ABSTRACT
Alteration in the cellular energy metabolism is a principal feature of tumors. An important role in modifying cancer cell metabolism belongs to the cancer-associated fibroblasts. However, the regulation of their interaction has been poorly studied to date. In this study we monitored the metabolic status of both cell types by using the optical redox ratio and the fluorescence lifetimes of the metabolic co-factors NAD(P)H and FAD, in addition to the intracellular pH and the hydrogen peroxide levels in the cancer cells, using genetically encoded sensors. In the co-culture of human cervical carcinoma cells HeLa and human fibroblasts we observed a metabolic shift from oxidative phosphorylation toward glycolysis in cancer cells, and from glycolysis toward OXPHOS in fibroblasts, starting from Day 2 of co-culturing. The metabolic switch was accompanied by hydrogen peroxide production and slight acidification of the cytosol in the cancer cells in comparison with that of the corresponding monoculture. Therefore, our HeLa-huFb system demonstrated metabolic behavior similar to Warburg type tumors. To our knowledge, this is the first time that these 3 parameters have been investigated together in a model of tumor-stroma co-evolution. We propose that determination of the start-point of the metabolic alterations and understanding of the mechanisms of their realization can open a new ways for cancer treatment.
KEYWORDS: co-culture of cancer cells and fibroblasts, energy metabolism, FLIM, genetically encoded sensors, hydrogen peroxide, intracellular pH, NAD(P)H and FAD, Warburg effect
Introduction
It is known that the main way to generate adenosine triphosphate (ATP) for providing living cells with energy is by oxidative phosphorylation (OXPHOS). However, in the case of cancer, an active role in cell metabolism belongs to anaerobic and aerobic glycolysis, or the “Warburg effect.” The use of the glycolytic metabolism even in the presence of oxygen is a principal metabolic feature of cancer cells.1 Rapidly proliferating cancer cells tend toward glycolysis because it provides intermediates for the biosynthesis of macromolecules as well as a fast production of ATP to support their high growth rate.2 It has been found that the glycolytic yield of ATP in most cancer cells is only 17 – 20%,3 and that an adequate level of OXPHOS is also needed for tumor growth.4 By contrast, slow-cycling cancer cells rely on OXPHOS and a mitochondrial supply of ATP.5 It has been suggested that the OXPHOS-dependent slow cycling phenotype may mediate multi-drug resistance.6 Therefore, aerobic glycolysis is not a universal feature of all cancer cells within any particular tumor.
An important role in modifying cancer cell metabolism belongs to the tumor stroma, and, especially, to cancer-associated fibroblasts (CAFs). Multiple studies report on the CAF's ability to promote tumorigenesis and to regulate cancer cell motility and stemness through the secretion of a number of growth factors, hormones and cytokines.7-10 Metabolic coupling between cancer cells and CAFs occupies a special role in these processes because metabolic reorganization underlies further changes leading to adaptation of the cancer cells. Interestingly, in some tumors CAFs undergo metabolic alterations and switch their metabolism to aerobic glycolysis when they interact with cancer cells, while the cancer cells, themselves, remain oxidative, the phenomenon being known as the “reverse Warburg effect.”11 In this case the CAFs secrete L-lactate and ketones into the intercellular space to “feed” the neoplastic cells. It is believed that the latter produce hydrogen peroxide to initiate oxidative stress in the CAFs and drive their aerobic glycolysis.12
It should be noted that there is a great diversity of metabolic phenotypes among tumors, and besides the above mentioned “Warburg type” and “reverse Warburg type,” there are tumors where both cancer cells and stroma are glycolytic or non–glycolytic.13
In general, the metabolism of cancer cells is more flexible compare to normal cells, which gives them the opportunity to survive in unfavorable conditions.1,6 Such factors as hypoxia, glucose deprivation, lactate accumulation and extracellular acidosis contribute a great deal to tumor progression through forming specific microenvironment and thus they determine, to a great extent, the metabolic phenotype.13 However the regulation of this metabolic flexibility has been poorly studied to date.
Cancer metabolism is considered as an attractive target for the development of new anti-cancer drugs.14 One of the promising strategies within this field is the metabolic uncoupling of the tumor stroma and the cancer cells.15 But the great degree of metabolic heterogeneity, both inter- and intratumoral, and the high plasticity of cancer cells significantly complicate the development of effective therapeutic strategies targeting metabolic pathways.
Here we analyze the metabolic interaction between cancer cells and fibroblasts in a co-culture model with a focus on the processes ongoing in the cancer cells. Three parameters related to metabolism were studied – the metabolic co-factors NAD(P)H and FAD, intracellular pH (pHi) and hydrogen peroxide concentrations. We hypothesized that the interaction of cancer cells with fibroblasts should lead to alterations in the cellular energy metabolism, resulting in changes of pHi as a consequence of the accumulation or depletion of metabolic by-products, mostly lactate, in the cell cytoplasm. Hydrogen peroxide is thought to act as a signal molecule in the metabolic coupling between cancer cells and the CAFs. To our knowledge, this work is the first attempt to trace the changes of these 3 aspects in living cancer cells in a model of tumor-stroma co-evolution.
Results
NAD(P)H and FAD in cancer cells and fibroblasts in mono- and co-culture
We analyzed NAD(P)H and FAD using 2-photon fluorescence microscopy and the fluorescence lifetime imaging (FLIM) technique. NAD(P)H and FAD are the main metabolic co-factors that work as electron acceptors and donors in the electron transport chain of mitochondria. Since these co-factors have a capability for fluorescence, and the fluorescence lifetimes differ for different states, the free or protein-bound, fluorescence intensity and lifetime measurements can be used to monitor the metabolic activity of the cells.16,17 It was established that, with an increased level of glycolysis, the NAD(P)H lifetimes, the contribution of protein-bound NAD(P)H and the fluorescence redox ratio (fluorescence intensity of FAD/NAD(P)H) decreased.18-20
For metabolic characterization of the cells the following parameters were analyzed: the optical redox ratio (FAD/NAD(P)H), the fluorescence lifetimes of NAD(P)H and FAD and the relative contribution of the components.
To estimate the overall cellular metabolic activity, ratiometric measurements of the fluorescence intensity of FAD to NAD(P)H were performed in the co-culture of cancer cells and fibroblasts and in the corresponding monocultures. Representative fluorescence and redox images are shown in Fig. 1 and Fig. 2. Monitoring of the redox ratio in the co-culture showed that in the cancer cells the redox ratio significantly decreased on Day 5 of co-culturing, indicating an increase in glycolytic activity. Meanwhile, the fibroblasts showed a gradual increase of their redox ratio from Day 1 to Day 5.
Figure 1.
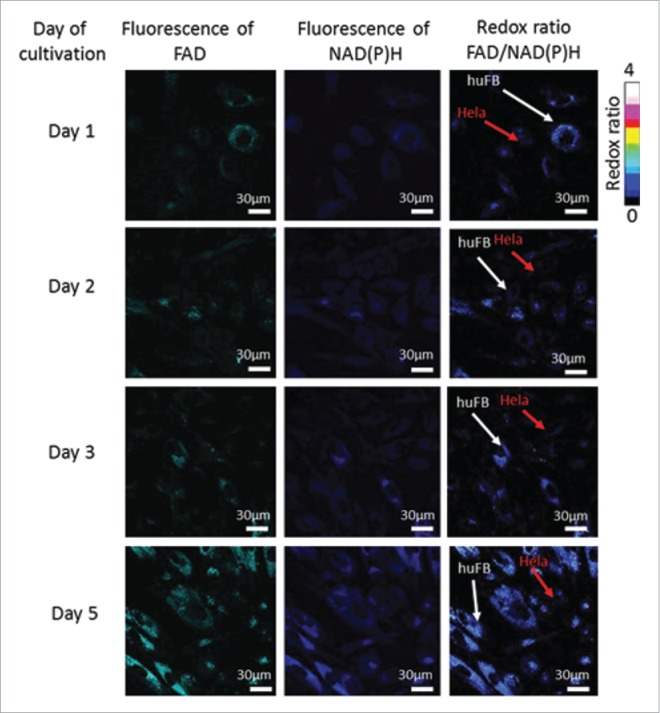
Fluorescence and optical redox ratio images of cancer cells and fibroblasts in co-culture. For NAD(P)H: excitation - 750 nm, detection - 455–500 nm, for FAD: excitation - 900 nm, detection - 500–550 nm. Image size is 213 × 213 μm (1024×1024 pixels).
Figure 2.
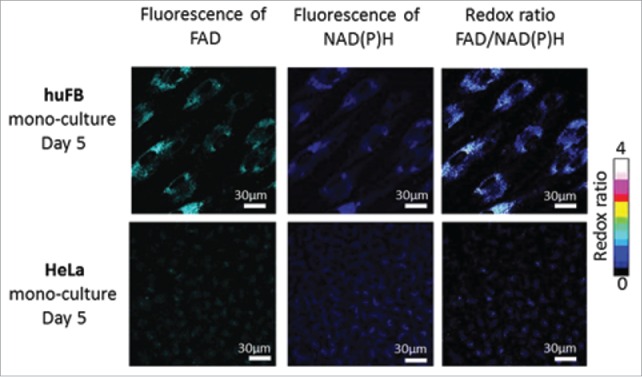
Fluorescence and optical redox ratio images of cancer cells and fibroblasts in mono-culture. For NAD(P)H: excitation - 750 nm, detection - 455–500 nm, for FAD: excitation - 900 nm, detection - 500–550 nm. Image size is 213 × 213 μm (1024×1024 pixels).
In the monocultures the redox ratio in the fibroblasts was higher than in the cancer cells but did not change during 5 days of cultivation in either cell type (1.4 vs. 0.35, p = 0.000001) (Fig. 3).
Figure 3.
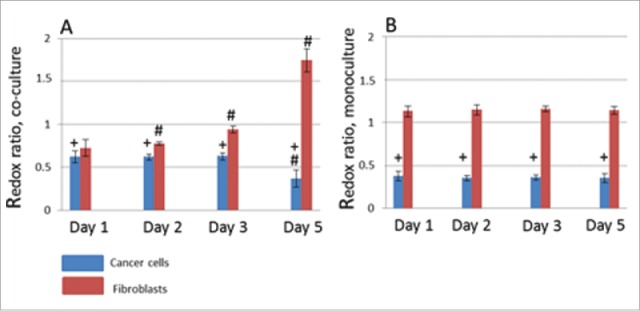
The optical redox ratio (FAD/NAD(P)H) in cancer cells (blue columns) and fibroblasts (red columns) in co-culture (A) and mono-cultures (B). M ± SD. # statistically significant difference from Day 1, p ≤ 0.005 + statistically significant difference from fibroblasts on the same day of cultivation, p ≤ 0.05
As expected for FLIM, the fluorescence decay curves for NAD(P)H and FAD were best fit to a double-exponential decay model, indicating the presence of 2 distinctly different lifetimes for the free and protein-bound forms of the co-factors. The fluorescence lifetimes of the free (τ1) and protein-bound (τ2) NAD(P)H were measured to be ∼0.4 and ∼2.6 ns, respectively. For the free (τ2) and protein-bound (τ1) the FAD fluorescence lifetimes were ∼2.7 and ∼0.4 ns. Fluorescence lifetimes were the same in both the cancer cells and fibroblasts and did not change during co-culturing (Table 1, p>0.05, the maximal р = 0.58).
Table 1.
Fluorescence lifetimes of NAD(P)H and FAD in mono- and co-cultures of cancer cells and fibroblasts. M ± SD.
| Mono-culture |
Co-culture |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HeLa |
huFB |
HeLa |
huFB |
||||||
| τ1, ns | τ2, ns | τ1, ns | τ2, ns | τ1, ns | τ2, ns | τ1, ns | τ2, ns | ||
| Day 1 | NAD(P)H | 0.45 ± 0.01 | 2.6 ± 0.1 | 0.43 ± 0.03 | 2.5 ± 0.5 | 0.48 ± 0.06 | 2.7 ± 0.1 | 0.45 ± 0.03 | 2.7 ± 0.1 |
| FAD | 0.42 ± 0.05 | 2.6 ± 0.1 | 0.41 ± 0.04 | 2.6 ± 0.1 | 0.42 ± 0.05 | 2.6 ± 0.1 | 0.4 ± 0.05 | 2.6 ± 0.1 | |
| Day 2 | NAD(P)H | 0.44 ± 0.02 | 2.6 ± 0.1 | 0.41 ± 0.01 | 2.6 ± 0.1 | 0.46 ± 0.02 | 2.7 ± 0.1 | 0.45 ± 0.03 | 2.7 ± 0.1 |
| FAD | 0.36 ± 0.04 | 2.5 ± 0.1 | 0.4 ± 0.04 | 2.8 ± 0.1 | 0.38 ± 0.03 | 2.6 ± 0.1 | 0.4 ± 0.04 | 2.7 ± 0.1 | |
| Day 3 | NAD(P)H | 0.43 ± 0.02 | 2.6 ± 0.1 | 0.41 ± 0.02 | 2.6 ± 0.1 | 0.44 ± 0.08 | 2.7 ± 0.1 | 0.44 ± 0.02 | 2.7 ± 0.1 |
| FAD | 0.4 ± 0.04 | 2.6 ± 0.1 | 0.4 ± 0.05 | 2.7 ± 0.1 | 0.39 ± 0.04 | 2.6 ± 0.1 | 0.4 ± 0.04 | 2.7 ± 0.1 | |
| Day 5 | NAD(P)H | 0.43± 0.2 | 2.5 ± 0.1 | 0.4 ± 0.03 | 2.6 ± 0.1 | 0.44 ± 0.03 | 2.6 ± 0.1 | 0.43 ± 0.02 | 2.6 ± 0.1 |
| FAD | 0.38 ± 0.05 | 2.6 ± 0.1 | 0.42 ± 0.04 | 2.8 ± 0.1 | 0.46 ± 0.07 | 2.8 ± 0.2 | 0.42 ± 0.04 | 2.8 ± 0.1 | |
The relative contribution of free NAD(P)H (α1) in cancer cells co-cultured with fibroblasts increased from 76.9 % on Day 1 to 79.45 % (p = 0.000001) on Day 2, and remained at that increased level during whole period of co-culturing. The relative contribution of free FAD (α2) in these cells increased on Day 3 from 27.9 to 32.4 % (p = 0.000000), and then did not change. The observed changes in the relative contributions of free NAD(P)H and FAD testify to an increased bias toward a glycolytic metabolism. By contrast, for the fibroblasts in the co-culture, the relative contributions of free NAD(P)H and FAD gradually decreased starting from Day 2, indicating a shift toward oxidative metabolism (Fig. 4). All cells in the population displayed the described changes.
Figure 4.
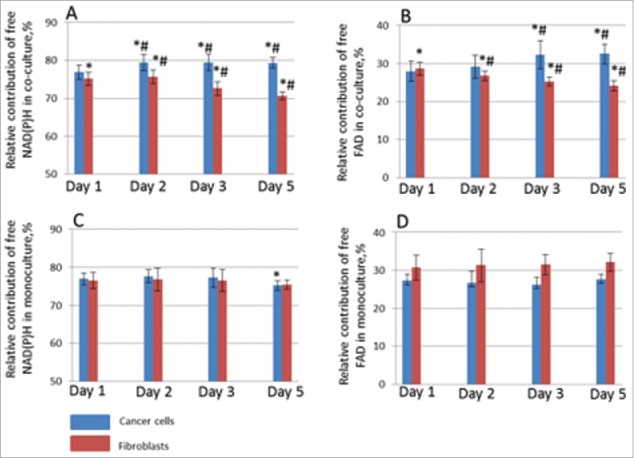
Relative contributions of metabolic cofactors in HeLa cells (blue columns) and fibroblasts (red columns). Free NAD(P)H (α1) in co-culture (A), free FAD (α1) in co-culture (B), free NAD(P)H (α1) in mono-culture (C), free FAD (α1) in mono-culture (D). * statistically significant difference from the same day in mono-culture, p ≤ 0.05 # statistically significant difference from Day 1, p ≤ 0.005
In monocultures of cancer cells and fibroblasts the relative contributions of the co-factors were fairly stable throughout the 5 days of cultivation (˜76% for free NAD(P)H and ˜30% for free FAD) without any statistical difference for the NAD(P)H and only a slight difference for FAD (p = 0.000011).
Therefore, both the optical redox ratio FAD/NAD(P)H and the FLIM measurements of the relative contributions of protein-bound and free NAD(P)H and FAD in cancer cells and fibroblasts showed similar changes in their cellular energy metabolism – a switch of the HeLa cells toward a glycolytic phenotype and a switch of the huFb toward OXPHOS as a result of co-cultivation.
pHi in cancer cells in mono- and co-culture
pHi in cancer cells was measured using the genetically encoded indicator, SypHer2. SypHer2 has 2 excitation peaks, at 420 nm and at 500 nm, and one emission peak at 516 nm. The excitation peak at 420 nm decreases with pH proportionally to the increase in the peak at 500 nm, thus allowing ratiometric measurement of pHi.21 Previously we have shown the possibility of assessing pHi distribution in cultured cancer cells and tumors using SypHer2.22 A more acidic pHi was recorded in the core of each tumor spheroid and in the center of the tumor nodule, presumably due to a hypoxia-induced increase in the use of the glycolytic metabolism and, as a consequence, the accumulation of lactate.
In monoculture of cancer cells stably expressing the sensor, the fluorescence ratio I500/I420 did not change with time, indicating a stable pHi (Fig. 5). On Day 1 of co-cultivation with fibroblasts, the SypHer2 ratio had already started to show a reduction compared to that in the monoculture (p = 0.028) because the pHi became more acidic. The lower SypHer2 ratio remained throughout further cultivation. Acidification of the cytosol was, likely, a consequence of a greater emphasis on the glycolytic metabolism in the cancer cells cultivated with fibroblasts compared with those in monoculture.
Figure 5.
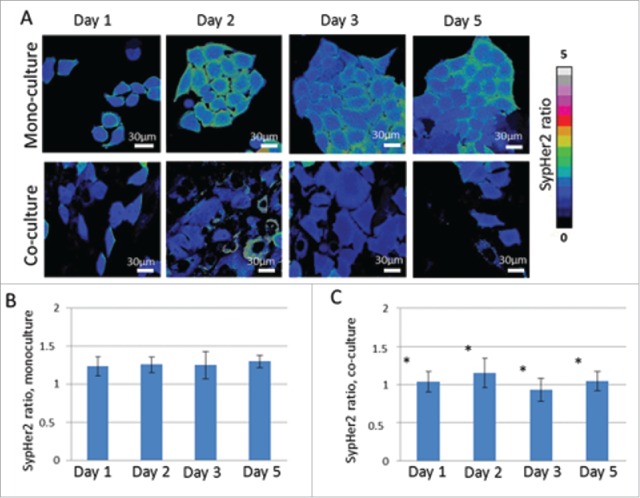
Analysis of pHi in cancer cells in mono-culture and in co-culture with fibroblasts using genetically encoded sensor SypHer2. Ratiometric images I500/I420 (A), SypHer2 ratio in mono-culture (B) and in co-culture (C) from Days 1 to 5 of culturing. * statistically significant difference from monoculture on the same day, p ≤ 0.05
H2O2 in cancer cells in mono- and co-culture
Hydrogen peroxide production in cancer cells was studied using the genetically encoded fluorescent probe, HyPer2, developed by Markvicheva et al.23 HyPer2 is a ratiometric (dual excitation) indicator with spectral characteristics similar to SypHer2. Measuring H2O2 with HyPer2 is based on the increase of the I500/I420 ratio upon H2O2 exposure. HyPer2 has been previously used to monitor the generation of H2O2 in cells activated by various growth factors.24
To test the hypothesis that hydrogen peroxide participates in a metabolic coupling between cancer and normal cells as a signal molecule, cancer cells stably expressing the H2O2- sensor HyPer2 were co-cultured with fibroblasts for 5 days. A monoculture of HeLa-HyPer2 cells imaged at the same time-points was used as a control. The relative amount of H2O2 was assessed by the ratio of fluorescence intensities excited at 500 nm and 420 nm (I500/I420). There were no differences between the I500/I420 values in the monoculture of cancer cells during the 5 days of cultivation (Fig. 6). In the co-culture, a 1.4-fold increase in the sensor signal occurred on Day 2 (p = 0.000001), and then it decreased to the base level matching that in the monoculture. This short flash of hydrogen peroxide production in the cancer cells testifies, most probably, to the function of hydrogen peroxide as a second messenger and oxidative stress trigger in the cancer cell-fibroblast interaction.
Figure 6.
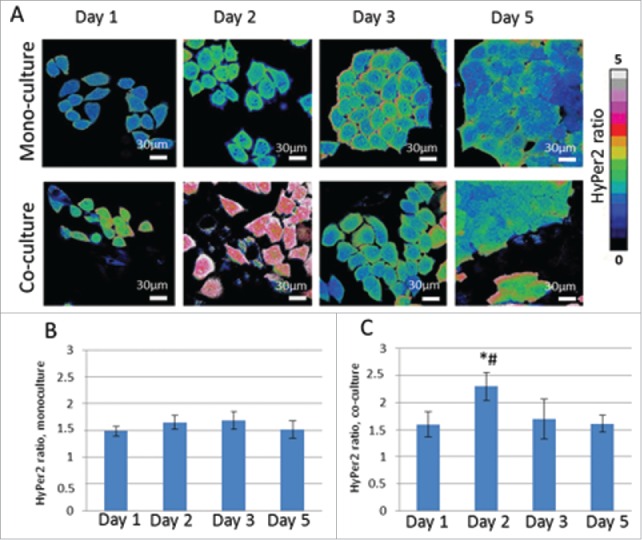
Analysis of hydrogen peroxide production in cancer cells in mono-culture and co-culture with fibroblasts using genetically encoded sensor HyPer2. Ratiometric images I500/I420 (A), SypHer2 ratio in mono-culture (B) and in co-culture (C) from Days 1 to 5 of culturing. * statistically significant difference from monoculture on the same day, p ≤ 0.05 # statistically significant difference from Day 1, p ≤ 0.005
Discussion
In this paper we investigated into metabolic changes that occur in cancer cells and fibroblast as a result of their interaction. In the co-culture of human cervical carcinoma cells, HeLa and human fibroblasts, we monitored an aspect of the metabolic status of both cell types by using the optical redox ratio FAD/ NAD(P)H and the fluorescence lifetimes of NAD(P)H and FAD, in addition to the intracellular pH and the hydrogen peroxide levels in the cancer cells, using genetically encoded sensors. We observed a metabolic shift from OXPHOS toward glycolysis in cancer cells, and from glycolysis to OXPHOS in fibroblasts, starting from Day 2 of co-culturing. The metabolic switch was accompanied by hydrogen peroxide production in the cancer cells on Day 2. The increased glycolytic metabolism of the cancer cells resulted in slight acidification of their cytosol in comparison with that of the corresponding monoculture. Therefore, our HeLa-huFb system demonstrated metabolic behavior similar to Warburg type tumors, where cancer cells use increased glycolytic and stromal cells tend toward increased non-glycolytic metabolism.
Previously, a glycolytic phenotype of cervical cancer had been demonstrated for cell cultures and human tumors. For example, Rossignol et al. reported that HeLa cells generate energy predominantly by glycolysis, but can change ATP generation to exclusively OXPHOS when the availability of glucose is limited.26 It was found by Herst et al. that HeLa cells that rely preferentially on OXPHOS in highly oxygenated conditions became more glycolytic upon hypoxia.1,27 Glycolytic metabolism of HeLa cells was also shown for tumor spheroids.28,29 Pinheiro et al.30 demonstrated the glycolytic phenotype of squamous cell carcinoma in biopsy samples. At the same time, fibroblasts retain a significant level of OXPHOS even if the glucose level is high.31,32
There are many methods for assessment of the cells metabolic status including direct methods such as measuring of the glucose uptake, lactate production and oxygen consumption.27 Indirect methods include immunocytochemistry or immunohistochemistry for metabolic markers (for example, the monocarboxylate transporters MCT1 and MCT4);30 evaluation of the key glycolytic enzymes (for example, hexokinase-II, phosphofructokinase and pyruvate kinase),28,29,33 and the microarray analysis of the glycolytic gene expression.34 However, all these methods are time-consuming. They require exogenous labels and do not allow to probe metabolic changes continuously and non-invasively. Moreover gene transcription and protein production do not fully reflect the metabolic flux.35,36
To date, multiphoton microscopy of the NAD(P)H and FAD redox states and fluorescence lifetimes provides an established technique for metabolic imaging in vitro and in vivo. Application of the method has been widely demonstrated in oncology for, not only fundamental, but also for pre-clinical and clinical studies of therapeutic agents and the development of diagnostic approaches.37 Major differences in the metabolic phenotypes of cancer and normal tissues allow to distinguish tumors at an early stage, as has been shown for basal cell carcinoma, head and neck squamous cell carcinoma, cervical intraepithelial neoplasia, melanoma and other tumors.38-41 However, to our knowledge, this highly sensitive and precise technique has never been used to analyze the energy metabolism during tumor-stroma co-evolution.
The optical redox ratio, the ratio of fluorescence intensities of FAD to NAD(P)H, is known to reflect the balance between oxidative phosphorylation and glycolysis. Cancer cells, where aerobic glycolysis predominates over OXPHOS, display a decreased redox ratio, as has been demonstrated for different cancer cell lines and tumors.42-44 In our study we detected a reduced ratio of FAD/NAD(P)H in cancer cells while it increased in fibroblasts in co-culture conditions, indicating metabolic shifts toward glycolysis and OXPHOS, respectively. This is, however, in contrast with a work by Scala et al.,45 who showed in vivo that not all tumor cells demonstrate the Warburg effect and the redox ratio (associated with the enhanced metabolic activity of tumor tissue) decreases with distance from the nearest vessel. In mono-cultures we observed an increased redox ratio in fibroblasts compared to cancer cells, which testifies to higher oxidative metabolism in the stromal cells. This result is consistent with the data by Ostrander et al, who showed that the MCF-10A and HMEC cell lines of normal mammary epithelial cells had an increased redox ratio when compared to 9 different breast cancer cell lines.43 Martinez-Outschoorn et al.25 also detected increased mitochondrial activity in fibroblasts when compared with the MCF-7 cell line in monoculture. Similar results were reported by R. de Andrade Natal et al.46 for biopsies of breast cancer patients.
Using FLIM we measured the fluorescence lifetimes and relative contribution of the short and long components of NAD(P)H and FAD in HeLa cells and human fibroblasts. Fluorescence lifetimes documented in the literature lie in the range 0.2-0.5 ns for free NAD(P)H, 1.8-2.7 ns for protein-bound NAD(P)H, 0.15-0.4 ns for protein-bound FAD and 2-2.8 ns for free FAD.17,47,48 Therefore, the values ∼0.4 ns and ∼2.6 ns for NAD(P)H, and ˜0.4 ns and ∼2.7 ns for FAD recorded in our study agree well with published data. We have found no differences in the fluorescence lifetimes of the NAD(P)H and FAD between cancer cells and fibroblasts and no changes in the process of co-cultivation in vitro. Although it is known that fluorescence lifetimes of the NAD(P)H and FAD depend on pH, and we detected slight acidification of the cytosol of cancer cells as result of metabolic changes, however this pHi alteration in cancer cells was, likely, insufficient to cause any changes in fluorescence lifetimes of the cofactors. The lifetime changes are normally observed when pH fluctuates in a wide range (5.0 – 9.0 pH units),49 whereas in our study pH changes did not exceed 0.2 pH units according to previous estimations.22
Another parameter to describe cellular energy metabolism by FLIM is the relative contribution of the fluorescence lifetime components corresponding to the free or protein-bound states of the co-factors. A decrease in the relative amount of protein-bound NAD(P)H (τ2) and FAD (τ1) is associated with an increased level of glycolysis. In our HeLa-huFb co-culture, a significant decrease in the contributions of protein-bound NAD(P)H and FAD was observed in the cancerous cells, whereas, in the fibroblasts, the contributions of protein-bound co-factors increased. Previously, in vivo, a decreased contribution of protein-bound NAD(P)H and FAD had been demonstrated for precancer development.50 A decreased amount of NAD(P)H and a decreased free/bound NAD(P)H ratio were also shown in oral squamous carcinoma cells when compared with non-malignant cells.20
Nevertheless, there have been very few studies in this field performed on the co-culture model. For example, a higher glucose uptake and increased lactate production in co-cultures of MCF-7 and CAFs compared with monocultures were demonstrated by Brauer et al., but in that work the cancerous and the normal cells were not separated.51 Martinez-Outschoorn et al.11,12,25 demonstrated the reversed Warburg effect (OXPHOS in cancer cells and glycolysis in fibroblasts) in co-cultures of MCF7 cancer cells and fibroblasts.
We analyzed pHi in cancer cells using the genetically-encoded sensor SypHer2 and revealed a minor acidification of the cytosol of cells co-cultured with fibroblasts. This observation corresponds well with metabolic shift to glycolysis shown by NAD(P)H and FAD fluorescence imaging: decreased pHi is associated with increased lactic acid production in conditions of increased glycolysis. ‘The combination of lactic acidosis and decreased cytosolic pH is a specific feature of cancer cells with high rates of glycolysis. It is known that the accumulation of lactate during tumor development may affect the pHi and drive metabolic changes.52 At the same time, for successful realization of glycolysis, an elevated pH level is required.53 We observed a difference between the pHi of cancer cells in mono- and in co-culture starting from the Day 1 of their co-cultivation with fibroblasts, and this pHi value remained unchanged throughout the whole period of the study. This small modification of pHi confirms that cytosolic pH is a quite stable parameter owing to the high buffering capacity of cells.54 However, the statistical significance of the pHi changes flags deep changes in intracellular processes in cancer cells during co-cultivation.
The ability of cancer cells to generate reactive oxygen species (ROS) is well documented.55,56 A number of studies have shown an increased level of hydrogen peroxide in cancer cells, which is essential for cancer development.12,56 To test if hydrogen peroxide participates in metabolic coupling between cancer cells and fibroblasts, we examined H2O2 production in cancer cells using the genetically encoded sensor HyPer2. We detected elevation of the sensor signal on Day 2 of co-cultivation, indicating a rise in the H2O2 concentration in the HeLa cells. Similar results were obtained by Martinez-Outschoorn et al.,12 who registered high level of ROS in MCF7 cancer cells on Day 2 of their co-culture with fibroblasts, and, using catalase, verified that the main component of the ROS was hydrogen peroxide. The authors suggested that through H2O2 secretion, initially glycolytic MCF7 cells shift adjacent fibroblasts toward the glycolytic state as a result of oxidative stress, so that the MCF7 cells become oxidative and the fibroblasts become glycolytic, displaying the “reverse Warburg effect.” Interestingly, on Day 5 H2O2 was generated mainly in the CAFs. It was suggested that H2O2 produced in the CAFs helps to maintain their glycolytic state. A principal difference from that work is that our HeLa-huFb co-culture system underwent the Warburg effect, as the cancer cells tended toward glycolytic metabolism and the fibroblasts – toward oxidative. This is consistent with the idea that cancer cells activate aerobic glycolysis because of a deviation of oxygen metabolism from the route that generates ATP (OXPHOS) to the route that produces O2·− and H2O2.57 We propose that the hydrogen peroxide secreted by cancer cells affects the adjacent fibroblasts where it activates the tricarboxylic acid cycle (TCA cycle) for its detoxification. This process was described by Mailloux et al.58 who demonstrated that TCA cycle acts as a universal oxidative defense mechanism, where a-ketoglutarate (KG) is a key detoxifier of ROS. The lactate eliminated from the cancer cells may act as a substrate in the TCA cycle.59 Since the conversion of lactate to pyruvate is accompanied by NAD(P)H production59 this could cause the high NAD(P)H fluorescence intensity detected in the fibroblasts in our study (Fig. 1). Measuring NAD(P)H fluorescence intensity in fibroblasts showed that it increased by 58.2% from 7562 ± 755 on Day 1 of co-cultivation to 12985 ± 738 on Day 5 (p = 0.0007). In monoculture of fibroblasts the intensity was 11470 ± 948 on Day 1 and 12800 ± 985 on Day 5 (p = 0.22).
Conclusions
In this study we combined, for the first time, imaging of NAD(P)H and FAD, with measurements of the levels of intracellular pH and hydrogen peroxide, to trace ongoing metabolic changes as a result of tumor-stroma interaction. Using co-cultures of HeLa cells and human fibroblasts to model the interaction, we observed a metabolic switch toward glycolytic metabolism in the HeLa cells and toward oxidative metabolism in the fibroblasts in the situation of co-cultivation, which was accompanied by both a reduction of pHi (acidification) and by H2O2 production in the cancer cells. The data about metabolic changes in tumor-stroma co-evolution are important for the development of anticancer drugs targeted on metabolic pathways in both cancer and adjacent stromal cells. Searching for the start-point of the metabolic alterations and an understanding of the mechanisms of their realization and regulation are promising fundamental tasks, opening new ways for cancer treatment.
Materials and methods
Cell cultures
HeLa Kyoto, human cervical carcinoma, stably expressing the pHi-sensor SypHer2 (HeLa-SypHer2) or the H2O2-sensor HyPer2 (HeLa-HyPer2), non-expressing counterpart and human skin fibroblasts (huFB) were used in the study. Genetically transfected cell lines were generated and characterized in the Institute of Bioorganic Chemistry Russian Academy of Science (Moscow, Russia). The huFB were obtained from the Koltzov Institute of Developmental Biology Russian Academy of Science (Moscow, Russia). For assessment of metabolic activity separate cell cultures that did not have fluorescent sensors were used.
The cells were cultured in DMEM containing 100 µg/ml penicillin, 100 µg/ml streptomycin sulfate and 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2.
The protocol for the co-culturing of cancer cells and fibroblasts was modified from that of ref. 25. For co-culturing, the huFB were plated in glass-bottom FluoroDishes in complete DMEM media without phenol red (Life Technologies) and then HeLa Kyoto cells were seeded within 4 hours. The total number of cells per dish in the co-culture was 1 × 105 with a 1:5 fibroblast-to-cancer cell ratio. In parallel, monotypic cultures were plated at the same quantity as in the corresponding co-culture. The day of plating was defined as a Day 0. The day after plating (Day 1), the medium was changed to DMEM with 5% FBS, and afterwards the medium was changed every other day. The cells were analyzed over a period of 5 days (Day 1, Day 2, Day 3 and Day 5).
Fluorescence microscopy and FLIM technique
Fluorescence confocal (one- and 2-photon) and FLIM images were obtained using an LSM 710 (Carl Zeiss, Germany) fluorescence laser-scanning microscope equipped with a femtosecond Ti:Sa laser with a repetition rate of 80 MHz and pulse duration of 140 fs, and an FLIM module based on time-correlated single photon counting - Simple Tau 152 TCSPC (Becker & Hickl GmbH, Germany). The fluorescence images were processed with ImageJ 1.39p software (NIH, USA). The analysis of the FLIM data was performed using SPCImage software (Becker & Hickl GmbH, Germany).
The images were acquired through a 40x, 1.2 NA water immersion objective. During image acquisition the cells were maintained at 37°C and 5% CO2.
NAD(P)H and FAD analysis
NAD(P)H and FAD fluorescences were excited at wavelengths of 750 nm or 900 nm and registered in the ranges 455-500 nm or 500-550 nm, respectively. The average power applied to the sample was ˜6 mW.
The fluorescence redox ratio was calculated as the ratio of the fluorescence intensity of FAD to that of NAD(P)H. The background signal, taken from an empty region of the image, was subtracted from the fluorescence images. The value of the ratio was analyzed for the cytoplasm of the cells. To selectively measure fluorescence in the cytoplasm 2-step analysis was used. For each image, a threshold was applied first, so that the much darker nucleus was below the threshold according to the work by Scala et.al.18 Then we selected ˜40x40 and ˜60x60 pixel zones in the cytoplasm of cancer cells and fibroblasts, correspondingly.
The method for NAD(P)H and FAD fluorescence lifetime measurements was adopted from that of ref. 18 and 20. 3 × 3 spatial pixel binning (binning factor 1) was used to obtain the decay profiles for ˜5000 photons in total. The lifetime decay curve of each pixel was fitted to a double-exponential decay model, where the short and long lifetime components (τ1 and τ2, respectively), and the relative contributions of the lifetime components (a1 and a2, where a1 + a2 = 100%) were estimated. The goodness of the fit, the χ2 value, was 1.2 ± 0.1 (single-sigma standard deviation). In a first approximation, for NAD(P)H the first (short, τ1) component can be attributed to its free, and the second (long, τ2) component to its protein-bound state. For FAD τ1 is attributed to the protein-bound and τ2 - to the free form. The absence of photobleaching was confirmed by monitoring the photon count rates throughout image acquisition. A threshold was applied so that the much darker nucleus was below the threshold.
Cancer cells and fibroblasts were identified by their cellular morphology. Cancer cells are much smaller than fibroblasts, have a round shape and large nucleus with a thick cytoplasmic layer. The fibroblasts have a typically spindle-shaped morphology, much greater size and a larger volume of cytoplasm compared to the cancer cells.
pHi and H2O2 imaging
Fluorescence of the genetically encoded sensors, SypHer2 and HyPer2, was excited at the wavelengths of 405 nm by diode laser and 488 nm by an argon laser with a diffraction grating. Emission was detected in the range 435–689 nm for excitation at 405 nm and at 509–689 nm for excitation at 488 nm. The background signal, taken from an empty region of the image, was subtracted from the measurements, and the ratio of emission intensity resulting from excitation at the 2 wavelengths was calculated (I500/I420).
Statistical analysis
The mean values (M) and standard deviations (SD) were calculated for the fluorescence lifetimes, the relative contributions of the long and short components of NAD(P)H and FAD, the redox ratio, and the SypHer2 and HyPer2 ratios. The data were compared using the Student's t-test and the one-way ANOVA with Fisher's post-hoc test, where appropriate. p ≤ 0.05 was considered statistically significant. The total number of cells used for mean value calculations was from 30 to 50 cells in 5-7 fields of view for each time-point. Final calculations were done from 3 independent samples.
Abbreviations
- ATP
Adenosine triphosphate
- CAFs
cancer-associated fibroblasts
- FAD
flavin adenine dinucleotide
- FLIM
fluorescence lifetime imaging
- NAD(P)H
Nicotinamide adenosine dinucleotide phosphate
- OXPHOS
oxidative phosphorylation
- pHi
intracellular pH
- ROS
reactive oxygen species
- TCA cycle
tricarboxylic acid cycle
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work has been financially supported by the Russian Science Foundation (grants No. 14-15-00646). The authors are grateful to Dr. Vladislav Shcheslavskiy (Becker & Hickle GmbH, Germany) for his help with FLIM and for his valuable discussions.
References
- [1].Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol Lett 2012; 4:1151-7; PMID:23226794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science 2001; 292:504-7; PMID:11283355; http://dx.doi.org/ 10.1126/science.1058079 [DOI] [PubMed] [Google Scholar]
- [3].Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun 2004; 313:459-65. [DOI] [PubMed] [Google Scholar]
- [4].Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol 2010; 30:1303-18; PMID:20100866; http://dx.doi.org/ 10.1128/MCB.01101-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Marusyk A, Polyak K. Tumor heterogeneity: Causes and consequences. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 2009; 1:105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Berridge MV, Herst PM, Tan AS. Metabolic flexibility and cell hierarchy in metastatic cancer. Mitochondrion 2010; 10:584-588; PMID:20709626; http://dx.doi.org/ 10.1016/j.mito.2010.08.002 [DOI] [PubMed] [Google Scholar]
- [7].Wang W, Li Q, Yamada T, Matsumoto K, Matsumoto I, Oda M, Watanabe G, Kayano Y, Nishioka Y, Sone S, et al.. Crosstalk to stromal fibroblasts induces resistance of lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors. Clin Cancer Res 2009; 15:6630-8; PMID:19843665; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-1001 [DOI] [PubMed] [Google Scholar]
- [8].Jedeszko C, Victor BC, Podgorski I, Sloane BF. Fibroblast hepatocyte growth factor promotes invasion of human mammary ductal carcinoma in situ. Cancer Res 2009; 69:9148-55; PMID:19920187; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matsumoto K, Nakamura T. Hepatocyte growth factor and the Met system as a mediator of tumor-stromal interactions. Int J Cancer 2006; 119:477-83; PMID:16453287; http://dx.doi.org/ 10.1002/ijc.21808 [DOI] [PubMed] [Google Scholar]
- [10].Cirri P, Chiarugi P. Cancer associated fibroblasts: the dark side of the coin. Am J Cancer Res 2011; 1(4):482-97; PMID:21984967 [PMC free article] [PubMed] [Google Scholar]
- [11].Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S, et al.. The reverse Warburg effect aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009; 8(23):3984-4001; PMID:19923890; http://dx.doi.org/ 10.4161/cc.8.23.10238 [DOI] [PubMed] [Google Scholar]
- [12].Lisanti MP, Martinez-Outschoorn UE, Lin Z, Pavlides S, Whitaker-Menezes D, Pestell RG, Howell A, Sotgia F. Hydrogen peroxide fuels aging, inflammation, cancer metabolism and metastasis. Cell Cycle 2011; 10(15):2440-49; PMID:21734470; http://dx.doi.org/ 10.4161/cc.10.15.16870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med 2014; 211(8):1503-23; PMID:25071162; http://dx.doi.org/ 10.1084/jem.20140692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vaupel P. Metabolic microenvironment of tumor cells: a key factor in malignant progression. Exp Oncol 2010; 32(3);125-7; PMID:21403604 [PubMed] [Google Scholar]
- [15].Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest 2013; 123(9):3685-92; PMID:23999443; http://dx.doi.org/ 10.1172/JCI69741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lakowitz J. Principles of fluorescence spectroscopy [Lakowitz, J. (ed.)] [63–95] (Springer-Verlag, 2006) [Google Scholar]
- [17].Guo HW, Chen CT, Wei YH, Lee OK, Gukassyan V, Kao FJ, Wang HW. Reduced nicotinamide adenine dinucleotide (NADH) fluorescence lifetime separates human mesenchymal stem cells from differentiated progenies. J Biomed Opt 2008; 13(5):050505; PMID:19021377; http://dx.doi.org/ 10.1117/1.2990752 [DOI] [PubMed] [Google Scholar]
- [18].Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. PNAS 2007; 104(49):19494-99; PMID:18042710; http://dx.doi.org/ 10.1073/pnas.0708425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walsh AJ, Cook RS, Manning HC, Hicks DJ, Lafontant A, Arteaga CL, Skala MC. Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res 2013; 73(20):6164-74; PMID:24130112; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rück A, Hauser C, Mosch S, Kalinina S. Spectrally resolved fluorescence lifetime imaging to investigate cell metabolism in malignant and nonmalignant oral mucosa cells. J Biomed Opt 2014; 19(9):096005; PMID:25202900; http://dx.doi.org/ 10.1117/1.JBO.19.9.096005 [DOI] [PubMed] [Google Scholar]
- [21].Matlashov ME, Bogdanova YA, Ermakova GA, Mishina NM, Ermakova YG, Nikitin ES, Balaban PM, Okabe S, Lukyanov SA, Enikolopov G, et al.. Fluorescent ratiometric pH indicator SypHer2: applications in neuroscience and regenerative biology. Biochim Biophys Acta Gen Subj 2015; 1850(11):2318-28; PMID:26259819; http://dx.doi.org/ 10.1016/j.bbagen.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shirmanova MV, Druzhkova IN, Lukina MM, Matlasov ME, Belousov VV, Snopova LB, Prodanetz NN, Dudenkova VV, Lukyanov SA, Zagaynova EV. Intracellular pH imaging in cancer cells in vitro and tumors in vivo using the new genetically encoded sensor SypHer2. BBA-Gen Subj 2015; 1850:1905-11; PMID:25964069; http://dx.doi.org/ 10.1016/j.bbagen.2015.05.001 [DOI] [PubMed] [Google Scholar]
- [23].Markvicheva KN, Bilan DS, Mishina NM, Gorokhovatsky AY, Vinokurov LM, Lukyanov S, Belousov VV. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorganic & Med Chem 2011; 19:1079-84; PMID:20692175; http://dx.doi.org/ 10.1016/j.bmc.2010.07.014 [DOI] [PubMed] [Google Scholar]
- [24].Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 2006; 3:281-6; PMID:16554833; http://dx.doi.org/ 10.1038/nmeth866 [DOI] [PubMed] [Google Scholar]
- [25].Martinez-Outschoorn UE, Lin Z, Trimmer C, Flomenberg N, Wang S, Pavlides S, Pestell RG, Howell A, Sotgia F, Lisanti MP. Cancer cells metabolically “fertilize” the tumor microenvironment with hydrogen peroxide, driving the Warburg effect. Implications for PET Imaging Hum Tumors Cell Cycle 2011; 10(15):2504-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 2004:64(1):985-93; PMID:14871829 [DOI] [PubMed] [Google Scholar]
- [27].Herst PM, Berridge MV. Cell surface oxygen consumption: a major contributor to cellular oxygen consumption in glycolytic cancer cell lines. Biochimica et Biophysica Acta 2007; 1767:170-7. [DOI] [PubMed] [Google Scholar]
- [28].Marín-Hernández A, Rodríguez-Enríquez S, Vital-González PA, Flores-Rodríguez FL, Macías-Silva M, Sosa-Garrocho M, Moreno-Sánchez R. Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J 2006; 273:1975-88; PMID:16640561; http://dx.doi.org/ 10.1111/j.1742-4658.2006.05214.x [DOI] [PubMed] [Google Scholar]
- [29].Rodríguez-Enríquez S, Gallardo-Pérez JC, Avilés-Salas A, Marín-Hernández A, Carre˜no-Fuentes L, Maldonado-Lagunas V, Moreno-Sánchez R. Energy metabolism transition in multi-cellular human tumor spheroids. J Cell Physiol 2008; 216:189-97; PMID:18264981; http://dx.doi.org/ 10.1002/jcp.21392 [DOI] [PubMed] [Google Scholar]
- [30].Pinheiro E, Garcia A, Morais-Santos F, Scapulatempo-Neto C, Mafra A, Steenbergen RDM, Boccardo E, Villa LL, Baltazar F, Longatto-Filho A. Lactate transporters and vascular factors in HPV-induced squamous cell carcinoma of the uterine cervix. BMC Cancer 2014; 14(751):2-12; PMID:24383423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Reitzer L, Wice B, Kennel D. Evidence that glutamine, not sugar, is the major energy source for cultured Hela cells. J Biol Chem 1979; 254:2669-76; PMID:429309 [PubMed] [Google Scholar]
- [32].Donnely M, Scheffler I. Energy metabolism in respiration deficient and wildtype Chinese hamster fibroblasts in culture. J Cell Physiol 1976; 89:9-52. [DOI] [PubMed] [Google Scholar]
- [33].Canesi L, Ciacci C, Betti M, Malatesta M, Gazzanelli G, Gallo G. Growth factors stimulate the activity of key glycolytic enzymes in isolated digestive gland cells from mussels (Mytilus galloprovincialis Lam.) through tyrosine kinase mediated signal transduction. Gen Comp Endocrinol 1999; 116(2):241-8; PMID:10562454; http://dx.doi.org/ 10.1006/gcen.1999.7366 [DOI] [PubMed] [Google Scholar]
- [34].Rodríguez-Enríquez S, Carreño-Fuentes L, Gallardo-Pérez JC, Saavedra E, Quezada H, Vega A, Marín-Hernández A, Olín-Sandoval V, Torres-Márquez ME, Moreno-Sánchez R. Oxidative phosphorylation is impaired by prolonged hypoxia in breast and possibly in cervix carcinoma. Int J Biochem Cell Biol 2010; 42(10):1744-51; PMID:20654728; http://dx.doi.org/ 10.1016/j.biocel.2010.07.010 [DOI] [PubMed] [Google Scholar]
- [35].ter Kuille BH, Westerhoff HV. Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett 2001; 500(3):169-71; PMID:11445079; http://dx.doi.org/ 10.1016/S0014-5793(01)02613-8 [DOI] [PubMed] [Google Scholar]
- [36].Bevalicqua A, Wilkinson SJ, Dimelow R, Murabito E, Rehman S, Nardelli M, van Eunen K, Rossell S, Bruggeman FJ, Blüthgen N, et al.. Vertical systems biology: from DNA to flux and back. SEB Exp Biol Ser 2008; 61:65-91; PMID:18709737 [PubMed] [Google Scholar]
- [37].Rajoria S, Zhao L, Intes X, Barroso M. FLIM-FRET for cancer applications. Curr Mol Imaging 2014; 3(2):144-61; PMID:26023359; http://dx.doi.org/ 10.2174/2211555203666141117221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shah AT, Beckler MD, Walsh AJ, Jones WP, Pohlmann PR, Skala MC. Optical metabolic imaging of treatment response in human head and neck squamous cell carcinoma. Plos One 2014:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun Y, Phipps JE, Meier J, Hatami N, Poirier B, Elson DS, Farwell DG, Marcu L. Endoscopic fluorescence lifetime imaging for in vivo intraoperative diagnosis of oral carcinoma. Microsc Microanal 2013. August; 19(4):791-8; PMID:23702007; http://dx.doi.org/ 10.1017/S1431927613001530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].De Beule PAA, Dunsby C, Galletly NP, Stamp GW, Chu AC, Anand U, Anand P, Benham CD, Naylor A, French PMW. A hyperspectral fluorescence lifetime probe for skin cancer diagnosis. Rev Sci Instrum 2007; 78:123101; PMID:18163714; http://dx.doi.org/ 10.1063/1.2818785 [DOI] [PubMed] [Google Scholar]
- [41].Dimitrov E, Riemann I, Ehlers A, Koehler MJ, Norgauer J, Elsner P, König K, Kaatz M. Spectral fluorescence lifetime detection and selective melanin imaging by multiphoton laser tomography for melanoma diagnosis. Exp Dermatol 2009; 18(6):509-15; PMID:19243426; http://dx.doi.org/ 10.1111/j.1600-0625.2008.00815.x [DOI] [PubMed] [Google Scholar]
- [42].Balu M, Zachary CB, Harris RM, Krasieva TB, König K, Tromberg BJ, Kelly KM. In vivo multiphoton microscopy of basal cell carcinoma. JAMA Dermatol 2015; 151(10):1068-74; PMID:25909650; http://dx.doi.org/ 10.1001/jamadermatol.2015.0453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chance B, Schoener B, Oshino R, Itshak F, Nakase Y. Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem 1979; 254:4764-71; PMID:220260 [PubMed] [Google Scholar]
- [44].Ostrander JH, McMahon CM, Lem S, Millon SR, Brown JQ, Seewaldt VL, Ramanujam N. Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res 2010. June 1; 70(11):4759-66; PMID:20460512; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Skala M., Fontanella . Longitudinal optical imaging of tumor metabolism and hemodynamics. J of Biomed Optics 2010; 15(1):011112-1-8; PMID:20210438; http://dx.doi.org/ 10.1117/1.3285584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].de Andrade Natal R, Pelegati VB, Bondarik C, Mendonça GR, Derchain SF, Lima CP, C. Cesar L, Sarian LO, Vassallo J. Increased metabolic activity detected by FLIM in human breast cancer cells with desmoplastic reaction: a pilot study. Proc. SPIE 2015; 9536:95360L [Google Scholar]
- [47].Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain MJ, Szabadkai G, Duchen MR. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 2014; 5:3936; PMID:24874098; http://dx.doi.org/ 10.1038/ncomms4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].van den Berg PAW, Feenstra KA, Mark AE, Berendsen HJC, Visser AJWG. Dynamic conformations of flavin adenine dinucleotide: simulated molecular dynamics of the flavin cofactor related to the time-resolved fluorescence characteristics. J Phys Chem 2002; 106:8858-69; http://dx.doi.org/ 10.1021/jp020356s [DOI] [Google Scholar]
- [49].Ogikubo S, Nakabayashi T, Adachi T, Islam Md S, Yoshizawa T, Kinjo M, Ohta N, Intracellular pH sensing using autofluorescence lifetime microscopy. J Phys Chem B 2011; 115:10385-90; PMID:21776989; http://dx.doi.org/ 10.1021/jp2058904 [DOI] [PubMed] [Google Scholar]
- [50].McGinty J, Galletly NP, Dunsby C, Munro I, Elson DS, Requejo-Isidro J, Cohen P, Ahmad R, Forsyth A, Thillainayagam AV, et al.. Wide-field fluorescence lifetime imaging of cancer. Biomed Opt Exp 2010:627-40; PMID:21258496; http://dx.doi.org/ 10.1364/BOE.1.000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Brauer HA, Makowski L, Hoadley KA, Casbas-Hernandez P, Lang LJ, Roman-Perez E, D'Arcy M, Freemerman AJ, Perou CM, Troester MA. Impact of tumor microenvironment and epithelial phenotypes on metabolism in breast cancer. Clin Cancer Res 2013; 19(3):571-85; PMID:23236214; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Aredia F, Scovassi AI. Multiple effects of intracellular pH modulation in cancer cells. Cancer Cell Microenv 2014; 1:e136 [Google Scholar]
- [53].Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer 2005; 5:786-95; PMID:16175178 [DOI] [PubMed] [Google Scholar]
- [54].Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer 2011; 11:671-7; PMID:21833026; http://dx.doi.org/ 10.1038/nrc3110 [DOI] [PubMed] [Google Scholar]
- [55].Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res 1991; 51:794-8; PMID:1846317 [PubMed] [Google Scholar]
- [56].Jezierska-Drutel A, Rosenzweig SA, Neumann CA. Role of oxidative stress and the microenvironment in breast cancer development and progression. Adv Cancer Res 2013; 119:107-25; PMID:23870510; http://dx.doi.org/ 10.1016/B978-0-12-407190-2.00003-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lopez-Lazaro M, The Warburg effect: why and how do cancer cells activate glycolysis in the presence of oxygen? Anti-Cancer Agents Med Chem 2008; 8:305-12; PMID:18393789; http://dx.doi.org/ 10.2174/187152008783961932 [DOI] [PubMed] [Google Scholar]
- [58].Mailloux RJ, Be´riault R, Lemire J, Singh R, Che´nier DR, Hamel RD, Appanna VD. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PlosOne 2007; 2(8):e690; PMID:17668068; http://dx.doi.org/ 10.1371/journal.pone.0000690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lemire J, Mailloux RJ, Appanna VD. Mitochondrial lactate dehydrogenase is involved in oxidative-energy metabolism in human astrocytoma cells (CCF-STTG1). Plos One 2008; 3(2):e1550; PMID:18253497; http://dx.doi.org/ 10.1371/journal.pone.0001550 [DOI] [PMC free article] [PubMed] [Google Scholar]


