ABSTRACT
Epigenetic mechanisms have not been characterized in ticks despite their importance as vectors of human and animal diseases worldwide. The objective of this study was to characterize the histones and histone modifying enzymes (HMEs) of the tick vector Ixodes scapularis and their role during Anaplasma phagocytophilum infection. We first identified 5 histones and 34 HMEs in I. scapularis in comparison with similar proteins in model organisms. Then, we used transcriptomic and proteomic data to analyze the mRNA and protein levels of I. scapularis histones and HMEs in response to A. phagocytophilum infection of tick tissues and cultured cells. Finally, selected HMEs were functionally characterized by pharmacological studies in cultured tick cells. The results suggest that A. phagocytophilum manipulates tick cell epigenetics to increase I. scapularis p300/CBP, histone deacetylase, and Sirtuin levels, resulting in an inhibition of cell apoptosis that in turn facilitates pathogen infection and multiplication. These results also suggest that a compensatory mechanism might exist by which A. phagocytophilum manipulates tick HMEs to regulate transcription and apoptosis in a tissue-specific manner to facilitate infection, but preserving tick fitness to guarantee survival of both pathogens and ticks. Our study also indicates that the pathogen manipulates arthropod and vertebrate cell epigenetics in similar ways to inhibit the host response to infection. Epigenetic regulation of tick biological processes is an essential element of the infection by A. phagocytophilum and the study of the mechanisms and principal actors involved is likely to provide clues for the development of anti-tick drugs and vaccines.
KEYWORDS: Anaplasma, epigenetics, histone modifying enzyme, histone, pathogen, tick
Introduction
It is increasingly recognized that intracellular pathogens manipulate the transcriptional programs of their host cells via epigenetic mechanisms, leading to radical effects, notably on stress and inflammatory responses.1-3 Indeed, such pathogens can be viewed as ‘epigenators’,4 by analogy to environmental factors, but acting within the cell.3 One such organism known to manipulate host cells is Anaplasma phagocytophilum (Rickettsiales: Anaplasmataceae), an emerging zoonotic pathogen transmitted by ticks of the genus Ixodes. The major A. phagocytophilum vector species are Ixodes scapularis in North America and Ixodes ricinus in Europe.5 A. phagocytophilum infects tick midgut, hemocytes and salivary glands and vertebrate host granulocytes causing human, canine, and equine granulocytic anaplasmosis and tick-borne fever of ruminants.6-13 To establish infection, A. phagocytophilum manipulates cell defense mechanisms in both ticks and vertebrate hosts by inhibition of cellular processes, such as apoptosis and the immune response.14-19
Recently, histone deacetylase 1 (HDAC1) upregulation was shown in A. phagocytophilum-infected granulocytes.20 Higher HDAC1 levels were associated with an increase in A. phagocytophilum survival, suggesting a mechanism for control of host cell gene expression and function based on epigenetic changes.20 Furthermore, recent studies showed that chromatin bound bacterial effector Ankyrin A (AnkA)21-23 recruits HDAC1 and modifies host gene expression, therefore supporting the role of A. phagocytophilum proteins secreted through the Type IV secretion system (a secretion system composed of a macromolecular complex that spans the bacterial inner and outer membranes and can also span the membrane of eukaryotic host cells used by Gram-negative bacteria for a variety of biological functions including the exchange of genetic material with other bacteria and the translocation of oncogenic DNA and effector proteins into eukaryotic host cells)24 in controlling host epigenetics and global DNA methylation.25,26 These results and the finding that A. phagocytophilum evolved common strategies for infection of ticks and vertebrate hosts suggest that the pathogen probably controls host cell epigenetics in both granulocytes and tick cells.13
However, the role of epigenetic mechanisms in tick cells is largely unknown.27 Recently, Kotsyfakis et al.28 annotated 34 transcripts as encoding candidate histone modifying proteins in the I. ricinus transcriptome but without further characterization. The I. scapularis tick genome that has been well characterized and therefore constitutes a good model for the study of tick-pathogen interactions.18,29 The objective of this study was to characterize the role of histones and histone modifying enzymes (HMEs) during A. phagocytophilum infection of I. scapularis. Among the enzyme families that write or erase modifications on histone tails, as well as other protein substrates, we focused on histone acetyltransferases (HATs),30 histone deacetylases (HDACs and Sirtuins),31,32 histone methyltransferases (HMTs)33 and histone demethylases (HDMs).34 We first identified in the I. scapularis genome/transcriptome and phylogenetically and structurally characterized 5 histones and 34 HMEs (HATs, HDACs, Sirtuins, HMTs, and HDMs) in comparison with similar proteins in other organisms. Then, we used transcriptomics and proteomics data to analyze the mRNA and protein levels of I. scapularis histones and HMEs in response to A. phagocytophilum infection of tick tissues and cultured cells. Finally, selected HMEs were functionally characterized by pharmacological studies in cultured tick cells. The results suggested that the manipulation of histones and HMEs by A. phagocytophilum is an epigenetic mechanism by which this pathogen simultaneously regulates several defense mechanisms, such as apoptosis, to increase infection and multiplication in tick cells.
Results and discussion
Identification of histones and HMEs in I. scapularis
I. scapularis histones
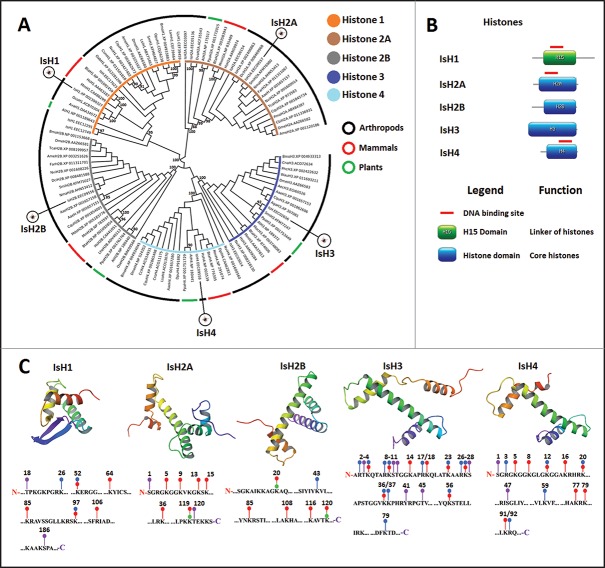
In the nucleus, DNA is wrapped into a complex known as chromatin by core histones H2A, H2B, H3, and H4 to form the nucleosome. Another linker histone, H1, binds the nucleosome at the entry and exit sites of the DNA to assemble higher order chromatin structures.35 Five putative histones were identified in the I. scapularis genome and transcriptome and classified as IsH1, IsH2A, IsH2B, IsH3, and IsH4 (Fig. 1A). All these histones except IsH4 were closely related to other tick histones from Ornithodoros coriaceus, Amblyomma variegatum, and Nuttalliella namaqua. The IsH4 was closely related to mammalian histone H4 from Homo sapiens, Mus musculus, and Bos taurus, and not to H4 from other arthropods (Fig. 1A). The analysis of the putative histones from I. scapularis showed typical domains (Fig. 1B) and tertiary fold structure with helix-turn-helix-turn-helix motifs (Fig. 1C) found in other organisms. Most key histone residues (lysine, tyrosine, serine, and threonine) that undergo posttranslational modifications were conserved between human and I. scapularis predicted homologs (Fig. 1C). Differential post-translational modifications by acetylation, methylation, phosphorylation, or ubiquitination of the conserved residues constitute an important component of the histone code.36 These results suggested that in I. scapularis, as in other organisms, gene regulation is affected by a combination of these histone modifications and, therefore, different chromatin states are defined by specific histone mark repertoires.37
Figure 1.
Characterization of I. scapularis histones. (A) Phylogenetic analysis of histone amino acid sequences. A neighbor joining phylogenetic tree was built using the amino acid sequences of the histones identified in I. scapularis (IsH1, IsH2A, IsH2B, IsH3, and IsH4) and their homologs in H. sapiens (Hs), M. musculus (Mm), B. taurus (Bta), D. melanogaster (Dme), Lycosa singoriensis (Lsi), A. variegatum (Ava), O. coriaceus (Oco), N. namaqua (Nma), Oreta pulchripes (Opu), Leptidea amurensis (Lam), Orussus abietinus (Oab), Leptidea juvernica (Lju), Bombyx mori (Bmo), Apis mellifera (Ame), Culex quinquefasciatus (Cqu), Aedes aegypti (Aae), Anopheles gambiae (Aga), Ceratosolen solmsi marchali (Csm), Nasonia vitripennis (Nvi), Fopius arisanus (Far), Cerapachys biroi (Cbi), Diaphorina citri (Dci), Penaeus monodon (Pmo), S. mimosarum (Smi), Tribolium castaneum (Tca), Latrodectus hesperus (Lhe), Musca domestica (Mdo), Oxyopes sertatus (Ose), P. h. corporis (Phc), Wasmannia auropunctata (Wau), Cybaeota nana (Cna), Lepeophtheirus salmonis (Lsa), Caligus rogercresseyi (Cro), C. clemensi (Ccl), Diadromus pulchellus (Dpu), Diprion pini (Dpi), Arabidopsis thaliana (At) and Physcomitrella patens (Ppa). Bootstrap values for internal branches and GenBank accession numbers are shown. (B) Major protein domains found in I. scapularis histones. (C) Predicted tertiary structure of I. scapularis histones with conserved positions of post-translational modifications. The modeled histone tertiary structures are shown color-coded from the N-terminus (red-orange) to the C-terminus (blue-purple). Selected amino acid positions are shown below each tertiary structure with post-translational modifications indicated with red ballstick, acetylation; blue ballstick, methylation; purple ballstick, phosphorylation; green bomb, ubiquitination.
I. scapularis HATs
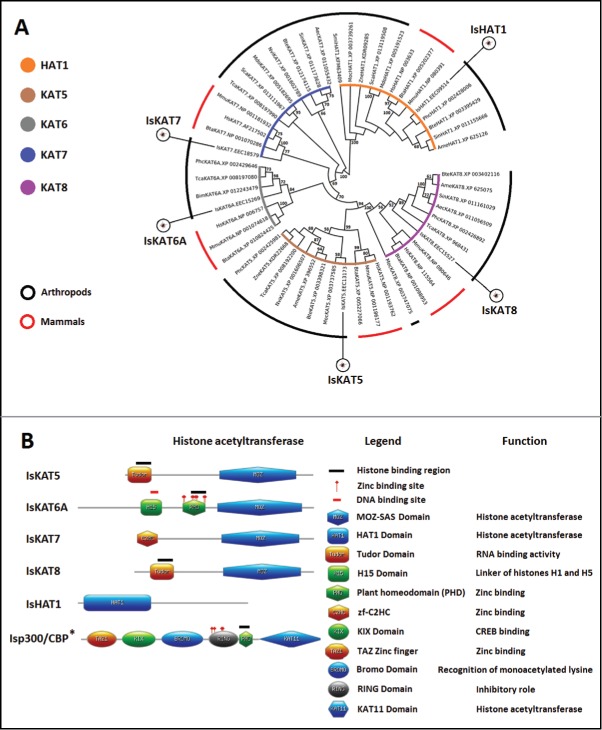
The HAT protein family contains “writer” enzymes that add acetyl groups to histones, while HDACs and Sirtuins are “eraser” enzymes that remove acetyl groups on or from histones, respectively.30-32 Histone acetylation is a dynamic process regulated by HATs that modulates the accessibility of DNA through alterations to the compaction of the chromatin and also regulates the binding of repressors and activators of gene transcription.38 Three major HAT families have been described in humans, including the GCN5/PCAF family (consisting of GCN5, PCAF, and related proteins), the p300/CBP family, and the MYST family, renamed in 2007 as lysine (K) acetyltransferase (KAT). According to this new classification, TIP60, MOZ, MORF, HBO1, and hMOF (MYST family) became KAT5, KAT6A, KAT6B, KAT7, and KAT8, respectively.3 From the I. scapularis genome and transcriptome data, we identified IsHAT1 (GCN5/PCAF family), Isp300/CBP (p300/CBP family), and several members of the MYST family (IsKATs) (Fig. 2A, Fig. 2B, and Supplementary File 1).
Figure 2.
Characterization of I. scapularis HATs. (A) Phylogenetic analysis of HAT amino acid sequences. A neighbor joining phylogenetic tree was built using the amino acid sequences of the HATs identified in I. scapularis (IsHAT1, IsKAT5, IsKAT6A, IsKAT7 and IsKAT8) and their homologs in H. sapiens (Hs), M. musculus (Mm), B. taurus (Bta), A. mellifera (Ame), Acromyrmex echinatior (Aec), Bombus terrestris (Bte), Bombus impatiens (Bim), M. domestica (Mdo), N. vitripennis (Nvi), P. h. corporis (Phc), Stomoxys calcitrans (Sca), Solenopsis invicta (Sin), T. castaneum (Tca), Zootermopsis nevadensis (Zne), M. occidentalis (Moc), and S. mimosarum (Smi). Bootstrap values for internal branches and GenBank accession numbers are shown. (B) Major protein domains found in I. scapularis HATs. *Additional characterization of Isp300/CBP is presented in Supplementary File 2: Figures S1A-S1C.
I. scapularis p300/CBP
The p300/CBP are transcriptional co-activator proteins playing a central role in coordinating and integrating multiple signal-dependent events with the transcription apparatus, allowing the appropriate level of gene activity to occur in response to diverse physiological stimuli.39 The phylogenetic analysis showed that Isp300/CBP is more closely related to similar proteins from chelicerates (i.e., Limulus Polyphemus and Stegodyphus mimosarum) than to either mammalian CBP or p300 or arthropod (i.e., Drosophila melanogaster) CBP proteins (Supplementary Figure 1A and Supplementary Fig. 1B). However, Isp300/CBP presented a domain organization similar to mammalian orthologs but without the third and fourth zinc-binding domain present in mammals, insects and other chelicerates (Supplementary File 1, Supplementary Figure 1A, and Supplementary Fig. 1C). The absence of the fourth zinc-binding domain was not due to incomplete Isp300/CBP annotation, as we found that the C-terminus is present (Supplementary Fig. 1C). These results suggested that Isp300/CBP has a deletion of the zinc-binding domains ZZ and TAZ2 with unknown functional implications.
I. scapularis HAT1
The IsHAT1 was the only representative of type B HATs identified in I. scapularis (Fig. 2A and Fig. 2B). A previous study identified a HAT in I. scapularis but did not classify it as IsHAT1.40 Type B HATs are distinguished by their substrate specificity and subcellular localization.41 HAT1 can be found in the cytoplasm and the nucleus with the ability to acetylate free, mainly newly synthesized histones H3 and H4 but not nucleosomal histones.41 IsHAT1 clustered together with HAT1 from another blood sucking arthropod, Pediculus humanus corporis and mammals such as H. sapiens, but not with HAT1 from other chelicerates, such as S. mimosarum spider and Metaseiulus occidentalis mite, that are closely related to ticks (Fig. 2A). The HAT1 domain with acetyltransferase activity in the human HAT1 was highly conserved in the I. scapularis homolog with 44% identity at the amino acids sequence level.
I. scapularis KAT
Four members of the MYST family were classified in I. scapularis as IsKAT5, IsKAT6A, IsKAT7, and IsKAT8 (Fig. 2A and Fig. 2B). Members of this family share the MYST (MOZ-SAS) domain, which confers the intrinsic acetyltransferase activity of these enzymes.30 The MYST domain was identified in all IsKATs (Fig. 2B) and their tertiary structure was highly conserved (Supplementary File 1). This protein family is extremely diverse in domain organization, complex formation, and biological function.30 Specifically, KAT6A (also known as MOZ) contains a PHD finger domain that is important for histone H3 binding, and this domain was identified in IsKAT6A (Fig. 2B). KAT6A acetylates H3 at lysine-9 and lysine-14,42,43 which were conserved in IsH3 (Fig. 1C). These results suggested a function for IsKATs similar to that described in other organisms.30
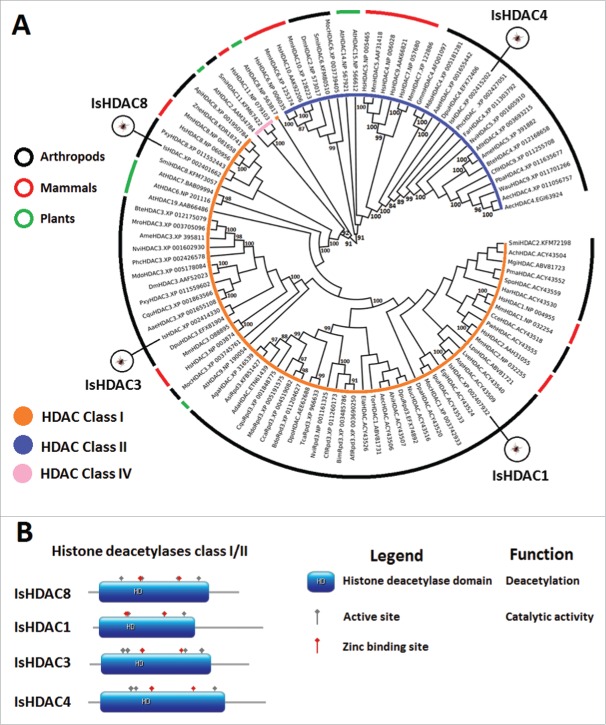
I. scapularis HDACs
From prokaryotes to mammals, HDACs are part of a large, evolutionarily conserved protein family.44 Human HDACs are organized into 4 classes, of which classes I, II, and IV are phylogenetically related, but class III (Sirtuins) is phylogenetically separated.31,32 HDAC class I contains 4 HDACs (HDAC1-3 and HDAC8), class II 6 HDACs (HDAC4-7, HDAC9 and HDAC10), and class IV only one HDAC (HDAC11).31 In I. scapularis, 3 class I HDACs (IsHDAC1, 3, and 8) and one class II HDAC (IsHDAC4) were identified (Fig. 3A, Figure 3B, and Supplementary File 1). A fragment of 144 amino acids (Supplementary Table 1) was found with 63% identity to human HDAC6 but was not included in further analyses because the fragment was too short for a more detailed characterization. In mammals, class I HDACs are ubiquitously expressed with a predominantly nuclear localization, but the class II HDAC4 can shuttle in and out of the nucleus.44 Class I HDACs are almost entirely comprised of a conserved deacetylase domain and have minimal N- and C-terminal regions, while HDAC4 possesses extensive N-terminus adapter domains.44 IsHDACs possess the typical active site and zinc/potassium binding sites (Fig. 3B and Supplementary File 1). Additionally, IsHDACs belonged to the α/β protein class with 7–8 β-strands at its core surrounded by several α-helices representing the Rossmann fold—a typical motif of nucleotide binding enzymes (Supplementary File 1).
Figure 3.
Characterization of I. scapularis HDACs. (A) Phylogenetic analysis of HDAC amino acid sequences. A neighbor joining phylogenetic tree was built using the amino acid sequences of the HDACs identified in I. scapularis (IsHDAC1, IsHDAC3, IsHDAC4 and IsHDAC8) and their homologs in H. sapiens (Hs), M. musculus (Mm), A. thaliana (At), Daphnia pulex (Dpu), C. quinquefasciatus (Cqu), Anopheles sinensis (Asi), Bactrocera dorsalis (Bdo), Ceratitis capitata (Cca), Anopheles darling (Ada), T. castaneum (Tca), Apis florea (Afl), B. impatiens (Bim), Camponotus floridanus (Cfl), M. domestica (Mdo), A. gambiae (Aga), Dendroctonus ponderosae (Dpo), N. vitripennis (Nvi), P. h. corporis (Phc), F. arisanus (Far), A. echinatior (Aec), Pogonomyrmex barbatus (Pba), W. auropunctata (Wau), A. aegypti (Aae), Glossina morsitans morsitans (Gmm), B. terrestris (Bte), A. mellifera (Ame), Z. nevadensis (Zne), Acyrthosiphon pisum (Api), Plutella xylostella (Pxy), Megachile rotundata (Mro), S. mimosarum (Smi), M. occidentalis (Moc), Amblyomma sp. (Asp), Cryptocellus centralis (Cce), L. polyphemus (Lpo), Prokoenenia wheeleri (Pwh), Hadrurus arizonensis (Har), Aphonopelma chalcodes (Ach), Eremocosta gigasella (Egi), Leiobunum verrucosum (Lve), Mastigoproctus giganteus (Mgi), Phrynus marginemaculatus (Pma), Dinothrombium pandorae (Dpa), Tanystylum orbiculare (Tor), Endeis laevis (Ela), Achelia echinata (Aec), Nymphon unguiculatum-charcoti (Nuc), Idiogaryops pumilis (Ipu), Stenochrus portoricensis (Spo), Ammothea hilgendorfi (Ahi), M. occidentalis (Moc), and S. mimosarum (Smi). Bootstrap values for internal branches and GenBank accession numbers are shown. (B) Major protein domains found in I. scapularis HDACs.
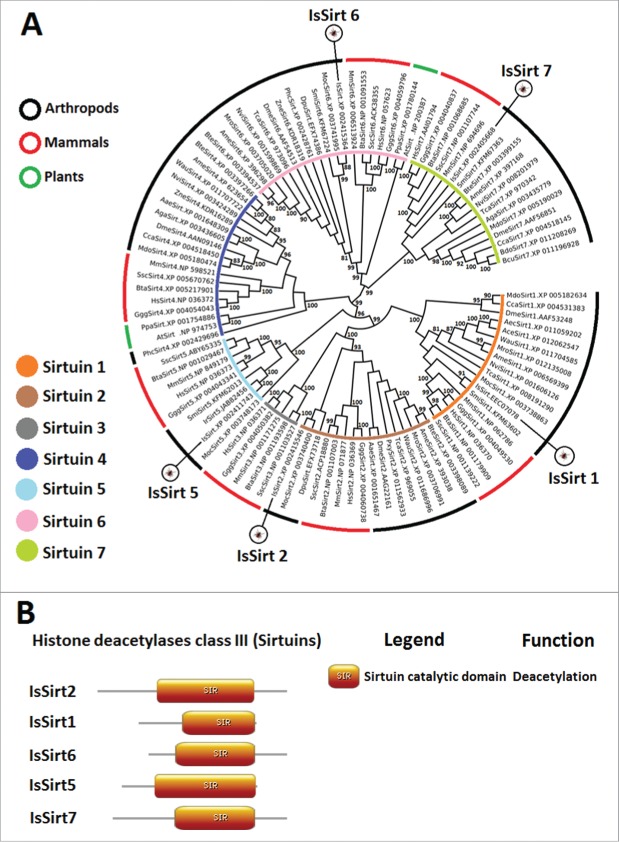
I. scapularis Sirtuins
Initially categorized as class III HDACs, Sirtuins are NAD+-dependent protein N-ε-acyl-lysine deacetylases that are phylogenetically and structurally distinct from class I, II, and IV HDACs. Sirtuins are involved in a wide spectrum of biological processes, such as transcriptional regulation, metabolism, aging, apoptosis, and DNA damage responses.45,46 There are 7 human Sirtuins, but a variable number of Sirtuins are found in other organisms.32 In parasitic pathogens, certain Sirtuins are essential for the survival and/or development of these organisms, determining their interest as drug targets.47 Sirtuins were also recently shown to be targets for host manipulation by parasites48 and bacteria.49 Of the 7 Sirtuins previously reported across all kingdoms of life,32 5 were identified and characterized in I. scapularis (IsSirt1, 2, 5–7) (Fig. 4A, Figure 4B, and Supplementary File 1). Not all organisms contain all 7 Sirtuins.32 For example, consistent with our results in I. scapularis, Sirt3 is absent in other arthropods32 (Fig. 4A). However, Sirt4, which is present in other arthropods such as D. melanogaster,32 was not found in I. scapularis (Fig. 4A). Like I. scapularis, the flatworm parasites Schistosoma mansoni and S. japonicum also lack Sirt3 and Sirt4.47,50 All Sirtuins including those identified in I. scapularis contain a conserved catalytic core domain composed of a NAD+-binding Rossmann fold domain and a smaller zinc-binding domain containing 4 highly conserved Cys residues (Fig. 4B and Supplementary File 1). The I. scapularis Sirtuins conform to the conserved tertiary structure of Sirtuins from other organisms (Supplementary File 1).51-55
Figure 4.
Characterization of I. scapularis Sirtuins. (A) Phylogenetic analysis of Sirtuin amino acid sequences. A neighbor joining phylogenetic tree was built using the amino acid sequences of the Sirtuins identified in I. scapularis (IsSirt1, IsSirt2, IsSirt5, IsSirt6 and IsSirt7) and their homologs in H. sapiens (Hs), M. musculus (Mm), B. taurus (Bta), Sus scrofa (Ssc), Gorila gorila gorila (Ggg), M. domestica (Mdo), S. mimosarum (Smi), C. capitata (Cca), A. echinatior (Aec), N. vitripennis (Nvi), Atta cephalotes (Ace), W. auropunctata (Wau), M. rotundata (Mro), A. mellifera (Ame), T. castaneum (Tca), A. aegypti (Aae), B. terrestris (Bte), D. pulex (Dpu), P. xylostella (Pxy), Z. nevadensis (Zne), P. h. corporis (Phc), A. gambiae (Aga), Bactrocera cucurbitae (Bcu), B. dorsalis (Bdo), M. occidentalis (Moc), A. thaliana (At) and P. patens (Ppa). Bootstrap values for internal branches and GenBank accession numbers are shown. (B) Major protein domains found in I. scapularis Sirtuins.
I. scapularis HMTs
Histone lysine methylation by HMTs regulates chromatin organization and, depending on the lysine residue that is targeted, either activates or represses gene expression.33 Based on structural features of their SET domain, HMTs are divided into 7 families that include the SUV39, EZH, SET1, SET2, PRDM, and SMYD families and other HMTs containing a SET domain. Only one HMT (DOT1L) does not contain a SET domain. 33,56 Of the 7 HMT families plus DOT1L that have been identified in humans33,56 we identified in I. scapularis members of the SET2 (IsSETD2, IsSETD4, IsSETD7), SUV39 (IsSETDB1-A, IsSETDB1-B, IsEHMT), and SMYD (IsSMYD3-5) families and the tick homolog of human DOT1L (IsDOT1L) (Fig. 5A, Figure 5B, and Supplementary File 1). All human SMYD family members contain a SET domain and a MYND zinc finger56 that were identified in IsSMYD4 and IsSMYD5 but not in IsSMYD3 (Fig. 5B and Supplementary File 1). However, the available IsSMYD3 sequence lacks the C-terminus, which may explain the absence of the SET domain and the MYND zinc finger (Fig. 5B). It was not possible to classify the IsEHMT as IsEHMT1 or IsEHMT2 because EHMTs from chelicerates (i.e., I. scapularis, S. mimosarum, and M. occidentalis) formed a cluster separate from mammal and insect EHMT1 and EHMT2 (Fig. 5A), suggesting that EHMT1 and EHMT2 possess amino acid sequence properties that may be taxa-specific. Nevertheless, the IsEHMT predicted tertiary structure was similar to the structure of human EHMT2 with Ankyrin, Pre-SET, and SET domains (Fig. 5B and Supplementary File 1).33
Figure 5.
Characterization of I. scapularis HMTs. (A) Phylogenetic analysis of HMT amino acid sequences. A neighbor joining phylogenetic tree was built using the amino acid sequences of the HMTs identified in I. scapularis (IsSETDB1-A, IsSETDB1-B, IsEHMT, IsSETD2, IsSETD4, IsSETD7, IsDOT1L, IsSMYD3, IsSMYD4, and IsSMYD5) and their homologs in H. sapiens (Hs), M. musculus (Mm), B. taurus (Bta), A. mellifera (Ame), A. echinatior (Aec), S. mansoni (Sma), B. terrestris (Bte), C. floridanus (Cfl), N. vitripennis (Nvi), P. h. corporis (Phc), M. occidentalis (Moc), S. invicta (Sin), D. pulex (Dpu), M. domestica (Mdo), Z. nevadensis (Zne), S. mimosarum (Smi), A. aegypti (Aae), O. abietinus (Oab), P. barbatus (Pba), and S. calcitrans (Sca). Bootstrap values for internal branches and GenBank accession numbers are shown. (B) Major protein domains found in I. scapularis HMTs.
I. scapularis HDMs
The HDMs are grouped into the Jumonji (JMJ) demethylase family or the LSD demethylase (lysine-specific histone demethylase) family that are mainly composed of LSD1 (also known as KDM1A) and LSD2 (also known as KDM1B).34 The 2 evolutionarily conserved LSD and JMJC histone demethylase families use different reaction mechanisms to establish demethylation.34 In humans, the LSD family has 2 members, LSD1 and LSD2, that use a flavin adenine dinucleotide (FAD)-dependent amine oxidation reaction to catalyze the demethylation of their substrate.34 All JMJC family members share a JMJC (Jumonji) domain that is essential in demethylase activity.34 The JMJCs are a multigenic family that in humans consists of 30 members, of which 18 have been shown to possess a dioxygenase reaction that depends on Fe(II) and α-ketoglutarate.34 Two copies of LSD1 were identified in I. scapularis (IsLSD1-A and IsLSD1-B), but no homolog for LSD2 was found (Fig. 6A and Fig. 6B). The two LSD1 shared 80% homology at the amino acid level, suggesting that they are paralogs with similar tertiary structures (Supplementary File 1). Both IsLSD1-A and IsLSD1-B possess SWIRM and amino oxidase domains (Fig. 6B) that are typical of human LSD1.34 IsLSD1-A and IsLSD1-B have 3 major differences: (i) IsLSD1-B is larger in the N-terminus than IsLSD1-A, (ii) IsLSD1-B does not possess a “spacer region” found in the amino oxidase domain of IsLSD1-A as well as in mammalian LSD1, and (iii) IsLSD1-A does not possess a NAD-binding domain present in the human LSD1 (Fig. 6C). These findings suggested that IsLSD1-A and IsLSD1-B might have different functions in I. scapularis. Six members of the JMJC demethylases were identified in I. scapularis (IsUTY, IsJARID2-A, IsJMJD1B, IsJARID1A, IsJMJD2B, and IsJARID2-B) (Fig. 6A and Fig. 6B). All I. scapularis JMJC demethylases possess the JMJC domain while other domains such as PHD and C5HC2 domains with zinc-binding activity were found in IsJARID1A and IsJMJD2B (Supplementary File 1). However, IsJMJD1B lacks the zinc-binding domain and only possesses the nickel-binding site. IsUTY presented the typical cluster of tetratricopeptide repeats (TRP) in the N-terminus and the JMJC domain at the C-terminus (Fig. 6C and Supplementary File 1). These results suggested that IsJMJDs are structurally and probably functionally conserved.
Figure 6.
Characterization of I. scapularis HDMs. (A) Phylogenetic analysis of HDM amino acid sequences. A neighbor joining phylogenetic tree was built using the amino acid sequences of the HDMs identified in I. scapularis (IsLSD1-A, IsLSD1-B, IsUTY, IsJARID1A, IsJARID2-A, IsJARID2-B, IsJMJD1B, and IsJMJD2B) and their homologs in H. sapiens (Hs), M. musculus (Mm), B. taurus (Bta), A. mellifera (Ame), B. terrestris (Bte), Harpegnathos saltator (Hsa), N. vitripennis (Nvi), P. h. corporis (Phc), S. invicta (Sin), T. castaneum (Tca), S. mimosarum (Smi), M. domestica (Mdo), and M. occidentalis (Moc). Bootstrap values for internal branches and GenBank accession numbers are shown. (B) Major protein domains found in I. scapularis HDMs.
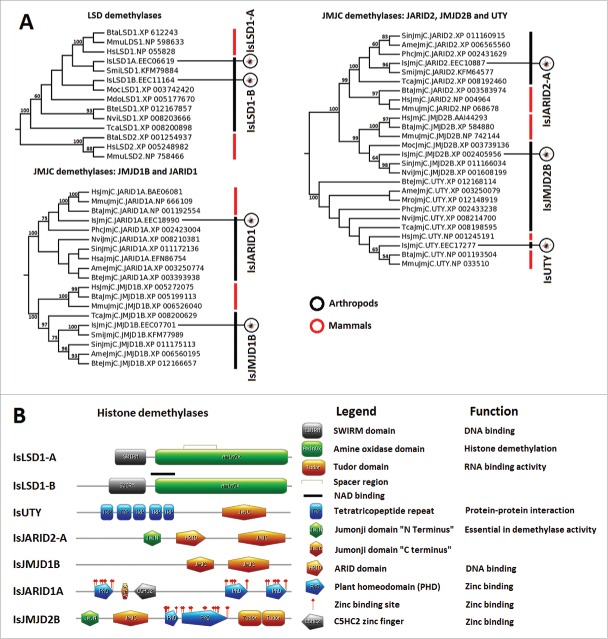
I. scapularis histone and HMEs mRNA and protein levels vary in response to A. phagocytophilum infection
The finding of histones and HMEs coding genes in I. scapularis suggested the existence of evolutionarily conserved regulatory mechanisms that have not been functionally characterized in ticks and other arthropod vectors of human and animal diseases. The tick response to A. phagocytophilum infection is largely regulated at the transcriptional level.18,19 Therefore, to functionally characterize the role of I. scapularis tick histones and HMEs in response to A. phagocytophilum infection, we first analyzed the quantitative transcriptomics and proteomics raw data deposited on the Dryad repository database, NCBI's Gene Expression Omnibus database and ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002181 and doi: 10.6019/PXD002181.18,19
The analysis of histone and HME mRNA and protein levels were conducted in uninfected versus A. phagocytophilum-infected samples in I. scapularis nymphs (N), adult female midguts (G) and salivary glands (SG), and ISE6 cultured cells that constitute a model for hemocytes infected with A. phagocytophilum.19 The results showed mRNA/protein-specific and tissue-specific differences between infected and uninfected samples (Fig. 7). At the mRNA level, differences were observed between N, G, SG, and ISE6 cells (Fig. 7). Differences at the mRNA level probably reflect tissue-specific responses to pathogen infection.18 At the protein level, many proteins were not detected by mass spectrometry but for the identified proteins, the results showed a low (31%) correlation between differential regulation at the mRNA and protein levels (Fig. 7). These results were similar to those obtained before for other genes and proteins in response to A. phagocytophilum infection suggesting that differences between mRNA and protein levels could be due to delay between mRNA and protein accumulation, which requires sampling at different time points and/or the role for posttranscriptional and posttranslational modifications in tick tissue-specific response to A. phagocytophilum infection.18,19 In fact, some histones and HMEs are regulated at the transcriptional levels while other are affected by posttranslational modifications.57
Figure 7.
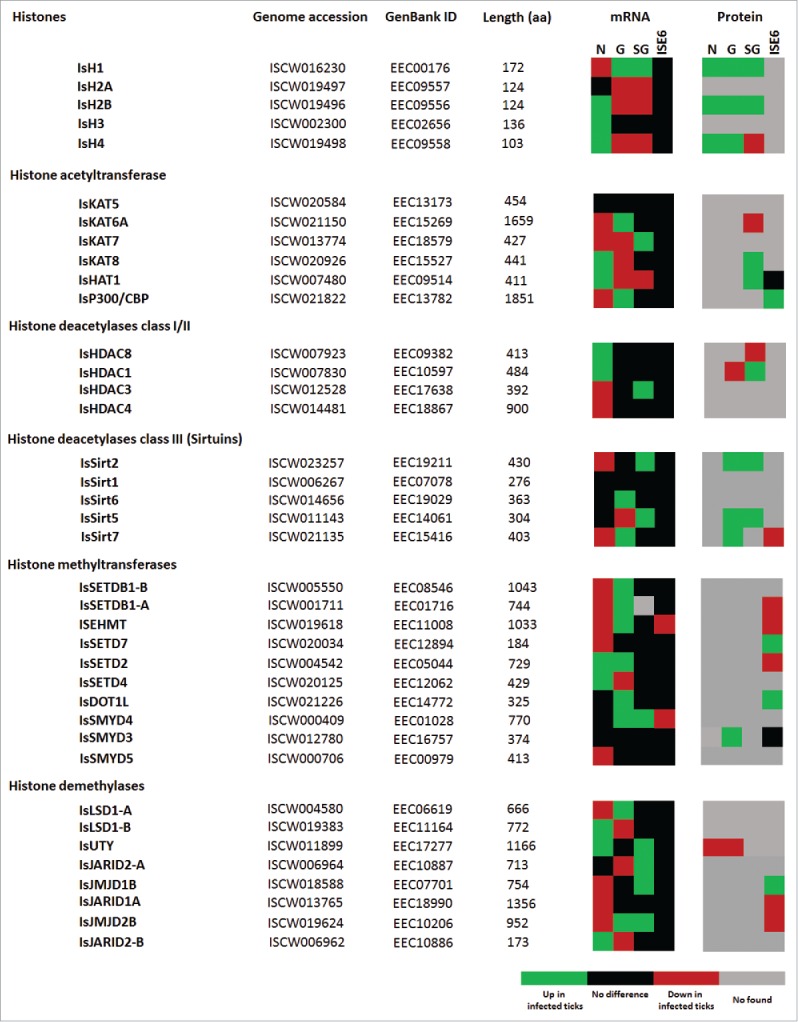
I. scapularis histone and HME mRNA and protein levels in response to A. phagocytophilum infection. Comparison of histone and HME mRNA and protein levels in I. scapularis nymphs (N), female midguts (G), female salivary glands (SG), and ISE6 cells (ISE6) in response to A. phagocytophilum infection. Transcriptomics and proteomics data were obtained from previously published datasets available in public repositories (see Methods).
Despite these general considerations, some results were functionally relevant for the characterization of tick-pathogen molecular interactions. Histone IsH1, IsH2A, and IsH4 protein levels were affected by A. phagocytophilum infection in tick N, G, and SG, while most histone mRNA levels varied in response to infection in these tissues (Fig. 7). Histones and their posttranslational modifications have key roles in chromatin remodeling and gene transcription with different roles in biological processes such as immunity.58 Therefore, higher histone protein levels in infected ticks probably reflected a tick response to infection with possible implications in the control of pathogen levels to promote tick survival.
The IsHDAC1 protein levels were lower in G and higher in SG in infected samples when compared to uninfected controls (Fig. 7). Garcia-Garcia et al.20 showed that HDAC1 expression and activity increase with A. phagocytophilum infection of human neutrophils, which is critical for pathogen survival. Upon A. phagocytophilum infection, downregulation of host defense genes also occurs along with HDAC1 binding and histone H3 deacetylation at their promoters.20 Recently, Mukherjee et al.59 showed that pathogen infection in insect results in the upregulation of HDACs to inhibit host immunity and facilitate infection. Therefore, as previously shown for other biological processes,14 the increase in HDAC1 levels in response to A. phagocytophilum in I. scapularis SG, insect, and human cells suggested that the pathogen manipulates arthropod and vertebrate host cell epigenetics in similar ways to inhibit host immune response and facilitate infection. Furthermore, considering the differential representation of IsHDCA1 proteins in tick G and SG (Fig. 7) and their role in cell apoptosis,60-62 these results suggested a compensatory mechanism by which these proteins regulate transcription and apoptosis in a tissue-specific manner in response to A. phagocytophilum infection. In infected tick SG, A. phagocytophilum may manipulate tick cells to increase IsHDCA1 levels, resulting in the inhibition of cell defense and apoptosis to facilitate infection, while lower IsHDAC1 levels in G may be a tick cell compensatory response to induce apoptosis to limit pathogen infection and promote tick survival. These results support the evolution of mechanisms by which A. phagocytophilum manipulates tick protective responses to facilitate infection but preserves tick fitness to guarantee survival of both pathogens and ticks.
The proteins IsSirt2, IsSirt5, and IsSirt7 were overrepresented in infected SG and/or G in A. phagocytophilum-infected ticks (Fig. 7). In SG, but not in G cells, IsSirt2 and IsSirt5 appeared to be regulated at the transcriptional level in response to infection (Fig. 7). In contrast, IsSirt7 levels were higher in tick G at both mRNA and protein levels (Fig. 7). The result in tick G cells was similar to that reported by Xie et al.,48 in which Cryptosporidium parvum (Apicomplexa) infection of human cells resulted in higher Sirt1 protein levels without changes in the hsirt1 mRNA levels. Recently, Eskandarian et al.49 showed that, in Listeria monocytogenes-infected cells, Sirt2 translocates to the nucleus, deacetylates histone H3 on lysine 18, and represses a subset of host genes to facilitate infection. All together, these results suggested that Sirtuins might play an essential role during host epigenetic manipulation by pathogens to facilitate infection in both vertebrate host and tick cells.
In tick SG, IsHAT1, and IsKAT8 protein levels increased in response to A. phagocytophilum infection (Fig. 7). A previous study proposed that histone acetylation by HATs mediates epigenetic regulation of transcriptional reprogramming in insects to inhibit host immune responses during infection,59 a mechanism that may be common in arthropods based on the results obtained here in ticks infected with A. phagocytophilum.
The expression and activity of HMTs have been linked to infection processes in mammals63,64 and plants.65 The bacterium Chlamydia trachomatis was found to target host histone methylation to ensure infection.64 In this study, we found that A. phagocytophilum infection increases mRNA levels of IsSETDB1-A, IsSETDB1-B, IsEHMT, IsSETD2, IsSMYD4, and IsDOT1L coding genes in tick G (Fig. 7), therefore suggesting a role for histone methylation during A. phagocytophilum infection in ticks.
HDMs also play an important role during pathogen infection. The inhibition of LSD1 and JMJD2 in mammalian cells results in heterochromatic suppression of a herpesvirus genome and blocks infection and reactivation both in vitro and in vivo.66 In rice, the expression of a JMJC demethylase was induced by stress signals and during pathogen infection resulting in increase resistance to the bacterial blight disease pathogen, Xanthomonas oryzae pathovar oryzae.67 Pathogen manipulation mediated by HDMs of the Kaposi's sarcoma-associated herpesvirus induces an overexpression of an RNA molecule that associates with demethylases UTX and JMJD3 to activate lytic replication of the virus.68 In ticks, we found that A. phagocytophilum infection modified the expression of HDM coding genes while the IsUTY protein was underrepresented in N and G (Fig. 7). As with other HMEs, these results suggested a possible implication of tick HDMs in response to A. phagocytophilum infection.
In ISE6 cells, the effect of A. phagocytophilum infection did not affect expression for most histone and HMEs coding genes (Fig. 7). Only a few histone and HMEs proteins were identified by proteomics analysis of ISE6 cells, providing evidence for the possible role of HMTs and HDMs during pathogen infection (Fig. 7).
Validation of transcriptomics and proteomics data for I. scapularis HMEs
The validation of transcriptomics and proteomics data was done on selected genes and proteins by real-time RT-PCR and immunofluorescence, respectively. However, although real-time RT-PCR is easy to perform to validate transcriptomics data, few antibodies are available that react with tick proteins for the validation of proteomics data. Herein, 10 I. scapularis HME coding genes were selected for real-time RT-PCR (Supplementary File 2). This analysis of mRNA levels in individual samples from infected and uninfected I. scapularis adult female G and SG and ISE6 cells corroborated transcriptomics results by demonstrating that gene up- or down-regulation was similar between transcriptomics and RT-PCR analyses for most samples (Supplementary File 2). As in previous experiments, the differences observed between the results of both analyses in tick G and SG could be attributed to intrinsic variation in gene expression and the fact that approximately 85% of the ticks used for transcriptomics were infected, while for RT-PCR all ticks were confirmed uninfected or infected with A. phagocytophilum before analysis.18,19
For the validation of proteomics data, 2 commercial antibodies were found reactive against IsSirt2 and IsHDAC1 and were used for immunofluorescence in I. scapularis G and SG tissue sections (Supplementary File 2). The results corroborated proteomics results by showing higher IsHDAC1 levels in infected tick SG (Supplementary File 2) and higher IsSirt2 levels in infected tick G and SG (Supplementary File 2).
Functional characterization of selected I. scapularis HME proteins
The characterization of the I. scapularis histone and HMEs mRNA and protein levels showed variations in response to A. phagocytophilum infection that suggested a functional role for these molecules during pathogen infection in ticks. To further characterize the function of selected HME proteins during A. phagocytophilum infection, infected ISE6 cells were left untreated or treated for 72 h with different pharmacological HME inhibitors or activators (Fig. 8A). The results showed that treatment with p300/CBP, HDAC, and Sirtuin inhibitors resulted in lower A. phagocytophilum infection levels, while activators of p300/CBP and Sirtuins resulted in higher infection levels when compared to untreated infected cells (Fig. 8A). Treatment with SMYD3, LSD1, and UTY inhibitors did not affect infection when compared to untreated controls (Fig. 8A). Therefore, the role of HMTs and HDMs during A. phagocytophilum infection of I. scapularis cells was not supported by functional studies, although these HME inhibitors may not be fully active in tick cells.
Figure 8.
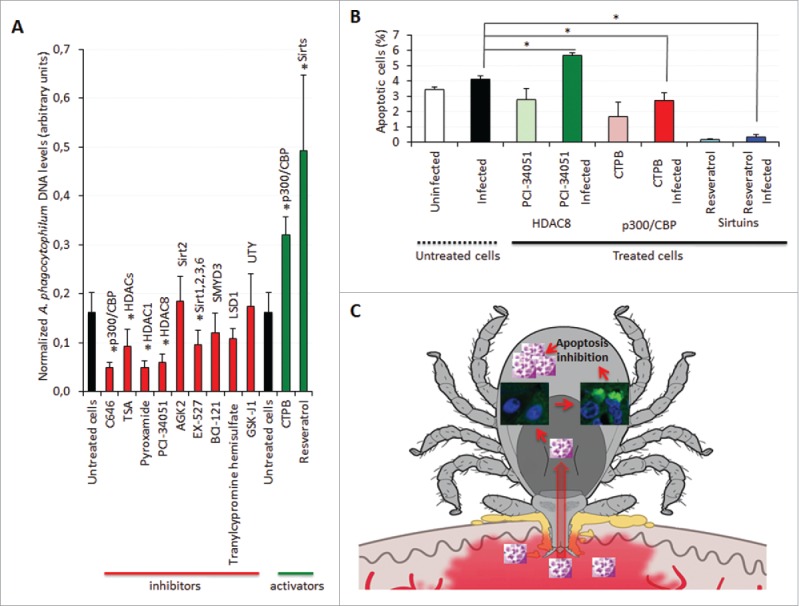
Characterization of the function of I. scapularis HMEs during A. phagocytophilum infection. (A) A. phagocytophilum DNA levels were determined in infected ISE6 tick cells untreated or treated for 72 h with C646 to inhibit p300/CBP, Trichostatin A (TSA) to inhibit HDACs, Pyroxamide to inhibit HDAC1, PCI-34051 to inhibit HDAC8, AGK2 to inhibit Sirt2, EX-527 to inhibit Sirt1/Sirt2/Sirt3/Sirt6, BCI-121 to inhibit SMYD3, Tranylcypromine hemisulfate to inhibit LSD1, GSK-J1 to inhibit UTY, CTPB to activate p300/CBP and Resveratrol to activate Sirtuins. Bacterial DNA levels were determined by msp4 real-time PCR normalizing against tick 16S rDNA. Results are shown as Ave+SD normalized Ct values and were compared between untreated and treated cells by Student's t-test with unequal variance (*P < 0.05; N = 4). (B) The percentage of apoptotic cells was determined by flow cytometry in uninfected and infected ISE6 tick cells untreated or treated for 72 h with PCI-34051 to inhibit HDAC8, CTPB to activate p300/CBP and Resveratrol to activate Sirtuins. Results are represented as Ave+SD and compared between untreated and treated infected cells by Student's t-test with unequal variance (*P <0.05; N = 4). (C) Schematic representation of the role of tick Sirtuins during A. phagocytophilum infection. A. phagocytophilum manipulates tick cell epigenetics to increase IsSirtuin levels resulting in the inhibition of cell apoptosis to facilitate pathogen infection and multiplication.
Intracellular bacteria such as A. phagocytophilum use different strategies to inhibit cell apoptosis in order to enhance their replication and survival.14 In I. scapularis, A. phagocytophilum inhibits tick cell apoptosis by different mechanisms in a tissue-specific manner.14,18,19 However, the mechanisms by which A. phagocytophilum modifies gene expression or protein levels to inhibit apoptosis are not known but likely involve secreted bacterial effector proteins or the effect of tick response to infection that also results in apoptosis inhibition facilitating pathogen infection and multiplication.69 It has been established that HMEs affect cell apoptosis.60-62 The results obtained here showed that IsHDAC8 is involved in the regulation of apoptosis in tick cells (Fig. 8B). The inhibition of IsHDAC8 resulted in higher percentage of apoptotic cells while the activation of Isp300/CBP and IsSirtuins resulted in lower percentage of apoptotic cells whether the cells were infected or not (Fig. 8B). As expected, the effect of HME inhibitors and activators on tick cell apoptosis correlated with the effect on A. phagocytophilum infection showing higher infection levels in cells with lower percentage of apoptotic cells (Fig. 8A and Fig. 8B).
These results support the view that A. phagocytophilum infection alters tick cell epigenetics to promote survival and replication. The mechanism by which A. phagocytophilum affects the epigenome of tick cells is unknown but, as shown in human cells, it is probably controlled by effector proteins secreted through the Type IV secretion system 23,25,26 or other secretion mechanisms.22
Conclusions
This is the first study on the global identification and characterization of tick histones and HMEs and their role in pathogen infection. We identified 5 histones and 34 HMEs in the tick disease vector, I. scapularis including HATs, HDACs, Sirtuins, HMTs, and HDMs. However, due to the incomplete genome sequence and assembly,29 additional HMEs could be present in I. scapularis. It is generally assumed that genes/proteins with similar sequence and structure have the same function in different organisms. However, the function described in model organisms like mammals and insects may be different in evolutionarily distant species such as ticks.70 The results obtained here show that A. phagocytophilum manipulates tick cell epigenetics to increase Isp300/CBP, IsHDACs, and IsSirtuin levels resulting in the inhibition of cell apoptosis to facilitate pathogen infection and multiplication (Fig. 8C). These results suggested that a compensatory mechanism might exist by which A. phagocytophilum manipulates tick HMEs to regulate transcription and apoptosis in a tissue-specific manner to facilitate infection but preserving tick fitness to guarantee survival of both pathogens and ticks.69 Similar to other biological processes involved in A. phagocytophilum infection,14 these results also suggested that the pathogen manipulates arthropod and vertebrate host cell epigenetics in similar ways to inhibit host responses and facilitate infection. Investigating gene expression by epigenetic regulation and the relevance of HMEs in tick biology and pathogen infection is essential to advance our knowledge of tick-pathogen molecular interactions with possible implications for the identification of new targets for anti-tick drug and vaccine development.
Materials and methods
Identification of histones and HMEs in I. scapularis
The I. scapularis transcriptome was searched with the following keywords: “histones,” “acetyltransferase,” “deacetylase,” “methyltransferase,” and “demethylase.” Collected hits were translated into protein sequences and domains searched using Pfam.71 To find the human homologs, collected I. scapularis hits were blasted against the H. sapiens database using the Blastp tool from BLAST72,73 and the sequences with the lowest E-value were selected. I. scapularis homologs found in the transcriptome were double-checked by searching the I. scapularis genome database using as queries the human homologs identified in the previous step. Domains identified with Pfam were double-checked using Conserved Domain74 from BLAST. Using the above strategy we could identify proteins at the protein family level, but it was not very accurate for identifying specific homologs. To identify specific homologs across relevant taxa we systematically searched mammal, arthropod and chelicerate databases using the Blastp tool from BLAST.72,73 Amino acid sequences collected from different mammals, arthropods and chelicerate species (indicated in the phylogenetic trees) were aligned with MAFFT (v7) configured for the highest accuracy using the scoring matrix 200PAM/k=2, alignment strategy MAFFT-FFT-NS-I, gap opening penalty 1.53 and offset value 0.123.75,76 Non-aligned regions were removed with Gblocks (v 0.91b).77 The neighbor joining (NJ) method, implemented in Molecular Evolutionary Genetics Analysis (MEGA, version 6) software,78 was used to obtain the best tree topologies. A proportion of Gamma distributed sites (G) were estimated in MEGA for each group of sequences. Reliability of internal branches was assessed using the bootstrapping method (1000 bootstrap replicates). Graphical representation and editing of the phylogenetic tree was performed with EvolView.79 All histones and HMEs identified in I. scapularis are summarized in Supplementary Table 1.
Tertiary structure prediction of I. scapularis histones and HMEs
To find the best homology templates for modeling the I. scapularis histones and HMEs tertiary structure we employed previously published methods.80 Briefly, a PSI-BLAST81 was performed with up to 5 iterations using the non-redundant database to compile the position-specific scoring matrix (PSSM). A subsequent PSI-BLAST was performed using the generated PSSM against the Protein DataBank (PDB;82) to obtain the structural template homologs for modeling. A multiple sequence alignment was performed using the default configuration of the MAFFT web-server.83 Structures were modeled using MODELLER84 with multiple templates and the models were evaluated using the protein model-qualifying servers QMEAN85 and RESPROX.86 All predicted models were edited to the sequences of the homolog structures or to unresolved secondary structures. Binding domains for HDACs and Sirtuins were produced by a structural superposition of the respective substrate bound human homolog using the Schrodinger's Maestro software package.87
Characterization of the I. scapularis histone and HMEs mRNA and protein levels in response to A. phagocytophilum infection
The quantitative transcriptomics and proteomics data for uninfected and A. phagocytophilum-infected I. scapularis N, G, SG, and ISE6 cells were obtained from previously published results18,19 and deposited at the Dryad repository database, NCBI's Gene Expression Omnibus database and ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD002181 and doi: 10.6019/PXD002181 and searched against identified I. scapularis histones and HMEs.
Determination of tick mRNA levels by real-time RT-PCR
The expression of selected genes was characterized using total RNA extracted from individual I. scapularis female G, SG, and ISE6 cells. Uninfected and A. phagocytophilum-infected samples were obtained as previously described.18,19 All ticks were confirmed as infected or uninfected by real-time PCR analysis of A. phagocytophilum msp4 DNA. Real-time RT-PCR was performed on RNA samples with gene-specific oligonucleotide primers (Supplementary Table 2) using the iScript One-Step RT-PCR Kit with SYBR Green and the iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA) following manufacturer's recommendations. A dissociation curve was run at the end of the reaction to ensure that only one amplicon was formed and that the amplicons denatured consistently in the same temperature range for every sample. The mRNA levels were normalized against tick cyclophilin and ribosomal protein S4 as described previously using the genNorm method (ddCT method as implemented by Bio-Rad iQ5 Standard Edition, Version 2.0).88 Normalized Ct values were compared between infected and uninfected tick samples by Student's t-test with unequal variance (P = 0.05; N = 3–17 biological replicates).
Immunofluorescence assay
Female ticks fed on A. phagocytophilum-infected and uninfected sheep and fixed with 4% paraformaldehyde in 0.2M sodium cacodylate buffer were embedded in paraffin and used to prepare sections on glass slides as previously described.18 The paraffin was removed from the sections through 2 washes in xylene and the sections were hydrated by successive 5 min washes with a graded series of 100%, 96%, and 65% ethanol and finally with distilled water. Next, the slides were treated with Proteinase K (Dako, Barcelona, Spain) for 7 min, washed with 0,1% PBS-Tween 20 (Sigma-Aldrich, St. Louis, MI, USA) and blocked with 2% bovine serum albumin (BSA; Sigma-Aldrich) in PBS-Tween 20 during 1h at room temperature. The slides were then incubated overnight at 4°C with rabbit anti-Sirt2 (ABIN1109013, Antibodies-online.com) and anti-HDAC-1 (ab1767, Abcam, Cambridge, UK) antibodies both diluted 1:1000 in 2% BSA/PBS-Tween 20. After 3 washes with PBS-Tween 20, the slides were incubated for 1h with goat-anti-rabbit IgG conjugated with FITC (Sigma-Aldrich) diluted 1:160 in 2% BSA/PBS-Tween 20. Finally, after 2 washes with PBS the slides were mounted on ProLong Diamond Antifade Mountant with DAPI reagent (Thermo Scientific™, Madrid, Spain). The sections were examined using a Leica SP2 laser scanning confocal microscope (Leica, Wetzlar, Germany) and IgGs from rabbit pre-immune serum were used as controls.
Pharmacological studies in cultured ISE6 tick cells
The I. scapularis embryo-derived tick cell line ISE6, provided by Ulrike Munderloh, University of Minnesota, USA, was cultured in L-15B300 medium as described previously,89 except that the osmotic pressure was lowered by the addition of one- fourth sterile water by volume. The ISE6 cells were infected with A. phagocytophilum (human NY18 isolate19) and maintained according to Munderloh et al.90 Uninfected and A. phagocytophilum-infected ISE6 cells were left untreated or treated for 72 h with 4 µM C646 (Sigma-Aldrich) to inhibit p300/CBP,91 5 µM Trichostatin A (Sigma-Aldrich) to inhibit HDACs,92 1 µM Pyroxamide (Abcam) to inhibit HDAC1,93 5 µM PCI-34051 (Sigma-Aldrich) to inhibit HDAC8,94 35 µM AGK2 (Sigma-Aldrich) to inhibit Sirt2,95 100 µM EX-527 (Sigma-Aldrich) to inhibit Sirt1/Sirt2/Sirt3/Sirt6,96 100 µM BCI-121 (Glixx laboratories, Southborough, MA, USA) to inhibit SMYD3,97 600 nM GSK-J1 (Sigma-Aldrich) to inhibit UTY,98 20 µM Tranylcypromine hemisulfate (Reaction Biology, Malvern, PA, USA) to inhibit LSD1,99 100 µM CTPB (Sigma-Aldrich) to activate p300/CBP100 and 50 µM Resveratrol (Sigma-Aldrich) to activate Sirtuins.101 After treatment, cells were harvested and used for Annexin V-FITC staining to detect cell apoptosis and for DNA extraction. A. phagocytophilum DNA levels were characterized by msp4 real-time PCR normalizing against tick 16S rDNA as described previously.88 Normalized Ct values were compared between untreated and treated cells by Student's t-test with unequal variance (P = 0.05; N = 4 biological replicates).
Annexin V-FITC staining to detect cell apoptosis after experimental infection with A. phagocytophilum
Approximately 5 × 105−1 × 106 uninfected and A. phagocytophilum-infected ISE6 tick cells were collected after different treatments. Apoptosis was measured by flow cytometry using the Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Immunostep, Salamanca, Spain) following the manufacturers protocols. The technique detects changes in phospholipid symmetry analyzed by measuring Annexin V (labeled with FITC) binding to phosphatidylserine, which is exposed in the external surface of the cell membrane in apoptotic cells. Cells were stained simultaneously with the non-vital dye propidium iodide (PI) allowing the discrimination of intact cells (Annexin V-FITC negative, PtdIns negative) and early apoptotic cells (Annexin V-FITC positive, PI negative). All samples were analyzed on a FAC-Scalibur flow cytometer equipped with CellQuest Pro software (BD Bio-Sciences, Madrid, Spain). The viable cell population was gated according to forward-scatter and side-scatter parameters. The percentage of apoptotic cells was compared between both treated and untreated infected and uninfected cells by Student's t-test with unequal variance (P = 0.05; N = 4 biological replicates).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to acknowledge José Ramón Marín Tébar (University of Castilla - La Mancha, Spain) for technical assistance with immunofluorescence. This research was supported by the Ministerio de Economia y Competitividad (Spain) grant BFU2011-23896 and the European Union (EU) Seventh Framework Program (FP7) ANTIGONE project number 278976. NA was funded by Ministerio de Economía y Competitividad, Spain. MV was supported by the Research Plan of the University of Castilla - La Mancha, Spain. RP received funding from the EU FP7 for research, technological development and demonstration under grant agreement number 602080 (A-ParaDDisE). The work of RP and ACC was also supported by institutional funds from the Center National de la Recherche Scientifique (CNRS), the Institut Pasteur de Lille and the Université de Lille 2. JJV was supported by the EU FP7 under grant agreement number 316304 and project LO1218, with financial support from the MEYS of the Czech Republic under the NPU I program.
Authors' contributions
ACC, JJV, and RP performed the identification and sequence analysis of histones and HMEs in I. scapularis. PA performed the pharmacological and apoptosis studies. NA performed the expression and immunofluorescence experiments. JF and ACC conceived and designed the study and wrote the manuscript. All authors have read and approved the final manuscript.
References
- 1.Gómez-Díaz E, Jordà M, Peinado MA, Rivero A. Epigenetics of host-pathogen interactions: the road ahead and the road behind. PLoS Pathog 2012; 8:e1003007; http://dx.doi.org/ 10.1371/journal.ppat.1003007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silmon de Monerri NC, Kim K. Pathogens hijack the epigenome: a new twist on host-pathogen interactions. Am J Pathol 2014; 184:897-911; PMID:24525150; http://dx.doi.org/ 10.1016/j.ajpath.2013.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheeseman K, Weitzman JB. Host-parasite interactions: an intimate epigenetic relationship. Cell Microbiol 2015; 17:1121-32; PMID:26096716; http://dx.doi.org/ 10.1111/cmi.12471 [DOI] [PubMed] [Google Scholar]
- 4.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev 2009; 23:781-3; PMID:19339683; http://dx.doi.org/ 10.1101/gad.1787609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13:297-311; PMID:22473470 [DOI] [PubMed] [Google Scholar]
- 6.de la Fuente J, Estrada-Peña A, Venzal JM, Kocan KM, Sonenshine DE. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci 2008; 13: 6938-46; PMID:18508706; http://dx.doi.org/ 10.2741/3200 [DOI] [PubMed] [Google Scholar]
- 7.Reichard MV, Roman RM, Kocan KM, Blouin EF, de la Fuente J, Snider TA, Heinz RE, West MD, Little SE, Massung RF. Inoculation of white-tailed deer (Odocoileus virginianus) with Ap-V1 or NY-18 strains of Anaplasma phagocytophilum and microscopic demonstration of Ap-V1 in Ixodes scapularis adults that acquired infection from deer as nymphs. Vector Borne Zoonotic Dis 2009; 9:565-8; PMID:18973438; http://dx.doi.org/ 10.1089/vbz.2008.0106 [DOI] [PubMed] [Google Scholar]
- 8.Sukumaran B, Narasimhan S, Anderson JF, DePonte K, Marcantonio N, Krishnan MN, Fish D, Telford SR, Kantor FS, Fikrig E. An Ixodes scapularis protein required for survival of Anaplasma phagocytophilum in tick salivary glands. J Exp Med 2006; 203:1507-17; PMID:16717118; http://dx.doi.org/ 10.1084/jem.20060208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of the genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol 2001; 51:2145-65; PMID:11760958; http://dx.doi.org/ 10.1099/00207713-51-6-2145 [DOI] [PubMed] [Google Scholar]
- 10.Rikihisa Y. Molecular pathogensis of Anaplasma phagocytophilum. Current Microbiol Rev 2011; 24:469-89; http://dx.doi.org/ 10.1128/CMR.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severo MS, Choy A, Stephens KD, Sakhon OS, Chen G, Chung DW, Le Roch KG, Blaha G, Pedra JH. The E3 ubiquitin ligase XIAP restricts Anaplasma phagocytophilum colonization of Ixodes scapularis ticks. J Infect Dis 2013; 208:1830-40; PMID:23901084; http://dx.doi.org/ 10.1093/infdis/jit380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuen S. Anaplasma phagocytophilum—the most widespread tick-borne infection in animals in Europe. Vet Res Commun 2010; 31:79-84; http://dx.doi.org/ 10.1007/s11259-007-0071-y [DOI] [PubMed] [Google Scholar]
- 13.Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum—a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol 2013; 3:31; PMID:23885337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Fuente J, Estrada-Peña A, Cabezas-Cruz A, Kocan KM. Anaplasma phagocytophilum uses common strategies for infection of ticks and vertebrate hosts. Trends Microbiol 2016; 24:173-80; http://dx.doi.org/ 10.1016/j.tim.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 15.Rikihisa Y. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat Rev Microbiol 2010; 8:328-39; PMID:20372158; http://dx.doi.org/ 10.1038/nrmicro2318 [DOI] [PubMed] [Google Scholar]
- 16.Rikihisa Y. Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet Parasitol 2010; 167:155-66; PMID:19836896; http://dx.doi.org/ 10.1016/j.vetpar.2009.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rikihisa Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev 2011; 24:469-89; PMID:21734244; http://dx.doi.org/ 10.1128/CMR.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayllón N, Villar M, Galindo RC, Kocan KM, Šíma R, López JA, Vázquez J, Alberdi P, Cabezas-Cruz A, Kopáček P, et al.. Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet 2015; 11:e1005120; http://dx.doi.org/ 10.1371/journal.pgen.1005120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villar M, Ayllón N, Alberdi P, Moreno A, Moreno M, Tobes R, Mateos-Hernández L, Weisheit S, Bell-Sakyi L, de la Fuente J. Integrated metabolomics, transcriptomics and proteomics identifies metabolic pathways affected by Anaplasma phagocytophilum infection in tick cells. Mol Cell Proteomics 2015; 14:3154-72; PMID:26424601; http://dx.doi.org/ 10.1074/mcp.M115.051938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Garcia JC, Barat NC, Trembley SJ, Dumler JS. Epigenetic silencing of host cell defense genes enhances intracellular survival of the rickettsial pathogen Anaplasma phagocytophilum. PLoS Pathog 2009; 5:e1000488; PMID:19543390; http://dx.doi.org/ 10.1371/journal.ppat.1000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol 2007; 9:2644-57; PMID:17587335; http://dx.doi.org/ 10.1111/j.1462-5822.2007.00985.x [DOI] [PubMed] [Google Scholar]
- 22.Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, Borjesson DL, Carlyon JA. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb Pathog 2010; 49:273-84; PMID:20600793; http://dx.doi.org/ 10.1016/j.micpath.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikihisa Y, Lin M, Niu H. Type IV secretion in the obligatory intracellular bacterium Anaplasma phagocytophilum. Cell Microbiol 2010; 12:1213-21; PMID:20670295; http://dx.doi.org/ 10.1111/j.1462-5822.2010.01500.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Z, Atmakuri K, Christie PJ. The ins and outs of bacterial type IV secretion substrates. Trends Microbiol 2003; 11:527-35; PMID:14607070; http://dx.doi.org/ 10.1016/j.tim.2003.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rennoll-Bankert KE, Garcia-Garcia JC, Sinclair SH, Dumler JS. Chromatin bound bacterial effector AnkA recruits HDAC1 and modifies host gene expression. Cell Microbiol 2015; 17:1640-52; PMID:25996657; http://dx.doi.org/ 10.1111/cmi.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinclair SH, Yegnasubramanian S, Dumler JS. Global DNA methylation changes and differential gene expression in Anaplasma phagocytophilum-infected human neutrophils. Clin Epigenetics 2015; 7:77; PMID:26225157; http://dx.doi.org/ 10.1186/s13148-015-0105-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamson SW, Browning RE, Budachetri K, Ribeiro JM, Karim S. Knockdown of selenocysteine-specific elongation factor in Amblyomma maculatum alters the pathogen burden of Rickettsia parkeri with epigenetic control by the Sin3 histone deacetylase corepressor complex. PLoS One 2013; 8:e82012; PMID:24282621; http://dx.doi.org/ 10.1371/journal.pone.0082012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotsyfakis M, Schwarz A, Erhart J, Ribeiro JM. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Sci Rep 2015; 5:9103; PMID:25765539; http://dx.doi.org/ 10.1038/srep09103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, et al.. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nature Commun 2016; 7:10507; http://dx.doi.org/ 10.1038/ncomms10507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang XJ. MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease. Biochim Biophys Acta 2015; 1853:1818-26; PMID:25920810; http://dx.doi.org/ 10.1016/j.bbamcr.2015.04.014 [DOI] [PubMed] [Google Scholar]
- 31.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006; 6:38-51; PMID:16397526; http://dx.doi.org/ 10.1038/nrc1779 [DOI] [PubMed] [Google Scholar]
- 32.Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells 2009; 28:407-15; PMID:19936627; http://dx.doi.org/ 10.1007/s10059-009-0169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol 2015; 16:499-513; PMID:26204160; http://dx.doi.org/ 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- 34.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13:297-311; PMID:22473470 [DOI] [PubMed] [Google Scholar]
- 35.Melters DP, Nye J, Zhao H, Dalal Y. Chromatin Dynamics in Vivo: A Game of Musical Chairs. Genes (Basel) 2015; 6:751-76; PMID:26262644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000; 403:41-5; PMID:10638745; http://dx.doi.org/ 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 37.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol 2010; 28:1057-68; PMID:20944598; http://dx.doi.org/ 10.1038/nbt.1685 [DOI] [PubMed] [Google Scholar]
- 38.Berger SL. The complex language of chromatin regulation during transcription. Nature 2007; 447:407-12; PMID:17522673; http://dx.doi.org/ 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- 39.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 2001; 114(Pt 13):2363-73; PMID:11559745 [DOI] [PubMed] [Google Scholar]
- 40.Hiragaki S, Suzuki T, Mohamed AA, Takeda M. Structures and functions of insect arylalkylamine N-acetyltransferase (iaaNAT); a key enzyme for physiological and behavioral switch in arthropods. Front Physiol 2015; 6:113; PMID:25918505; http://dx.doi.org/ 10.3389/fphys.2015.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007; 26:5319-28; PMID:17694075; http://dx.doi.org/ 10.1038/sj.onc.1210602 [DOI] [PubMed] [Google Scholar]
- 42.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, et al.. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol 2008; 28:6828-43; PMID:18794358; http://dx.doi.org/ 10.1128/MCB.01297-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell 2009; 17:674-86; PMID:19922872; http://dx.doi.org/ 10.1016/j.devcel.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 44.Mihaylova MM, Shaw RJ. Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol Metab 2013; 24:48-57; PMID:23062770; http://dx.doi.org/ 10.1016/j.tem.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herranz D, Muñoz-Martin M, Cañamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 2010; 1:3; PMID:20975665; http://dx.doi.org/ 10.1038/ncomms1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaeberlein M. Lessons on longevity from budding yeast. Nature 2010; 464:513-9; PMID:20336133; http://dx.doi.org/ 10.1038/nature08981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lancelot J, Cabezas-Cruz A, Caby S, Marek M, Schultz J, Romier C, Sippl W, Jung M, Pierce RJ. Schistosome sirtuins as drug targets. Future Med Chem 2015; 7:765-82; PMID:25996069; http://dx.doi.org/ 10.4155/fmc.15.24 [DOI] [PubMed] [Google Scholar]
- 48.Xie H, Lei N, Gong AY, Chen XM, Hu G. Cryptosporidium parvum induces SIRT1 expression in host epithelial cells through downregulating let-7i. Hum Immunol 2014; 75:760-5; PMID:24862934; http://dx.doi.org/ 10.1016/j.humimm.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eskandarian HA, Impens F, Nahori MA, Soubigou G, Coppée JY, Cossart P, Hamon MA. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 2013; 341:1238858; PMID:23908241; http://dx.doi.org/ 10.1126/science.1238858 [DOI] [PubMed] [Google Scholar]
- 50.Lancelot J, Caby S, Dubois-Abdesselem F, Vanderstraete M, Trolet J, Oliveira G, Bracher F, Jung M, Pierce RJ. Schistosoma mansoni sirtuins: characterization and potential as chemotherapeutic targets. PLoS Negl Trop Dis 2013; 7:e2428; PMID:24069483; http://dx.doi.org/ 10.1371/journal.pntd.0002428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol 2001; 8:621-5; PMID:11427894; http://dx.doi.org/ 10.1038/89668 [DOI] [PubMed] [Google Scholar]
- 52.Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, et al.. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure 2007; 15:377-389; PMID:17355872; http://dx.doi.org/ 10.1016/j.str.2007.02.002 [DOI] [PubMed] [Google Scholar]
- 53.Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, et al.. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem 2009; 284:24394-405; PMID:19535340; http://dx.doi.org/ 10.1074/jbc.M109.014928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem 2011; 286:14575-87; PMID:21362626; http://dx.doi.org/ 10.1074/jbc.M111.218990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X, Allison D, Condon B, Zhang F, Gheyi T, Zhang A, Ashok S, Russell M, MacEwan I, Qian Y, et al.. The 2.5Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem 2013; 56:963-9; PMID:23311358; http://dx.doi.org/ 10.1021/jm301431y [DOI] [PubMed] [Google Scholar]
- 56.Völkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie 2007; 89:1-20; PMID:16919862; http://dx.doi.org/ 10.1016/j.biochi.2006.07.009 [DOI] [PubMed] [Google Scholar]
- 57.Zou C, Mallampalli RK. Regulation of histone modifying enzymes by the ubiquitin-proteasome system. Biochim Biophys Acta 2014; 1843:694-702; PMID:24389248; http://dx.doi.org/ 10.1016/j.bbamcr.2013.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen R, Kang R, Fan XG, Tang D. Release and activity of histone in diseases. Cell Death Dis 2014; 5:e1370; http://dx.doi.org/ 10.1038/cddis.2014.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee K, Fischer R, Vilcinskas A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front Zool 2012; 9:25; PMID:23035888; http://dx.doi.org/ 10.1186/1742-9994-9-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang AG, Kim SU, Lee SH, Kim SK, Seo SB, Yu DY, Lee DS. Histone deacetylase 1 contributes to cell cycle and apoptosis. Biol Pharm Bull 2005; 28:1966-70; PMID:16204956; http://dx.doi.org/ 10.1248/bpb.28.1966 [DOI] [PubMed] [Google Scholar]
- 61.Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci U S A 2012; 109:E481-9; PMID:22223663; http://dx.doi.org/ 10.1073/pnas.1118403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakrabarti A, Oehme I, Witt O, Oliveira G, Sippl W, Romier C, Pierce RJ, Jung M. HDAC8: a multifaceted target for therapeutic interventions. Trends Pharmacol Sci 2015; 36:481-92; PMID:26013035; http://dx.doi.org/ 10.1016/j.tips.2015.04.013 [DOI] [PubMed] [Google Scholar]
- 63.Imai K, Togami H, Okamoto T. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem 2010; 285:16538-45; PMID:20335163; http://dx.doi.org/ 10.1074/jbc.M110.103531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 2010; 6:e1000995; PMID:20657819; http://dx.doi.org/ 10.1371/journal.ppat.1000995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berr A, McCallum EJ, Alioua A, Heintz D, Heitz T, Shen WH. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol 2010; 154:1403-14; PMID:20810545; http://dx.doi.org/ 10.1104/pp.110.161497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, et al.. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 2014; 6:265ra169; PMID:25473037; http://dx.doi.org/ 10.1126/scitranslmed.3010643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li T, Chen X, Zhong X, Zhao Y, Liu X, Zhou S, Cheng S, Zhou DX. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 2013; 25:4725-36; PMID:24280387; http://dx.doi.org/ 10.1105/tpc.113.118802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rossetto CC, Pari G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 2012; 8:e1002680; PMID:22589717; http://dx.doi.org/ 10.1371/journal.ppat.1002680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de la Fuente J, Villar M, Cabezas-Cruz A, Estrada-Peña A, Ayllón N, Alberdi P. Tick-host-pathogen interactions: conflict and cooperation. PLoS Pathog 2016, in press; PMID:26808628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ayllón N, Naranjo V, Hajdušek O, Villar M, Galindo RC, Kocan KM, Alberdi P, Šíma R, Cabezas-Cruz A, Rückert C, et al.. Nuclease Tudor-SN is involved in tick dsRNA-mediated RNA interference and feeding but not in defense against flaviviral or Anaplasma phagocytophilum rickettsial infection. PLoS One 2015; 10:e0133038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al.. Pfam: the protein families database. Nucleic Acids Res 2014; 42:D222-30; PMID:24288371; http://dx.doi.org/ 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712; http://dx.doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 73.Madden TL, Tatusov RL, Zhang J. Applications of network BLAST server. Methods Enzymol 1996; 266:131-41; PMID:8743682; http://dx.doi.org/ 10.1016/S0076-6879(96)66011-X [DOI] [PubMed] [Google Scholar]
- 74.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al.. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015; 43:D222-6; PMID:25414356; http://dx.doi.org/ 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 2005; 33:511-8; PMID:15661851; http://dx.doi.org/ 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katoh K, Standley D. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772-80; PMID:23329690; http://dx.doi.org/ 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-52; PMID:10742046; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 78.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/ 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H, Gao S, Lercher M, Hu S, Chen W. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 2012; 40:W569-72; PMID:22695796; http://dx.doi.org/ 10.1093/nar/gks576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valdés JJ, Schwarz A, Cabeza de Vaca I, Calvo E, Pedra JH, Guallar V, Kotsyfakis M. Tryptogalinin is a tick Kunitz serine protease inhibitor with a unique intrinsic disorder. PLoS One 2013; 8:e62562; http://dx.doi.org/ 10.1371/journal.pone.0062562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-402; PMID:9254694; http://dx.doi.org/ 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res 2000; 28:235-42; PMID:10592235; http://dx.doi.org/ 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 2008; 9:286-98; PMID:18372315; http://dx.doi.org/ 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- 84.Webb B, Sali A. Comparative Protein Structure Modeling Using Modeller. Curr Protoc Bioinformatics 2014; 47:1-32; PMID:25199789 [DOI] [PubMed] [Google Scholar]
- 85.Benkert P, Tosatto SCE, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins 2008; 71:261-77; PMID:17932912; http://dx.doi.org/ 10.1002/prot.21715 [DOI] [PubMed] [Google Scholar]
- 86.Berjanskii M, Zhou J, Liang Y, Lin G, Wishart DS. Resolution-by-Proxy: A Simple Measure for Assessing and Comparing the Overall Quality of NMR Protein Structures. J Biomol NMR 2012; 53:167-80; PMID:22678091; http://dx.doi.org/ 10.1007/s10858-012-9637-2 [DOI] [PubMed] [Google Scholar]
- 87.Schrödinger Maestro v9.1 New York 2010 [Google Scholar]
- 88.Ayllón N, Villar M, Busby AT, Kocan KM, Blouin EF, Bonzón-Kulichenko E, Galindo RC, Mangold AJ, Alberdi P, Pérez de la Lastra JM, et al.. Anaplasma phagocytophilum inhibits apoptosis and promotes cytoskeleton rearrangement for infection of tick cells. Infect Immun 2013; 81:2415-25; http://dx.doi.org/ 10.1128/IAI.00194-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J Parasitol 1994; 80:533-43; PMID:8064520; http://dx.doi.org/ 10.2307/3283188 [DOI] [PubMed] [Google Scholar]
- 90.Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, et al.. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol 1999; 37:2518-24; PMID:10405394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu XY, Huang CS, Li Q, Chang RM, Song ZB, Zou WY, Guo QL. p300 exerts an epigenetic role in chronic neuropathic pain through its acetyltransferase activity in rats following chronic constriction injury (CCI). Mol Pain 2012; 8:84; PMID:23176208; http://dx.doi.org/ 10.1186/1744-8069-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roy S, Tenniswood M. Site-specific acetylation of p53 directs selective transcription complex assembly. J Biol Chem 2007; 282:4765-71; PMID:17121856; http://dx.doi.org/ 10.1074/jbc.M609588200 [DOI] [PubMed] [Google Scholar]
- 93.Kutko MC, Glick RD, Butler LM, Coffey DC, Rifkind RA, Marks PA, Richon VM, LaQuaglia MP. Histone deacetylase inhibitors induce growth suppression and cell death in human rhabdomyosarcoma in vitro. Clin Cancer Res 2003; 9:5749-55; PMID:14654560 [PubMed] [Google Scholar]
- 94.Lopez G, Bill KL, Bid HK, Braggio D, Constantino D, Prudner B, Zewdu A, Batte KLD, Pollock RE. HDAC8, a potential therapeutic target for the treatment of malignant peripheral nerve sheath tumors (MPNST). PLoS One 2015; 10:e0133302; PMID:26200462; http://dx.doi.org/ 10.1371/journal.pone.0133302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nie H, Chen H, Han J, Hong Y, Ma Y, Xia W, Ying W. Silencing of SIRT2 induces cell death and a decrease in the intracellular ATP level of PC12 cells. Int J Physiol Pathophysiol Pharmacol 2011; 3:65-70; PMID:21479103 [PMC free article] [PubMed] [Google Scholar]
- 96.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol 2006; 26:28-38; PMID:16354677; http://dx.doi.org/ 10.1128/MCB.26.1.28-38.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peserico A, Germani A, Sanese P, Barbosa AJ, di Virgilio V, Fittipaldi R, Fabini E, Bertucci C, Varchi G, Moyer MP, et al.. A SMYD3 Small-Molecule Inhibitor Impairing Cancer Cell Growth. J Cell Physiol 2015; 230:2447-60; PMID:25728514; http://dx.doi.org/ 10.1002/jcp.24975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al.. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012; 488:404-8; PMID:22842901; http://dx.doi.org/ 10.1038/nature11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lynch JT, Cockerill MJ, Hitchin JR, Wiseman DH, Somervaille TC. CD86 expression as a surrogate cellular biomarker for pharmacological inhibition of the histone demethylase lysine-specific demethylase 1. Anal Biochem 2013; 442:104-6; PMID:23911524; http://dx.doi.org/ 10.1016/j.ab.2013.07.032 [DOI] [PubMed] [Google Scholar]
- 100.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem 2003; 278:19134-40; PMID:12624111; http://dx.doi.org/ 10.1074/jbc.M301580200 [DOI] [PubMed] [Google Scholar]
- 101.Wallenborg K, Vlachos P, Eriksson S, Huijbregts L, Arnér ES, Joseph B, Hermanson O. Red wine triggers cell death and thioredoxin reductase inhibition: effects beyond resveratrol and SIRT1. Exp Cell Res 2009; 315:1360-71; PMID:19268663; http://dx.doi.org/ 10.1016/j.yexcr.2009.02.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.