Abstract
Acute kidney injury (AKI) is one of the leading causes of in-hospital morbidity and mortality, particularly in critically ill patients. Although our understanding of AKI at the molecular level remains limited due to its complex pathophysiology, recent advances in both quantitative and spatial mass spectrometric approaches offer new opportunities to assess the significance of renal metabolomic changes in AKI models. In this study, we evaluated lipid changes in early ischemia-reperfusion (IR)-related AKI in mice by using sequential window acquisition of all theoretical spectra (SWATH)-mass spectrometry (MS) lipidomics. We found a significant increase in two abundant ether-linked phospholipids following IR at 6 h postinjury, a plasmanyl choline, phosphatidylcholine (PC) O-38:1 (O-18:0, 20:1), and a plasmalogen, phosphatidylethanolamine (PE) O-42:3 (O-20:1, 22:2). Both of these lipids correlated with the severity of AKI as measured by plasma creatinine. In addition to many more renal lipid changes associated with more severe AKI, PC O-38:1 elevations were maintained at 24 h post-IR, while renal PE O-42:3 levels decreased, as were all ether PEs detected by SWATH-MS at this later time point. To further assess the significance of this early increase in PC O-38:1, we used matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS) to determine that it occurred in proximal tubules, a region of the kidney that is most prone to IR injury and also rich in the rate-limiting enzymes involved in ether-linked phospholipid biosynthesis. Use of SWATH-MS lipidomics in conjunction with MALDI-IMS for lipid localization will help in elucidating the role of lipids in the pathobiology of AKI.
Keywords: AKI, acute kidney injury, IR, ischemia-reperfusion, lipidomics, SWATH-MS, sequential window acquisition of all theoretical spectra mass spectrometry, MALDI-IMS, matrix-assisted laser desorption ionization imaging mass spectrometry
acute kidney injury (aki) is a major cause of morbidity and mortality in critically ill patients and can progress to chronic kidney disease and dialysis dependency (4, 7, 22, 23, 40). Although the overall in-hospital mortality rate is up to 50%, it can exceed 75% in critically ill patients and the prognosis of AKI still remains relatively poor (45). Ischemia-reperfusion (IR) is one of the major causes of AKI clinically, and so it is widely used in animal models to study AKI. Evidence shows that kidney injury and cell death can be caused during ischemic stress with depletion of ATP, activation of enzymes including phospholipases, proteases, and reactive oxygen species (ROS), generating enzymes like nitric oxide synthase that occur as early as 2 h after IR (8, 15, 18). Since the kidney is the major organ for excreting metabolic end products, its injury can cause changes in the renal metabolic profile, and understanding its influence on kidney function will shed light on new potential diagnostic markers and therapeutic targets in AKI (17, 21, 34, 43). Advances in functional genomics and proteomics have fostered progress in our understanding of the pathogenic mechanisms involved and identified some novel protein biomarkers and candidate therapeutic targets in plasma, urine, and tissues which precede changes in commonly used clinical markers of renal dysfunction like plasma creatinine (6, 14, 18, 29, 41). Comparatively little, however, is known regarding the role of lipids in pathogenic mechanisms of AKI (1, 5, 39).
Major lipids like glycerophospholipids and sphingolipids have an important role in membrane bilayer structure, energy storage, and cell signaling and provide functional support to membrane proteins (36). Prior studies have shown that various forms of AKI, including IR, induce cellular membrane instability, causing lipid dysfunction and abnormal lipid accumulation within the kidney, which may be either protective or toxic to the kidney depending on the nature of the lipid species and the time course of the injury (13, 25, 46). For example, ether-linked phosphatidylcholines (PC) and phosphatidylethanolamines (PE) have been implicated in a range of developmental and pathogenic processes related to tissue injury, where they are enriched in proximal tubules of the kidney and can modulate membrane turnover, as well as protect against oxidative stress by reducing sensitivity to ROS in the case of plasmalogens that are characterized by the presence of a vinyl ether linkage of the sn1 alkyl chain (3, 10). Ether lipids can also be hydrolyzed by phospholipase A2 (PLA2) enzymes during ischemic injury, leading to the accumulation of fatty acid precursors of inflammatory oxylipids that may lead to irreversible cell injury within an hour of the insult (3, 24). Lipid oxidation has also been negatively correlated with IR-induced AKI (16). Thus there is a need to apply experimental approaches capable not only of identifying changes in a broad range of lipids within the renal lipidome that correlate with the severity of AKI but also of helping to determine whether these changes occur in areas of the kidney that are principally affected by AKI. In other words, both quantitative and spatial information on renal lipid changes are required to identify those lipid changes that are most likely to be pathophysiologically relevant and/or of greatest potential biomarker utility.
Although targeted mass spectrometry (MS) approaches have been used to identify new lipids which may have causative roles in AKI (31, 47), the significance of those lipids identified is often not clear. Furthermore, targeted MS approaches suffer from underrepresentation of major lipid classes within any single MS assay, necessitating the use of many independent MS approaches to obtain a picture of renal lipid changes. To overcome this limitation, we utilized an untargeted sequential window acquisition of all theoretical spectra (SWATH)-MS method (35) to identify novel lipid changes early following IR-induced kidney injury and select for those that correlated with AKI severity. This data-independent acquisition MS method combines the advantages of high throughput shotgun lipidomics with the high reproducibility of selected reaction-monitoring MS approaches. Using matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS), we then sought to localize those lipids that changed in the kidney in proportion to AKI severity early after IR to specific renal structures known to be affected by IR to further assess their potential significance.
MATERIALS AND METHODS
Animals.
Male C57BL/6 mice (10–12 wk old) were used for experiments. Animals were fed a standard diet and allowed free access to water. The Institutional Animal Care and Use Committee of the University of Alabama at Birmingham approved all the animal work.
Renal IR surgery.
For anesthetization, mice (n = 6) were induced using 2.5% isoflurane by inhalation and then maintained with 1.5% isoflurane by inhalation during sham or IR surgery. Under aseptic precautions, a right nephrectomy was performed via a right flank incision. A similar incision was made in the left flank, and the left renal pedicle was exposed, secured, and clamped with a microserrefine atraumatic vascular clamp (18055-05, Fine Science Tools, Foster City, CA) for 30 min. Blanching of the entire kidney ensured loss of blood flow. During this period, the kidney was kept moist, using sterile gauze soaked in saline. At the end of ischemia, the clamp was removed to allow reperfusion, which was confirmed visually, and the kidney was returned to the abdominal cavity in its original position. An additional group of animals (n = 6) underwent sham surgery in which a right nephrectomy was performed and the left renal pedicle was exposed, but was not clamped. Mice were euthanized with a lethal dose of pentobarbital sodium (0.4 ml, 50 mg/kg), and blood was collected for creatinine levels, which was determined using liquid chromatography-tandem MS (LC-MS/MS) performed at the UAB-UCSD O'Brien Center Bioanalytical Core facility.
Tissue processing.
One-half of each mouse kidney was weighed and homogenized in 500 μl PBS-butylated hydroxytoluene (BHT; 2,6-Di-tert-butyl-4-methylphenol, B1378, Sigma) for 30 s followed by sonication for 10 s using a sonic dismembrator (FB-50, Fisher Scientific). Homogenates were stored under argon at −80°C until extraction. Tissue weight was normalized between animals, and lipids were extracted by the Bligh-Dyer method, dried down under argon, and stored at −80°C until MS analysis. The other half of the kidney was embedded in 10% gelatin blocks for MALDI-IMS as described below.
SWATH-MS.
Lipid extracts were resuspended in 500 μl 2:1 methanol:chloroform containing 5 mM ammonium acetate. Samples were infused into a 5600 Triple-TOF mass spectrometer (Sciex) by isocratic flow at 7 μl/min using a 500-μl Hamilton gas tight syringe. For syringe cleaning, before and in between samples, the direct infusion syringe was cleaned with multiple solvents. The solvent wash steps included two flushes with 100% methanol, two flushes with 100% acetonitrile, two flushes with 100% isopropyl alcohol, and two flushes with 100% direct infusion solvent. Calibration standards for direct infusion analyses were provided by Sciex. The APCI Positive Calibration Solution (4460131) and APCI Negative Calibration Solution (4460134) were utilized. The syringe was cleaned in between samples by washing for 10 min with the direct infusion solvent. Spectra were collected over the range 200-1,200 m/z. A 250-ms high-resolution time-of-flight (TOF) scan was acquired first, followed by a series of high-sensitivity collision-induced dissociation (CID) product ion scans starting at 200 m/z and increasing 1 size step/1 Da mass through 1,200 m/z. The collision energy per step was 35 eV, and a collision energy spread of 15 eV was applied per product ion scan. Parameters for instrumentation, such as curtain gas, GS1, GS2, spray voltage, and temperature were 20.00, 15.00, 5,500.00 (positive ion mode)/4,500 (negative ion mode), and 400°C, respectively. This yielded fragmentation into product ions to assist in identification of specific lipid classes. Two technical replicates were performed per sample in positive ion mode.
The acquired TOF MS and MS/MS data were processed with LipidView 1.2 software (Sciex, Concord, Ontario, Canada). TOF precursor scans were tethered to product ion TOF MS/MS scans for identification of lipids with a mass tolerance of 5 mDa and deisotoping of the spectra. A mass tolerance of 0.01 Da was used for product ion scans with smoothing of individual spectra. The signal-to-noise threshold was set to 3 with a minimum intensity cut off of 10 counts to be accepted, and lipid search parameters were set to allow for up to 6 double bonds and lyso-species variants.
Hydroxyoctadecadienoic acid/hydroxyeicosatetraenoic acid measurement.
For renal hydroxyoctadecadienoic acid/hydroxyeicosatetraenoic acid (HODE/HETE) measurement, animals were perfused through the left ventricle with 20 ml PBS containing 50 μM BHT (Sigma-Aldrich, St. Louis, MO). The kidney was removed, weighed, and homogenized in 2 ml PBS/BHT. Homogenates were stored in 500-μl aliquots under argon at −80°C before analysis. Lipids were extracted from 500-μl homogenate aliquots and analyzed by LC-MS/MS exactly as described in Black et al. (2) using a Shimadzu prominence HPLC system and an AB/Sciex API-4000 Q TRAP mass spectrometer.
Tissue sectioning and matrix application for MALDI-IMS.
Mouse kidneys were embedded in 10% gelatin (G1890, wt/vol, aqueous, Sigma-Aldrich), frozen in liquid nitrogen-cooled isopentane, and stored at −80°C until use. Coronal cryosections (14 μM) were cut at −20°C and thaw-mounted onto indium tin oxide (ITO)-coated slides (CG-81IN-S115, Delta Technologies). A digital image of the slide with cryosections was then acquired at 2,600-dpi resolution against a black background using a Canon Lide 210 scanner. Alternating sections from each block were placed on conventional microscope slides for immunofluorescence staining.
Matrix (2,5-dihydroxybenzoic acid; DHB; 149357-25G, Sigma-Aldrich) was applied to each slide via sublimation using a vacuum sublimation apparatus comprising an Edwards RV12 vacuum pump with pirani vacuum gauge and digital display (Edwards Vacuum) attached to a glass sublimation chamber (8023, Ace Glassware). The slide was affixed to the bottom of the condenser unit of the sublimation chamber using double-sided carbon conductive tape (77802, Electron Microscopy Sciences), and 300 mg of DHB was evenly distributed on the bottom of the chamber. The chamber was attached to the vacuum apparatus via a cold trap (Z120790, Sigma-Aldrich). The pressure was allowed to reach 0.05 Torr while a slurry of dry ice and ethanol in the cold trap was maintained. Ice water was kept between 5 and 10°C in the condenser unit of the sublimation chamber, which was placed on top of a heated sand bath kept at 140°C for 10 min. After returning to ambient temperature and pressure, the matrix-coated slides were removed from the chamber and subjected to MALDI-IMS.
MALDI-TOF imaging and MS/MS.
Slides with DHB-coated tissue sections were placed into an MTP Slide Adaptor II (Bruker Daltonics, Billerica, MA) and analyzed on an AutoFlex Speed MALDI-TOF/TOF mass spectrometer (Bruker Daltonics). Positive ion images were obtained in reflector mode over the mass range m/z 60-1,600 using a spatial resolution of 20 μM raster step size. The sample rate was 2.00 GS/s, and the detector gain was 16.3×. Laser size 1_minimum was used, and laser intensity was kept at 100% with the global attenuator set to 30%. The frequency was 1,000 Hz with 100 laser shots added per spot. Calibration was performed using a mixture of the following lipids: LPC (16:0), LPC (18:0), LPC (20:0), PC (16:0/18:1), PC (16:0/18:2), and PC (16:0/20:4) all purchased from Avantipolar Lipids (Alabaster). MALDI images were processed using FlexImaging 4.0 software (Bruker Daltonics). Average spectra were normalized to the total ion current (TIC).
MALDI-TOF MS/MS fragment ion spectra were collected using a low-mass LIFT method in positive ion mode over the mass range m/z 40–950. The sample rate was 1.00 GS/s, and the detector gain was 3.8×. Laser size 4_large was used at a frequency of 200 Hz, and laser intensity was 77%. The method was calibrated using the peptide angiotensin II (8208241, Bruker). The PCIS window was set to 4 Da on either side of the parent ion. Three spectra were summed together per MS/MS run, with 500 laser shots collected per spectrum. The resulting MS/MS spectra were loaded into FlexAnalysis software (Bruker Daltonics) to subtract the baseline and label the peaks. MS/MS spectra were interpreted using LIPID MAPS to match precursor ion masses and interpret fragmentation patterns.
Immunofluorescence staining.
For immunofluorescence staining, sections collected onto conventional microscope slides were fixed with acetone, blocked with blocking buffer (2% normal horse serum, 2% normal goat serum in PBS) for 1 h, followed by incubation with rat anti-PECAM1 and fluorescein-labeled Lotus tetragonolobus lectin (Vector Laboratories, CA) for 2 h at room temperature. Following washes with PBS, sections were incubated in secondary antibody conjugated to Alexa Fluor 555 for 30 min at room temperature. Following washes with PBS, slides were mounted using Vectashield Hard Set Mounting Medium with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories).
Statistics.
Statistical analysis was performed using SigmaStat (Systat Software, San Jose, CA). Student's t-test was used for single comparisons, and the Pearson product moment test was used for correlation analyses. For all statistical analyses, P < 0.05 was considered significant.
RESULTS
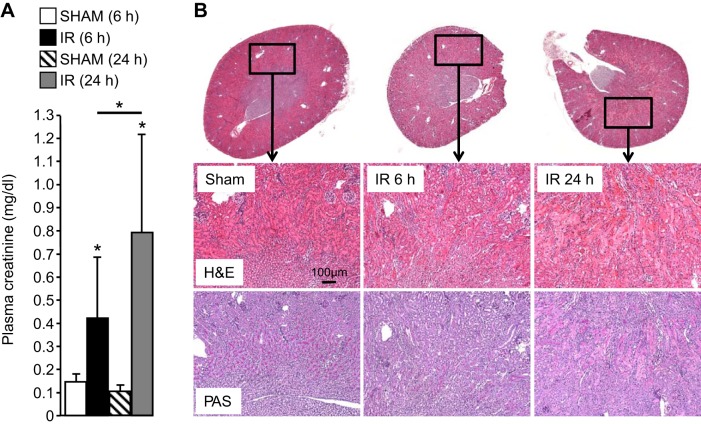
We reasoned that many lipids would be changed at 24 h post-IR as a result of the excessive injury and inflammation at this time point (12) and that most therefore may not play significant roles in the initial pathology of AKI. To address this, we chose an additional early time point assuming that fewer lipids would be changed with IR and those that increase are more likely to exhibit a causal relationship with AKI severity. Compared with sham mice, plasma creatinine levels in IR mice were significantly higher at 6 and 24 h after reperfusion (P < 0.001) (Fig. 1A). Histological features of tubular necrosis, loss of brush borders and cast formation, were evident at 24 h (Fig. 1B) in the kidneys of animals undergoing IR, but not in the kidneys from animals undergoing sham surgery. Importantly, renal injury as assessed by these parameters was less severe at 6 h compared with 24 h.
Fig. 1.
A: plasma creatinine concentrations in control (sham) and mice undergoing renal ischemia-reperfusion (IR) at the indicated time points (n = 6). B: hematoxylin/eosin (H&E)- and periodic acid-Schiff (PAS)-stained kidney sections from the indicated control (sham) and IR mice. Note higher plasma creatinine and more pronounced histological features of acute kidney injury (AKI) at 24 h compared with 6 h. *P < 0.05.
Ether-linked phospholipids are increased early in IR-induced AKI.
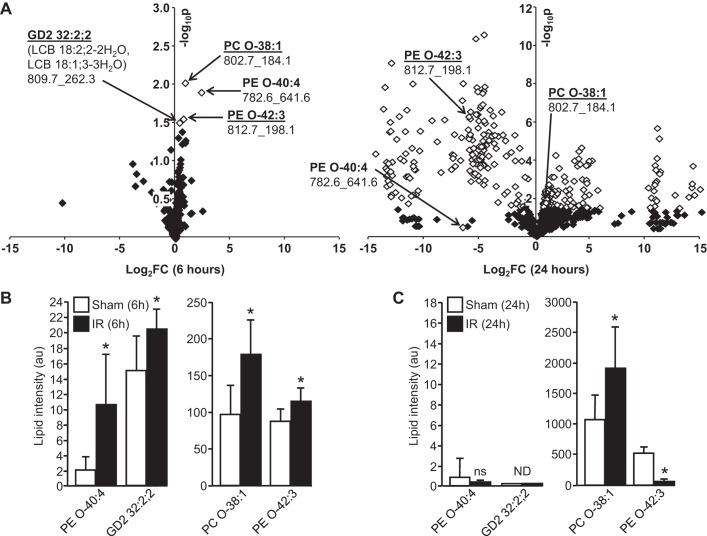
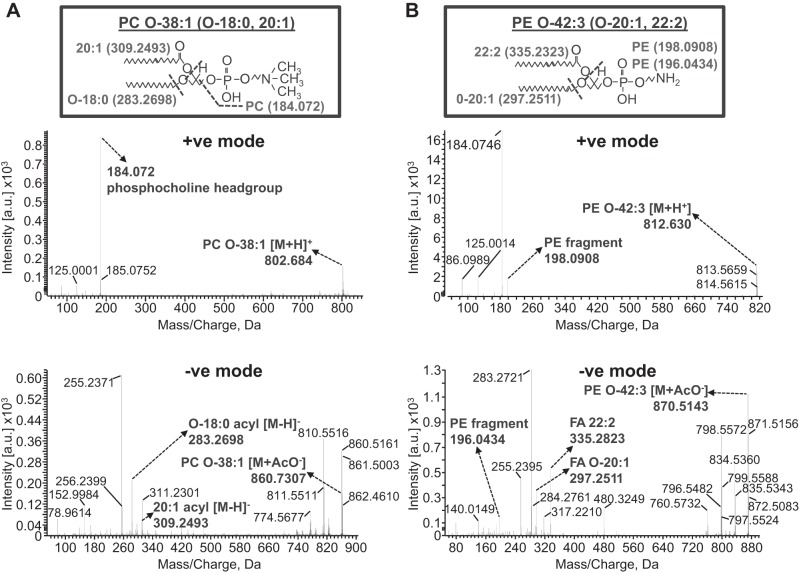
We performed SWATH-MS analysis on kidney homogenates from IR and sham mice at both the 6- and 24-h time points after surgery. Together with the accurate parent lipid mass, MS/MS analysis in positive and negative modes provided identification of the parent lipid and their acyl chain distribution, respectively. Although numerous lipids were detected in positive mode at both time points, only four were changed (all increases) to a statistically significant extent at 6 h after IR (of a total 938) (Fig. 2A, left). Interestingly, of these four lipids that were increased significantly at 6 h after IR, three were identified as ether-linked phospholipids (one an abundant PC: PC O-38:1, and two PEs: an abundant PE O-42:3 and a minor PE O-40:4) (Fig. 2A). SWATH-MS/MS analysis in positive mode and negative modes produced ions which allowed the phospholipid headgroup and acyl chain designations, respectively, to be made for the two abundant ether lipids as follows: PC O-38:1 (PC O-18:0, 20:1) and PE O-42:3 (PE O-20:1, 22:2) (Fig. 3, A and B). PC O-38:1 is a plasmanyl choline (Fig. 3A), while PE O-42:3 is a plasmalogen by virtue of the vinyl linkage of the 20:1 sn1 alkyl chain (Fig. 3B).
Fig. 2.
Sequential window acquisition of all theoretical spectra (SWATH) mass spectrometry (MS) of renal lipids following IR-induced kidney injury. A: volcano plots showing relative changes (log2 fold-change on x-axis) in renal lipids with intensity values >10 (i.e., 3× signal-to-noise ratio) at 6 (left) and 24 h (right) IR. Fold-changes that are statistically significant are marked as white symbols (>1.301 −log10 P value on y-axis is statistically significant). For each lipid named, numbers below correspond to parent ion mass followed by tandem MS (MS/MS) fragment ion mass used for identification. B: intensities of lipids found to be significantly changed at 6-h IR relative to sham controls. C: intensities of lipids in B at 24 h. See Tables 1–5 for list of all lipid changes at 24 h post-IR. *P < 0.05.
Fig. 3.
SWATH-MS/MSALL in positive mode (top) and negative mode (bottom) showing the ion fragments used to identify PC O-38:1 (PC O-18:0/20:1; A) and PE O-42:3 (O-20:1, 22:2; B). For PC O-38:1, the protonated parent ion mass in positive mode is 802.684 m/z (expected mass 802.668 m/z), and 184.072 m/z (expected mass 184.074 m/z) is the phosphocholine fragment ion. In negative mode, the parent ion acetate adduct ion mass is 860.731 m/z (expected mass 860.675 m/z), and two fragment ions corresponding to 18:0 and 20:1 acyl chains are 283.270 m/z (expected mass 283.264 m/z) and 309.249 m/z (expected mass 309.279 m/z), respectively. B: for PE O-42:3 (O-20:1, 22:2), the protonated parent ion mass in positive mode is 812.630 m/z (expected mass 812.653 m/z), and 198.0908 m/z (expected mass 198.10 m/z) is the PE fragment (196.0434 m/z in negative mode spectrum below). In negative mode, the parent ion acetate adduct ion mass is 870.5143 m/z (expected mass 870.560 m/z), and two fragment ions corresponding to O-20:1 and 22:2 acyl chains are 297.2511 m/z (expected mass 297.301 m/z) and 335.2823 m/z (expected mass 335.296 m/z), respectively.
A minor ganglioside (glycospgingolipids enriched in membrane lipid rafts with sialic acid residues linked to the sugar chain), GD2 32:2;2, was also increased with IR at 6 h, but not detected at 24 h in either sham or IR mice (Fig. 2). As anticipated, many more lipids were changed to a statistically significant extent at 24 h after IR (36% of a total of 1,088) (Fig. 2A, right). All statistically significant lipid changes detected at 24 h post-IR are shown in Tables 1–5. Interestingly, while the abundant PC O-38:1 remained elevated in IR kidneys at 24 h compared with the 24-h sham group, the low-abundance PE O-40:4 was present at comparably low levels in kidneys of both IR and sham mice at 24 h (Fig. 2, B and C). PE O-42:3, on the other hand, was present at high levels but was decreased at 24 h compared with its sham control group (Fig. 2C), even though this lipid was increased relative to its sham control group at 6 h post-IR (Fig. 2B). Interestingly, all ether-linked PEs and PEs detected by SWATH-MS 24 h post-IR were reduced with AKI (Tables 1–3).
Table 1.
Phosphatidylcholine (PC) and phosphatidylethanolamine (PE) species changed to a statistically significant extent 24 h post-IR-induced AKI
| Lipid | MS/MS, m/z | Intensity Change | Fold- Change | Lipid | MS/MS, m/z | Intensity Change | Fold- Change |
|---|---|---|---|---|---|---|---|
| PC 34:1 | 760.6_184.1 | 659.1→1295.1 | +2 | PE 34:1 | 718.6_198.1 | 62.3→1.8 | −34.6 |
| PC 34:4 | 754.6_184.1 | 1082.2→2,014.7 | +1.9 | PE 36:0 | 748.6_198.1 | 9900.4→155.8 | −63.6 |
| PC 34:6 | 750.6_184.1 | 940.4→244.1 | −3.9 | PE 36:1 | 746.6_198.1 | 3229.9→31.6 | −102.2 |
| PC 36:0 | 790.6_184.1 | 418.1→1290.5 | +3.1 | PE 38:0 | 776.6_198.1 | 1057.6→14.9 | −71.0 |
| PC 36:1 | 788.7_184.1 | 398.6→1461.5 | +3.7 | PE 38:1 | 774.6_198.1 | 267.5→1.5 | −178.3 |
| PC 36:2 | 786.6_184.1 | 927.0→2635.3 | +2.8 | PE 38:4 | 768.7_198.1 | 273.8→11.6 | −23.6 |
| PC 36:3 | 784.6_184.1 | 1584.5→3037.3 | +1.9 | PE 38:6 | 764.6_198.1 | 81.1→ND | − |
| PC 36:4 | 782.6_184.1 | 698.4→1290.8 | +1.9 | PE 40:0 | 804.7_198.1 | 189.4→3.6 | −52.6 |
| PC 38:2 | 814.6_184.1 | 3757.9→6117.2 | +1.6 | PE 40:1 | 802.7_198.1 | 723.0→1.0 | −723 |
| PC 38:4 | 810.7_184.1 | 4265.8→7589.2 | +1.8 | PE 40:2 | 800.7_198.1 | 1089.5→3.4 | −320.4 |
| PC 38:5 | 808.7_184.1 | 1265.6→2983.7 | +2.4 | PE 40:3 | 798.7_198.1 | 304.8→11.0 | −27.7 |
| PC 38:6 | 806.7_184.1 | 877.4→1505.9 | +1.7 | PE 42:0 | 832.7_198.1 | 190.4→3.8 | −50.1 |
| PC 40:2 | 842.7_184.1 | 1641.7→2940.4 | +1.8 | PE 42:3 | 826.7_198.1 | 2551.5→55.9 | −45.6 |
| PC 40:4 | 838.7_184.1 | 1623.9→3683.5 | +2.3 | PE 42:4 | 824.7_198.1 | 999.9→30.6 | −32.7 |
| PC 42:4 | 866.7_184.1 | 1335.2→2610.5 | +2.0 | PE 42:5 | 822.7_198.1 | 245.0→14.8 | −16.6 |
| PC 42:5 | 864.7_184.1 | 3081.6→5870.6 | +1.9 | PE 42:6 | 820.7_198.1 | 177.8→11.9 | −14.9 |
| PC 42:6 | 862.7_184.1 | 5128.4→7823.7 | +1.5 | PE 44:0 | 860.7_198.1 | 307.7→22.4 | −13.7 |
| PC 44:1 | 900.7_184.1 | 1402.1→2530.1 | +1.8 | PE 44:1 | 858.7_198.1 | 340.5→47.2 | −7.2 |
| PC 44:6 | 890.7_184.1 | 4260.8→6964.1 | +1.6 | PE 44:2 | 856.7_198.1 | 435.2→28.2 | −15.4 |
| PC 46:1 | 928.8_184.1 | 1072.0→1759.3 | +1.6 | PE 44:3 | 854.7_198.1 | 763.8→18.8 | −40.6 |
| PC 48:5 | 948.8_184.1 | 1184.5→1913.6 | +1.6 | PE 44:4 | 852.7_198.1 | 438.6→15.8 | −27.8 |
| PC 48:6 | 946.8_184.1 | 1154.4→1793.8 | +1.6 | PE 44:5 | 850.7_198.1 | 196.4→5.1 | −38.5 |
| PE 34:0 | 720.6_198.1 | 285.0→4.1 | −69.5 | PE 44:6 | 848.7_198.1 | 201.1→7.2 | −27.9 |
| PE 46:0 | 888.7_198.1 | 810.2→28.4 | −28.5 | PE 48:0 | 916.8_198.1 | 612.8→28.4 | −21.6 |
| PE 46:2 | 884.7_198.1 | 526.6→17.9 | −29.4 | PE 48:1 | 914.8_198.1 | 445.7→14.5 | −30.7 |
| PE 46:3 | 882.8_198.1 | 253.2→6.7 | −37.8 | PE 48:2 | 912.8_198.1 | 802.8→14.2 | −56.5 |
| PE 46:4 | 880.8_198.1 | 422.0→10.6 | −39.8 | PE 48:3 | 910.7_198.1 | 447.0→15.9 | −28.1 |
| PE 46:5 | 878.8_198.1 | 862.7→29.0 | −29.8 | PE 48:5 | 906.8_198.1 | 764.7→21.5 | −35.6 |
| PE 46:6 | 876.7_198.1 | 1408.2→54.5 | −25.8 | PE 48:6 | 904.8_198.1 | 1069.4→38.8 | −27.6 |
IR, ischemia-reperfusion; AKI, acute kidney injury; tandem mass spectrometry (MS/MS), m/z: parent ion m/z followed by fragment ion m/z used for lipid identification; intensity change, average lipid intensity sham→average lipid intensity IR. Reductions are highlighted in bold. Note that all statistically significant changes in PEs 24 h post-IR were reductions.
Table 5.
Glycosphingolipid species changed to a statistically significant extent 24 h post-IR-induced AKI
| Lipid | MS/MS, m/z | Intensity Change | Fold-Change |
|---|---|---|---|
| Hex3Cer 26:3;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 924.8_262.2 | ND→18.7 | + |
| Hex3Cer 26:4;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 922.8_262.2 | 1.7→16.2 | +9.5 |
| Hex3Cer 27:3;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 954.8_262.2 | ND→12.3 | + |
| Hex3Cer 27:4;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 952.8_262.2 | ND→13.5 | + |
| Hex3Cer 28:0;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 974.8_262.2 | 2.5→19.7 | +7.9 |
| Hex3Cer 28:1;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 972.8_262.2 | 0.8→17.5 | +21.9 |
| Hex3Cer 29:2;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 984.8_264.2 | 3.7→16.2 | +4.4 |
| Hex3Cer 30:0;3 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 986.8_264.2 | 4.6→13.6 | +3.0 |
| Hex3Cer 30:1;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 984.8_262.2 | 3.1→13.8 | +4.5 |
| Hex3Cer 30:4;3 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 978.8_264.2 | 1.6→39.2 | +24.5 |
| Hex3Cer 30:4;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 978.8_262.2 | 2.3→41.3 | +18.0 |
| Hex3Cer 32:0;3 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 1014.8_264.2 | 34.8→17.7 | −2.0 |
| Hex3Cer 32:4;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 1006.9_262.2 | 1.9→32.1 | +16.9 |
| Hex3Cer 32:4;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 1022.9_262.2 | 1.1→15.7 | +14.3 |
| Hex3Cer 34:2;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 1054.9_262.2 | 0.3→17.6 | +58.7 |
| Hex3Cer 38:1;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 1113.0_264.2 | 14.5→4.4 | −3.3 |
| Hex3Cer 38:2;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 1111.0_264.2 | 21.8→12.4 | −1.8 |
| Hex3Cer 38:4;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 1107.0_264.2 | 20.6→6.6 | −3.1 |
| Hex3Cer 39:3;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 1122.9_264.2 | 18.1→2.4 | −7.5 |
| Hex3Cer 40:1;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 1141.0_264.2 | 20.7→5.6 | −3.7 |
| Hex2Cer 26:0;3 (LCB 18:0;2-2H2O) | 768.7_266.2 | ND→19.6 | + |
| Hex2Cer 26:3;2 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 746.6_262.3 | 1.0→18.7 | +18.7 |
| Hex2Cer 30:3;2 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 802.7_262.3 | 3.3→74.5 | +22.6 |
| Hex2Cer 30:3;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 834.7_262.3 | 20.3→8.6 | −2.4 |
| Hex2Cer 32:3;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 862.7_262.3 | 18.4→1.4 | −13.1 |
| Hex2Cer 32:3;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 862.7_264.3 | ND→19.0 | +19 |
| Hex2Cer 34:1;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 878.8_262.3 | ND→16.2 | +16.2 |
| Hex2Cer 34:1;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 894.7_262.3 | ND→17.6 | +17.6 |
| Hex2Cer 36:3;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 894.7_262.3 | ND→17.6 | +17.6 |
| Hex2Cer 36:3;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 918.8_264.2 | 18.4→9.0 | −2.0 |
| Hex2Cer 39:4;2 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 926.8_262.2 | ND→16.5 | + |
| Hex2Cer 40:2;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 960.8_262.2 | 1.0→19.7 | +19.7 |
| Hex2Cer 40:3;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 974.8_262.2 | 0.9→31.1 | +34.6 |
| Hex2Cer 41:1;3 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 976.8_264.2 | 9.2→39.4 | +4.3 |
| HexCer 33:2;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 716.6_264.3 | ND→13.5 | + |
| Cer 28:1;2 (LCB 18:1;2-H2O, LCB 18:0;3-2H2O) | 454.3_282.3 | ND→18.5 | + |
| CerP 26:1;4 (LCB 18:1;2-H2O, LCB 18:0;3-3H2O) | 538.4_264.3 | ND→200.7 | + |
| CerP 44:1;3 (LCB 18:2;2-H2O, LCB 18:1;3-3H2O) | 774.6_262.3 | 1.4→25.1 | +17.9 |
| CerPE 36:3;4 (LCB 18:2;2-H2O, LCB 18:1;3-3H2O) | 717.6_262.3 | 2.7→29.1 | +10.8 |
| CerPE 38:1;3 (LCB 18:2;2-H2O, LCB 18:1;3-3H2O) | 733.6_262.3 | ND→11.7 | + |
| SGalCer 26:0;3 (LCB 18:0;2-2H2O) | 686.5_266.3 | 353.3→6.3 | −56.1 |
| SGalCer 26:1;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 700.5_264.3 | ND→58.2 | + |
| SGalCer 29:3;4 (LCB 18:1;2-2H2O, LCB 18:0;3-2H2O) | 738.6_282.3 | ND→15.8 | + |
| SGalCer 29:4;3 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 720.6_264.2 | 4.0→24.3 | +6.1 |
| SGalCer 30:3;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 752.6_262.3 | 17.5→ND | − |
| SGalCer 30:4;2 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 718.6_262.3 | 24.5→349.3 | +14.3 |
| SGalCer 30:4;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 750.6_262.3 | 71.1→2.2 | +32.3 |
| SGalCer 31:1;4 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 770.6_262.3 | 14.2→ND | − |
| SGalCer 32:0;3 (LCB 18:0;2-2H2O) | 770.6_266.3 | 130.8→2.0 | −65.4 |
| SGalCer 32:0;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 786.6_282.3 | 26.0→1.2 | −21.7 |
| SGalCer 34:0;3 (LCB 18:0;2-2H2O) | 798.6_266.3 | 79.4→ND | − |
| SGalCer 34:0;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 814.7_282.3 | 31.7→ND | − |
| SGalCer 38:4;2 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 830.7_262.3 | 2.2→100.2 | +45.6 |
| SGalCer 40:0;4 (LCB 18:1;2-2H2O, LCB 18:0;3-3H2O) | 898.8_264.2 | 6.0→26.4 | +4.4 |
| SGalCer 40:1;3 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 880.8_262.3 | ND→23.6 | + |
| SGalCer 44:4;2 (LCB 18:2;2-2H2O, LCB 18:1;3-3H2O) | 914.8_262.2 | 2.9→19.2 | +6.6 |
Cer, ceramide; Hex3Cer, trihexosylceramide; Hex2Cer, dihexosylceramide; CerP, ceramide 1-phosphate; CerPE, ceramide phosphoethanolamine; SGalCer, 3-O-sulfo galactosylceramide; LCB, long-chain base. Text in parentheses shows possible LCBs based on fragment ion m/z. Reductions are highlighted in bold.
Table 3.
Lysophosphatidylcholine (LPC), ether-linked LPC (LPC O), lysophosphatidylethanolamine (LPE), and ether-linked LPE (LPE O) species changed to a statistically significant extent 24 h post-IR-induced AKI
| Lipid | MS/MS, m/z | Intensity Change | Fold- Change | Lipid | MS/MS, m/z | Intensity Change | Fold- Change |
|---|---|---|---|---|---|---|---|
| LPC 18:0 | 524.4_184.1 | 718.1→1325.2 | +1.85 | LPC O-22:5 | 556.4_184.1 | 110.2→ND | − |
| LPC 20:0 | 552.4_184.1 | 103.1→ND | − | LPC O-22:6 | 554.1_184.1 | 207.7→ND | − |
| LPC 20:4 | 544.4_184.1 | 58.5→ND | − | LPC O-24:1 | 592.4_184.1 | 20.3→142.4 | +7.02 |
| LPC 26:6 | 624.4_184.1 | 16.4→160.0 | +9.76 | LPC O-24:2 | 590.4_184.1 | 22.2→173.0 | +7.79 |
| LPC O-14:0 | 454.3_184.1 | 24.1→50.8 | +2.11 | LPC O-24:3 | 588.5_184.1 | 119.4→379.2 | +3.18 |
| LPC O-16:0 | 482.4_184.1 | 353.1→800.4 | +2.27 | LPC O-24:6 | 582.4_184.1 | 77.7→ND | − |
| LPC O-16:2 | 478.3_184.1 | 43.2→ND | − | LPC O-28:6 | 638.5_184.1 | ND→80.2 | + |
| LPC O-20:0 | 538.4_184.1 | 266.4→39.0 | −6.83 | LPE 16:0 | 454.3_313.3 | 30.9→190.3 | +6.16 |
| LPC O-20:3 | 532.4_184.1 | 102.7→ND | − | LPE 18:0 | 482.4_341.3 | 53.2→307.3 | +5.78 |
| LPC O-20:4 | 530.4_184.1 | 94.7→ND | − | LPE 22:0 | 538.4_198.1 | 443.9→29.7 | −14.95 |
| LPC O-22:0 | 566.4_184.1 | 104.0→ND | − | LPE 24:6 | 568.4_198.1 | 91.6→ND | − |
| LPC O-22:1 | 564.4_184.1 | 49.3→ND | − | LPE O-20:0 | 496.3_198.1 | 185.5→14.5 | −12.79 |
| LPC O-22:3 | 560.4→184.1 | 111.2→39.7 | +2.80 | LPE O-26:3 | 574.4_198.1 | 21.1→ND | − |
| LPC O-22:4 | 558.4_184.1 | 114.0→ND | − |
Reductions are highlighted in bold.
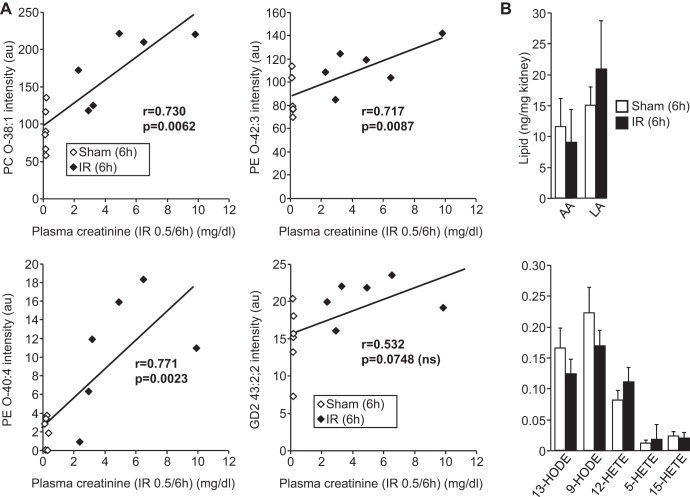
A positive correlation was seen between kidney PC O-38:1 levels and AKI severity at 6 h as measured by plasma creatinine concentration (r = 0.730, P < 0.0062). Similar positive associations were present for PE O-42:3 (r = 0.717, P = 0.0087) and PE O-40:4 (r = 0.771, P = 0.0023) (Fig. 4A). However, the increase in renal GD2 43:2;2 levels at 6 h did not show a statistically significant association with plasma creatinine (r = −0.532, P = 0.0748). This suggests that early increases in ether phospholipids are directly related to the pathology of AKI in this IR model.
Fig. 4.
Increases in ether lipids correlate with severity of AKI and occur independently of lipid oxidation. A: correlation analysis (Pearson product moment) between intensity of ether lipids and plasma creatinine concentrations in control (sham) and IR mice. Note that increases in GD2 32:2;2 do not correlate with the extent of renal injury as measured by plasma creatinine. B: levels of the indicated oxidized fatty acids [hydroxyoctadecadienoic acid (HODES) and hydroxyeicosatetraenoic acid (HETEs)] and their precursors [arachidonic acid (AA) and linoleic acid (LA)] in kidneys from the indicated control (sham) and IR mice.
Early changes in renal lipids following IR occur independently of lipid oxidation.
Ether lipids can be hydrolyzed by PLA2 enzymes during ischemic injury, leading to the accumulation of unsaturated linoleic and arachidonic acids and their oxidized derivatives (HODE and HETE acids, respectively) (42), many of which exhibit inflammatory properties that may lead to irreversible cell injury (3, 16, 24). We therefore measured levels of HODEs and HETEs, as well as their fatty acid precursors, in kidneys of sham and IR animals at 6 h. No statistically significant differences in major HODEs and HETEs or linoleic and arachidonic acids were detected (Fig. 4B), suggesting that phospholipid hydrolysis and fatty acid oxidation are not responsible for the lipid changes observed.
Localization of ether lipid increases in IR to proximal tubules by MALDI tissue imaging.
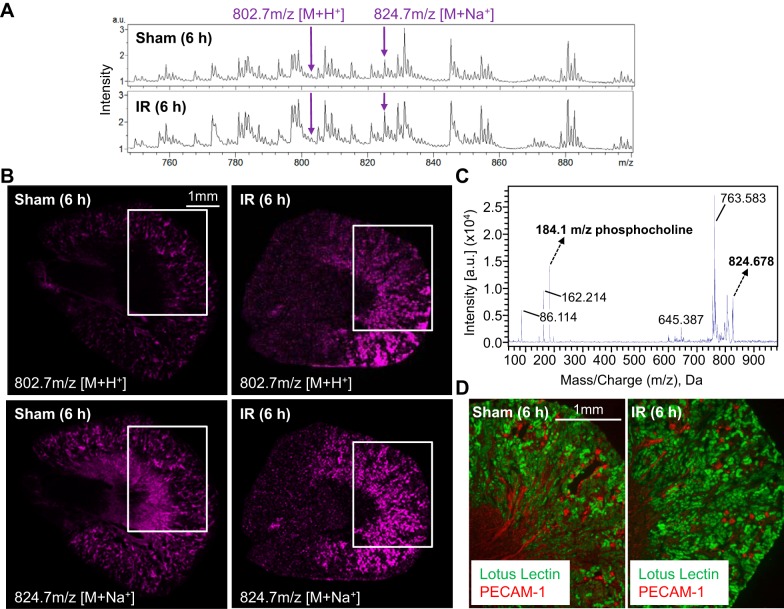
PC O-38:1 was the most abundant of the ether phospholipids increased early in AKI (6 h) (Fig. 2, A and B) and also the only one remaining elevated compared with sham controls at 24 h (Fig. 2, C and D). To provide additional evidence for its significance in addition to its correlation with AKI severity (Fig. 4A), we performed MALDI-IMS in positive mode to determine whether PC O-38:1 localized to specific renal structures commonly affected by IR (Fig. 5). The spatial distribution of PC O-38:1 was largely restricted to the corticomedullary area of the kidney, and immunofluorescence staining of adjacent tissue sections to those taken for MALDI imaging with L. tetragonolobus lectin (for proximal tubules) and anti-PECAM1 antibody (for endothelial cells) confirmed a distribution predominantly localized to proximal tubules rather than blood vessels (which is consistent with generation of the lipid in situ, as opposed to entry of the lipid from the circulation) (Fig. 5C). Similarly to the positive mode SWATH-MS/MS analysis (Fig. 3A), MALDI- MS/MS in positive mode on the corticomedullary area of sham and IR kidney cryosections gave the 184.1-m/z phosphocholine headgroup fragment ion expected for the PC O-38:1 sodium adduct (Fig. 5D).
Fig. 5.
Increases in PC O-38:1 with AKI are predominantly localized to proximal tubules. A: representative matrix-assisted laser desorption ionization-imaging MS (IMS) spectra from 6-h sham control and 6-h IR kidneys. Arrows denote ions at 802.7 and 824.7 m/z consistent with protonated [(M+H+)] PC O-38:1 and sodiated [(M+Na+)] PC O-38:1, respectively. B: positive ion mode MALDI-IMS images of coronal kidney sections from 6-h sham and 6-h IR mice showing the distribution of the minor 802.7-m/z protonated [(M+H+)] PC O-38:1 and the more abundant 824.7-m/z sodiated [(M+Na+)] PC O-38:1. C: positive mode MS/MS analysis of 824.7-m/z (M+Na+) ion showing a 184.1-m/z fragment indicative of the PC headgroup in PC O-38:1 D: Lotus tetragonolobus lectin staining of proximal tubules in coronal kidney sections adjacent to those in A demonstrates that PC O-38:1 is most abundant in proximal tubular areas. White boxes in B mark the areas corresponding to those shown on adjacent sections stained with L. tetragonolobus lectin in D.
DISCUSSION
In this study, we used SWATH-MS to demonstrate that 6 h after renal IR in mice, the relative abundance of a small number of ether-linked phospholipids is significantly increased in the kidney compared with control animals (Fig. 2) independently of major changes in lipid oxidation (Fig. 4B). Importantly, renal levels of these ether phospholipids correlated with plasma creatinine as a marker of AKI severity (Fig. 4A). This suggests that they might play a causal or protective role in AKI and/or may prove to be effective early biomarkers for AKI. With respect to the latter possibility, it will be important to develop more sensitive targeted LC-MS/MS methods to determine whether any of the lipid changes we found are manifested in urine.
Lipid MALDI-IMS in combination with immunofluorescence staining of adjacent kidney cryosections revealed a predominant localization of the most abundant ether lipid increased (PC O-38:1) to the proximal tubules of the kidney, where rate-limiting peroxisomal acyltransferase and synthase enzymes for ether lipid biosynthesis are located (10) (Fig. 5). Indeed, this region of the kidney is particularly rich in peroxisomes and is also known to be more prone to developing ischemia-related injury (19, 26, 44). Thus spatially restricted ether lipid production in the proximal tubules of the kidney may play important roles early in AKI. However, some glomerular PC O-38:1 may also be present that could be detected using greater spatial resolution for MALDI-IMS than the 20 μm used in this study.
Ether-linked PC and PE species differ from the more common diacyl PCs and PEs in their utilization of a fatty alcohol, as opposed to a fatty acid, at the sn1 position (ether vs. ester sn1 linkage) (20). Elevated levels of ether lipids have been widely reported in cancer tissues, and the discovery of important biological activities of distinctive ether-like lipids such as platelet-activating factor (PAF) (9, 37) greatly stimulated interest in these lipids. Notably, deficiency of ether-linked phospholipids in general is associated with numerous peroxisomal disorders including Zellweger's, and impaired peroxisomal ether lipid synthesis has been linked to defects in membrane trafficking and increased apoptosis (10, 38). Peroxisomal ether lipid synthesis has also been shown to be important for sustaining membrane phospholipid composition and cell viability (19). Our finding of elevations in select ether-linked phospholipids early in IR-induced kidney injury and the putative localization of the most abundant (PC O-38:1) to proximal tubules therefore provides important new information on the specific types of ether-linked phospholipids that are involved in AKI.
Twenty-four hours following AKI induction (a time point commonly used to study mechanisms of AKI in this model), there were considerably more lipid changes associated with AKI (Fig. 2B and Tables 1–4), likely because of the greater number and complexity of cellular and molecular processes related to injury, inflammation, and tissue repair occurring at this later time. In addition, of the two abundant ether phospholipids increased at 6 h (PC O-38:1 and PE O-42:3), only PC O-38:1 was still elevated compared with control animals at 24 h, with the plasmalogen PE O-42:3 reduced at this later time point (Fig. 2). Although plasmalogens and other ether phospholipids can be hydrolyzed by calcium-independent PLA2 enzymes at early time points in renal injury, their rapid turnover can result in maintenance of high levels which may be required to preserve normal membrane function and to confer protective antioxidant effects that prevent irreversible cell injury (28, 30). This property may be of particular importance in AKI, where the accumulation of myeloperoxidase (MPO) activity derived from inflammatory leukocytes and ROS in the kidney can cause irreversible damage (27). Indeed, “plasmalogen-deficient” mouse models succumb to oxidative stress, tissue injury, and peroxisomal dysfunction (10, 32, 33). Sustained increases in renal PC O-38:1 levels up to 24 h following IR-induced AKI may therefore be significant in this respect, while the later reductions in the plasmalogen PE O-42:3 may reflect its “consumption” at later time points in IR due to abundant ROS generation and excessive damage to proximal tubules that are rich in the peroxisomal enzymes required for the synthesis of plasmalogens and other ether lipids (3). Indeed, all ether PEs detected 24 h post-IR were similarly reduced (Table 2), strongly supporting this notion. This suggests that preventing ether PE reductions or restoring their levels (perhaps through modification of key enzymes) could provide some therapeutic benefit in AKI.
Table 4.
Sphingomyelin (SM) species changed to a statistically significant extent 24 h post-IR-induced AKI
| Lipid | MS/MS, m/z | Intensity Change | Fold-Change | Lipid | MS/MS, m/z | Intensity Change | Fold-Change |
|---|---|---|---|---|---|---|---|
| SM 26:0;4 | 625.5_184.1 | ND→169.4 | + | SM 39:4;2 | 767.7_184.1 | 2118.6→424.5 | −5.00 |
| SM 26:1;4 | 623.4_184.1 | ND→285.5 | + | SM 40:0;3 | 805.7_184.1 | 580.8→1484.3 | +2.56 |
| SM 32:2;4 | 705.6_184.1 | 1160.2→3841.1 | +3.31 | SM 40:0;4 | 821.6_184.1 | 785.2→1583.3 | +2.02 |
| SM 32:3;3 | 687.5_184.1 | 689.3→19.2 | −35.90 | SM 40:1;3 | 803.7_184.1 | 886.3→1603.7 | +1.81 |
| SM 32:3;4 | 703.6_184.1 | 116.3→4380.6 | +37.7 | SM 40:3;2 | 783.6_184.1 | 1670.6→3774.5 | +2.26 |
| SM 32:4;3 | 685.6_184.1 | ND→182.5 | + | SM 42:0;3 | 833.7_184.1 | 593.8→1129.9 | +1.90 |
| SM 32:4;4 | 701.6_184.1 | 39.7→655.0 | +16.50 | SM 42:3;4 | 843.7_184.1 | 1524.4→3444.7 | +2.26 |
| SM 34:0;3 | 721.6_282.3 | 1.5→40.2 | +26.8 | SM 42:4;2 | 809.7_184.1 | 441.7→3768.9 | +8.53 |
| SM 34:1;3 | 719.6_262.3 | 21.9→303.3 | +13.85 | SM 43:0;4 | 863.7_184.1 | 3370.3→5603.2 | +1.66 |
| SM 34:2;3 | 717.6_184.1 | 168.7→354.8 | +2.10 | SM 43:1;4 | 861.7_184.1 | 5080.8→7635.9 | +1.50 |
| SM 34:4;4 | 729.6_184.1 | ND→65.4 | + | SM 43:4;2 | 823.7_184.1 | 1765.7→3678.6 | +2.08 |
| SM 34:5;4 | 743.6_184.1 | 98.5→ND | − | SM 44:3;2 | 839.7_184.1 | 3620.0→5410.1 | +1.5 |
| SM 35:0;4 | 751.6_184.1 | 621.1→211.4 | −2.94 | SM 45:0;4 | 891.7_184.1 | 1907.2→3507.7 | +1.84 |
| SM 35:3;4 | 745.6_184.1 | 40.7→132.5 | +3.26 | SM 45:1;4 | 889.7_184.1 | 3747.7→5730.3 | +1.53 |
| SM 36:0;3 | 749.6_184.1 | 299.0→74.7 | −4.00 | SM 45:3;4 | 885.7_184.1 | 848.1→2141.5 | +2.53 |
| SM 36:0;4 | 765.6-184.1 | 216.9→456.3 | +2.10 | SM 45:4;2 | 851.7_184.1 | 1176.3→2286.0 | +1.94 |
| SM 36:1;4 | 763.6_184.1 | 227.0→44.9 | −5.06 | SM 45:4;3 | 867.7_184.1 | 1083.3→1967.6 | +1.82 |
| SM 36:4;4 | 757.6_184.1 | 248.3→809.6 | +3.26 | SM 45:4;4 | 883.7_184.1 | 116.0→2077.5 | +17.91 |
| SM 38:0;4 | 793.6_184.1 | 850.3→1674.8 | +1.97 | SM 46:4;2 | 865.7_184.1 | 1388.5→2391.1 | +1.72 |
| SM 38:2;4 | 789.7_184.1 | 562.4→2039.4 | +3.63 | SM 47:0;2 | 887.7_184.1 | 598.1→1557.9 | +2.61 |
| SM 38:3;2 | 755.6_184.1 | 914.5→1802.2 | +1.97 | SM 47:2;4 | 915.8_184.1 | 965.9→1804.7 | +1.87 |
| SM 38:4;4 | 785.7_184.1 | 723.5→1443.8 | +2.00 | SM 48:2;3 | 913.8_184.1 | 772.9→1423.7 | +1.84 |
| SM 39:0;2 | 775.6_184.1 | 390.7→1003.4 | +2.57 | SM 48:2;4 | 929.8_184.1 | 773.8→1335.3 | +1.73 |
Reductions are highlighted in bold.
Table 2.
Ether-linked PC and ether-linked PE species changed to a statistically significant extent 24 h post-IR-induced AKI
| Lipid | MS/MS, m/z | Intensity Change | Fold-Change | Lipid | MS/MS, m/z | Intensity Change | Fold-Change |
|---|---|---|---|---|---|---|---|
| PC O-20:5 | 542.4_184.1 | 75.7→ND | − | PC O-46:2 | 912.8_184.1 | 1296.8→2044.6 | +1.6 |
| PC O-20:6 | 540.4_184.1 | 72.8→ND | − | PC O-48:0 | 944.8_184.1 | 1059.4→1867.8 | +1.8 |
| PC O-30:4 | 684.5_184.1 | ND→175.1 | + | PC O-48:1 | 942.8_184.1 | 1224.9→2333.8 | +1.9 |
| PC O-32:1 | 718.6_184.1 | 815.4→3278.8 | +4.0 | PC O-48:2 | 940.8_184.1 | 1777.2→3636.6 | +2.1 |
| PC O-32:6 | 708.5_184.1 | 32.7→2.9 | −11.3 | PC O-48:3 | 938.8_184.1 | 1707.0→3629.3 | +2.1 |
| PC O-34:3 | 742.6_184.1 | 117.8→30.1 | −3.9 | PE O-26:3 | 588.4_198.1 | 38.1→1.8 | −21.2 |
| PC O-34:4 | 740.6_184.1 | 70.0→24.6 | −2.9 | PE O-34:0 | 706.5_198.1 | 81.4→ND | |
| PC O-36:0 | 776.6_184.1 | 227.4→520.6 | +2.3 | PE O-34:1 | 718.6_198.1 | 62.3→1.8 | −34.6 |
| PC O-36:1 | 774.6_184.1 | 263.2→950.9 | +3.6 | PE O-36:0 | 734.6_198.1 | 1080.5→9.5 | −113.7 |
| PC O-36:3 | 770.6_184.1 | 1583.1→757.4 | −2.1 | PE O-36:1 | 732.6_198.1 | 130.9→2.9 | −45.1 |
| PC O-36:4 | 768.7_184.1 | 2618.3→1422.4 | −1.8 | PE O-36:6 | 722.6_198.1 | 26.3→ND | − |
| PC O-36:5 | 766.6_184.1 | 810.3→469.7 | −1.7 | PE O-38:1 | 760.6_198.1 | 85.6→ND | − |
| PC O-38:0 | 804.7_184.1 | 708.9→1266.8 | +1.8 | PE O-38:5 | 752.6_198.1 | 78.1→2.1 | −37.2 |
| PC O-38:1 | 802.7_184.1 | 1068.9→1915.5 | +1.8 | PE O-40:0 | 790.7_198.1 | 55.0→ND | |
| PC O-38:2 | 800.7_184.1 | 1044.7→1614.4 | +1.6 | PE O-40:1 | 788.7_198.1 | 127.6→2.0 | −63.8 |
| PC O-38:4 | 796.7_184.1 | 985.0→1548.1 | +1.6 | PE O-40:2 | 786.6_198.1 | 226.5→1.4 | −161.8 |
| PC O-38:5 | 794.7_184.1 | 739.6→1470.4 | +2.0 | PE O-40:3 | 784.6_198.1 | 418.8→2.3 | −182.1 |
| PC O-38:6 | 792.7_184.1 | 583.5→1064.1 | +1.8 | PE O-40:4 | 782.6_641.6 | 0.83→ND | − |
| PC O-40:5 | 822.7_184.1 | 927.1→2110.9 | +2.3 | PE O-40:6 | 778.6_198.1 | 145.4→0.4 | −363.5 |
| PC O-42:0 | 860.7_184.1 | 3957.8→6326.0 | +1.6 | PE O-42:0 | 818.7_198.1 | 198.3→7.1 | −28.0 |
| PC O-44:0 | 888.7_184.1 | 3229.0→5976.6 | +1.9 | PE O-42:1 | 816.7_198.1 | 296.8→2.5 | −118.7 |
| PC O-44:1 | 886.7_184.1 | 1066.5→2880.9 | +2.7 | PE O-42:2 | 814.6_198.1 | 257.7→3.5 | −73.6 |
| PC O-46:1 | 914.8_184.1 | 1093.2→2015.6 | +1.8 | PE O-42:3 | 812.6_198.1 | 516.3→7.4 | −69.8 |
| PE O-42:4 | 810.7_198.1 | 244.1→1.4 | −174.4 | PE O-46:2 | 870.7_198.1 | 582.6→17.7 | −32.9 |
| PE O-42:5 | 808.7_198.1 | 80.3→8.9 | −9.0 | PE O-46:3 | 868.8_198.1 | 817.7→21.2 | −38.6 |
| PE O-42:6 | 806.7_198.1 | 76.1→9.0 | −8.5 | PE O-46:4 | 866.7_198.1 | 611.1→14.8 | −41.3 |
| PE O-44:0 | 848.7_198.1 | 201.1→7.2 | −2.8 | PE O-46:5 | 864.7_198.1 | 188.5→3.6 | −52.4 |
| PE O-44:1 | 844.7_198.1 | 1167.3→24.1 | −48.4 | PE O-46:6 | 862.7_198.1 | 274.7→9.4 | −29.2 |
| PE O-44:2 | 842.7_198.1 | 1165.8→29.1 | −40.1 | PE O-48:0 | 902.8_198.1 | 789.7→30.8 | −25.6 |
| PE O-44:3 | 840.7_198.1 | 2050.0→44.5 | −46.1 | PE O-48:1 | 900.7_198.1 | 302.9→10.7 | −28.3 |
| PE O-44:4 | 838.7_198.1 | 1047.9→25.7 | −40.1 | PE O-48:2 | 898.8_198.1 | 392.0→12.2 | −32.1 |
| PE O-44:6 | 834.7_198.1 | 223.5→8.5 | −26.3 | PE O-48:4 | 894.7_198.1 | 1061.0→19.3 | −55.0 |
| PE O-46:0 | 874.7_198.1 | 985.7→33.4 | −29.5 | PE O-48:5 | 892.8_198.1 | 973.8→27.9 | −34.9 |
| PE O-46:1 | 872.7_198.1 | 589.5→16.1 | −36.6 | PE O-48:6 | 890.7_198.1 | 1066.1→37.4 | −28.5 |
ND: not detected. Phospholipids that were increased at 6 h post-IR are in italics. Reductions are highlighted in bold. Note that all statistically significant changes in ether-linked PEs 24 h post-IR were reductions.
In conclusion, our study demonstrates that an untargeted SWATH-MS lipidomics approach in conjunction with MALDI imaging can be affordably used to provide quantitative and spatial information with which to narrow those changes in the renal lipidome associated with AKI to those lipids that have the greatest potential to be of pathophysiological significance by virtue of their correlation with AKI severity as well as their localization to renal sites known to be principally affected by AKI. Similarly, using a combination of quantitative and spatial MS approaches, Grove et al. (11) identified lipid changes associated with diabetic renal pathology. In addition to the use of different matrices for MALDI-IMS in this study, a separate MALDI Fourier transform ion cyclotron resonance MS system was used to obtain MS/MS spectra for diagnostic fragmentation analysis with improved mass resolution. Refinements to MALDI-IMS such as this, together with greater spatial resolution and the use of alternative matrices, will undoubtedly increase the number of candidate lipids that can be identified in this way and thus further our understanding of the molecular mechanisms in AKI. Determining whether those renal lipids identified early in AKI can be correlated with similar lipid changes in urine or plasma could also lay the foundation for the development of novel biomarkers for both diagnostic and prognostic benefits earlier in the course of AKI.
GRANTS
This work was supported by a Novel Research Project grant from the Lupus Research Institute to J. H. Kabarowski and National Institutes of Health Grants KO1 DK103931 (to S. Bolisetty) and RO1 DK059600 (to A. Agarwal).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: S.R., K.B.W., D.G., S. Barnes, A.A., and J.H.K. provided conception and design of research; S.R., K.B.W., L.W., B.C., S. Bolisetty, and J.H.K. performed experiments; S.R., K.B.W., L.W., and J.H.K. analyzed data; S.R., K.B.W., S. Barnes, and J.H.K. interpreted results of experiments; S.R., K.B.W., and J.H.K. prepared figures; S.R., K.B.W., and J.H.K. drafted manuscript; S.R., K.B.W., and J.H.K. edited and revised manuscript; S.R., K.B.W., L.W., B.C., S. Bolisetty, D.G., S. Barnes, A.A., and J.H.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the University of Alabama at Birmingham (UAB) Animal Resources Program Comparative Pathology Laboratory for assistance with histology and the UAB-UCSD O'Brien Center for Acute Kidney Injury Research (P30 DK079337) for creatinine measurements.
REFERENCES
- 1.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 69: 89–95, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Black LL, Srivastava R, Schoeb TR, Moore RD, Barnes S, Kabarowski JH. Cholesterol-independent suppression of lymphocyte activation, autoimmunity, and glomerulonephritis by apolipoprotein A-I in normocholesterolemic lupus-prone mice. J Immunol 195: 4685–4698, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta 1822: 1442–1452, 2012. [DOI] [PubMed] [Google Scholar]
- 4.Clermont G, Acker CG, Angus DC, Sirio CA, Pinsky MR, Johnson JP. Renal failure in the ICU: comparison of the impact of acute renal failure and end-stage renal disease on ICU outcomes. Kidney Int 62: 986–996, 2002. [DOI] [PubMed] [Google Scholar]
- 5.Devarajan P. Emerging biomarkers of acute kidney injury. Contrib Nephrol 156: 203–212, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Devarajan P. Proteomics for biomarker discovery in acute kidney injury. Semin Nephrol 27: 637–651, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Druml W. Long term prognosis of patients with acute renal failure: is intensive care worth it? Intensive Care Med 31: 1145–1147, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Edelstein CL, Ling H, Schrier RW. The nature of renal cell injury. Kidney Int 51: 1341–1351, 1997. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira MA, Barcelos LS, Teixeira MM, Bakhle YS, Andrade SP. Tumor growth, angiogenesis and inflammation in mice lacking receptors for platelet activating factor (PAF). Life Sci 81: 210–217, 2007. [DOI] [PubMed] [Google Scholar]
- 10.Gorgas K, Teigler A, Komljenovic D, Just WW. The ether lipid-deficient mouse: tracking down plasmalogen functions. Biochim Biophys Acta 1763: 1511–1526, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Grove KJ, Voziyan PA, Spraggins JM, Wang S, Paueksakon P, Harris RC, Hudson BG, Caprioli RM. Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J Lipid Res 55: 1375–1385, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull TD, Kamal AI, Boddu R, Bolisetty S, Guo L, Tisher CC, Rangarajan S, Chen B, Curtis LM, George JF, Agarwal A. Heme oxygenase-1 regulates myeloid cell trafficking in AKI. J Am Soc Nephrol 26: 2139–2151, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson AC, Stahl A, Zager RA. Triglyceride accumulation in injured renal tubular cells: alterations in both synthetic and catabolic pathways. Kidney Int 67: 2196–2209, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Kashani K, Al-Khafaji A, Ardiles T, Artigas A, Bagshaw SM, Bell M, Bihorac A, Birkhahn R, Cely CM, Chawla LS, Davison DL, Feldkamp T, Forni LG, Gong MN, Gunnerson KJ, Haase M, Hackett J, Honore PM, Hoste EA, Joannes-Boyau O, Joannidis M, Kim P, Koyner JL, Laskowitz DT, Lissauer ME, Marx G, McCullough PA, Mullaney S, Ostermann M, Rimmele T, Shapiro NI, Shaw AD, Shi J, Sprague AM, Vincent JL, Vinsonneau C, Wagner L, Walker MG, Wilkerson RG, Zacharowski K, Kellum JA. Discovery and validation of cell cycle arrest biomarkers in human acute kidney injury. Crit Care 17: R25, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kribben A, Wieder ED, Wetzels JF, Yu L, Gengaro PE, Burke TJ, Schrier RW. Evidence for role of cytosolic free calcium in hypoxia-induced proximal tubule injury. J Clin Invest 93: 1922–1929, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulah E, Tascilar O, Acikgoz S, Tekin IO, Karadeniz G, Can M, Gun B, Barut F, Comert M. Oxidized LDL accumulation in experimental renal ischemia reperfusion injury model. Ren Fail 29: 409–415, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Ling XB, Mellins ED, Sylvester KG, Cohen HJ. Urine peptidomics for clinical biomarker discovery. Adv Clin Chem 51: 181–213, 2010. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Yan S, Ji C, Dai W, Hu W, Zhang W, Mei C. Metabolomic changes and protective effect of (L)-carnitine in rat kidney ischemia/reperfusion injury. Kidney Blood Press Res 35: 373–381, 2012. [DOI] [PubMed] [Google Scholar]
- 19.Lodhi IJ, Wei X, Yin L, Feng C, Adak S, Abou-Ezzi G, Hsu FF, Link DC, Semenkovich CF. Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab 21: 51–64, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnusson CD, Haraldsson GG. Ether lipids. Chem Phys Lipids 164: 315–340, 2011. [DOI] [PubMed] [Google Scholar]
- 21.Makris K, Kafkas N. Neutrophil gelatinase-associated lipocalin in acute kidney injury. Adv Clin Chem 58: 141–191, 2012. [DOI] [PubMed] [Google Scholar]
- 22.Mehta RL, Pascual MT, Soroko S, Savage BR, Himmelfarb J, Ikizler TA, Paganini EP, Chertow GM, and Program to Improve Care in Acute Renal Disease. Spectrum of acute renal failure in the intensive care unit: the PICARD experience. Kidney Int 66: 1613–1621, 2004. [DOI] [PubMed] [Google Scholar]
- 23.Metnitz PG, Krenn CG, Steltzer H, Lang T, Ploder J, Lenz K, Le Gall JR, Druml W. Effect of acute renal failure requiring renal replacement therapy on outcome in critically ill patients. Crit Care Med 30: 2051–2058, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Molitoris BA, Simon FR. Renal cortical brush-border and basolateral membranes: cholesterol and phospholipid composition and relative turnover. J Membr Biol 83: 207–215, 1985. [DOI] [PubMed] [Google Scholar]
- 25.Naito M, Bomsztyk K, Zager RA. Renal ischemia-induced cholesterol loading: transcription factor recruitment and chromatin remodeling along the HMG CoA reductase gene. Am J Pathol 174: 54–62, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallone TL, Silldorff EP, Turner MR. Intrarenal blood flow: microvascular anatomy and the regulation of medullary perfusion. Clin Exp Pharmacol Physiol 25: 383–392, 1998. [DOI] [PubMed] [Google Scholar]
- 27.Portilla D. Role of fatty acid beta-oxidation and calcium-independent phospholipase A2 in ischemic acute renal failure. Curr Opin Nephrol Hypertens 8: 473–477, 1999. [DOI] [PubMed] [Google Scholar]
- 28.Portilla D, Creer MH. Plasmalogen phospholipid hydrolysis during hypoxic injury of rabbit proximal tubules. Kidney Int 47: 1087–1094, 1995. [DOI] [PubMed] [Google Scholar]
- 29.Portilla D, Schnackenberg L, Beger RD. Metabolomics as an extension of proteomic analysis: study of acute kidney injury. Semin Nephrol 27: 609–620, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Portilla D, Shah SV, Lehman PA, Creer MH. Role of cytosolic calcium-independent plasmalogen-selective phospholipase A2 in hypoxic injury to rabbit proximal tubules. J Clin Invest 93: 1609–1615, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reis A, Rudnitskaya A, Chariyavilaskul P, Dhaun N, Melville V, Goddard J, Webb DJ, Pitt AR, Spickett CM. Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res 56: 413–422, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiss D, Beyer K, Engelmann B. Delayed oxidative degradation of polyunsaturated diacyl phospholipids in the presence of plasmalogen phospholipids in vitro. Biochem J 323: 807–814, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodemer C, Thai TP, Brugger B, Kaercher T, Werner H, Nave KA, Wieland F, Gorgas K, Just WW. Inactivation of ether lipid biosynthesis causes male infertility, defects in eye development and optic nerve hypoplasia in mice. Hum Mol Genet 12: 1881–1895, 2003. [DOI] [PubMed] [Google Scholar]
- 34.Sigdel TK, Klassen RB, Sarwal MM. Interpreting the proteome and peptidome in transplantation. Adv Clin Chem 47: 139–169, 2009. [DOI] [PubMed] [Google Scholar]
- 35.Simons B, Kauhanen D, Sylvanne T, Tarasov K, Duchoslav E, Ekroos K. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites 2: 195–213, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramaniam S, Fahy E, Gupta S, Sud M, Byrnes RW, Cotter D, Dinasarapu AR, Maurya MR. Bioinformatics and systems biology of the lipidome. Chem Rev 111: 6452–6490, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun L, He Z, Ke J, Li S, Wu X, Lian L, He X, He X, Hu J, Zou Y, Wu X, Lan P. PAF receptor antagonist Ginkgolide B inhibits tumourigenesis and angiogenesis in colitis-associated cancer. Int J Clin Exp Pathol 8: 432–440, 2015. [PMC free article] [PubMed] [Google Scholar]
- 38.Thai TP, Rodemer C, Jauch A, Hunziker A, Moser A, Gorgas K, Just WW. Impaired membrane traffic in defective ether lipid biosynthesis. Hum Mol Genet 10: 127–136, 2001. [DOI] [PubMed] [Google Scholar]
- 39.Thongboonkerd V. Proteomic analysis of renal diseases: unraveling the pathophysiology and biomarker discovery. Expert Rev Proteomics 2: 349–366, 2005. [DOI] [PubMed] [Google Scholar]
- 40.Uchino S, Kellum JA, Bellomo R, Doig GS, Morimatsu H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, Tolwani A, Ronco C,.Beginning, and ending supportive therapy for the kidney. I. Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA 294: 813–818, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Uehara T, Horinouchi A, Morikawa Y, Tonomura Y, Minami K, Ono A, Yamate J, Yamada H, Ohno Y, Urushidani T. Identification of metabolomic biomarkers for drug-induced acute kidney injury in rats. J Appl Toxicol 34: 1087–1095, 2014. [DOI] [PubMed] [Google Scholar]
- 42.Waddington EI, Croft KD, Sienuarine K, Latham B, Puddey IB. Fatty acid oxidation products in human atherosclerotic plaque: an analysis of clinical and histopathological correlates. Atherosclerosis 167: 111–120, 2003. [DOI] [PubMed] [Google Scholar]
- 43.Wei Q, Xiao X, Fogle P, Dong Z. Changes in metabolic profiles during acute kidney injury and recovery following ischemia/reperfusion. PLoS One 9: e106647, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitehouse T, Stotz M, Taylor V, Stidwill R, Singer M. Tissue oxygen and hemodynamics in renal medulla, cortex, and corticomedullary junction during hemorrhage-reperfusion. Am J Physiol Renal Physiol 291: F647–F653, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Yang F, Zhang L, Wu H, Zou H, Du Y. Clinical analysis of cause, treatment and prognosis in acute kidney injury patients. PLoS One 9: e85214, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zager RA, Andoh T, Bennett WM. Renal cholesterol accumulation: a durable response after acute and subacute renal insults. Am J Pathol 159: 743–752, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao YY, Vaziri ND, Lin RC. Lipidomics: new insight into kidney disease. Adv Clin Chem 68: 153–175, 2015. [DOI] [PubMed] [Google Scholar]