1). Depletion of endogenous xCdt1 led to loss of Mcm loading, whereas chromatin loading of Orc and Cdc6 proteins increased (Fig. 2A, lane 3).30 Addition of the wild-type Cdt1 protein to the Cdt1-depleted extracts restored Mcm2 loading in a dose-dependent manner (Fig. 2A, lanes 4–6). However, R-S and R-C mutants showed significantly impaired Mcm2 loading activity (Fig. 2A, lanes 7–12). In contrast, the RR-AA mutant restored the Mcm2 loading more efficiently than the wild-type; chromatin loading was significantly restored even at 0.6 nM by RR-AA but not by the wild-type (Fig. 2A, lanes 6 and 15). The chromatin binding of PCNA, DNA polymerase α and DNA polymerase ϵ at 75 min was restored 0.6 nM by RR-AA more efficiently than by the wild-type (Fig. 2A, lanes 21 and 30), consistent with the effect on Mcm loading. These results suggest that these arginine residues of Cdt1 play critical roles in its licensing activity.
Figure 2.
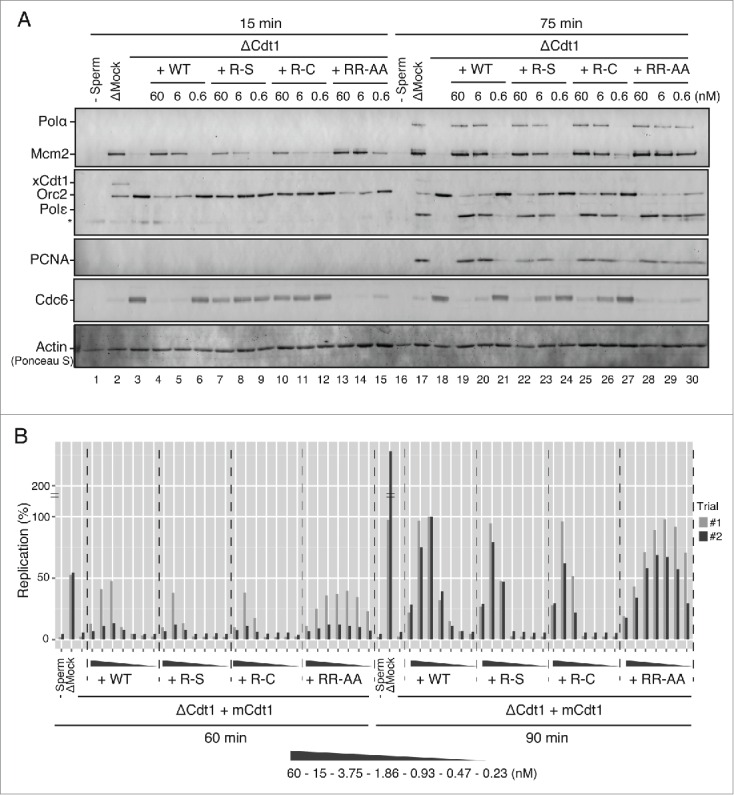
Licensing and replication activities of various mutant Cdt1 proteins. (A) Western blot analysis of Mcm2, Orc2, Cdc6, Cdt1, pol α, pol ϵ, and PCNA on chromatin in the egg extract at 15 min and 75 min. S-phase entry occurs at about 40 min under the condition employed. The extracts were either mock-depleted or Cdt1-depleted in combination with buffer or wild-type/mutant Cdt1 proteins (60, 6 and 0.6 nM). Asterisk indicates a non-specific band. Actin bands stained with Ponceau S are shown as loading control to show chromatin recovery. (B) DNA replication at 60 min and 90 min in either mock-depleted extract or Cdt1-depleted extract supplemented with buffer or wild-type/mutant Cdt1 in the range of 60 nM to 0.23 nM. The replication efficiency is shown by setting the value of [α-32P]dATP incorporation in the presence of WT mCdt1 at 90 min as 100%. Two independent experiments were carried out with very similar results.
The differential efficacy of mutant Cdt1 proteins in Mcm-loading was recapitulated also in DNA replication assay. A Cdt1-depleted extract did not replicate sperm chromatin, but DNA replication could be restored by addition of the wild-type Cdt1 protein (Fig. 2B). Consistent with previous study,31 Cdt1 has an optimum concentration for DNA replication; sufficient DNA replication was not observed at a low concentration due to inefficient Mcm loading, whereas the elongation of nascent DNA strand is inhibited at a higher concentration. Cdt1 added at the concentration from 3.7 to 15 nM was optimum for DNA replication. In contrast, R-S and R-C mutants showed impaired DNA replication at the concentration below 3.75 nM, consistent with its weak licensing activity. In contrast, the RR-AA mutant stimulated DNA replication more efficiently than the wild type, especially at low concentrations (1.86–0.23 nM) (Fig. 2B). These results indicate that the arginine finger-like mutant of Cdt1 has more potent licensing and DNA replication activities than the wild-type but that the R223 mutations decrease these activities.
We next evaluated the role of Geminin in functions of these Cdt1 mutants by codepleting Geminin. To inhibit the licensing activity of endogenous xCdt1, approximately 100 nM of Geminin was required.8,30,32,33 In mock depletion, Xenopus egg extract was supplemented with sufficient recombinant Geminin (400 nM) to inhibit Mcm loading onto the chromatin (Fig. 3A, compare lanes 1 and 2). An equal amount of Cdt1 protein (wild-type or mutant) was added to Cdt1 and Geminin double-depleted extract (Fig. 3A, lanes 4, 6, 8 and 10). We found that RR-AA could load Mcm more efficiently than the wild type Cdt1 in the absence of added Geminin, whereas the R-S or R-C mutant could not. The presence of recombinant Geminin did not completely inhibit the licensing activity of mutant Cdt1 RR-AA (Fig. 3A, compare lanes 5 and 11). Titration of Geminin in Cdt1-depleted extracts indicate that RR-AA could load Mcm on chromatin even at a high concentration of Geminin (120 nM) (Fig. 3B, lanes 11–16), while Mcm loading was inhibited at 40 nM of added Geminin with the wild-type Cdt1 (Fig. 3B, lanes 4-9). Geminin added at a concentration as high as 600 nM, under which the Mcm loading was completely inhibited in the egg extracts, did not completely suppress Mcm loading in the presence of the RR-AA mutant Cdt1 (Fig. 3C, lane10). This result suggests that RR-AA mutant is refractory to the licensing inhibition by Geminin. To evaluate whether the elevated licensing activity of the RR-AA mutant can be explained by its resistance to Geminin, we carefully compared the licensing activity of wild-type and the mutant in the absence of Geminin. In xCdt1 and Geminin double-depleted extract, RR-AA activated the Mcm2 loading significantly more effectively than the wild type at high (10 nM) and low (3.3 nM) concentrations (Fig. 3C, lanes 14–25; 3 independent experiments. See also Fig. S3). Since an additional mutation in RR-AA and the wild-type is the same, the altered function in RR-AA is most likely mediated by the amino acid changes at the 2 arginine residues. This result suggests that the arginine finger-like mutant of Cdt1 can exert hyper-licensing activity even in a Geminin-independent manner.
Figure 3.
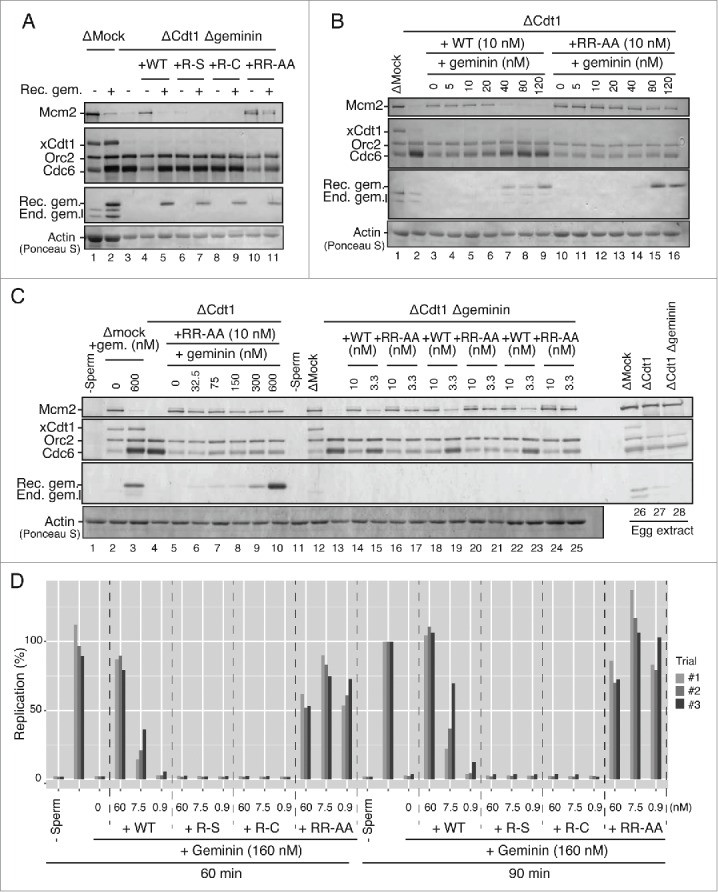
Licensing and replication activities of various mutant Cdt1 proteins in the presence of Geminin. (A) Western blot analysis of Mcm2, Orc2, Cdc6, Cdt1, and Geminin on chromatin in the egg extract that was either mock-depleted or Cdt1/Geminin double-depleted in the presence of 6 nM wild-type or mutant Cdt1 (R-S, R-C, and RR-AA) proteins with (+) or without (−) Geminin (400 nM). (B) Western blot analysis of indicated proteins in the Cdt1-depleted extract supplemented with 10 nM wild-type or RR-AA Cdt1 protein and Geminin protein at the indicated concentrations. (C) Western blot analysis of indicated proteins in the Cdt1-depleted extract in the presence of various amounts of Geminin (lanes 4–10) or Cdt1/Geminin double-depleted extract supplemented with 10 and 3.3 nM wild-type or RR-AA proteins (lanes 13–25; 3 independent experiments). The licensing activity of the mutant Cdt1/RR-AA is not significantly inhibited by addition of Geminin at a very high concentration. (D) The ability of the wild-type and mutant Cdt1 to restore DNA replication that has been inhibited by an excess Geminin protein was examined. Xenopus egg extract was supplemented with 160 nM Geminin, which resulted in complete inhibition of DNA replication. Then wild-type or mutant Cdt1 proteins were added at the indicated concentrations and incubated in 60 and 90 min, respectively. The replication efficiency is shown by setting the value of [α-32P]dATP incorporation in the absence of mCdt1 and Geminin at 90 min as 100.
The licensing activities of mutant Cdt1 proteins were further examined by more carefully analyzing their ability to counteract the inhibition of DNA replication by Geminin. Addition of Geminin (160 nM) completely inhibited DNA replication (Fig. 3D). Under this condition, addition of the wild-type Cdt1 at 60 nM restored DNA replication activity to the level without Geminin, while only a low or very little DNA replication was observed in the presence of the wild-type Cdt1 at 7.5 nM or 0.9 nM, respectively. On the other hand, R-S and R-C Cdt1 could not rescue the DNA replication even at 60 nM. In contrast, efficient DNA replication was observed with RR-AA even at 0.9 nM. Taken together, these results are consistent with the conclusion that the R-S and R-C mutants are significantly impaired in licensing and replication activities, whereas RR-AA is hyperactive. The latter mutant appears to be insensitive to inhibition by Geminin, but is also able to stimulate the licensing in a manner independent of Geminin. We conclude that all the 3 arginine residues of Cdt1 play important roles in both licensing and replication.
Stimulation of helicase and DNA-binding activities of Mcm by Cdt1 protein
We previously reported that Cdt1 stimulates Mcm helicase.23 In order to understand how the arginine mutants affect the licensing and replication activities, we next examined the mutant Cdt1 proteins in DNA helicase assays using Mcm4/6/7. We first confirmed that the stimulation of Mcm4/6/7 helicase activity is correlated with the Cdt1 protein amount in glycerol gradient fractions (Fig. 4A). The intensities of the full-length bands in fractions #13 of the wild-type and mutant Cdt1 proteins in glycerol gradient fractionation were quantified and an equal amount (as a full-length polypeptide) was used for helicase and DNA binding assays of the Mcm4/6/7 complex. As shown in Figure 4B, all the Cdt1 mutants (R-S, R-C and RR-AA) have exhibited similar extent of stimulation of both Mcm4/6/7 helicase and single stranded DNA-binding activities (Fig. 4B, left and right; Fig. S4A). Since Cdt1 alone could not form clear protein-DNA complexes in gel-shift assays under this condition (Fig. 4B right, lanes 15–18), the biotin-labeled single stranded and forked DNAs were prepared and the pull-down assay were carried out using streptavidin beads. We show that each Cdt1 mutant protein efficiently binds to both single-stranded and forked DNA (Fig. 4C and 4D, lanes 10–13), indicating that all the mutant proteins bind to DNA with affinity similar to that of the wild-type. Nitrocellulose filter DNA binding assays using labeled single-stranded and double-stranded DNA as well as the single-stranded and double-stranded DNA cellulose binding assays also showed that DNA-binding activities of the wild-type and 3 mutant Cdt1 proteins are similar (data not shown and Fig. S4B).
Figure 4.
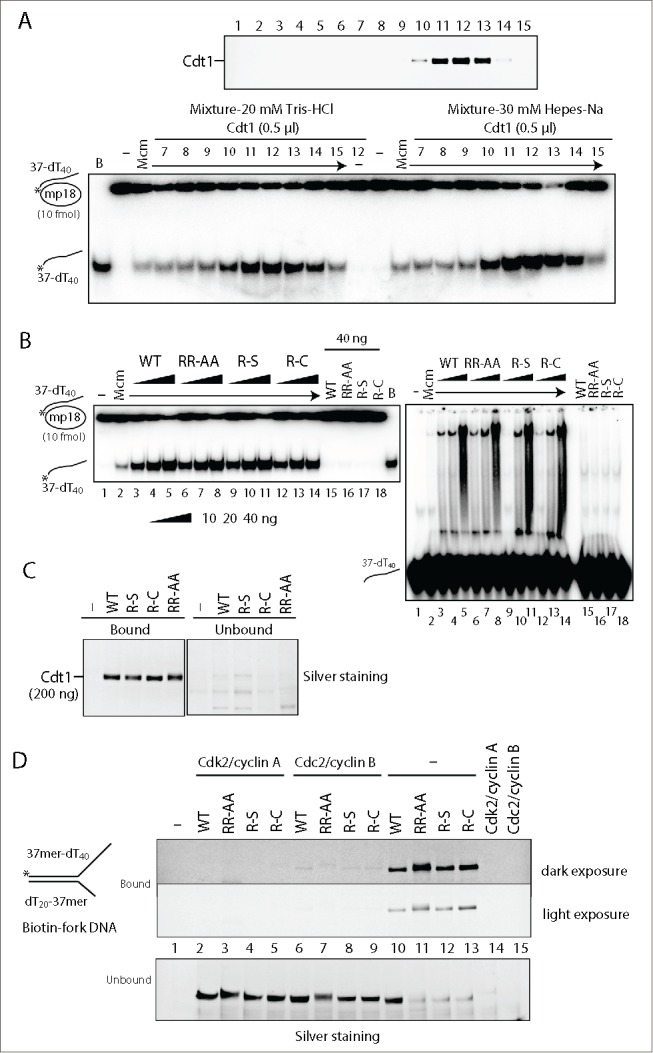
Effect of Cdt1 arginine mutations on the helicase and DNA-binding activities of Mcm. (A) The wild-type Cdt1 protein (0.5 μl) from the glycerol gradient fractions (silver staining pattern shown in the upper panel) was added to DNA helicase assays of Mcm4/6/7 (50 ng) conducted in reaction buffers containing Tris-HCl or Hepes-Na (lower panel). (B) The fractions #13 of wild-type or mutant Cdt1 protein in the second glycerol gradient fractionation were quantified and equal amounts (10, 20, and 40 ng) were added to the helicase and DNA-binding assays of Mcm4/6/7 complex (50 ng). (C and D) Pull-down assays using a biotinylated oligonucleotide. An equal amount of the wild-type or mutant Cdt1 (200 ng) was incubated with 5′-biotinylated oligonucleotide (37mer-dT40, 50 pmole) (C) or 5′-biotinylated Y-fork (37mer-dT40/dT20-37mer, 40 pmole) (D) at 30°C for 30 min. The bound proteins were collected by using streptavidin-coated M-280 magnetic, and were analyzed on 4–20% SDS-PAGE, followed by silver staining.
Defective complex formation of mutant Cdt1 (R-S and R-C) and Mcm
It has been known that the licensing involves the recruitment of Mcm2∼7 to chromatin by its direct interaction with Cdt1.11,23 We and others reported that mutations in the C-terminal domain of Cdt1 reduced or abolished its interaction with Mcm, thereby inactivating chromatin loading and DNA replication activities.23-26 Therefore, we next set out to evaluate the interaction between Cdt1 and Mcm complex. The purified Mcm2 alone, Mcm4/6/7 or Mcm2∼7 complex was mixed with various Cdt1 (containing a Flag-tag), and immunoprecipitation using anti-Flag antibody agarose beads, or anti-Cdt1 antibody coupled with protein G Dynabeads were performed. We found that the affinity of wild type or each mutant Cdt1 with Mcm2 protein was similar (Fig. 5A, upper). Similarly, interaction with Mcm4/6/7 or Mcm2∼7 was not affected by the mutation (Fig. 5A, lower panels). These results indicate that the mutant Cdt1 proteins physically interact with Mcm with similar affinity to that of the wild-type.
Figure 5.
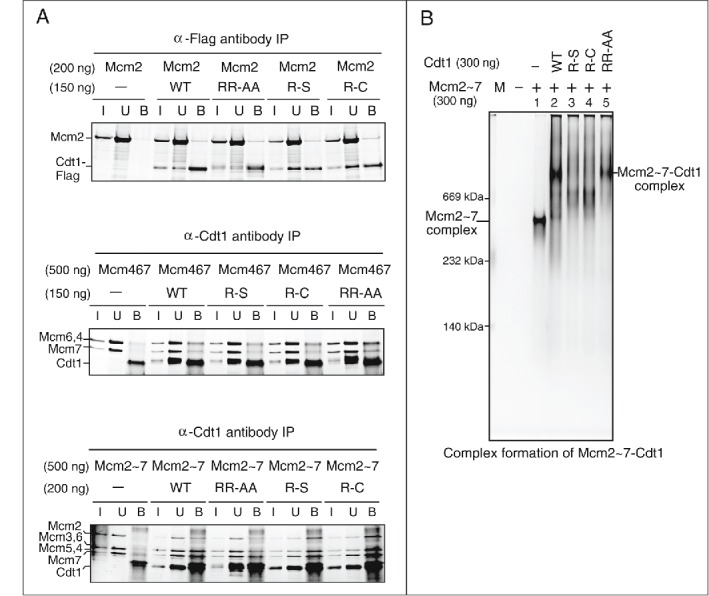
Physical interactions of Cdt1 with Mcm complexes. (A) Co-immunoprecipitation of Mcm and Cdt1. Co-immunoprecipitation assays were performed using anti-Flag M2 antibody agarose beads (upper; Mcm2), anti-Cdt1 antibody (middle and lower panels; Mcm4/6/7 or Mcm2∼7 complex) coupled with Dynabead protein G. All of them were mixed with indicated proteins in buffer, and washed 4 times after 1 hr incubation. Bound proteins were eluted with 200 μg/ml Flag peptide (upper) or 0.1 M citric acid, pH 2.2 (middle and lower). The samples were run on SDS-PAGE, followed by silver staining. I: Input (7 % of the input); U: unbound (upper, 40 % of the unbound; middle, 20 % of the unbound; lower, 25 % of the unbound); B, bound. * indicates a non-specific band. (B) Complex formation of Mcm2∼7-Cdt1 was examined on a native gel. The indicated amount of Mcm2∼7 and Cdt1 proteins were mixed and electrophoresed on a 5% native polyacrylamide gel and stained with silver.
We then analyzed the complex formation using native gel electrophoresis. Mcm2∼7 migrates as a complex about 600 kDa, consistent with it being a hexameric complex (Fig. 5B, lane 1). Upon addition of Cdt1, the band of the Mcm2∼7 complex disappears and a slow-migrating band composed presumably of Mcm2∼7 and Cdt1 was detected (Fig. 5B, lane 2). Actually, the slow-migrating band was run on a second dimension SDS-PAGE, the result of which confirmed that it consisted with Mcm2∼7 and Cdt1 (data not shown). The RR-AA mutant forms a similar complex with Mcm2∼7 (Fig. 5B, lanes 2 and 5), whereas both R-S and R-C generated bands migrating between Mcm2∼7 and Mcm2∼7-Cdt1 complexes (Fig. 5B, lanes 3–4). Thus, R-S and R-C, although physically interacting with Mcm, may form an “incomplete” complex with Mcm, thus resulting in deficiency in licensing and DNA replication.
Phosphorylation of Cdt1
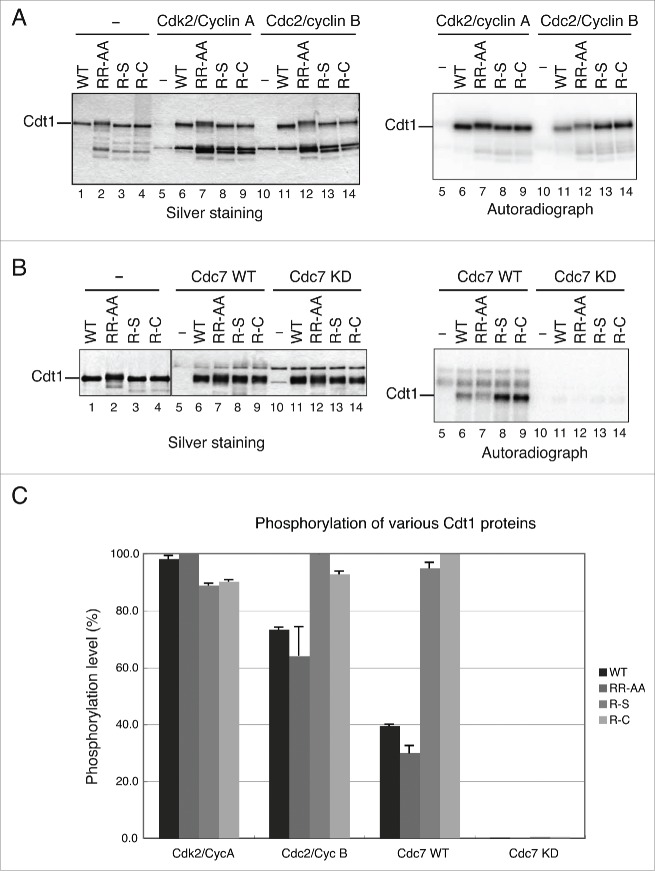
It was previously reported that Cdt1 is dephosphorylated in the absence of active Cdc2, which resulted in its rebinding to chromatin in G2/M phase. Under this condition, Mcm is hypophosphorylated and its release from chromatin in G2 phase is severely inhibited.20,34 It has also been reported that Cdt1 associates with Cdc7, and chromatin-bound Cdt1 is regulated in a Cdc7-dependent manner.21 Specifically, Cdt1 is dissociated form chromatin during S phase progression by the kinase activity of the Cdc7 complex. Thus, Cdk and Cdc7 regulate the activity of Cdt1. We first examined the phosphorylation of Cdt1 by these kinases in vitro. Equal amounts of the wild-type and mutant Cdt1 proteins were subjected to kinase assays using various kinases (Fig. 6A and B, left, lanes 1–4). Phosphorylation levels of Cdt1 mutants by Cdk2/Cyclin A were indistinguishable to that of the wild-type Cdt1 (Fig. 6A right, lanes 6–9). However, the phosphorylation of R-S and R-C with Cdc2/Cyclin B was significantly more vigorous than that of wild-type (Fig. 6A, lanes 11–14). Consistent with previous report, Cdt1 protein was phosphorylated by Cdc7/Dbf4 but not by the kinase-defective Cdc7 kinase (KD), although the phosphorylation level was lower than that by Cdk2/Cyclin A (Fig. 6B and data not shown). Interestingly, the phosphorylation levels of R-S and R-C mutants by Cdc7 were significant higher than that of the wild-type, whereas that of RR-AA was somewhat lower (Fig. 6B right, compare lanes 7–9 to lane 6). We have conducted mass spectrometry analyses of the phosphorylated residues in the in vitro phosphorylated Cdt1 wild-type and mutant proteins. The ratio of phosphorylated peptides to total peptides in the R-S mutant, phosphorylated by Cdc2/CyclinB1 or Cdc7/ASK, increased by 60 % or 25 %, respectively, compared to the non-treated sample. In contrast, it decreased in the RR-AA mutant phosphorylated by Cdc2 or Cdc7 by 15 % or 25 %, respectively (data not shown), consistent with the results of SDS-PAGE gel. The increased phosphorylation of R-S and R-C by Cdc7 kinase probably accelerates release of Cdt1 from chromatin, resulting in reduced Mcm chromatin loading as well as reduced DNA synthesis. In contrast, the ablation of Cdc7-mediated phosphorylation of RR-AA may inhibit removal of Cdt1 from chromatin during S phase progression, and might result in increase of Mcm-loading and DNA synthesis. Thus, we speculate that combined effect of Cdc2 and Cdc7 kinase on the mutant Cdt1 may be related to their phenotypes.
Figure 6.
Phosphorylation of the wild-type and mutant Cdt1 proteins by various kinases in vitro. (A) Standard reaction mixtures for in vitro phosphorylation by Cdk2/Cyclin A and Cdc2/Cyclin B kinase complex were incubated with wild-type and mutant Cdt1 proteins at the same concentration for 1 hr at 37°C, and products were analyzed on 5–20% SDS-PAGE. The gel was stained with silver (left panel) and dried for autoradiogram (right panel). (B) In vitro phosphorylation assays were conducted with Cdc7/Dbf4 kinase (Cdc7 WT) and Cdc7 kinase dead mutant (Cdc7 KD). The reaction products were analyzed on 5–20% SDS-PAGE, which was silver-stained (left panel), dried, and autoradiographed (right panel). (C) The levels of Cdt1 phosphorylation in the autoradiograms in A and B were quantified by Fuji image analyzer, expressed as relative values (the maximum level of kinase activities taken as 100) and were plotted.
Since CDK activities are involved in inhibiting Cdt1 association with chromatin,20,35 the effect of Cdk2/Cyclin A and Cdc2/Cyclin B on DNA binding activity was examined. We found that Cdt1 phosphorylated by Cdk2/Cyclin A could not bind to forked DNA (Fig. 4D, lanes 2–5), consistent with previous report.20 Similarly, phosphorylation by Cdc2/Cyclin B greatly reduced DNA binding (Fig. 4D, lanes 6–8). In addition, phosphorylation by Cdc7/ASK also weakened the affinity to DNA (Fig. S5). Therefore, the hyper-phosphorylation of R-S and R-C by Cdk and Cdc7 would reduce their association with chromatin, leading to reduced Mcm loading and DNA replication, consistent with above results.
Altered interaction between the RR-AA Cdt1 and Geminin
To ensure the regulated licensing during cell cycle, Geminin plays an important role by directly binding to Cdt1 and thereby inhibiting its licensing activity.9,10 The results in Xenopus egg extracts indicated that RR-AA mutant is refractory to inhibition by Geminin. Therefore, interaction with Geminin was investigated using purified GST-tagged Geminin and various mutants of Cdt1. Pull-down assay were carried out using anti-Flag antibody agarose beads or glutathione sepharose beads (Fig. 7A). Wild type and mutant Cdt1 showed identical level of Geminin binding in both assays (Fig. 7A), suggesting that overall affinity of Geminin to Cdt1 was not affected by the mutations.
Figure 7.
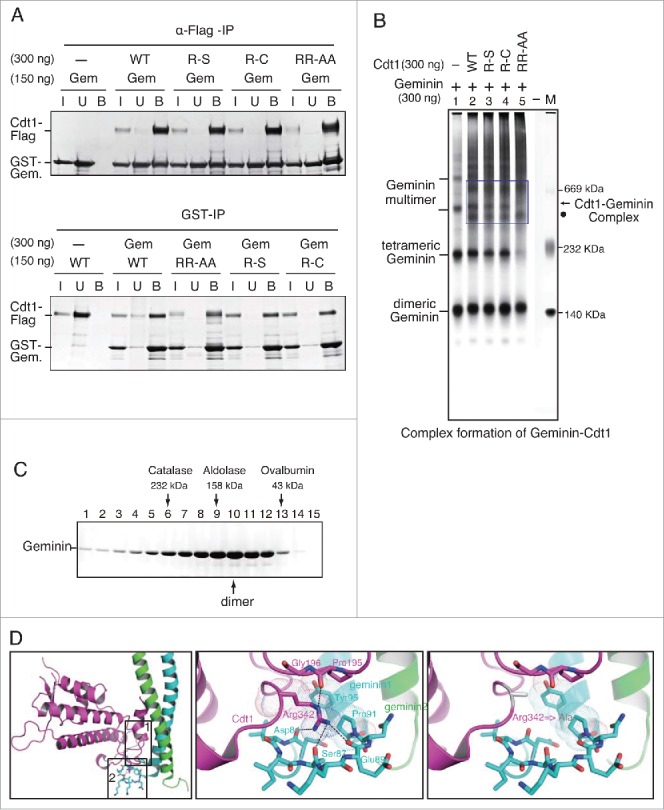
Physical interactions of Geminin and Cdt1. (A) Pull down assays of Geminin and Cdt1. The purified GST-tagged Geminin were mixed with Flag-tagged Cdt1, and immunoprecipitation was performed using anti-Flag M2 antibody agarose beads (upper) or glutathione beads (lower). The associated proteins collected on beads were subjected to SDS-PAGE, followed by silver staining. I: Input (7 % of the input); U: unbound (upper, 25 % of the unbound; lower, 33 % of the unbound); B, bound. (B) The complex formation of Geminin-Cdt1 was examined on a native gel. Indicated proteins were mixed and electrophoresed on a 5% native polyacrylamide gel and stained with silver. The bands in the square boxes indicate the Cdt1-Geminin complexes. The arrow and dot indicate different complexes of Cdt1-Geminin, respectively. (C) Partially purified GST-Geminin protein was fractionated by 15–35% glycerol gradient centrifugation at 45,000 rpm for 16 hr. Proteins in the fractions were analyzed by SDS-PAGE and stained with silver. The glycerol gradient fractionation of purified Geminin protein showed that the Geminin mainly forms a dimeric complex. (D) Effect of Arg342,Arg346-Ala substitution of Cdt1 on its interaction with Geminin. See Discussion for details.
Next, we examined the complex formation between Geminin and various Cdt1 mutants on a native protein gel (Fig. 7B). Although glycerol gradient fractionation of purified Geminin protein showed that it was broadly distributed from 50 kDa through 300 kDa, with a peak around the size of a dimeric complex (Fig. 7C), native gel electrophoresis showed that Geminin forms multiple high molecular-weight complexes appearing as a ladder (Fig. 7B, lane 1), consistent with the structural studies showing that Geminin could form dimers and tetramers.36,37 In the presence of Geminin and Cdt1 proteins, several new distinct bands that were not detected with Geminin alone were detected. There are no differences between the wild-type and R-S/R-C mutant Cdt1 proteins in the patterns of complex formation (Fig. 7B, lanes 3 and 4). In contrast, we found that RR-AA formed a fast-migrating band (representing a dimeric Geminin complex) but the band representing a tetramer of Geminin complex was largely gone (Fig. 7B, lane 5; Fig. S6A, lane 5). Instead, 2 slow migrating bands (dot and star) were detected by anti-Cdt1 and -GST (the tag for Geminin) antibodies, suggesting they contain both RR-AA and Geminin proteins (Fig. S6A and S6B, lane 5). In addition, slow-migrating bands (arrow) of Cdt1 were generated between the wild-type and R-S/R-C mutant Cdt1 proteins, but not in RR-AA mutant in which a more slow-migrating band (star) appeared instead (Fig. 7B; Fig. S6B, compare lanes 2–4 to lane 5). These results suggest that the RR-AA mutant could form a larger complex with Geminin. Since the effect of Geminin on the licensing activity could vary depending on the states of the complex18, the results could be consistent with the fact that RR-AA mutant is refractory to inhibition by Geminin.
Discussion
Here we report crucial arginine residues on Cdt1 that differentially affect DNA replication, licensing and complex formation with Geminin and Mcm (Table 1). Serine or cysteine substitution of 223th arginine (R-S or R-C) resulted in reduced DNA replication/licensing activity, whereas alanine substitution of 342nd and 346th arginines constituting an arginine finger-like structure (RR-AA) resulted in enhanced activity. We show here that 223th arginine is required for formation of the functional Mcm-Cdt1 complex and that 342nd and 346th arginines are involved in formation of a larger inhibitory complex with Geminin. Our results also indicate that arginine mutations affect the extent of phosphorylation of Cdt1 by Cdc2-CyclinB or Cdc7-ASK. Thus, conserved arginine residues of Cdt1 play crucial roles in generating proper complexes with Mcm, Geminin and also possibly with regulatory kinases, thereby profoundly affecting its licensing activity.
Table 1.
Summary of the biochemical properties of mutants of Cdt1 protein.
| WT | RR-AA | R-S | R-C | ||
|---|---|---|---|---|---|
| DNA replication | ++ | ++++ | + | + | |
| Licensing activity | ++ | ++++ | + | + | |
| Complex formation with Mcm2˜7 | ++ | ++ | − | − | |
| Complex formation with Geminin | ++ | − | ++ | ++ | |
| Stimulation of MCM helicase | ++ | ++ | ++ | ++ | |
| DNA-binding activity | ++ | ++ | ++ | ++ | |
| Kinase activity | Cdk2/Cyclin A | ++ | ++ | ++ | ++ |
| Cdc2/Cyclin B | ++ | +/++ | +++ | +++ | |
| Cdc7/Dbf4 | ++ | + | +++ | +++ | |
The assays were performed as described in Materials and Methods. The activity of the wild type Cdt1 is indicated as ++ in each assay; the activities of mutant Cdt1 proteins are shown by the number of plus signs. No activity is shown as a minus sign.
The function of Cdt1 mutants correlates with conformational change with Geminin or Mcm
It has been proposed that a Cdt1-Geminin complex can exist in 2 distinct states, ‘permissive’ and ‘inhibitory’ complexes.18 The results reported here support this hypothesis. The RR-AA Cdt1 mutant can form a complex with Geminin that is larger than that generated by the wild-type, and is hyperactive in DNA replication and in recruitment of the Mcm complex to chromatin. Consistent with the previous proposal, the RR-AA Cdt1 is competent for licensing and replication in the presence of a high level of Geminin (Fig. 3). The Cdt1(RR-AA)-Geminin, forming preferentially a permissive complex, can recruit Mcm to chromatin even at a very low concentration.
It was previously reported that 210th arginine in human Cdt1 (present at the Cdt1:Geminin interface; corresponding to R223 in the mouse Cdt1) is crucial for Cdt1 licensing activity and DNA replication.18 Consistent with this, we show here that the R-S/R-C Cdt1 are severely impaired in DNA replication and Mcm loading activities. Our results further show that the R-S/R-C Cdt1 are not able to generate a functional complex with Mcm2∼7, while interaction with Geminin is not affected. This suggests an unexpected possibility that the tertiary interface of Cdt1 with Geminin may be involved in interaction with Mcm. The structural information indicated that charge complementarity between the C-terminal segments of Cdt1 and Mcm6 is essential for Cdt1-Mcm interaction.24-26 Our results suggest that similar charged interactions at the central domains of Cdt1 also are important for functional Cdt1-Mcm interaction as well as for licensing. This is consistent with previous deletion analyses of Cdt1 which indicate that the central domain containing R223 is crucial for its functions.17,22,27
Effect of arginine mutations on phosphorylation
CDK negatively regulates pre-RC assembly by inhibiting Mcm loading and re-replication. Cdt1 protein accumulates in the nucleus during G1 phase, leading to Mcm2∼7 complex loading on chromatin. In S phase, Cdt1 activity is restrained by Geminin stabilization and Cdt1 degradation.3,38 In the G2/M phases, Cdt1 starts to accumulate again, but cannot interact with chromatin because of the inhibitory phosphorylation mediated by mitotic kinase. A recent study has shown that the mutant Cdt1 deficient in mitotic phosphorylation re-associates with chromatin in G2/M phase, resulting in unscheduled DNA licensing and re-replication.19 This indicates that the proper phosphorylation of Cdt1 in G2/M phase is critical for regulated licensing. It was reported before that the kinase activity of the Cdc7 complex is also required for Cdt1 dissociation from chromatin at the onset of S phase.21 Thus, both Cdk and Cdc7 would regulate chromatin binding of Cdt1. We found that R-S/R-C Cdt1 are phosphorylated significantly more efficiently by Cdc2/Cyclin B and by Cdc7 in vitro. This suggests a possibility that R-S/R-C Cdt1 may be hyperphosphorylated and that its association with chromatin may be inhibited, resulting in reduced licensing. In contrast, RR-AA is poorly phosphorylated by Cdc7 in vitro, which may result in hypophosphorylation of Cdt1. This would facilitate reassociation of Cdt1 to chromatin in G2/M phase and would also be inhibitory for its release from chromatin at the onset of S phase, leading to increased licensing and DNA replication in Xenopus egg extracts. This would also explain the Geminin-independent replication stimulatory effect of the RR-AA mutant.
The DNA-binding domain of Cdt1 has been proposed to contain the N-terminal and middle domains.11,14 The arginine mutations did not affect the affinity to DNA (Fig. 4B & C). On the other hand, the phosphorylation of Cdt1 by Cdk dramatically inhibits its affinity to DNA (Fig. 4D). Thus, the efficacy of Cdk-mediated phosphorylation of Cdt1 may have profound effect on its association with DNA, which may ultimately affect its Mcm loading activity.
Structural consideration of RR-AA mutation
Crystallographic studies suggest that Cdt1 interact with Geminin through 3 interfaces to form a complex.12,18 As shown in Figure 7D, Arg342 (loop-2 of Cdt1; secondary interaction) has a van der Waals contact with Tyr95 and Pro91 of Geminin. Arg342 interacts also with Pro195 and Gly196 of the upper loop (loop-1 of Cdt1; primary interaction) of Cdt1, the major Geminin binding interface (Fig. 7D, middle and right panels). Carbonyl group of Pro195 makes hydrogen bonding with Arg342, and Gly196 is in contact with the aliphatic part of Arg342 (loop-2; Fig. 7D, middle panel). In addition, the Arg346 also interacts with Glu89 of Geminin. Substitution of Arg342 and Arg346 with alanine results in loss of the primary and secondary interactions as well as that of hydrogen bonding (Fig. 7D, right panel), which would extensively affect the normal conformation of the Geminin-Cdt1 complex. This finding supports the notion that differences in the stoichiometry of Cdt1 and Geminin can cause the formation of different Cdt1-Geminin complexes with different effect.15-18,30
We conclude that the complex formation of functionally proficient Cdt1-Geminin and Cdt1-Mcm and proper phosphorylation of Cdt1 are required for the regulated spatial and temporal chromatin loading of the Mcm and DNA replication.
Materials and methods
Cloning of wild-type and mutant forms of Cdt1 genes into baculovirus transfer vectors
Site-directed mutagenesis of the Cdt1 genes was conducted with the QuikChange site-directed mutagenesis kit (Agilent Technologies). The oligonucleotides 5′- CGACCAGCTGACCGCATGGCATCCGGCATTCAATGTGGACG-3′, 5′-GAGCATGCTCCACAATAGCTCTGAGACTGTGACC-3′, and 5′-GAGCATGCTCCACAATTGCTCTGAGACTGTGACC-3′ were used as the primers to clone the full-length Cdt1 coding regions containing R342R346-AA, R223-S and R223-C Cdt1 mutants, respectively, in pFastbac1.
Antibodies
The antibodies against following Xenopus proteins were described previously; anti-Xenopus Mcm2 antibody,39 antibodies for Xenopus Cdt1, Cdc6, Orc2 and Geminin,30 anti-Xenopus Pol ϵ antibody,40 anti-Pol α antibody.41 Anti-PCNA antibody (PC 10) was purchased from Sigma Aldrich.
Purification of recombinant proteins
Recombinant wild-type or various mutant forms of Cdt1 protein, expressed in insect cells, were purified by Ni-NTA and anti-Flag antibody beads. The peak fractions were further fractionated on 15∼35% glycerol gradient which was run at 38,000 rpm for 17 hr, as previously described.23 To remove contaminating nucleases, further purification was carried out as follows. The fractions from 10 to 14 in glycerol gradient were pooled and treated with Benonase (final 1 U/ml). The samples were bound to Ni-sepharose for 1 hr at 4°C and thoroughly washed with buffer containing 5 mM ATP and 10 mM Mg-acetate. Then, Cdt1 proteins were eluted with elution buffer containing the 0.5 M imidazole, 2 mM ATP, 2 mM Mg-acetate, and 100 mM Na-acetate. The eluates were further fractionated by second glycerol gradient centrifugation.
The highly purified recombinant mouse Mcm4/6/7 and Mcm2∼7 protein complexes were prepared from insect cells as previously described.42 The recombinant GST-Geminin protein was purified from E. coli cells by glutathione sepharose beads according to the manufacture's protocol. GST-Geminin was further fractionated by 15-35% glycerol gradient centrifugation at 45,000 rpm for 16 hr. The glycerol gradient fractionation indicated that GST-Geminin mainly formed a dimer. Each fraction was subjected to SDS-PAGE (5–20% polyacrylamide) to detect the proteins by silver staining.
Pull-down experiments
Pull-down experiments were performed using anti-Flag M2 antibody agarose beads (Sigma), glutathione beads or anti-Cdt1 antibody (Santa Cruz Biotechnology) coupled to Dynabeads protein G (Life Technologies). Indicated proteins were mixed in PBS buffer (Cdt1-Mcm interaction) or buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM sodium acetate, 5 mM Mg-acetate, 1 mM ATP, 1 mM EDTA, 10% glycerol, 0.01% Triton X, and proteinase inhibitor (Cdt1-Geminin interaction). After 1 hr incubation, beads were washed 3 times with the above buffer, and then once with PBS buffer. Bound proteins were eluted with 200 μg/ml Flag peptide or 0.1 M citric acid (pH 2.2). The samples were run on SDS-PAGE, followed by silver staining.
Complex formation analysis
The complex formation of Mcm2∼7-Cdt1 and Geminin-Cdt1 were examined in a native gel as follows. 300 ng each of Mcm2∼7 and Cdt1 or Geminin and Cdt1 were mixed on ice in a buffer containing of 20 mM Tris-HCl (pH 7.5), 10 mM creatine-phosphate, 5 mM Mg-acetate, 5 mM DTT, and 0.01% Triton X-100, and then electrophoresed on a 5% Tris-glycine native polyacrylamide gel containing 5% glycerol, followed by staining with silver or western-blotting. The proteins that had been electrophoresed under a non-denaturing condition were transferred to a membrane after the gel was incubated in 20 mM Tris-150 mM glycine-0.1% SDS for 10 min. The membrane was then treated with 50 mM Tris pH7.5-2% SDS-0.8% 2-mercaptoethanol at 50°C for 30 min, washed in TBST (Tris-buffered saline, 0.1% Tween 20) and then incubated with anti-Cdt1 antibody (Santa Cruz Biotechnology). The membrane was then washed in TBST buffer and incubated in anti-GST antibody to detect Geminin protein. The protein markers, thyroglobulin (669 kDa), catalase (232 kDa) and lactate dehehydrogenase (140 kDa), were run side by side.
In vitro phosphorylation
The wild type and mutant Cdt1 proteins (100 ng) were incubated with Cdk2/CyclinA, Cdc2/CyclinB (purchased from Carna Biosciences, Inc.), or Cdc7/ASK complex at 37°C for 30 min in a reaction mixture consisting of 20 mM Tris-HCl (pH 7.5), 30 mM NaCl, 10 mM MgCl2, 1 mM ATP, and 0.01% Triton X-100, 10 mM 2-glycerophosphate, 1 µM Calyculin A in the presence of [γ-32P]ATP. The phosphorylated proteins were analyzed by SDS-polyacrylamide gel electrophoresis, followed by silver staining. The radioactivity on the same gel was detected by a Bio-Image analyzer (BAS 2500; Fuji).
DNA-binding assays using biotinylated oligonucleotide
Wild-type or mutant Cdt1 (200 ng) and 5′-biotinylated oligonucleotides were incubated at 30°C for 30 min in a buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM sodium acetate, 10 mM Mg(CH3COO)2, 1 mM DTT, 1 mM ATP, 5 mM NaF; 1 mM NaVO4, and 0.01% Triton X-100. Streptavidin-coated M-280 magnetic beads was added to the mixture and incubated for 30 min at 4°C. After the removal of the supernatant, the beads were collected, washed with the buffer, and were analyzed on 4–20% SDS-PAGE.
The coupled reactions of phosphorylation and DNA binding analysis in Figure 4D were conducted as follows. After phosphorylation of Cdt1 with Cyclin A/Cdk2 and Cdc2/CyclinB for 30 min at 30°C in the above buffer, a mixture were diluted into above binding buffer and biotin-labeled fork DNA (37mer-dT40/dT20-37mer) were added, and the incubation was continued for 30 min at 30°C. And then, the streptavidin-coated M-280 magnetic beads (Dynal Biotech ASA) were added to the mixture and incubated for 30 min at 4°C. After the removal of the supernatant (unbound), the beads (bound) were washed 4 times in a buffer (400 μl) containing 20 mM Tris (pH 7.5), 100 mM Na-Acetate, 1 mM DTT, 0.01% Triton X-100, 5 mM NaF, 1 mM NaVO4 and 10% glycerol, re-suspended in SDS sample buffer, and were analyzed on 4–20% gradient PAGE, followed by silver staining.
Gel-shift and helicase assays
DNA helicase and DNA-binding activities were examined basically as previously described.42
Replication assays
Replication assays contained 10 μl of interphase extract with Flag-peptide-eluted mCdt1 preparations or control buffer. Demembranated Xenopus sperm nuclei were added at a final concentration of 2,500 nuclei/μl. DNA synthesis was monitored by [α-32P]dATP incorporation after 60 min or 90 min at 23°C. After the incubation of sperm nuclei, the samples (4 μl) were removed and added to the 10 μl of a reaction-stop buffer (0.25 mg/ml proteinase K, 8 mM EDTA, 80 mM Tris–HCl (pH 8.0), 0.13% (w/v) phosphoric acid, 10% (w/v) Ficoll, 5% (w/v) SDS, and 0.2% (w/v) bromophenol blue). After incubation at 37°C for 1 hr, the samples were diluted by addition of 40 μl of TE (50 mM Tris-HCl (pH 8.0), 1 mM EDTA) and spotted onto a Whatman DE81 cellulose chromatography paper (GE Healthcare). The filter paper was washed with 0.5 M sodium phosphate buffer (pH 6.8), deionized water and then 100% ethanol. Amount of 32P on the filter paper was measured by a scintillation counter.
Chromatin-binding assays in xenopus egg extracts
To isolate the chromatin fraction, sperm chromatin (2500 nuclei/μl) was incubated with interphase extracts of Xenopus eggs (20 μl) at 23°C for indicated time in the figure or legends. Chromatin fraction was isolated as described previously.43
Mass spectrometer analyses of Cdt1 phosphorylated in vitro
About 2 μg of Cdt1 protein (wild-type and mutants), either non-treated or phosphorylated by Cdc2/CyclinB or Cdc7/ASK, was treated with trypsin and concentrated by IMAC-C18 (immobilized metal ion affinity chromatography) to improve the detection sensitivity of phosphorylated molecules. Each fraction was analyzed via LCMS/MS on Triple TOF 5600 Plus (AB Sciex) mass spectrometer. Analysis of the data with ProteinPilot 4.5beta software (AB Sciex) identified 600-1000 peptides with a 95% confidence at a false discovery rate of 1%. No phosphorylation was identified on the T24 and S61 residues.
Supplementary Material
Abbreviations
- Cdc7 kinase
Cell division cycle 7-related protein kinase
- Cdk
cyclin-dependent kinase
- Cdt1
Cdc10-dependent transcript 1
- Mcm
mini-chromosome maintenance
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Katsuhiko Kamada (RIKEN Discovery Research Institute, Japan) for structural analyses of interaction of Cdt1 (wild-type and RR-AA) with Geminin. Naoko Kakusho and Rino Fukatsu for excellent technical assistance. We thank the members of our laboratories for helpful discussions.
References
- [1].Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 2010; 79:89-130; PMID:20373915; http://dx.doi.org/ 10.1146/annurev.biochem.052308.103205 [DOI] [PubMed] [Google Scholar]
- [2].Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div 2006; 1:22; PMID:17042960; http://dx.doi.org/ 10.1186/1747-1028-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 2003; 278:30854-8; PMID:12840033; http://dx.doi.org/ 10.1074/jbc.C300251200 [DOI] [PubMed] [Google Scholar]
- [4].Nishitani H, Lygerou Z. DNA replication licensing. Front Biosci 2004; 9:2115-32; PMID:15353274; http://dx.doi.org/ 10.2741/1315 [DOI] [PubMed] [Google Scholar]
- [5].Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al.. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. Embo J 2006; 25:1126-36; PMID:16482215; http://dx.doi.org/ 10.1038/sj.emboj.7601002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 2006; 8:84-90; PMID:16362051; http://dx.doi.org/ 10.1038/ncb1346 [DOI] [PubMed] [Google Scholar]
- [7].Sugimoto N, Kitabayashi I, Osano S, Tatsumi Y, Yugawa T, Narisawa-Saito M, Matsukage A, Kiyono T, Fujita M. Identification of novel human Cdt1-binding proteins by a proteomics approach: proteolytic regulation by APC/CCdh1. Mol Biol Cell 2008; 19:1007-21; PMID:18162579; http://dx.doi.org/ 10.1091/mbc.E07-09-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998; 93:1043-53; PMID:9635433; http://dx.doi.org/ 10.1016/S0092-8674(00)81209-X [DOI] [PubMed] [Google Scholar]
- [9].Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000; 290:2309-12; PMID:11125146; http://dx.doi.org/ 10.1126/science.290.5500.2309 [DOI] [PubMed] [Google Scholar]
- [10].Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol 2001; 3:107-13; PMID:11175741; http://dx.doi.org/ 10.1038/35055000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yanagi K, Mizuno T, You Z, Hanaoka F. Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem 2002; 277:40871-80; PMID:12192004; http://dx.doi.org/ 10.1074/jbc.M206202200 [DOI] [PubMed] [Google Scholar]
- [12].Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, Enomoto T, Tada S, Kim Y, Cho Y. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature 2004; 430:913-7; PMID:15286659; http://dx.doi.org/ 10.1038/nature02813 [DOI] [PubMed] [Google Scholar]
- [13].Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem 2001; 276:44905-11; PMID:11555648; http://dx.doi.org/ 10.1074/jbc.M105406200 [DOI] [PubMed] [Google Scholar]
- [14].Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PI, Lygerou Z. Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. Embo J 2007; 26:1303-14; PMID:17318181; http://dx.doi.org/ 10.1038/sj.emboj.7601597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lutzmann M, Maiorano D, Mechali M. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J 2006; 25:5764-74; PMID:17124498; http://dx.doi.org/ 10.1038/sj.emboj.7601436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xouri G, Dimaki M, Bastiaens PI, Lygerou Z. Cdt1 interactions in the licensing process: a model for dynamic spatiotemporal control of licensing. Cell Cycle 2007; 6:1549-52; PMID:17598984; http://dx.doi.org/ 10.4161/cc.6.13.4455 [DOI] [PubMed] [Google Scholar]
- [17].Kisielewska J, Blow JJ. Dynamic interactions of high Cdt1 and geminin levels regulate S phase in early Xenopus embryos. Development 2012; 139:63-74; PMID:22096080; http://dx.doi.org/ 10.1242/dev.068676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, et al.. Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing. Proc Natl Acad Sci U S A 2009; 106:19807-12; PMID:19906994; http://dx.doi.org/ 10.1073/pnas.0905281106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Coulombe P, Gregoire D, Tsanov N, Mechali M. A spontaneous Cdt1 mutation in 129 mouse strains reveals a regulatory domain restraining replication licensing. Nat Commun 2013; 4:2065; PMID:23817338; http://dx.doi.org/ 10.1038/ncomms3065 [DOI] [PubMed] [Google Scholar]
- [20].Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, Fujita M. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem 2004; 279:19691-7; PMID:14993212; http://dx.doi.org/ 10.1074/jbc.M313175200 [DOI] [PubMed] [Google Scholar]
- [21].Ballabeni A, Zamponi R, Caprara G, Melixetian M, Bossi S, Masiero L, Helin K. Human CDT1 associates with CDC7 and recruits CDC45 to chromatin during S phase. J Biol Chem 2009; 284:3028-36; PMID:19054765; http://dx.doi.org/ 10.1074/jbc.M803609200 [DOI] [PubMed] [Google Scholar]
- [22].Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. Embo J 2011; PMID:22045335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].You Z, Masai H. Cdt1 forms a complex with the minichromosome maintenance protein (MCM) and activates its helicase activity. J Biol Chem 2008; 283:24469-77; PMID:18606811; http://dx.doi.org/ 10.1074/jbc.M803212200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jee J, Mizuno T, Kamada K, Tochio H, Chiba Y, Yanagi K, Yasuda G, Hiroaki H, Hanaoka F, Shirakawa M. Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins. J Biol Chem 2010; 285:15931-40; PMID:20335175; http://dx.doi.org/ 10.1074/jbc.M109.075333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu C, Wu R, Zhou B, Wang J, Wei Z, Tye BK, Liang C, Zhu G. Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res 2012; 40:3208-17; PMID:22140117; http://dx.doi.org/ 10.1093/nar/gkr1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wei Z, Liu C, Wu X, Xu N, Zhou B, Liang C, Zhu G. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem 2010; 285:12469-73; PMID:20202939.15653632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ferenbach A, Li A, Brito-Martins M, Blow JJ. Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res 2005; 33:316-24; PMID:15653632; http://dx.doi.org/ 10.1093/nar/gki176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev 2000; 14:1765-76; PMID:10898791 [PMC free article] [PubMed] [Google Scholar]
- [29].Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem 2005; 280:27420-30; PMID:15901724; http://dx.doi.org/ 10.1074/jbc.M502764200 [DOI] [PubMed] [Google Scholar]
- [30].Ode KL, Fujimoto K, Kubota Y, Takisawa H. Inter-origin cooperativity of geminin action establishes an all-or-none switch for replication origin licensing. Genes Cells 2011; 16:380-96; PMID:21426446; http://dx.doi.org/ 10.1111/j.1365-2443.2011.01501.x [DOI] [PubMed] [Google Scholar]
- [31].Tsuyama T, Watanabe S, Aoki A, Cho Y, Seki M, Enomoto T, Tada S. Repression of nascent strand elongation by deregulated Cdt1 during DNA replication in Xenopus egg extracts. Mol Biol Cell 2009; 20:937-47; PMID:19064889; http://dx.doi.org/ 10.1091/mbc.E08-06-0613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem 2001; 2:15; PMID:11737877; http://dx.doi.org/ 10.1186/1471-2091-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Waga S, Zembutsu A. Dynamics of DNA binding of replication initiation proteins during de novo formation of pre-replicative complexes in Xenopus egg extracts. J Biol Chem 2006; 281:10926-34; PMID:16497662; http://dx.doi.org/ 10.1074/jbc.M600299200 [DOI] [PubMed] [Google Scholar]
- [34].Fujita M, Yamada C, Tsurumi T, Hanaoka F, Matsuzawa K, Inagaki M. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J Biol Chem 1998; 273:17095-101; PMID:9642275; http://dx.doi.org/ 10.1074/jbc.273.27.17095 [DOI] [PubMed] [Google Scholar]
- [35].Ballabeni A, Melixetian M, Zamponi R, Masiero L, Marinoni F, Helin K. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J 2004; 23:3122-32; PMID:15257290; http://dx.doi.org/ 10.1038/sj.emboj.7600314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Okorokov AL, Orlova EV, Kingsbury SR, Bagneris C, Gohlke U, Williams GH, Stoeber K. Molecular structure of human geminin. Nat Struct Mol Biol 2004; 11:1021-2; PMID:15378034; http://dx.doi.org/ 10.1038/nsmb835 [DOI] [PubMed] [Google Scholar]
- [37].Thepaut M, Maiorano D, Guichou JF, Auge MT, Dumas C, Mechali M, Padilla A. Crystal structure of the coiled-coil dimerization motif of geminin: structural and functional insights on DNA replication regulation. J Mol Biol 2004; 342:275-87; PMID:15313623; http://dx.doi.org/ 10.1016/j.jmb.2004.06.065 [DOI] [PubMed] [Google Scholar]
- [38].Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev 2004; 18:673-86; PMID:15075294; http://dx.doi.org/ 10.1101/gad.1180204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J 1997; 16:3320-31; PMID:9214647; http://dx.doi.org/ 10.1093/emboj/16.11.3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells 2006; 11:993-1007; PMID:16923121; http://dx.doi.org/ 10.1111/j.1365-2443.2006.00998.x [DOI] [PubMed] [Google Scholar]
- [41].Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. Embo J 1998; 17:5699-707; PMID:9755170; http://dx.doi.org/ 10.1093/emboj/17.19.5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].You Z, De Falco M, Kamada K, Pisani FM, Masai H. The Mini-Chromosome Maintenance (Mcm) Complexes Interact with DNA Polymerase alpha-Primase and Stimulate Its Ability to Synthesize RNA Primers. PLoS One 2013; 8:e72408; PMID:23977294; http://dx.doi.org/ 10.1371/journal.pone.0072408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J 2003; 22:2526-35; PMID:12743046; http://dx.doi.org/ 10.1093/emboj/cdg238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.