ABSTRACT
The human APOBEC3 family of DNA-cytosine deaminases comprises 7 members (A3A-A3H) that act on single-stranded DNA (ssDNA). The APOBEC3 proteins function within the innate immune system by mutating DNA of viral genomes and retroelements to restrict infection and retrotransposition. Recent evidence suggests that APOBEC3 enzymes can also cause damage to the cellular genome. Mutational patterns consistent with APOBEC3 activity have been identified by bioinformatic analysis of tumor genome sequences. These mutational signatures include clusters of base substitutions that are proposed to occur due to APOBEC3 deamination. It has been suggested that transiently exposed ssDNA segments provide substrate for APOBEC3 deamination leading to mutation signatures within the genome. However, the mechanisms that produce single-stranded substrates for APOBEC3 deamination in mammalian cells have not been demonstrated. We investigated ssDNA at replication forks as a substrate for APOBEC3 deamination. We found that APOBEC3A (A3A) expression leads to DNA damage in replicating cells but this is reduced in quiescent cells. Upon A3A expression, cycling cells activate the DNA replication checkpoint and undergo cell cycle arrest. Additionally, we find that replication stress leaves cells vulnerable to A3A-induced DNA damage. We propose a model to explain A3A-induced damage to the cellular genome in which cytosine deamination at replication forks and other ssDNA substrates results in mutations and DNA breaks. This model highlights the risk of mutagenesis by A3A expression in replicating progenitor cells, and supports the emerging hypothesis that APOBEC3 enzymes contribute to genome instability in human tumors.
KEYWORDS: APOBEC3, ATR kinase, cytosine deamination, cell cycle checkpoint, DNA replication, DNA replication stress, single-stranded DNA
Introduction
The human AID/APOBEC3 family of DNA-cytosine deaminases comprises 7 apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins, A3A through A3H, and the related activation-induced deaminase (AID). The AID/APOBEC3 enzymes function within the adaptive and innate immune systems by mutating host and pathogen DNA.1,2 AID deaminates immunoglobulin (Ig) genes in germinal center B cells to promote antibody diversity through class-switch recombination and somatic hypermutation.3,4 APOBEC3 enzymes play a role in innate immunity as viral restriction factors, in part by inducing mutations in viral genomes.5-7 AID/APOBEC3 family members also inhibit retrotransposition of genetic elements including long terminal repeat (LTR) and non-LTR (LINE-1) elements.8-11
Although these deaminases have important physiologic functions in innate defense, their DNA mutator activity also threatens host genome integrity.12-14 Deamination by AID outside of the Ig loci leads to off-target mutations and recurrent IgH-Myc translocations that drive B cell malignancies.15,16 Similarly, somatic mutation clusters consistent with APOBEC3 activity have been recognized in tumor genomes,17-20 suggesting deamination of the cellular genome by aberrant activity of APOBEC3 enzymes. The role of APOBEC3 enzymes in generating these mutational signatures in cancer genomes is further supported by the finding of clustered mutations in genomes of yeast that express ectopic APOBEC3 cytosine deaminases.21-25 APOBEC3 enzymes function exclusively on ssDNA substrates,8,26,27 and the mutation clusters may arise from highly processive deaminase activity on transiently exposed ssDNA. Within the cellular genome, ssDNA is exposed during several frequent cellular processes: transcription, recombination, and replication. ssDNA exposed during transcription is a well-defined target for AID deamination, which occurs at transcription bubbles and R-loops during antibody diversification.28,29 Additionally, resection of double-stranded DNA breaks (DSBs) during DNA repair exposes ssDNA that is susceptible to mutations caused by APOBECs.22,23 Recent studies involving whole-genome sequencing suggest that APOBEC-induced mutagenesis occurs primarily within lagging strand templates arising during DNA replication. This has been demonstrated by ectopic expression of human APOBEC3 enzymes in model organisms as well as analysis of cancer genomes.30-33 Although these studies suggest ssDNA at replication forks is vulnerable to deamination by APOBECs, this has not been established experimentally in mammalian cells.
In this study we investigate the potential for A3A to deaminate cellular ssDNA exposed during replication. A3A is among the most potent of the human APOBEC3 deaminases, and can restrict nuclear DNA viruses and retrotransposition events.8,34–36 We recently showed that the mechanism for A3A inhibition of L1 retrotransposition occurs by deamination of ssDNA transiently exposed during L1 integration.9 We have also previously shown that A3A can act on the cellular genome to induce DNA breaks that activate the DNA damage response (DDR).37 The DDR orchestrates the recognition and repair of DNA damage, and is activated by both DNA breaks and stalled replication.38,39 The protein kinases Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad3-related (ATR) are essential regulators of DDR signaling and activate transducers via phosphorylation of specific marks.40 ATM is primarily responsive to DSB formation, which results in auto-phosphorylation at serine 1981.41 Activated ATM then signals via phosphorylation of histone variant H2AX at serine 139 and checkpoint kinase 2 (Chk2) at threonine 68. This signaling cascade halts cell cycle progression at the G1/S and intra-S checkpoints to allow for repair of DSBs.42-44 In a parallel arm of the DDR, ATR signaling is activated by replication stress after Replication Protein A (RPA) binds exposed ssDNA.38,45–47 Checkpoint kinase 1 (Chk1) is activated by ATR via phosphorylation at serine 345 and serine 317.48 ATR- and Chk1-dependent signals are essential for activation of the G2/M checkpoint and contribute to intra-S checkpoint activation.47,49 These signaling events ensure that repair of genomic lesions and high-fidelity DNA replication occur prior to mitosis.
Our previous work demonstrated that DNA breaks caused by A3A lead to ATM-mediated signaling and activation of the G1/S checkpoint.37 We now investigate the hypothesis that A3A-induced cell cycle arrest may also occur through activation of the replication checkpoint via ATR in response to deamination of replicating DNA. In the present study, we show that A3A activates ATR signaling leading to G2/M arrest in ATM-deficient cells. We also show that A3A expression leads to increased DNA damage signaling upon replication stress, and accumulation of genomic uracil during DNA replication. Finally, our data show that A3A induces less DNA damage in quiescent cells compared to cycling cells. Our findings demonstrate that DNA replication provides ssDNA substrate that is susceptible to deamination by A3A. Thus, aberrant A3A activity leading to genomic damage may specifically impact rapidly replicating cells such as undifferentiated progenitors. Taken together, our data contribute to a model in which A3A deaminates transiently exposed ssDNA at replication forks and explain the somatic cytosine mutations.
Results
A3A activates the DNA replication checkpoint
To determine the DDR signaling pathways that are activated in response to A3A expression, we examined protein phosphorylation and cell cycle progression in stable cell lines generated to express A3A from an inducible promoter. A3A was expressed by treatment with doxycycline in either osteosarcoma (U2OS) or hepatoma (HepaRG) cell lines harboring inducible A3A genes. We then used Western blotting to examine checkpoint signaling with phospho-specific antibodies to proteins involved in the DDR (Fig. 1A). In both HepaRG and U2OS cell lines, A3A expression was accompanied by robust phosphorylation of the histone variant H2AX at serine 139 (γH2AX), a marker for DSBs and an early DDR signaling event.50 As previously reported,37 we observed activation of ATM kinase as demonstrated by phosphorylation of S1981, as well as phosphorylation of the downstream checkpoint kinase Chk2 on residue T68.42 In addition, we detected phosphorylation of checkpoint kinase Chk1 at S317 and S345 residues, which are known substrates for the ATR kinase.48 These data suggest that both ATM and ATR signaling pathways are activated as a result of A3A expression.
Figure 1.
ATR signaling is activated by A3A expression. (A) A3A activates DNA damage checkpoints. Inducible cell lines were treated with doxycycline for 24 hours and analyzed by Western blot using antibodies to HA, γH2AX, phosphorylated Chk2 (T68), phosphorylated Chk1 (S345 and S317), phosphorylated ATM (S1981), Chk2, Chk1, ATM, and ATR. GAPDH was used as a loading control. (B) A3A expression results in cell cycle arrest. Inducible cells lines were treated with doxycycline for 24 or 36 hours, then pulsed with BrdU for 40 minutes prior to harvest. Cells were fixed, stained with anti-BrdU antibody and 7-AAD, and analyzed by flow cytometry. The percentage of cells in each gate are indicated. Accompanying chart shows the fraction of cells in G1, S, and G2/M phase at 36 hours post-induction. (C) A3A activates ATR signaling in ATM-deficient cells. Inducible AT22IJE-A3A cells with mutant ATM or wild-type ATM were treated with doxycycline to induce A3A expression and analyzed by Western blot using antibodies to HA, γH2AX, phosphorylated Chk1 (S345 and S317), Chk1, phosphorylated ATM (S1981), ATM, and ATR. (D) Cell-cycle progression following A3A induction in AT22IJE cells with mutant ATM or wild-type ATM was determined by propidium iodide staining. Accompanying chart shows the fraction of cells in G1, S, and G2/M phase at 24 hours after treatment, which was similar to results obtained 48 hours after treatment. Results are representative of 3 independent experiments.
Since DDR signaling impacts cell cycle progression, we next examined the effect of A3A expression on the cell cycle in the two inducible cell lines. Cells were induced to express A3A, and cell cycle progression was assessed by flow cytometry after BrdU incorporation and staining. Both HepaRG and U2OS cell lines exhibited cell cycle arrest after 24 hours of A3A expression, which persisted at 36 hours (Fig. 1B). HepaRG cells were arrested in early S phase, as expected for ATM-mediated activation in response to DNA damage.37 In contrast, A3A expression in the U2OS cell line resulted in arrest predominantly in G2/M. We hypothesized that the G1/S and intra-S checkpoints may be insufficient in U2OS cells, and thus the G2/M arrest observed is mediated by ATR activation and signaling.
To determine the impact of A3A on ATR-mediated checkpoint activation, we generated stable lines with inducible A3A expression in the AT22IJE cell line derived from a patient with Ataxia-Telangiectasia which encodes a truncated ATM protein that is rapidly degraded.51 We compared the effect of A3A expression in an AT22IJE cell line without functional ATM (ATM-) to AT22IJE cells with an ectopic, constitutively expressed wild-type ATM (ATM+). We induced A3A expression with doxycycline in these cell lines and assessed phosphorylation of DDR proteins (Fig. 1C) and cell cycle progression by flow cytometry after propidium iodide staining (Fig. 1D). As expected, ATM was detected by Western blotting in ATM+ cells, and exhibited increased phosphorylation upon A3A expression whereas no ATM was detectable in ATM- cells (Fig. 1C). Similar to our observations in HepaRG and U2OS cells, A3A expression in both AT22IJE cell lines resulted in phosphorylation of H2AX and Chk1. To confirm that phosphorylation of Chk1 was dependent on ATR signaling, we used a small molecule inhibitor of ATR kinase activity (VE822)52 to inhibit ATR signaling in AT22IJE cells induced to express A3A. Upon treatment with the ATR inhibitor, both cell types exhibited decreased phosphorylation of Chk1 (Fig. S1), thus indicating that Chk1 activation is mediated by ATR in response to A3A expression. Following A3A induction, ATM- cells arrested at the G2/M checkpoint whereas ATM+ cells arrested in G1/S (Fig. 1D). These data indicate that when ATM signaling is intact, A3A expression results in G1/S arrest. However, in the absence of functional ATM, cell cycle arrest occurs in G2/M. Together these findings show that A3A activates checkpoints mediated by both ATR and by ATM.
Replication stress renders cells vulnerable to A3A-induced DNA damage
We hypothesized that activation of the ATR-mediated DNA replication checkpoint reflects the capacity for A3A to disrupt cellular DNA replication, which ultimately leads to cell cycle arrest. Based on this hypothesis, we investigated whether replication stress could potentiate genomic damage induced by A3A (Fig. 2). U2OS-A3A cells were treated with hydroxyurea (HU) to limit deoxyribonucleotide production, thus inhibiting replicative polymerase progression and generating ssDNA at the fork through uncoupling of MCM2-7 helicase and replicative polymerases.53 The HU treatment was restricted to 4 hours to allow for stalled replication without irreparable collapse of forks and genome damage.54 To investigate the impact of A3A on cells during replication stress and following recovery from transient replication stress, we examined cells that were pulsed with HU and then cultured for an extended period without HU (see schematic in Fig. 2A). For each treatment condition, cells were either treated with doxycycline (+dox) to induce A3A expression or not treated (-dox). Doxycycline dose was titrated to induce a low level of A3A expression that generated minimal DNA damage in the absence of HU treatment (Fig. S2). We observed that HU treatment alone was sufficient to cause H2AX phosphorylation, which is indicative of DNA damage independent of A3A expression. However, this damage was resolved after a period of recovery in the absence of A3A (Fig. 2B-D). In contrast, Western blotting showed the H2AX phosphorylation mark persisted through the recovery period in A3A-expressing cells (Fig. 2B). Immunofluorescent imaging of cells recovering from HU treatment also showed a significantly increased number of A3A-expressing cells exhibiting γH2AX staining as compared to uninduced cells (Fig. 2C-D). HU treatment of cells induced to express the catalytically inactive A3A-C106S mutant yielded no increase in H2AX phosphorylation following HU recovery (Fig. S3). These findings indicate that replication stress renders genomic DNA more susceptible to damage by A3A.
Figure 2.
Replication stress potentiates A3A-induced DNA damage. (A) Schematic of hydroxyurea (HU) treatment schedules. U2OS-A3A cells were cultured in the presence (+dox) or absence (-dox) of low-dose doxycycline (0.01 μg/mL) for 24 hours in each condition. Cells were treated with 5 mM HU for 4 hours to cause a transient replication arrest followed by removal of HU to allow for recovery of replication. Conditions included: no HU (top), HU treatment 4 hours prior to harvest (middle), HU treatment for 4 hours followed by removal of HU and 16 hours of recovery time (bottom). (B) Replication arrest leads to phosphorylation of H2AX in cells expressing A3A. Western blot analysis of cells treated as in (A) using antibodies to HA and γH2AX and phosphorylated Chk1 (S345). GAPDH was used as a loading control. (C) Analysis of γH2AX staining in U2OS-A3A cells cultured without HU, with HU, or with HU followed by recovery. Cells were fixed and stained with anti-γH2AX antibody, and analyzed by confocal microscopy. Nuclei were stained with DAPI. Scale bar is 50 μm. (D) Quantification of cells with γH2AX staining in (C). Error bars are SD. Statistical analysis was performed using a paired 2-tail t test. Results are representative of 3 independent experiments.
A3A-induced deamination and DNA damage are increased during DNA replication
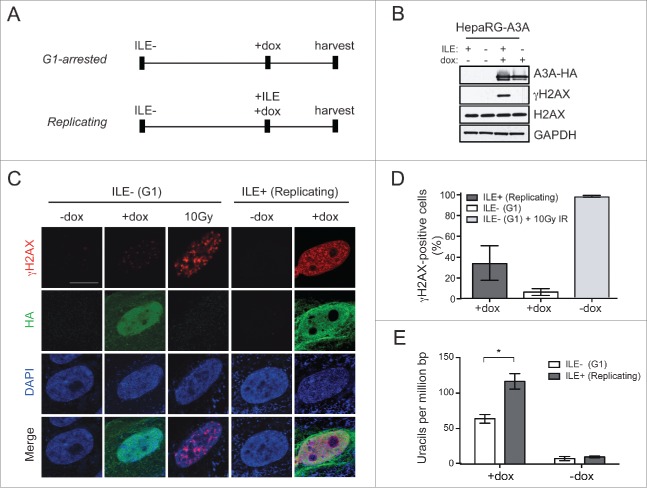
To determine the impact of A3A expression on the cellular genome during DNA replication we investigated the vulnerability of cells to A3A activity at different stages of the cell cycle. We compared DNA damage caused by A3A in cell populations enriched in either G1 or S-G2 phases. HepaRG-A3A cells were synchronized in G1 by depleting media of isoleucine (ILE).55 Untreated cells and those induced to express A3A remained largely in G1 phase following ILE depletion (Fig. S4A). After maintaining cultures in ILE-depleted media for 40 hours, cells were cultured in complete media to stimulate S phase entry and were simultaneously induced with doxycycline to express A3A (see schematic in Fig. 3A). After 20 hours of recovery in complete media, cells were enriched in S phase and the majority were replicating regardless of A3A expression (Fig. S4B-C). We then used Western blotting and microscopy to examine γH2AX in cells induced to express A3A in different stages of the cell cycle. By Western blotting, we observed robust H2AX phosphorylation upon A3A expression in S-G2 phase cells. H2AX phosphorylation was undetectable when A3A was expressed in cells synchronized in G1 phase (Fig. 3B). This result was confirmed by immunofluorescent staining of HepaRG cells induced to express A3A, in which more cells exhibited γH2AX staining during replication as compared to those synchronized in G1 phase (Fig. 3C-D). As a control we demonstrated the capability of these cells to mount a DDR following exposure to ionizing radiation (Fig. 3C-D). These observations suggest that cells are particularly susceptible to the activity of A3A during DNA replication.
Figure 3.
A3A-induced cytosine deamination and DNA damage occur primarily during DNA replication. (A) Schematic of cellular synchronization assay. HepaRG-A3A cells were synchronized in G1 phase by culture in isoleucine (ILE)-deficient media for 40 hours. Cells were then induced with doxycycline and cultured for 20 hours either in ILE-deficient media to remain in G1, or ILE-containing media to reach S phase. (B) Expression of A3A leads to H2AX phosphorylation in S phase cells. Western blot analysis of cells treated as in (A) using antibodies to HA and γH2AX. GAPDH was used as a loading control. (C) Analysis of γH2AX staining in HepaRG-A3A cells in G1 or S phase. Cells were treated as in (A) and additionally, HepaRG cells arrested in G1 by culture in ILE-deficient media were irradiated at a dose of 10 Gy. Cells were fixed and stained with anti-HA and anti-γH2AX antibodies, and analyzed by confocal microscopy. Nuclei were stained with DAPI. Scale bar is 10 μm. (D) Quantification of cells with γH2AX staining in (C). (E) Genomic uracil incorporation is increased in replicating cells that express A3A. Genomic DNA was harvested from HepaRG-A3A cells treated as in (A) and incorporated uracils were converted to abasic sites, labeled with Cy5-streptavidin, and transferred to a nylon membrane. Genomic DNA was analyzed for uracil content by phosphorimager and uracil quantitation was determined using a validated set of uracil-containing standards. Error bars are SD. Statistical analysis was performed using an unpaired 2-tail t test. Results are representative of 3 independent experiments.
To determine whether DDR signaling due to A3A correlated with cytosine deamination, genomic DNA was collected from cell lysates and the quantity of uracil in genomes from G1 and S-G2 phase cells was measured as previously described.56 In cells induced to express A3A, genomic uracil quantity in replicating cells was double that of resting cells (Fig. 3E). These data show that cytosine deamination by A3A is more prevalent in replicating cells, and are consistent with the hypothesis that ssDNA at replication forks is a substrate for A3A-mediated DNA damage.
Our data thus far suggest that A3A-induced DNA damage is inflicted upon replicating DNA. Therefore we hypothesized that non-dividing cells should incur minimal DNA damage following A3A expression when compared to actively dividing cells. To test this hypothesis, we examined doxycycline-treated HepaRG-A3A cells in both quiescent and actively cycling states (Fig. 4). Cellular quiescence was achieved by serum deprivation, and confirmed by decreased BrdU and EdU incorporation upon examination by flow cytometry and microscopy (Fig. S5). Cells were examined by microscopy after doxycycline treatment, and we observed that γH2AX was significantly increased in cycling cells compared to quiescent cells (Fig. 4A). As a control, both cell populations were exposed to ionizing radiation and examined for immunofluorescent staining of γH2AX (Fig. 4A). We quantified the number of γH2AX-positive cells and found similar proportions in both irradiated cell types (Fig. 4B). Immunoflourescent staining results were confirmed by Western blotting, which showed that A3A expression correlated with a large increase in γH2AX protein in actively cycling cells (cultured in 10% FBS), whereas minimal γH2AX was detected in serum-deprived cells (Fig. 4C). These experiments show that quiescent cells were less susceptible to DNA damage induced by A3A. Based on these data, we propose that ssDNA exposed at replication forks serves as substrate for damage by A3A (see model in Fig. 4D).
Figure 4.
Quiescent cells are less susceptible than cycling cells to DNA damage by A3A. (A) Non-replicating cells do not accumulate phosphorylated H2AX upon expression of A3A. HepaRG-A3A cells were cultured in media containing 0.1% FBS for 48 hours to arrest cell division or 10% FBS to allow cell cycling. Cells were treated with (+dox) or without (-dox) doxycycline for 24 hours or with 10 Gy γ-irradiation 1 hour prior to being fixed and stained with anti-HA and anti-γH2AX antibodies. Cells were analyzed by confocal microscopy. Nuclei were stained with DAPI. Scale bar is 50 μm. (B) Quantification of cells with γH2AX staining in (A). Error bars are SD. Statistical analysis was performed using an unpaired 2-tail t test. (C) Lysates from HepaRG cells treated as in (A) were analyzed by Western blot using antibodies to HA, γH2AX. GAPDH was used as a loading control. Results are representative of 3 independent experiments. (D) Proposed ssDNA substrates of A3A-induced deamination leading to viral restriction and cellular DNA damage.
Discussion
APOBEC3 enzymes act on ssDNA substrates, and have been shown to deaminate cytosines in reverse-transcribed viral cDNA, viruses with DNA genomes, or transiently exposed ssDNA during retrotransposon integration (see Fig. 4D). Although cellular regulation of APOBEC3 enzyme activity is unknown, current literature suggests that overexpressed or misregulated APOBEC3 enzymes are also capable of mutating the cellular genome.18,37,57 Genome editing events consistent with APOBEC3 activity have been associated with predictable mutational patterns identified in cancer genomes characterized by regional clusters of somatic mutations termed kataegis.17,19,20,58,59 These mutation clusters likely arise from the action of APOBEC3 enzymes on long stretches of ssDNA. In vitro assays have also demonstrated that APOBEC3 enzymes can bind short ssDNA substrates, and suggest that shorter substrates are susceptible to deamination.26 Seplyarskiy, et al. recently provided a biological correlate to these in vitro data by identifying non-clustered APOBEC signature mutations in tumor genome sequences.32 Thus, the degree of genomic damage caused by APOBEC3 enzymes is in part limited by the availability and length of exposed ssDNA. Consequently, identification of the cellular ssDNA substrates with which APOBEC3 enzymes interact is critical to understanding their capacity to damage the cellular genome.
Mutation of cellular DNA by AID, a member of the APOBEC3 family, is transcription-dependent and occurs on the non-transcribed ssDNA template.28,60,61 Similarly, A3A has been reported to deaminate the non-transcribed strand in models of in vitro transcription,26,27 although less efficiently than AID62 and these data have not been recapitulated in mammalian cells. Strand-coordinated clusters of mutations found in cancer genomes have been reported to be co-localized with DNA breakpoints, and postulated to represent APOBEC3 activity at resected ends of DSBs.23 Additionally, expression of A3A and A3B in yeast can produce clusters of break-associated mutations indicative of deamination of resected DSBs.22 The ssDNA at replication forks has been suggested as an additional substrate that is susceptible to APOBEC3 deamination based on recent data from genome sequencing analyses32,33 and model organism systems.30,31 These studies point to the potential for APOBEC3 enzymes to deaminate various cellular substrates, however the capacity for A3A to damage ssDNA during replication has not been previously demonstrated in mammalian cells.
Our data are the first to show in human cells that replicating DNA is highly susceptible to deamination by A3A, whereas quiescent genomes incur minimal damage when A3A is expressed. Furthermore, these data show that cellular processing of uracils created by A3A leads to DNA damage signaling indicative of DNA breaks and replication arrest. Together, these data suggest that ssDNA exposed transiently at replication forks during S phase is a substrate for A3A-induced DNA damage and checkpoint activation.
Our findings contribute to a paradigm for the interaction between A3A and cellular DNA that results in genome instability. A3A-induced deamination is dependent on the interaction of A3A with ssDNA exposed at the replication fork, resected ends at DSBs, or transcription bubbles, and these interactions result in cellular DNA damage and activation of the DNA damage response (Fig. 4D). While this manuscript was under review, several groups published complementary data suggesting APOBEC3 deamination of ssDNA at replication forks in various computational examinations of tumor genome sequences32,33 and model organism evaluations utilizing ectopic APOBEC3 expression.30,31 Notably, Hoopes, et al demonstrated preferential deamination of the lagging strand template in replicating yeast exposed to human A3A and A3B.31 These findings were echoed in a genome sequence analysis of E. coli by Bhagwat, et al. following exposure to the C-terminal deaminase domain of A3G over many generations of replication.30 Additionally, analysis of nearly 4000 whole-genome and whole-exome sequenced cancers by Seplyarskiy, et al. indicated enrichment of APOBEC3 signature mutations on lagging strand templates.32 Together with our findings, this growing body of data reveals the susceptibility of ssDNA exposed during replication to mutagenesis by APOBEC3 enzymes.
We previously showed that A3A expression results in DSBs that activate the DNA damage checkpoint via ATM signaling.37 We now show that A3A also activates the replication checkpoint through ATR activation. In the absence of functional ATM or an intact G1/S checkpoint, A3A expression resulted in activation of ATR signaling and G2/M arrest. ATR activation occurs in response to RPA-bound ssDNA,63 thus these signaling events indicate that A3A acts on cytosines in exposed ssDNA. Deaminated cytosines are repaired by the base excision repair (BER) pathway, during which the uracil base is removed by uracil-DNA glycosylase (UNG) and the resulting abasic site is replaced.64 Error-prone or interrupted processing of deaminated cytosine can lead to base substitutions and DNA breaks.65 Extensive deamination events at replication forks may overwhelm or outpace the cellular BER capacity, thus creating prolonged abasic sites and dysfunctional replication forks. Cells subjected to polymerase stalling and ssDNA generation at the fork through HU treatment exhibited persistent DNA damage signaling when A3A was expressed. By deaminating cytosines in replicating templates, A3A interrupts the replication machinery, and may elicit further ssDNA substrate through stalled replication or collapse of replication forks. This process may create DSBs, and resulting end resection or break-induced replication would then expose additional ssDNA, ultimately leading to a destabilized genome.
APOBEC3 enzymes have been proposed to contribute to oncogenesis.13,17,18 Here we report that A3A expression has a minimal effect on quiescent cells in contrast to actively cycling cells that incur deamination and DNA damage. These data suggest that non-replicating cells may be relatively protected from mutagenesis by A3A, but that A3A poses a threat to genomic integrity of replicating progenitors. This raises the possibility that proliferating immature progenitor cells may be at greater risk for mutations and damage in the context of misregulated A3A activity, potentially leading to genome instability and cellular transformation. Alternatively, A3A may contribute to somatically acquired mutations in previously transformed cells. Replicating DNA is vulnerable to genomic errors, and recovery from replication stress is dependent on intact cell cycle checkpoints and DNA repair mechanisms. Hallmarks of transformed cells include deficiency of essential replication checkpoint components, DNA repair defects, and uncontrolled replication.66 We found that cells exposed to A3A during stalled replication exhibited persistent DNA damage signaling upon recovery from replication stress, suggesting inadequate repair of genomic damage. Robust A3A expression in replicating cancer cells may lead to mutations that, in the absence of intact checkpoint activation and DNA repair mechanisms, contribute to further genome instability and clonal evolution of tumors. Our findings also raise the possibility that A3A activity within cancer cells could be exploited to induce genome toxicity and cellular lethality as a form of cancer therapy.
Materials and methods
Cell lines
U2OS cells were purchased from the American Tissue Culture Collection (ATCC) and maintained in Dulbecco's Modified Eagle's Medium (DMEM) containing 100 U/mL of penicillin and 100 μg/mL of streptomycin, and supplemented with 10% fetal bovine serum (FBS). ATM-deficient and ATM-complemented AT22IJE cells were a gift from Y. Shiloh (Tel Aviv University) and were cultured in similar media. HepaRG cells were a gift from R. Everett (University of Glasgow) and were grown as previously described.67 Inducible HepaRG-A3A and U2OS-A3A cells were constructed using a dual vector system with pLKO-EGFP-TetR and pLKO-TetO-A3A as previously described.37,67 To construct inducible AT22IJE-A3A cell lines, HA-tagged A3A cDNA was inserted into an entry vector with the CMV promoter and upstream Tetracycline Response Element (pEN_Tmcs). A pSLIK-A3A plasmid was generated by Gateway cloning of the pEN_Tmcs-A3A plasmid into a pSLIK lentivirus vector with a constitutively expressed neomycin resistance gene and tetracycline transactivator (rTA)68 to generate a single vector system of doxycycline-inducible expression of A3A-HA. Production of lentiviral particles was achieved by transfection of 293T cells as previously described.67 Following transduction with the pSLIK-A3A lentivirus, cells were selected in 1 μg/mL neomycin. After selection, cells were maintained in 0.5 μg/mL neomycin. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Cell-cycle analysis
HepaRG, U2OS, and AT22IJE cells were grown in the presence or absence of doxycycline (0.1-2 μg/ml, Clontech), fixed in 70% ice-cold ethanol, washed in PBS, and resuspended in staining solution containing 20 μg/ml propidium iodide (Sigma) and 200 μg/ml RNAse A (Roche). For BrdU incorporation, cells were stained using the APC BrdU Flow Kit (BD Bioscience) according to the manufacturer's instructions. Data were collected using an Accuri C6 Flow Cytometer (BD Bioscience) and analyzed by FlowJo software (Version 10.0.8). Experiments were each performed 3 times, and at least 10,000 cells were analyzed per sample.
Western blots, ATR inhibition, and immunofluorescence
For Western blotting, lysates were prepared by harvesting cells in 1x lithium dodecyl sulfate sample buffer (Novex) and 10% 1 M dithiothreitol (DTT, Sigma) and boiling for 10 minutes, then run on bis-tris gels and transferred to a nitrocellulose membrane (Amersham). For ATR inhibition experiments, the small molecule ATR inhibitor VE-822 (Vertex Pharmaceuticals) was dissolved in dimethyl sulfoxide (DMSO) to make a stock solution. Cells were co-treated with doxycycline and VE-822 (100-150 nM) for 24 hours prior to harvest. Controls were treated with an equal volume of vehicle (DMSO). For immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 10 minutes. Nuclei were visualized by staining with 4,6-diamidino-2-phenylindole (DAPI, Sigma). Images were acquired using a Zeiss LSM 710 confocal microscope. Quantification of cell staining was performed on at least 200 cells per condition using ImageJ. Representative images are shown.
Antibodies
Commercially available antibodies used in this study were obtained from Cell Signaling (H2AX, Chk2, Chk2-P-T68, Chk1-P-S345, Chk1-P-S317, γH2AX), Millipore (γH2AX), Epitomics (ATM), Abcam (ATM-P-S1981, ATR), Santa Cruz (HA, Chk1, Tubulin), GeneTex (GAPDH), and Biolegend (HA). Fluorescent secondary antibodies were from Invitrogen. Secondary antibodies for Western blotting included goat anti-rabbit IgG and goat anti-mouse IgG (Jackson ImmunoResearch).
Cell cycle synchronization, replication stress, and quiescence
To synchronize in G1 phase, cells were cultured in isoleucine-free DMEM (ILE-) for 40 hours.55 Cells were released from isoleucine-block by replacing the media with DMEM containing isoleucine (ILE+) for 20 hours to obtain a population enriched for S-G2 phase. To induce replication stress, cells were treated with 5 mM hydroxyurea (HU, Sigma) for 4 hours. For recovery experiments, cells were released into fresh media following treatment with 5 mM HU for 4 hours. To achieve quiescence, HepaRG cells were grown to confluence, then incubated in DMEM supplemented with 0.1% FBS and 1% pen/strep for 48 hours prior to evaluation by Western blot or immunofluorescence.69 To evaluate for DNA replication, cultured cells grown on coverslips were pulsed with 10 μM EdU (Invitrogen) for 40 minutes. Cells were fixed in 4% paraformaldehyde and permeabilized in 1% Triton X-100. After washing, they were incubated in click reaction cocktail containing 10 mM sodium ascorbate (Sigma), 2 mM CuSO4 (Sigma), and 10 μM AlexaFluor 488 Azide (Invitrogen) for 45 minutes. Cells were then washed and incubated in DAPI for 5 minutes. Cells were imaged and analyzed as described above.
Statistical analysis
Standard deviations and p values were generated in Graphpad Prism 6 using paired and unpaired 2-tail t tests.
Uracil quantification assay
Uracil quantitation was performed as previously described.56 Briefly, genomic DNA was extracted and treated with E. coli uracil DNA glycosylase (New England BioLabs) and an aldehyde-reactive probe (Dojindo Laboratories). The DNA samples were spotted onto a positively charged nylon membrane (Immobilon-Ny; Millipore) and membranes were then incubated in a solution containing streptavidin-Cy5 (GE Healthcare). A uracil-containing oligonucleotide duplex served as a uracil standard, and several dilutions of this DNA were processed alongside genomic DNA samples. Fluorescent imaging was achieved using a Typhoon 9210 Phosphorimager. Cy5 fluorescence was analyzed by ImageJ software, and the intensities of the uracil-containing duplexes were used to generate a standard calibration plot of Cy5 fluorescence versus uracil quantity. The numbers of uracils in the genomic DNA samples were determined by interpolating their fluorescence intensities in the calibration plot. These amounts were normalized for the amount of DNA loaded for each sample to calculate the number of uracils per 106 bp (1 million bp = ∼1.05×10−15g).
Supplementary Material
Acknowledgments
We thank members of the Weitzman Lab for insightful discussions and input, especially Milan Savani and Alexandra Wells for technical assistance. We are grateful to Yossi Shiloh and Roger Everett for generously sharing reagents. We thank Florin Tuluc for advice on flow cytometry. We thank Eric Brown and Rahul Kohli for critical reading of the manuscript.
Funding
The authors declare no competing financial interests. A.M.G. was supported as an Eagles Fly for Leukemia Scholar, by a Young Investigator Award from the Alex's Lemonade Stand Foundation, by a fellowship grant from the Canuso Family Foundation, and by the National Institutes of Health (K12 CA076931). D.C.A. was supported in part by T32 CA115299 and F32 GM112414. S.S. was supported by a Thomas C. Rumble Fellowship from Wayne State University. Research in Bhagwat lab was supported by a grant to A.S.B. from the National Institutes of Health (GM57200). Research on APOBEC enzymes in the Weitzman lab was supported by grants to M.D.W. from the National Institutes of Health (CA181259 and CA185799), an award from the W.W. Smith Charitable Trust, and funds from The Children's Hospital of Philadelphia.
References
- [1].Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 2002; 79:285-96; PMID:11863358; http://dx.doi.org/ 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- [2].Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 2005; 22:367-77; PMID:15496550; http://dx.doi.org/ 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- [3].Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 1999; 274:18470-6; PMID:10373455; http://dx.doi.org/ 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- [4].Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annual Rev Biochem 2007; 76:1-22; http://dx.doi.org/ 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- [5].Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002; 418:646-50; PMID:12167863; http://dx.doi.org/ 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- [6].Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell 2003; 113:803-9; PMID:12809610; http://dx.doi.org/ 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- [7].Harris RS, Dudley JP. APOBECs and virus restriction. Virology 2015; 479-480:131-45; PMID:25818029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 2006; 16:480-5; PMID:16527742; http://dx.doi.org/ 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- [9].Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife 2014; 3:e02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA 2006; 103:8780-5; PMID:16728505; http://dx.doi.org/ 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 2006; 281:22161-72; PMID:16735504; http://dx.doi.org/ 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- [12].Narvaiza I, Landry S, Weitzman MD. APOBEC3 proteins and genomic stability: the high cost of a good defense. Cell Cycle 2012; 11:33-8; PMID:22157092; http://dx.doi.org/ 10.4161/cc.11.1.18706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med 2015; 21:274-84; PMID:25820175; http://dx.doi.org/ 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- [14].Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med 2003; 197:1173-81; PMID:12732658; http://dx.doi.org/ 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM, et al.. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell 2009; 16:232-45; PMID:19732723; http://dx.doi.org/ 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Greisman HA, Lu Z, Tsai AG, Greiner TC, Yi HS, Lieber MR. IgH partner breakpoint sequences provide evidence that AID initiates t(11;14) and t(8;14) chromosomal breaks in mantle cell and Burkitt lymphomas. Blood 2012; 120:2864-7; PMID:22915650; http://dx.doi.org/ 10.1182/blood-2012-02-412791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al.. Mutational processes molding the genomes of 21 breast cancers. Cell 2012; 149:979-93; PMID:22608084; http://dx.doi.org/ 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al.. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013; 494:366-70; PMID:23389445; http://dx.doi.org/ 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al.. Signatures of mutational processes in human cancer. Nature 2013; 500:415-21; PMID:23945592; http://dx.doi.org/ 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al.. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499:214-8; PMID:23770567; http://dx.doi.org/ 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lada AG, Stepchenkova EI, Waisertreiger IS, Noskov VN, Dhar A, Eudy JD, Boissy RJ, Hirano M, Rogozin IB, Pavlov YI. Genome-Wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genet 2013; 9:e1003736; PMID:24039593; http://dx.doi.org/ 10.1371/journal.pgen.1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2013; 2:e00534; PMID:23599896; http://dx.doi.org/ 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al.. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 2012; 46:424-35; PMID:22607975; http://dx.doi.org/ 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lada AG, Dhar A, Boissy RJ, Hirano M, Rubel AA, Rogozin IB, Pavlov YI. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biol Direct 2012; 7:47; discussion; PMID:23249472; http://dx.doi.org/ 10.1186/1745-6150-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chan K, Sterling JF, Roberts SA, Bhagwat AS, Resnick MA, Gordenin DA. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS genetics 2012; 8:e1003149; PMID:23271983; http://dx.doi.org/ 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mitra M, Hercik K, Byeon IJ, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon CH, Charlton LM, Nam G, et al.. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res 2014; 42:1095-110; PMID:24163103; http://dx.doi.org/ 10.1093/nar/gkt945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pham P, Landolph A, Mendez C, Li N, Goodman MF. A biochemical analysis linking APOBEC3A to disparate HIV-1 restriction and skin cancer. J Biol Chem 2013; 288(41):29294-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 2003; 422:726-30; PMID:12692563; http://dx.doi.org/ 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- [29].Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol 2003; 4:442-51; http://dx.doi.org/ 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- [30].Bhagwat AS, Hao W, Townes JP, Lee H, Tang H, Foster PL. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc Natl Acad Sci USA 2016; 113:2176-2181; PMID:26839411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hoopes JI, Cortez LM, Mertz TM, Malc EP, Mieczkowski PA, Roberts SA. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Reports 2016; 14:1273-1282; PMID:26832400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Seplyarskiy VB, Soldatov RA, Popadin KY, Antonarakis SE, Bazykin GA, Nikolaev SI. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res 2016; 25:174-182; http://dx.doi.org/ 10.1101/gr.197046.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, Rheinbay E, Kim J, Maruvka YE, Braunstein LZ, et al.. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 2016; 164:538-49; PMID:26806129; http://dx.doi.org/ 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res 2006; 34:89-95; PMID:16407327; http://dx.doi.org/ 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 2008; 320:230-3; PMID:18403710; http://dx.doi.org/ 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- [36].Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol 2015; 89:688-702; PMID:25355878; http://dx.doi.org/ 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep 2011; 12:444-50; PMID:21460793; http://dx.doi.org/ 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature 2000; 408:433-9; PMID:11100718; http://dx.doi.org/ 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- [39].Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004; 432:316-23; PMID:15549093; http://dx.doi.org/ 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- [40].Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 2013; 5:a012716; PMID:24003211; http://dx.doi.org/ 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421:499-506; PMID:12556884; http://dx.doi.org/ 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- [42].Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 2000; 60:5934-6; PMID:11085506. [PubMed] [Google Scholar]
- [43].Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 2001; 410:842-7; PMID:11298456; http://dx.doi.org/ 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- [44].Shiloh Y. The ATM-Mediated DNA-Damage response: taking shape. Trends Biochem Sci 2006; 31:402-10; PMID:16774833; http://dx.doi.org/ 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- [45].Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40:179-204; PMID:20965415; http://dx.doi.org/ 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair 2007; 6:953-66; PMID:17531546; http://dx.doi.org/ 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- [47].Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616-27; PMID:18594563; http://dx.doi.org/ 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 2001; 21:4129-39; PMID:11390642; http://dx.doi.org/ 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al.. Chk1 is an essential kinase that is regulated by ATR and required for the G(2)/M DNA damage checkpoint. Genes Devel 2000; 14:1448-59; PMID:10859164; http://dx.doi.org/ 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858-68; PMID:9488723; http://dx.doi.org/ 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- [51].Ziv Y, Jaspers NG, Etkin S, Danieli T, Trakhtenbrot L, Amiel A, Ravia Y, Shiloh Y. Cellular and molecular characteristics of an immortalized ataxia-telangiectasia (group AB) cell line. Cancer Res 1989; 49:2495-501; PMID:2539904. [PubMed] [Google Scholar]
- [52].Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, Vallis KA, Hammond EM, Olcina MM, Gillies McKenna W, et al.. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis 2012; 3:e441; PMID:23222511; http://dx.doi.org/ 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014; 16:2-9; PMID:24366029; http://dx.doi.org/ 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 2010; 37:492-502; PMID:20188668; http://dx.doi.org/ 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cifuentes E, Croxen R, Menon M, Barrack ER, Reddy GP. Synchronized prostate cancer cells for studying androgen regulated events in cell cycle progression from G1 into S phase. J Cell Physiol 2003; 195:337-45; PMID:12704643; http://dx.doi.org/ 10.1002/jcp.10317. [DOI] [PubMed] [Google Scholar]
- [56].Shalhout S, Haddad D, Sosin A, Holland TC, Al-Katib A, Martin A, Bhagwat AS. Genomic uracil homeostasis during normal B cell maturation and loss of this balance during B cell cancer development. Mol Cell Biol 2014; 34:4019-32; PMID:25154417; http://dx.doi.org/ 10.1128/MCB.00589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A, Vogel RI, Nikas JB, Law EK, Brown WL, et al.. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res 2013; 73:7222-31; PMID:24154874; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al.. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013; 45:970-6; PMID:23852170; http://dx.doi.org/ 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, et al.. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 2015; 47(9):1067-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol 2003; 4:452-6; PMID:12692548; http://dx.doi.org/ 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- [61].Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res 2003; 31:2990-4; PMID:12799424; http://dx.doi.org/ 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Love RP, Xu H, Chelico L. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J Biol Chem 2012; 287:30812-22; PMID:22822074; http://dx.doi.org/ 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 2003; 100:13827-32; PMID:14605214; http://dx.doi.org/ 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol 2013; 5:a012583; PMID:23545420; http://dx.doi.org/ 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Krokan HE, Saetrom P, Aas PA, Pettersen HS, Kavli B, Slupphaug G. Error-free vs. mutagenic processing of genomic uracil–relevance to cancer. DNA Repair 2014; 19:38-47; PMID:24746924; http://dx.doi.org/ 10.1016/j.dnarep.2014.03.028. [DOI] [PubMed] [Google Scholar]
- [66].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [67].Everett RD, Parsy ML, Orr A. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol 2009; 83:4963-77; PMID:19264778; http://dx.doi.org/ 10.1128/JVI.02593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al.. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci USA 2006; 103:13759-64; PMID:16945906; http://dx.doi.org/ 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Langan TJ, Chou RC. Synchronization of mammalian cell cultures by serum deprivation. Methods Mol Biol 2011; 761:75-83; PMID:21755442; http://dx.doi.org/ 10.1007/978-1-61779-182-6_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.