Abstract
Vaccines represent one of the greatest contributions of the scientific community to global health. Yet, many pathogens remain either unchallenged or inadequately hindered by commercially available vaccines. Respiratory viruses pose distinct and difficult challenges due to their ability to rapidly spread, adapt, and modify the host immune response. Considerable research has been directed to understand the role of respiratory virus immunomodulatory proteins and how they influence the host immune response. We review here efforts to develop next-generation vaccines through targeting these key immunomodulatory genes in influenza virus, coronaviruses, respiratory syncytial virus, measles virus, and mumps virus.
Keywords: Respiratory Viruses, Next-generation Vaccines, Pathogenesis, Immunology, Respiratory Syncytial Virus, Coronavirus, Influenza, Measles, Mumps
Introduction
For much of the past century, vaccine development has focused on direct attenuation or inactivation of the pathogen. This approach, albeit quite successful for a large number of human pathogens, is not completely amenable to all. To be effective, vaccines must have the following properties: (1) high immunogenicity, (2) be able to induce lasting memory, (3) be genetically stable, (4) be physically or thermally stable, and most importantly, (5) be safe. Finding the proper balance between these properties has proven quite difficult to develop effective vaccine preparations to many respiratory viruses. Though effective and historically successful vaccines currently exist for several respiratory viruses, the efficacy of several of these current vaccines is marginal and there remains an unmet need for many other respiratory viruses.
Live-attenuated vaccines have a long history of success and are generally more immunogenic than non-replicating vaccines types due to persistence of antigen that mimics infection. However, the classical attenuation process by serial passage can be lengthy because it may be challenging to achieve a proper balance of safety and potency and because of the possibility for reversions or compensatory mutations. Many current efforts to design live-attenuated vaccines have focused on ways to suppress the nonstructural and immunomodulatory proteins of respiratory viruses by more genetically stable genetic modifications such as deletion or codon deoptimization. The major advantage to these approaches is that they permit recovery of a genetically stable infectious virus, while often attenuating and boosting the immune response. We review here efforts targeting immunomodulatory genes towards design of next generation live-attenuated vaccines to influenza virus, coronaviruses, respiratory syncytial virus, measles virus, and mumps virus.
Influenza
Influenza viruses are pleomorphic, enveloped viruses, which encode single-strand negative-sense segmented RNA genomes. Influenza viruses remain one of the most prevalent respiratory pathogens in circulation and have been responsible for millions of deaths annually. The 1918 Spanish flu pandemic, which resulted in many millions of deaths worldwide, highlights the potential of the virus to be highly pathogenic and cause severe disease [1]. There are three primary serotypes based on genetic variations in the surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA): influenza A (IAV), influenza B (IBV), and influenza C (ICV). Current influenza vaccine preparations consist of either inactivated or live-attenuated trivalent or quadrivalent preparations of antigens from two IAV and either one or two IBV strains. ICV is far less common than IAV or IBV and is associated with more mild disease. However, seasonal variations due to a variety of factors including genetic drift and incongruities between the antigens of the vaccine and the dominant strains within a flu season have resulted in marginal efficacy of the influenza vaccine at best and the need for annual vaccinations. The availability of more immunogenic live-attenuated influenza vaccine options opens the door to the potential for genetic modifications, which may improve duration and efficacy of protective immunity. Despite existing vaccines, continued efforts are being made to continue to improve influenza vaccine design through incorporation of better understanding of the virus and its relationship to the host.
The genome of influenza consists of 7 or 8 negative-strand segments that encode at least 11 proteins, with potentially several additional protein variants due to ribosomal frameshifts, alternative splicing, and truncations having been identified [2,3] (Figure 1). Segments PB2, HA, NP, and NA are monocistronic and are not known to code for alternative spliced or truncated protein variants. Segment PB1, which encodes a subunit of the viral polymerase, is also known to be translated into a couple protein variants: an N-terminal truncation of PB1 called PB1-N40 and an alternatively spliced PB1-F2 protein [4,5]. Segment PA, which encodes an additional subunit of the viral polymerase complex, is known to express at least three alternative forms, including a protein called PA-X, which is formed via a ribosomal frameshift [6,7]. In addition, segments 7 and 8 each encode for two protein variants. Segment 7 encodes for the proteins M1 and M2, which function in packaging/assembly and formation of proton ion channels that are important during multiple phases of the virus cycle, respectively. Segment 8 encodes for the nonstructural proteins NS1 and NS2. NS1 functions directly in host cell modulation through a variety of mechanisms and represents a primary target of modifications for vaccine design (Table 1). NS2, also known as nuclear export protein (NEP), facilitates export of RNP complexes from the nucleus following assembly [8,9].
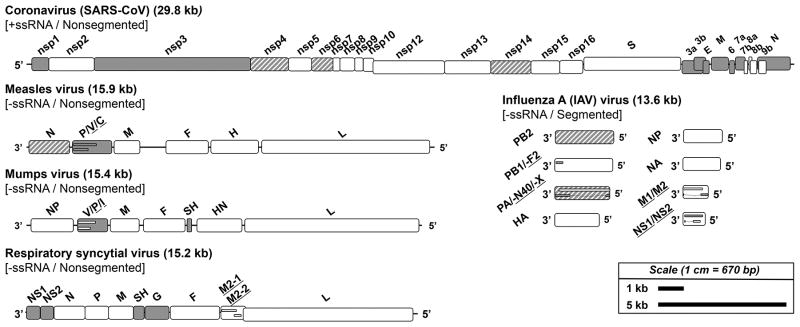
Figure 1. Genomic organization and immunomodulatory genetic elements of select respiratory virus pathogens.
The genomes of a coronavirus (SARS-CoV), measles virus, mumps virus, RSV, and influenza virus (IAV) are shown to scale with key genes and proteins identified. Genes and proteins directly involved in immunomodulation are shown in gray and those indirectly involved in immunomodulation are shown in gray stripes. For genes or segments encoding multiple overlapping proteins, small inset bars are shown to represent these products and the associated protein names are underlined and listed in order for the genes shown top to bottom.
Table 1. Overview of mechanisms of respiratory virus immunomodulatory activity.
Different mechanisms are described for host immunomodulatory activity including the proteins for influenza, coronavirus, RSV, measles, and mumps.
| Host Disruption | Virus: Proteins | Refs |
|---|---|---|
| Interferon Signaling |
Influenza: NS1, PB1-F2 Coronavirus: nsp1, nsp3, M, N, E, and several accessory proteins RSV: NS1, NS2, G Measles: V, C Mumps: V |
3, 15 – 19, 50, 68, 71 – 75, 118, 122, 123 |
| Host Transcription/Translation |
Influenza: NS1, PB2, PA, PA-X Coronavirus: nsp1, and several accessory proteins Measles: N, P, C |
3,10, 11, 21, 22, 30, 50, 118, 120 |
| Apoptosis, Cell Cycle Control, and Cell Stress Pathways |
Influenza: NS1, PB1-F2, PA-X Coronavirus: nsp1, E RSV: NS1, NS2, SH Measles: V, P, N Mumps: V, SH |
3, 5, 26, 30 – 32, 50 – 53, 68, 71 – 75, 82, 118, 121 – 123 |
| Inflammation |
Influenza: M2, PB1-F2 Coronavirus: nsp1, nsp3, N, E, and several accessory proteins RSV: NS1, NS2, SH, G Measles: V, C Mumps: V |
3,12, 26 – 29, 50, 51, 56, 68, 71 – 75, 82, 90, 91, 118, 122, 123 |
| Ubiquitination |
Influenza: NS1 Coronavirus: nsp3 |
3,18, 50, 122, 123 |
There are several immunomodulatory genes identified for influenza including PB1-F2, PA-X, M2, and NS1 (Table 1). Several other influenza genes are known to interfere with normal cell activity and indirectly impact host immune function. PB2 and PA affect host cell protein synthesis by facilitating cap snatching from host cell mRNAs and indirectly suppress the host antiviral response [10,11]. Influenza M2 ion channel activity is known to activate inflammasomes through activation of the NOD-like receptor, NLRP3 [12]. Inhibition of M2 activity early during influenza infection prevents uncoating and replication, consequently, it remains a key target for antivirals and both subunit and chimeric vaccines [13,14]. NS1 is the primary immunomodulatory protein of influenza and is known to inhibit TLR signaling through several mechanisms, disrupt host cell ubiquitination, alter host cell transcription by inhibiting activation and blocking nuclear translocation of several transcription factors, suppress host translation, and induce PI3K signaling [15–22]. Suppression of NS1 has been a common approach for generation of new live-attenuated vaccine candidates. Early studies involving either deletion or truncation of NS1 resulted in attenuated candidates that still provide protection in mice [23]. A novel mechanism for modifying the levels of this protein for vaccine design has recently been described through the use of codon deoptimization. Codon usage deoptimization was first described as a mechanism of virus gene modulation in 2006 as a genetically stable mechanism to reduce poliovirus protein synthesis without altering the protein amino acid sequence [24]. This approach involves substituting the least used codons based on host cell codon usage bias into the coding sequence of the viral gene. This approach was recently employed to lower the gene expression of NS1 and NS2 resulting in attenuated strains that are protective to homologous and heterologous challenge [25].
PB1-F2 and PA-X were more recently identified and have been shown to greatly impact the host response during influenza infection. PB1-F2 was discovered in 2001 and is translated as a result of an alternative reading frame on the PB1 segment and has been shown to induce apoptosis, activate the inflammatory pathway, and enhance the pathogenesis of secondary bacterial infections [26–29]. PA-X is a small peptide, which plays a role in suppression of host translation, and has been identified as a key regulator of influenza virulence by regulating inflammatory and apoptotic pathways of the host during infection [6,30–32]. Recently, deletion of PA-X was shown to alter the pathogenicity of the 1918 H1N1 pandemic virus [6,32]. PB1-F2 and PA-X may represent new intriguing targets for next-generation vaccines.
Coronaviruses
Coronaviruses are positive-sense, single-strand RNA viruses that are associated with a wide spectrum of upper and lower respiratory illnesses fluctuating in severity from the common cold to severe acute respiratory disease. Similar to RSV, there are no vaccines available and antivirals such as ribavirin have limited to no efficacy [33,34]. There are currently six known human coronaviruses. The recent epidemics of severe acute respiratory syndrome (SARS) coronavirus (2002 – 2003) and the ongoing epidemic of Middle East respiratory syndrome (MERS) coronavirus (2012 – present) are associated with mortality rates of approximately 10% and 36%, respectively [35]. These epidemics highlight the need for an effective vaccine and therapeutics. Modern advances in studying the virus in applicable animal models have provided new insight into pathogenesis of the virus and represent stable platform for testing of live-attenuated candidates [36–40]. Despite a high clinical and economic burden, there remain several challenges, which continue to hinder vaccine development including the potential for rapid reversion and recombination in live-attenuated vaccine candidates and unclear coronavirus biology.
The genome of coronaviruses, which ranges in size from 27 to 32 kb, represent the largest known RNA virus genomes [41]. Coronaviruses encode up to 16 nonstructural proteins (nsps), at least 7 structural proteins and a diverse set of accessory proteins that vary in form and function between strains [42] (Figure 1). The function of many of the accessory proteins for coronaviruses remains unclear or unknown. Coronaviruses encode many known immunomodulatory proteins. Nsp1, nsp3, membrane (M), nucleocapsid (N), envelope (E), and many accessory proteins have been shown to play key roles in host immune modulation (Table 1). In addition, nsps 3, 4, and 6 are actively involved in host membrane modifications forming virus-specific double membrane vesicles and convoluted membrane structures that facilitate replication complex formation [43–45]. These induced membrane structures play a key role in evading activation of host toll-like receptors by shielding double-strand RNA during replication. Though not a conventional immunomodulatory protein, recently, nsp14 was identified as having an exonuclease RNA proofreading activity and is responsible for conferring resistance of the virus to mutagenesis and nucleoside-analog based therapeutics [46–48]. While development of vaccine candidates has been largely focused on optimized expression of the spike (S) attachment glycoprotein, there have been several novel strategies targeting immunomodulatory elements of the coronavirus genome in development of potential live-attenuated vaccines.
Deletion of the open-reading frames 4, 5a, 7a, and HE have resulted in attenuation in a murine coronavirus, however these mutations had no appreciable effect on virulence of SARS-CoV [49,50]. However, considerable energy has been invested in generating a recombinant live-attenuated vaccine targeting E. The coronavirus envelope (E) protein is a small integral membrane protein that has been well studied and has many identified functions including roles in assembly, budding, and host cell modifications [50–52]. Very little E is incorporated into coronavirus virions, with as few as 20 copies per virion, however there is a high concentration of E detected within the host cell during virus replication [53–55]. Despite its various functions, E is dispensable for virus replication [42,56–59]. A SARS-CoV vaccine containing an E deletion has been generated and is highly attenuated in hamsters exhibiting less detected viral antigen by lung histopathology and lower viral titers [50,51,56,60]. An entirely new direction in coronavirus vaccine design has focused on modifying the replication fidelity of the virus. Introduction of a deletion to the exonuclease domain of nsp14 has resulted in an attenuated virus, which has an increased mutational frequency, attenuation in vivo, and surprisingly retains the exonuclease deletion during serial passaging [61,62]. The discovery of the protective nature of nsp14 to nucleoside-analog based therapeutics presents a new target for design of recombinant live-attenuated candidates.
Respiratory Syncytial Virus
Respiratory syncytial virus (RSV) is a leading cause of lower respiratory disease in young children [63,64]. To date, there remain no RSV vaccines available and present monoclonal antibody prophylaxis options are not feasible in most populations [65]. RSV represents a significant challenge to vaccine design for several reasons. First, the highest clinical burden is associated with infants under the age of 1, which have immune systems that remain compromised [63,64,66]. Second, the virus is known to encode several proteins that facilitate innate immune evasion, and this may contribute to natural infection failing to induce long-lasting immunity that prevents re-infection. Third, early efforts at designing a formalin-inactivated RSV vaccine ended in failure resulting in enhanced disease and mortality among infants contracting RSV following vaccinations [67,68]. The regrettable events surrounding these early efforts have hampered vaccine efforts. Lastly, there remain no animal models that fully recapitulate human RSV pathogenesis or RSV vaccine-enhanced disease. Unfortunately, traditional attenuation methods to dampen virus replication have yet to result in a promising balance of safety and immunogenicity. RSV reverse genetics system was developed in 1995, and other groups generated similar systems [69,70]. These reagents have permitted the development of next-generation vaccines that integrate current knowledge of RSV pathogenesis, immune evasion mechanisms, phylogenetic diversity, and immunogenicity.
The approximately 15 kb genome of RSV consists of 10 genes encoding 11 proteins (Figure 1) [68]. The genome encodes at least four established immunomodulatory proteins: nonstructural proteins 1 (NS1) and 2 (NS2), attachment glycoprotein (G), and the small hydrophobic protein (SH) (Table 1). The two nonstructural and non-essential proteins (NS1 and NS2) have been shown to suppress host innate immunity by interferon suppression, activate pro-survival pathways, which antagonize apoptosis, and are expressed in high quantities early in infection [71–75]. Early efforts to delete these proteins to generate vaccine candidates resulted in either over-attenuation (ΔNS1) or under-attenuated candidates (ΔNS2) [76,77]. Site-directed mutagenesis of key functional residues of NS1 and NS2 presents an additional challenge because very little is known of which determinants of the proteins are functional relevant and RSV, like many other respiratory viruses, is genetically instable. Although the mechanism remains unclear, codon-deoptimization has been successfully applied to RSV. Codon deoptimization of NS1 and NS2 lowered detectable NS1 and NS2 protein levels and subsequently increased STAT2 protein levels compared to wild-type RSV [78]. This approach resulted in a virus that was attenuated compared to wild-type in primary human cell cultures, but induced slightly higher neutralizing antibodies. These data suggest that modification of RSV NS proteins by codon-deoptimization may represent a new strategy to developing next-generation RSV live-attenuated vaccine candidates. In addition to the RSV nonstructural proteins, the small hydrophobic protein (SH) and the attachment glycoprotein (G) and have also been shown to play several functional roles including modulating the host response.
RSV SH is a short transmembrane protein, which is structurally similar to a class of small viroporins and has been shown to increase membrane permeability when expressed in bacteria and forms multimeric pore complexes in artificial membranes [79–81]. The function of SH in RSV biology remains largely unclear, however it has been shown to affect apoptosis and inhibit TNF-α signaling [82]. Deletion of SH protein results in minor attenuation in mice and chimpanzees, yet incorporation of the deletion had no apparent attenuation in humans in a live-attenuated vaccine candidate [83–85]. RSV G is a heavily glycosylated protein which has been shown to play a major role in RSV attachment through engagement with host glycosaminoglycans (GAGs) [86,87]. In coordination with the fusion (F) glycoprotein, RSV G is also believed to help facilitate cell entry. During infection in vivo, the majority of induced antibodies are targeted to RSV F and G. Yet, RSV G antibodies are generally far less efficient at neutralization and protection than F antibodies [88]. Furthermore, RSV G exists in two distinct forms, a membrane-bound surface exposed structure and a shorter secreted form. The secreted RSV G functions as an immune decoy and reduces antibody efficacy [89]. Several studies have indicated that G also modulates the innate immune response through structural mimicry and inhibition of TLR activation [90,91]. However, studies involving deletion of the RSV G gene have demonstrated that G protein is not required of RSV infection [92–95]. Development of RSV vaccines targeting RSV G have mostly consisted of expressing whole forms of the protein in a component vaccine or in chimeric strains (such as Sendai Virus or PIV) for its potential to induce protective antibodies [96–99]. However, the inability to delete G without over attenuating the virus as a vaccine candidate has made altering the immunomodulatory activity of RSV G difficult. Despite being associated with less immunogenicity than RSV F, RSV G vaccines have shown promise as potential targets of neutralizing antibodies. One study demonstrated using a G subunit vaccine that protective immunity could be induced to both RSV A and RSV B subgroups without enhanced pulmonary pathology [100,101]. Due in large part to the inability of RSV natural infection to induce long-lasting protective immunity, development of next generation live-attenuated RSV vaccines will likely need to identify new mechanisms to suppress or alter the activity of the viral immunomodulatory genes as well as promoting more protective immunogenic antigens.
Measles and Mumps
Measles and mumps viruses, like RSV, are members of the family Paramyxoviridae and are enveloped, negative-strand RNA viruses that are associated with highly contagious disease which may be fatal. Yet, the development of measles and mumps vaccines have reduced the incidence of measles and mumps worldwide 99.9% and 95.9%, respectively, since introduction of the vaccines [102]. The first vaccines for measles and mumps were passaged live-attenuated viruses developed in the 1950’s and 1960’s by John Enders and Maurice Hilleman, respectively [103,104]. The current MMR vaccine preparation includes live-attenuated strains based on these two strains as well as a live-attenuated strain of rubella virus [105]. However, there remains a need for further improving the efficacy of the measles and mumps vaccines. Studies have shown that the measles vaccine has noticeable levels of vaccine failure with 2 to 10% of children receiving two doses of the vaccine failing to develop protective immunity [106–109]. Historically, the live-attenuated Jeryl Lynn strain of mumps vaccine has been quite safe and effective over the last 50 years, however recent outbreaks, even among highly vaccinated populations, have prompted questions about whether the vaccine can be improved [110–114]. The recent re-emergence of measles and mumps in nations that have had historically low or undetectable levels of infections and the inability to eradicate the viruses has promoted renewed efforts to improve the efficacy of the current MMR vaccine preparation [105,115–117].
Measles virus has a 16 kb nonsegmented, negative-sense RNA genome which consists of 6 genes (N, P/V/C, M, F, HN, and L) that encode for six structural proteins and two nonstructural proteins (Figure 1) [118]. Mumps virus has a similar genomic organization, encoding seven genes (N, V/P, M, F, SH, H, and L). There are three measles proteins, which are directly associated with immunomodulatory activity during infection: P, C, and V (Table 1) [118]. In addition, the nucleocapsid (N) protein has also been shown in measles virus to indirectly modify the host response through inhibition of translation and the capacity to induce apoptosis [119,120]. Numerous new vaccine preparations have been made to improve the safety and immunogenicity of the H, F, and N proteins, however, novel vaccine strains targeting immunomodulatory genes have been largely absent [121]. Much work is currently being done to understand genetic differences between children, which may enhance or suppress the immune response to measles vaccination [107]. These data in combination with developments in reverse genetic approaches may open the door to novel live-attenuated vaccine preparations. Mumps has two primary proteins involved in immunomodulatory activity: V and SH. One recent mumps vaccine candidate employs a dual deletion of V and SH genes and results in attenuation, immunogenicity, and genetic stability [122–124]. Development on improved measles and mumps next generation vaccines is likely to focus on ways of modifying the expression or activity of these genes to optimize development of protective immunity.
Expert Commentary and Five-year View
The development of vaccines to respiratory viruses such as smallpox, measles, mumps, rubella and more recently, influenza, has transformed the landscape of medical practice and contributed substantially to near doubling of life expectancy over the last 100 years. However, the methods for classical attenuation of live-attenuated viruses such as passaging or cold adapting are not directly amenable to more prevalent modern respiratory viruses such as RSV, for which there remain no current commercially available vaccines. To develop safe and effective vaccines to the most pressing respiratory challenges, we envision novel approaches combining advancements in reverse genetics with a more detailed understanding of the biology of the virus to be the cornerstone of the next generation of respiratory virus vaccines. A common strategy to enhance and improve the host response is modifying the immunomodulatory activities of viral genes. We have discussed here several novel strategies, which have provided new tools to tweak and adjust the function of immunomodulatory elements of respiratory viruses for vaccine design.
Deletions or generation of vaccines based on other genetic backgrounds have presented a new opportunity to remove more virulent or pathogenic elements of respiratory virus systems and optimize expression of antigenic factors. Modifications to viral codon usage represent a new approach that has rapidly been expanded as a mechanism for suppression of viral protein expression without deletion. Over the last few years, codon-optimization and deoptimization have been successfully employed as described earlier for poliovirus, RSV, and influenza, as well as for hepatitis A, porcine reproductive and respiratory syndrome virus (PRRSV), and LCMV [24,78,125–128]. The mechanism of codon-deoptimization for protein suppression remains largely speculative, but is believed to consist of a combination of modifying translational kinetics and altering RNA secondary structure. As described earlier, the nonstructural proteins of RSV are known to be dispensable for viral replication and immunomodulatory, however deletion either over- or under-attenuates the virus for vaccine design. Codon-deoptimization is likely to be expanded as an approach for development of next-generation vaccines to address these issues by altering protein function through a genetically stable mechanism.
Vaccines will remain the foundation for modern advancements in public health. We expect that over the next five years, the development of next-generation live-attenuated vaccines will focus primarily on not just improvements to antigenic display and optimizing the immunogenicity of pathogens, but also suppressing the immunomodulatory elements of the virus that limit necessary elements of the host response to establish optimal protective immunity.
Key Issues.
Respiratory viruses continue to pose many new challenges for vaccine design and development.
Influenza virus vaccine remains marginally effective from season to season and fails to provide broadly-protective immunity. Several new immunomodulatory proteins of influenza have been identified and new techniques are being made available, which may result in a new generation of influenza vaccines.
Despite a clear potential for pandemic disease, no vaccines remain available for coronaviruses. A number of immunomodulatory genes for coronaviruses have been identified and novel strategies including targeting viral replication fidelity represent new directions in vaccine development.
RSV remains a major respiratory pathogen for young children and currently no prophylactic options are available for prevention. Codon deoptimization represents a potentially new strategy for RSV gene modulation, which has been shown to be effective for immunomodulatory proteins NS1 and NS2. Combining this approach with alterations to changes to other key regulatory genes may provide the basis for a novel RSV live-attenuated vaccine.
Future development of respiratory virus vaccines will likely target combining changes to immunomodulatory genes with improvements in reverse genetics. Codon modifications represent a key new development that may allow catered modifications of specific immunomodulatory genes in vaccine development.
Acknowledgments
This work was supported by NIH grants R01AI087798 (MLM), U19 AI095227 (MLM), and T32 AI074492 (CCS) and the Emory Children’s Center for Childhood Infections and Vaccines (CCIV).
References
- 1.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 2.Szewczyk B, Bienkowska-Szewczyk K, Krol E. Introduction to molecular biology of influenza a viruses. Acta Biochim Pol. 2014;61:397–401. [PubMed] [Google Scholar]
- 3**.Dash P, Thomas PG. Host detection and the stealthy phenotype in influenza virus infection. Curr Top Microbiol Immunol. 2015;386:121–147. doi: 10.1007/82_2014_412. This article provides a great review of influenza biology and host-pathogen interactions including key immunomodulatory elements and their role in pathogenesis. [DOI] [PubMed] [Google Scholar]
- 4.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 6.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. 2013;87:2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolff T, O’Neill RE, Palese P. NS1-Binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 11.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 12.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinto LH, Lamb RA. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst. 2007;3:18–23. doi: 10.1039/b611613m. [DOI] [PubMed] [Google Scholar]
- 14.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 15.Talon J, Horvath CM, Polley R, Basler CF, Muster T, et al. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, et al. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 17.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Li M, Zheng H, Muster T, Palese P, et al. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig S, Wang X, Ehrhardt C, Zheng H, Donelan N, et al. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 22.Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 23.Talon J, Salvatore M, O’Neill RE, Nakaya Y, Zheng H, et al. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc Natl Acad Sci USA. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, et al. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. This article describes the original basis for codon-deoptimization as it was applied to poliovirus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nogales A, Baker SF, Ortiz-Riano E, Dewhurst S, Topham DJ, et al. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J Virol. 2014;88:10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005;1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Goffic R, Leymarie O, Chevalier C, Rebours E, Da Costa B, et al. Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. PLoS Pathog. 2011;7:e1002202. doi: 10.1371/journal.ppat.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, et al. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 2010;6:e1001014. doi: 10.1371/journal.ppat.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desmet EA, Bussey KA, Stone R, Takimoto T. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol. 2013;87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Liu X. Crucial role of PA in virus life cycle and host adaptation of influenza A virus. Med Microbiol Immunol. 2015;204:137–149. doi: 10.1007/s00430-014-0349-y. [DOI] [PubMed] [Google Scholar]
- 32.Hu J, Mo Y, Wang X, Gu M, Hu Z, et al. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol. 2015;89:4126–4142. doi: 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiou HE, Liu CL, Buttrey MJ, Kuo HP, Liu HW, et al. Adverse effects of ribavirin and outcome in severe acute respiratory syndrome: experience in two medical centers. Chest. 2005;128:263–272. doi: 10.1378/chest.128.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller MP, Dresser L, Raboud J, McGeer A, Rea E, et al. Adverse events associated with high-dose ribavirin: evidence from the Toronto outbreak of severe acute respiratory syndrome. Pharmacotherapy. 2007;27:494–503. doi: 10.1592/phco.27.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middle East respiratory syndrome coronavirus (MERS-CoV) World Health Organization (WHO); 2015. [Last accessed 21 June 2015]. Available at: http://www.who.int/mediacentre/factsheets/mers-cov/en/ [Google Scholar]
- 36.Roberts A, Lamirande EW, Vogel L, Jackson JP, Paddock CD, et al. Animal models and vaccines for SARS-CoV infection. Virus Res. 2008;133:20–32. doi: 10.1016/j.virusres.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel LN, Roberts A, Paddock CD, Genrich GL, Lamirande EW, et al. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV) Vaccine. 2007;25:2173–2179. doi: 10.1016/j.vaccine.2006.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts A, Deming D, Paddock CD, Cheng A, Yount B, et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agnihothram S, Yount BL, Jr, Donaldson EF, Huynh J, Menachery VD, et al. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. MBio. 2014;5:e00047–00014. doi: 10.1128/mBio.00047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enjuanes L, Dediego ML, Alvarez E, Deming D, Sheahan T, et al. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. 2008;133:45–62. doi: 10.1016/j.virusres.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knoops K, Barcena M, Limpens RW, Koster AJ, Mommaas AM, et al. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beachboard DC, Anderson-Daniels JM, Denison MR. Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications. J Virol. 2015;89:2080–2089. doi: 10.1128/JVI.02776-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013;4 doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith EC, Blanc H, Surdel MC, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith EC, Denison MR. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr Opin Virol. 2012;2:519–524. doi: 10.1016/j.coviro.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Haan CA, Masters PS, Shen X, Weiss S, Rottier PJ. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Regla-Nava JA, Castano-Rodriguez C, et al. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. This article provides a great review of the known immunomodulatory genes of coronaviruses with a focus on targeting E in a vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Regla-Nava JA, Alvarez E, et al. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7:e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieto-Torres JL, Dediego ML, Alvarez E, Jimenez-Guardeno JM, Regla-Nava JA, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu DX, Inglis SC. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin XJ, Liu DX, Wang HC, Zhou Y. A study on the mutagenicity of 102 raw pharmaceuticals used in Chinese traditional medicine. Mutat Res. 1991;260:73–82. [PubMed] [Google Scholar]
- 55.Westerbeck JW, Machamer CE. A Coronavirus E Protein is Present in Two Distinct Pools with Different Effects on Assembly and the Secretory Pathway. J Virol. 2015 doi: 10.1128/JVI.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeDiego ML, Alvarez E, Almazan F, Rejas MT, Lamirande E, et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dediego ML, Pewe L, Alvarez E, Rejas MT, Perlman S, et al. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almazan F, DeDiego ML, Sola I, Zuniga S, Nieto-Torres JL, et al. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4:e00650–00613. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ortego J, Escors D, Laude H, Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts A, Vogel L, Guarner J, Hayes N, Murphy B, et al. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J Virol. 2005;79:503–511. doi: 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, et al. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med. 2012;18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009;360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamal-Bahl S, Doshi J, Campbell J. Economic analyses of respiratory syncytial virus immunoprophylaxis in high-risk infants: a systematic review. Arch Pediatr Adolesc Med. 2002;156:1034–1041. doi: 10.1001/archpedi.156.10.1034. [DOI] [PubMed] [Google Scholar]
- 66.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 67.Kim HW, Leikin SL, Arrobio J, Brandt CD, Chanock RM, et al. Cell-mediated immunity to respiratory syncytial virus induced by inactivated vaccine or by infection. Pediatr Res. 1976;10:75–78. doi: 10.1203/00006450-197601000-00015. [DOI] [PubMed] [Google Scholar]
- 68.Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011;162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, et al. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, et al. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology. 2012;434:129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bitko V, Shulyayeva O, Mazumder B, Musiyenko A, Ramaswamy M, et al. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-kappaB-dependent, interferon-independent mechanism and facilitate virus growth. J Virol. 2007;81:1786–1795. doi: 10.1128/JVI.01420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin H, Cheng X, Traina-Dorge VL, Park HJ, Zhou H, et al. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine. 2003;21:3647–3652. doi: 10.1016/s0264-410x(03)00426-2. [DOI] [PubMed] [Google Scholar]
- 73.Lo MS, Brazas RM, Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol. 2005;79:9315–9319. doi: 10.1128/JVI.79.14.9315-9319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected] J Virol. 2004;78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swedan S, Musiyenko A, Barik S. Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J Virol. 2009;83:9682–9693. doi: 10.1128/JVI.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, et al. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin H, Zhou H, Cheng X, Tang R, Munoz M, et al. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology. 2000;273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- 78.Meng J, Lee S, Hotard AL, Moore ML. Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. MBio. 2014;5:e01704–01714. doi: 10.1128/mBio.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Collins PL, Mottet G. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J Gen Virol. 1993;74(Pt 7):1445–1450. doi: 10.1099/0022-1317-74-7-1445. [DOI] [PubMed] [Google Scholar]
- 80.Perez M, Garcia-Barreno B, Melero JA, Carrasco L, Guinea R. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology. 1997;235:342–351. doi: 10.1006/viro.1997.8696. [DOI] [PubMed] [Google Scholar]
- 81.Carter SD, Dent KC, Atkins E, Foster TL, Verow M, et al. Direct visualization of the small hydrophobic protein of human respiratory syncytial virus reveals the structural basis for membrane permeability. FEBS Lett. 2010;584:2786–2790. doi: 10.1016/j.febslet.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the respiratory syncytial virus small hydrophobic protein. J Virol. 2007;81:8361–8366. doi: 10.1128/JVI.02717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bukreyev A, Whitehead SS, Murphy BR, Collins PL. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, et al. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis. 2005;191:1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- 86.Levine S, Klaiber-Franco R, Paradiso PR. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68(Pt 9):2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 87.Feldman SA, Hendry RM, Beeler JA. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol. 1999;73:6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olmsted RA, Elango N, Prince GA, Murphy BR, Johnson PR, et al. Expression of the F glycoprotein of respiratory syncytial virus by a recombinant vaccinia virus: comparison of the individual contributions of the F and G glycoproteins to host immunity. Proc Natl Acad Sci USA. 1986;83:7462–7466. doi: 10.1073/pnas.83.19.7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bukreyev A, Yang L, Fricke J, Cheng L, Ward JM, et al. The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J Virol. 2008;82:12191–12204. doi: 10.1128/JVI.01604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tripp RA, Jones LP, Haynes LM, Zheng H, Murphy PM, et al. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat Immunol. 2001;2:732–738. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- 91.Polack FP, Irusta PM, Hoffman SJ, Schiatti MP, Melendi GA, et al. The cysteine-rich region of respiratory syncytial virus attachment protein inhibits innate immunity elicited by the virus and endotoxin. Proc Natl Acad Sci USA. 2005;102:8996–9001. doi: 10.1073/pnas.0409478102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plattet P, Plemper RK. Envelope protein dynamics in paramyxovirus entry. MBio. 2013;4 doi: 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McLellan JS, Ray WC, Peeples ME. Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol. 2013;372:83–104. doi: 10.1007/978-3-642-38919-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Teng MN, Whitehead SS, Collins PL. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology. 2001;289:283–296. doi: 10.1006/viro.2001.1138. [DOI] [PubMed] [Google Scholar]
- 95.Techaarpornkul S, Collins PL, Peeples ME. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology. 2002;294:296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- 96.Tang RS, Spaete RR, Thompson MW, MacPhail M, Guzzetta JM, et al. Development of a PIV-vectored RSV vaccine: preclinical evaluation of safety, toxicity, and enhanced disease and initial clinical testing in healthy adults. Vaccine. 2008;26:6373–6382. doi: 10.1016/j.vaccine.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 97.Schmidt AC, Wenzke DR, McAuliffe JM, St Claire M, Elkins WR, et al. Mucosal immunization of rhesus monkeys against respiratory syncytial virus subgroups A and B and human parainfluenza virus type 3 by using a live cDNA-derived vaccine based on a host range-attenuated bovine parainfluenza virus type 3 vector backbone. J Virol. 2002;76:1089–1099. doi: 10.1128/JVI.76.3.1089-1099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones B, Zhan X, Mishin V, Slobod KS, Surman S, et al. Human PIV-2 recombinant Sendai virus (rSeV) elicits durable immunity and combines with two additional rSeVs to protect against hPIV-1, hPIV-2, hPIV-3, and RSV. Vaccine. 2009;27:1848–1857. doi: 10.1016/j.vaccine.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Falsey AR, Walsh EE, Capellan J, Gravenstein S, Zambon M, et al. Comparison of the safety and immunogenicity of 2 respiratory syncytial virus (rsv) vaccines--nonadjuvanted vaccine or vaccine adjuvanted with alum--given concomitantly with influenza vaccine to high-risk elderly individuals. J Infect Dis. 2008;198:1317–1326. doi: 10.1086/592168. [DOI] [PubMed] [Google Scholar]
- 100.Power UF, Plotnicky H, Blaecke A, Nguyen TN. The immunogenicity, protective efficacy and safety of BBG2Na, a subunit respiratory syncytial virus (RSV) vaccine candidate, against RSV-B. Vaccine. 2003;22:168–176. doi: 10.1016/s0264-410x(03)00570-x. [DOI] [PubMed] [Google Scholar]
- 101.Plotnicky H, Siegrist CA, Aubry JP, Bonnefoy JY, Corvaia N, et al. Enhanced pulmonary immunopathology following neonatal priming with formalin-inactivated respiratory syncytial virus but not with the BBG2NA vaccine candidate. Vaccine. 2003;21:2651–2660. doi: 10.1016/s0264-410x(03)00055-0. [DOI] [PubMed] [Google Scholar]
- 102.Roush SW, Murphy TV Vaccine-Preventable Disease Table Working G. Historical comparisons of morbidity and mortality for vaccine-preventable diseases in the United States. JAMA. 2007;298:2155–2163. doi: 10.1001/jama.298.18.2155. [DOI] [PubMed] [Google Scholar]
- 103.Katz SL. John F. Enders and measles virus vaccine--a reminiscence. Curr Top Microbiol Immunol. 2009;329:3–11. doi: 10.1007/978-3-540-70523-9_1. [DOI] [PubMed] [Google Scholar]
- 104.Buynak EB, Hilleman MR. Live attenuated mumps virus vaccine. 1. Vaccine development. Proc Soc Exp Biol Med. 1966;123:768–775. doi: 10.3181/00379727-123-31599. [DOI] [PubMed] [Google Scholar]
- 105.Betakova T, Svetlikova D, Gocnik M. Overview of measles and mumps vaccine: origin, present, and future of vaccine production. Acta Virol. 2013;57:91–96. doi: 10.4149/av_2013_02_91. [DOI] [PubMed] [Google Scholar]
- 106.Haralambieva IH, Ovsyannikova IG, O’Byrne M, Pankratz VS, Jacobson RM, et al. A large observational study to concurrently assess persistence of measles specific B-cell and T-cell immunity in individuals following two doses of MMR vaccine. Vaccine. 2011;29:4485–4491. doi: 10.1016/j.vaccine.2011.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haralambieva IH, Ovsyannikova IG, Pankratz VS, Kennedy RB, Jacobson RM, et al. The genetic basis for interindividual immune response variation to measles vaccine: new understanding and new vaccine approaches. Expert Rev Vaccines. 2013;12:57–70. doi: 10.1586/erv.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Poland GA, Jacobson RM, Thampy AM, Colbourne SA, Wollan PC, et al. Measles reimmunization in children seronegative after initial immunization. JAMA. 1997;277:1156–1158. [PubMed] [Google Scholar]
- 109.Pannuti CS, Morello RJ, Moraes JC, Curti SP, Afonso AM, et al. Identification of primary and secondary measles vaccine failures by measurement of immunoglobulin G avidity in measles cases during the 1997 Sao Paulo epidemic. Clin Diagn Lab Immunol. 2004;11:119–122. doi: 10.1128/CDLI.11.1.119-122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pagano JS, Levine RH, Sugg WC, Finger JA. Clinical trial of new attenuated mumps virus vaccine (Jeryl Lynn strain): preliminary report. Prog Immunobiol Stand. 1967;3:196–202. [PubMed] [Google Scholar]
- 111.Weibel RE, Stokes J, Jr, Buynak EB, Leagus MB, Hilleman MR. Jeryl Lynn strain live attenuated mumps virus vaccine. Durability of immunity following administration. JAMA. 1968;203:14–18. [PubMed] [Google Scholar]
- 112.Young ML, Dickstein B, Weibel RE, Stokes J, Jr, Buynak EB, et al. Experiences with Jeryl Lynn strain live attenuated mumps virus vaccine in a pediatric outpatient clinic. Pediatrics. 1967;40:798–803. [PubMed] [Google Scholar]
- 113.Marin M, Quinlisk P, Shimabukuro T, Sawhney C, Brown C, et al. Mumps vaccination coverage and vaccine effectiveness in a large outbreak among college students--Iowa, 2006. Vaccine. 2008;26:3601–3607. doi: 10.1016/j.vaccine.2008.04.075. [DOI] [PubMed] [Google Scholar]
- 114.Barskey AE, Glasser JW, LeBaron CW. Mumps resurgences in the United States: A historical perspective on unexpected elements. Vaccine. 2009;27:6186–6195. doi: 10.1016/j.vaccine.2009.06.109. [DOI] [PubMed] [Google Scholar]
- 115.Poland GA, Jacobson RM. The re-emergence of measles in developed countries: time to develop the next-generation measles vaccines? Vaccine. 2012;30:103–104. doi: 10.1016/j.vaccine.2011.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smits G, Mollema L, Hahne S, de Melker H, Tcherniaeva I, et al. Seroprevalence of mumps in The Netherlands: dynamics over a decade with high vaccination coverage and recent outbreaks. PLoS One. 2013;8:e58234. doi: 10.1371/journal.pone.0058234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mahamud A, Fiebelkorn AP, Nelson G, Aguon A, McKenna J, et al. Economic impact of the 2009–2010 Guam mumps outbreak on the public health sector and affected families. Vaccine. 2012;30:6444–6448. doi: 10.1016/j.vaccine.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 118.Rima BK, Duprex WP. New concepts in measles virus replication: getting in and out in vivo and modulating the host cell environment. Virus Res. 2011;162:47–62. doi: 10.1016/j.virusres.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 119.Sato H, Masuda M, Kanai M, Tsukiyama-Kohara K, Yoneda M, et al. Measles virus N protein inhibits host translation by binding to eIF3-p40. J Virol. 2007;81:11569–11576. doi: 10.1128/JVI.00570-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bhaskar A, Bala J, Varshney A, Yadava P. Expression of measles virus nucleoprotein induces apoptosis and modulates diverse functional proteins in cultured mammalian cells. PLoS One. 2011;6:e18765. doi: 10.1371/journal.pone.0018765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121*.de Vries RD, Stittelaar KJ, Osterhaus AD, de Swart RL. Measles vaccination: new strategies and formulations. Expert Rev Vaccines. 2008;7:1215–1223. doi: 10.1586/14760584.7.8.1215. This article offers a great review of current measles vaccination strategies and next generation improvements on vaccine formulations. [DOI] [PubMed] [Google Scholar]
- 122.Xu P, Chen Z, Phan S, Pickar A, He B. Immunogenicity of novel mumps vaccine candidates generated by genetic modification. J Virol. 2014;88:2600–2610. doi: 10.1128/JVI.02778-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu P, Luthra P, Li Z, Fuentes S, D’Andrea JA, et al. The V protein of mumps virus plays a critical role in pathogenesis. J Virol. 2012;86:1768–1776. doi: 10.1128/JVI.06019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu P, Li Z, Sun D, Lin Y, Wu J, et al. Rescue of wild-type mumps virus from a strain associated with recent outbreaks helps to define the role of the SH ORF in the pathogenesis of mumps virus. Virology. 2011;417:126–136. doi: 10.1016/j.virol.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cheng BY, Ortiz-Riano E, Nogales A, de la Torre JC, Martinez-Sobrido L. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J Virol. 2015;89:3523–3533. doi: 10.1128/JVI.03401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ni YY, Zhao Z, Opriessnig T, Subramaniam S, Zhou L, et al. Computer-aided codon-pairs deoptimization of the major envelope GP5 gene attenuates porcine reproductive and respiratory syndrome virus. Virology. 2014;450–451:132–139. doi: 10.1016/j.virol.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 127.Aragones L, Guix S, Ribes E, Bosch A, Pinto RM. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog. 2010;6:e1000797. doi: 10.1371/journal.ppat.1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]