Significance
Although two-component systems are a ubiquitous means of rapid bacterial adaptation to changing environments, identification of the specific signals detected by sensor kinases can be challenging. Also, little is known about the diverse, poorly characterized family of sensor kinases that detect intramembrane signals. We show that the major type IV pilin, PilA, is an inhibitory intramembrane ligand for the PilS sensor kinase that controls pilA expression and we characterize the mechanism of signal transduction. Because the conserved N-terminal domain of PilA alone can repress pilA expression, peptides corresponding to this short region could have potential as therapeutic agents to suppress type IV Pili (T4P) biogenesis.
Keywords: type IV pili, two-component system, transcriptional regulation, Pseudomonas aeruginosa, Geobacter sulfurreducens
Abstract
Type IV pili are important virulence factors for many pathogens, including Pseudomonas aeruginosa. Transcription of the major pilin gene—pilA—is controlled by the PilS-PilR two-component system in response to unknown signals. The absence of a periplasmic sensing domain suggested that PilS may sense an intramembrane signal, possibly PilA. We suggest that direct interactions between PilA and PilS in the inner membrane reduce pilA transcription when PilA levels are high. Overexpression in trans of PilA proteins with diverse and/or truncated C termini decreased native pilA transcription, suggesting that the highly conserved N terminus of PilA was the regulatory signal. Point mutations in PilA or PilS that disrupted their interaction prevented autoregulation of pilA transcription. A subset of PilA point mutants retained the ability to interact with PilS but could no longer decrease pilA transcription, suggesting that interaction between the pilin and sensor kinase is necessary but not sufficient for pilA autoregulation. Furthermore, PilS’s phosphatase motif was required for the autoregulation of pilA transcription, suggesting that under conditions where PilA is abundant, the PilA–PilS interaction promotes PilR dephosphorylation and thus down-regulation of further pilA transcription. These data reveal a clever bacterial inventory control strategy in which the major subunit of an important P. aeruginosa virulence factor controls its own expression.
A wide variety of bacteria, including the opportunistic pathogen Pseudomonas aeruginosa, use type IV Pili (T4P) for attachment to surfaces and host tissues, biofilm formation, DNA uptake, and twitching motility (1–4). T4P are retractile surface appendages comprised predominantly of thousands of subunits of the major pilin protein, PilA (5), which is rapidly polymerized and depolymerized by a complex assembly machine. Also incorporated into T4P are small amounts of the minor pilins FimU, PilV, PilW, PilX, and PilE (P. aeruginosa nomenclature), which prime pilus assembly (6, 7).
PilA and the minor pilins share a similar lollipop-like topology, with a highly conserved, hydrophobic N-terminal α-helix packed against a variable C-terminal antiparallel β-sheet (8, 9). The first ∼24 residues of mature pilins anchor the subunits in the inner membrane until they are polymerized by the assembly machinery (10). When pili are retracted, subunits are disassembled from the base of the pilus and returned to the inner membrane for reuse in subsequent cycles of extension. In P. aeruginosa, high levels of PilA in the inner membrane lead to decreased pilA transcription, whereas depletion of PilA inner membrane pools significantly elevates pilA transcription (11, 12). These data suggest that pilA expression is responsive to levels of intracellular PilA, although the sensory mechanism is unknown.
pilA transcription in P. aeruginosa and many other species is dependent on the PilS-PilR two-component regulatory system (13–16). Two-component systems (TCSs) allow bacteria to rapidly detect and adapt to changes in their environment (17). The sensor kinase (SK) detects physical or chemical signals, typically via an extracytoplasmic domain flanked by two transmembrane (TM) segments (17). On detection of an activating signal, the protein undergoes autophosphorylation on a conserved His residue in the cytoplasmic kinase domain. The phosphate is transferred to a cytoplasmic response regulator, which regulates gene expression in response to the stimulus (18).
In the PilS-PilR TCS, PilR is the cytoplasmic response regulator that activates pilA transcription (19). Its cognate SK PilS is atypical, with six TM segments connected by very short loops and no obvious external signal input domain (20). When PilS is absent, pilA transcription is significantly reduced, whereas loss of PilR abrogates pilA transcription (13). Interestingly, overexpression of full-length PilS also decreases pilin expression, whereas expression of its cytoplasmic kinase domain alone leads to constitutive pilA transcription, a common result of decoupling sensor kinases from their signal input domains (19, 21, 22).
Recently, a conserved E/DxxN/T phosphatase motif adjacent to the phosphorylated His was identified in the HisKA family of SKs to which PilS belongs, indicating that many SKs can have dual kinase and phosphatase activities to fine tune regulation (23). PilS has a canonical ExxN motif at position 320–323, beside the H319 phosphorylation site. Coupled with the observation that PilS overexpression reduces pilA transcription, the presence of this motif suggests that PilS could have intrinsic phosphatase activity on phospho-PilR.
Among the most significant challenges in the TCS field is identification of the specific signal(s) to which SKs respond. The observation that pilA transcription is inversely correlated with levels of PilA in the inner membrane, coupled with the unusual six TM topology of PilS, suggested that it recognizes an intramembrane signal, possibly PilA itself. Here, we show that direct intramembrane interactions between the conserved, hydrophobic N terminus of PilA and one or more TM of PilS down-regulate pilA transcription, and define the sequence elements on PilA involved in interaction and regulation. Based on analysis of PilS mutants, we propose that PilA–PilS interactions likely maintain PilS in a phosphatase state when membrane pools of PilA are high, providing a sensitive feedback mechanism for pilin inventory control.
Results
Overexpression of Heterologous Pilins Reduces PilA Levels by Decreasing pilA Transcription.
P. aeruginosa strains carry one of five pilA alleles (groups I–V) (24), encoding pilins that vary in size and sequence of the C-terminal domain (9), plus the presence or absence of posttranslational modifications (25, 26) and accessory proteins (25, 27). However, the first ∼24 amino acids of mature pilins are highly conserved (Fig. S1), whereas the C-terminal domains are diverse (24). In contrast, PilS in P. aeruginosa strains is invariant (28), regardless of the strain’s pilin type. These data—plus PilS’s lack of an extramembranous signal recognition domain—suggested that the highly conserved hydrophobic N terminus of PilA could be the PilS ligand. To test whether diverse pilins could suppress PilA expression in a heterologous strain, V5-epitope tagged pilins of groups I, III, IV, and V were overexpressed in strain PAK (group II). Intracellular levels of native PAK PilA were monitored using PAK PilA-specific antisera. Overexpression of each of the heterologous pilins from an inducible vector reduced native PilA levels by >50% (Fig. 1A).
Fig. S1.
Major and minor pilins have conserved N termini. An amino acid sequence alignment of residues 1–24 of the mature major and minor pilins overexpressed in Fig. 1 shows high sequence identity with the native PAK PilA sequence. Heterologous PilAs from other sources are near identical in this domain, whereas minor pilins are more divergent. *Identical residue between the sequence and that of PAK PilA in a given position.
Fig. 1.
Overexpression of heterologous pilins reduces native PilA levels. (A) V5 epitope tagged major pilins from PA1244 (group I), PA14 (group III), PA5196 (group IV), PA1457 (group V), and the G. sulfurreducens PilA were overexpressed in the PAK (group II) strain. Heterologous PilAs overexpressed to similar extents (as demonstrated by α-V5 Western blot) reduced native PilA levels. (B) In the absence of the TM segments of PilS (pilScyt), overexpression of group III PilA no longer reduces native pilin levels. (C) Of the minor pilins—FimU, PilV, and PilE—only PilE caused a reduction in native pilin levels. For all blots, the flagellin band was used as a loading control. Numbers represent relative expression of native PilA in recombinant strains compared the empty vector control, as measured by densitometry and a one-way ANOVA statistical test (n = 3). (D) PilA and minor pilins FimU and PilE were overexpressed in PAK+pMS402 ppilA, and relative luminescence as a function of pilA promoter activity was recorded. Mean and SEM of three independent experiments are shown.
Geobacter sulfurreducens (GS), which regulates pilA transcription using a similar PilSR TCS (29), has unusually short type IV pilins that retain the characteristic conserved hydrophobic N-terminal α-helix, but lack a globular C-terminal domain. This architecture is thought to promote efficient electron transfer through pilus “nanowires” (30). The first 24 residues of the mature GS pilin are 92% identical to those of P. aeruginosa PilA (Fig. S1). When the GS pilin was overexpressed in P. aeruginosa, native pilin expression was reduced to the same extent as with heterologous P. aeruginosa pilins (Fig. 1A), showing that the N-terminal segment of a pilin is sufficient for repression of PilA expression. This autoregulatory phenotype is dependent on the TM segments of PilS. The chromosomal copy of pilS was replaced with a version encoding an N-terminal truncation lacking the TM segments: residues 3–176 (PilScyt). Although this form of PilS supported near-WT levels of PilA expression, overexpression of the heterologous group III PilA in this background failed to reduce native PilA levels (Fig. 1B).
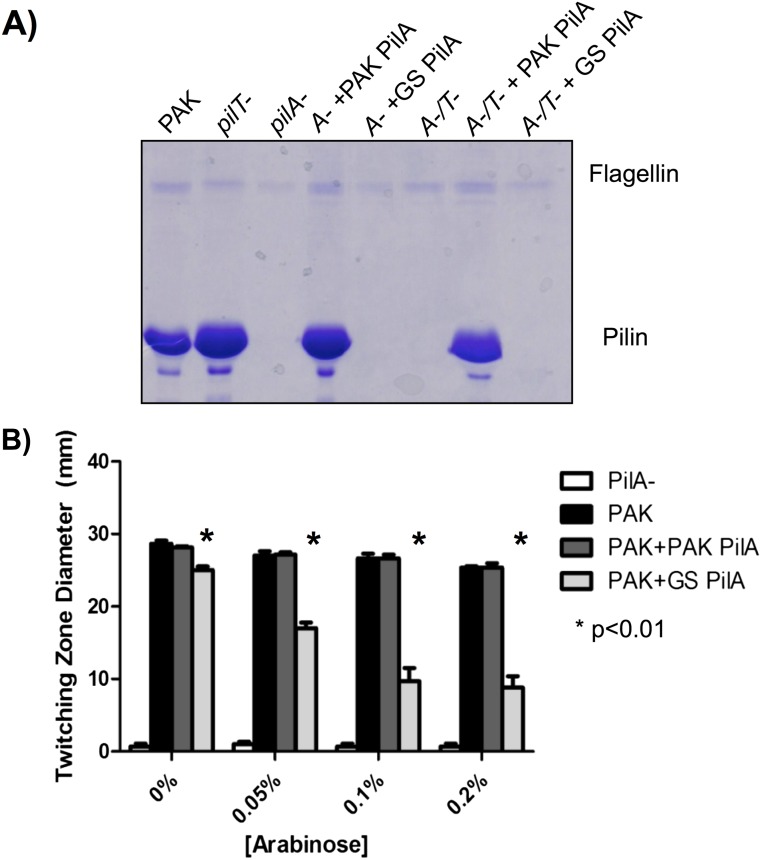
Although overexpression of heterologous pilins in the presence of WT PilS reduced native PilA levels, T4P function was unaffected if the heterologous subunits were competent for assembly. GS can assemble P. aeruginosa pilins (31), but the reverse was not true, even when pilus retraction was blocked to maximize the capture of assembled pili (Fig. S2A). When the assembly-incompetent GS pilins were expressed from an inducible vector in P. aeruginosa, twitching motility decreased in an inducer concentration-dependent manner (Fig. S2B).
Fig. S2.
Overexpression of nonfunctional pilins can impair normal T4P function. (A) SDS/PAGE of a sheared surface protein preparation examining surface piliation after complementation of a PAK pilA deletion with either the native or G. sulfurreducens (GS) pilA in both a retraction-proficient and -deficient (pilT−) background. G. sulfurreducens PilA cannot be assembled, even under retraction-deficient conditions, whereas complementation with PAK PilA can fully restore surface piliation in both the presence and absence of normal pilus retraction. (B) Results of a twitching stab assay of PAK strains overexpressing either PAK PilA or GS PilA at increasing levels of arabinose induction. Overexpression of the functional PAK pilin does not affect twitching motility, regardless of the level of induction. However, overexpression of a pilin that cannot be assembled, like GS PilA, can impair twitching motility at higher concentrations.
To address whether overexpression of other pilin-like proteins with divergent N termini could impact PilA levels, each of the minor pilins—FimU, PilV, PilW, PilX, or PilE—was overexpressed in strain PAK. However, the levels of PilW and PilX were lower than those of other minor pilins and they were therefore excluded. Of the three remaining minor pilins, only PilE reduced native PilA levels, to ∼60% of the vector control. Neither FimU nor PilV had an effect on PilA levels when overexpressed (Fig. 1C). Thus, specific sequences in the pilin N terminus are required for pilA regulation.
To determine if PilA autoregulation occurred at the level of transcription, the pilA promoter was cloned upstream of a luciferase (lux) reporter (pMS402-ppilA). pilA promoter activity was monitored in PAK carrying the pBADGr vector or pBADGr-pilA, pBADGr-pilE—both of which reduced native pilin levels when overexpressed—or pBADGr-fimU, which had no effect on PilA levels (Fig. 1D). In agreement with the Western blot data in Fig. 1C, overexpression of FimU had no impact on pilA transcription. However, pilA transcription was significantly decreased in both PilA and PilE overexpression strains (Fig. 1D). Due to the inherent leakiness of the pBADGr promoter, differences in pilA transcription as a result of protein overexpression can be seen even at t = 0, when expression from pBADGr is first induced with arabinose. Together, these data suggest that the conserved N termini of PilA and PilE, but not FimU, contain the appropriate sequence information needed for down-regulation of pilA transcription.
PilA and PilS Interact Directly in the Inner Membrane.
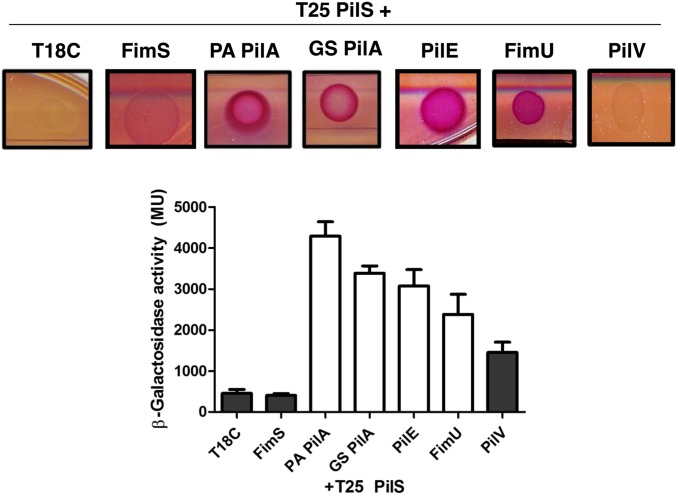
We next tested our hypothesis that direct PilA–PilS interaction led to decreased pilA transcription, using a bacterial adenylate cyclase two-hybrid (BACTH) assay (32). T18-pilin fusions of PAK PilA, G. sulfurreducens PilA, and minor pilins PilE, FimU, and PilV were coexpressed with T25-PilS in Escherichia coli BTH 101, and potential interactions were detected by monitoring β-galactosidase activity. PilA from P. aeruginosa and G. sulfurreducens, as well as the minor pilin, PilE, interacted with PilS (Fig. 2; P < 0.01), correlating with their ability to reduce pilA transcription when overexpressed in P. aeruginosa. Unexpectedly, FimU, which had no effect on pilA transcription, interacted with PilS, suggesting that interaction and regulation are separable phenotypes. Based on the results of a one-way ANOVA analysis (P > 0.05) and a negative result in the McConkey plate assay, PilV did not interact with PilS (Fig. 2, gray bars).
Fig. 2.
PilA and select minor pilins interact with PilS. T25-PilS interacts with PilA from both P. aeruginosa and G. sulfurreducens, PilE, and to a lesser extent, FimU, each tagged with the T18 domain of adenylate cyclase. PilV did not interact with PilS in either the McConkey agar plate assay or the β-galactosidase activity assay (gray bars). β-Galactosidase activity resulting from protein–protein interactions was measured by ONPG-hydrolysis as described in Materials and Methods. FimS was used as a membrane-bound negative control for PilS interactions.
E5 and P22 of PilA Are Important for Interaction and Regulation.
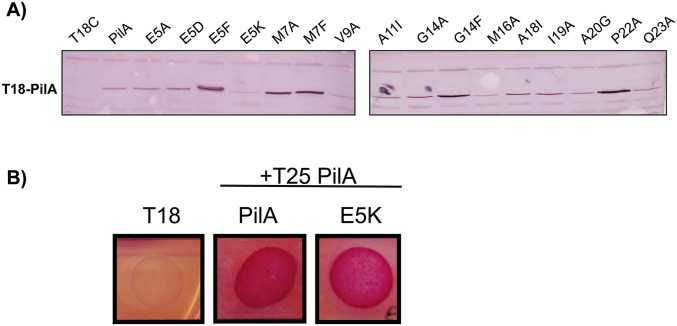
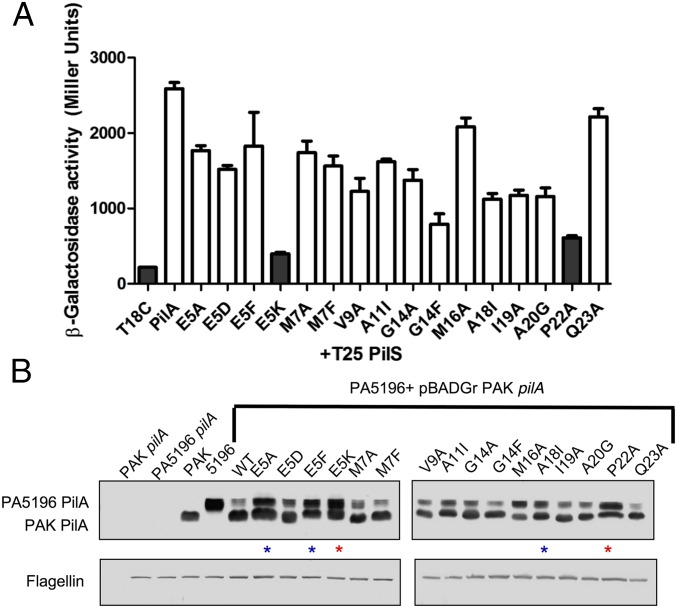
The ability of PilA, PilE, and FimU to interact with PilS, even though FimU did not repress pilA transcription, implied that specific pilin N-terminal residues were important for discrimination of the regulatory signal. To dissect the residues required for both the PilA–PilS interaction and subsequent regulation of pilA transcription, a series of point mutations was generated in the PAK T18-PilA fusion and the inducible pBADGr-pilA construct, respectively. We targeted residues E5, A20, and P22, important for a number of T4P-related functions (8, 15, 33–36), plus additional residues conserved between PilA and PilE, but not FimU (Fig. S1), suggesting that they could be regulatory positions. In general, the native residue was substituted to Ala, but E5A, E5D, E5F, and E5K substitutions were also made to establish which R-group characteristics were important at this position. The stability of each mutant T18-PilA fusion was verified by α-PilA Western blot (Fig. S3). Although its expression is slightly decreased compared with the other T18-PilA fusions, T18-PilA E5K is present in high enough abundance to support interaction with WT T25-PilA (Fig. S3B). Only substitutions at E5 or P22 disrupted the PilA–PilS interaction in the BACTH assay (Fig. 3A). Interestingly, only PilE and FimU have a P22 residue, supporting a role for a kinked N-terminal helix in pilin–PilS interactions. A positive charge at position E5 was nonpermissive, as E5K, but not E5A, E5F, or E5D, abolished the PilA–PilS interaction (Fig. 3A).
Fig. S3.
T18-PilA mutant fusions are stable. (A) α-PAK PilA (1:7,500 dilution) Western blot was performed to determine the stability of all T18-PilA point mutants generated in E. coli DH5α. Although there is some difference in protein levels, each of the point mutants are present and lower abundance mutants do not necessarily affect β-galactosidase output, as mutations such as PilA M16A or Q23A are at low levels but interact robustly with PilS in the BACTH assay. (B) Despite being present at a lower abundance than most other T18-PilA variants, PilA E5K is still present in high enough abundance to reveal interactions in the BACTH assay where they are present.
Fig. 3.
PilA residues E5 and P22 are important for the PilA–PilS interaction and PilA autoregulation. Point mutations in the conserved N terminus of PAK PilA were generated using site-directed mutagenesis. (A) T18-PilA fusions coexpressed with T25-PilS in the β-galactosidase liquid assay were tested for interaction. PilA E5K and P22A fail to interact with PilS (gray bars, P < 0.01). (B) The same set of PAK PilA point mutants were overexpressed in strain PA5196 (group IV), and both PAK mutant and native PA5196 PilA levels were detected by Western blot with α-PAK and α-PA5196 PilA-specific antibodies. Overexpression of PAK WT and most mutant PilA derivatives decreased PA5196 PilA levels, whereas E5A, E5F, and M16A interacted with PilS, but their overexpression resulted in near-WT levels of native PilA (blue stars), and E5K and P22A transformants did not interact with PilS and also had near-WT levels of PA5196 PilA (red stars).
The effects of PilA point mutant overexpression were also tested directly in P. aeruginosa. To more easily differentiate between plasmid and chromosomally encoded pilins, PAK PilA point mutants were overexpressed in strain PA5196 (group IV). Because PA5196 pilins are glycosylated with d-arabinofuranose (25, 26), they are of larger mass than those of PAK, allowing the two to be readily separated on 18% SDS/PAGE. Coincubation of Western blots with non–cross-reactive α-PAK PilA and α-PA5196 PilA antibodies allowed for the simultaneous detection of both pilins. PAK PilA point mutants were all stably expressed in PA5196 (Fig. 3B, lower band). Overexpression of most PAK PilA derivatives reduced native pilin levels to the same extent as the WT pilin, whereas PAK PilA E5K and P22A failed to reduce PA5196 PilA levels (Fig. 3B, upper band), consistent with their inability to interact with PilS. Notably, PAK PilA E5A and E5F could interact with PilS (Fig. 3A) but had no effect on PA5196 PilA expression. The M16A and A18I point mutants had intermediate phenotypes, in that their overexpression reduced levels of PA5196 PilA, but to a lesser extent than the WT PAK pilin. Together, the data suggest that the PilA–PilS interaction is necessary but not sufficient for modulating PilA expression levels and that specific intermolecular contacts that depend on PilA N-terminal conformation (P22) and charge (E5) are required for proper signal transduction.
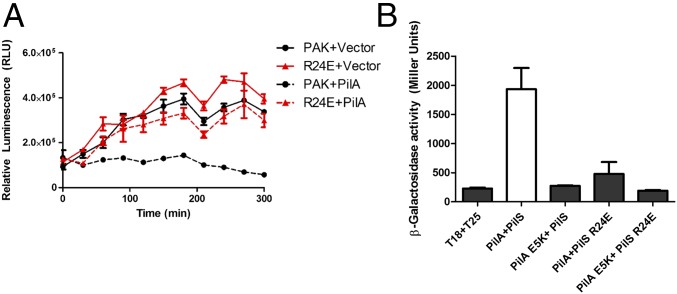
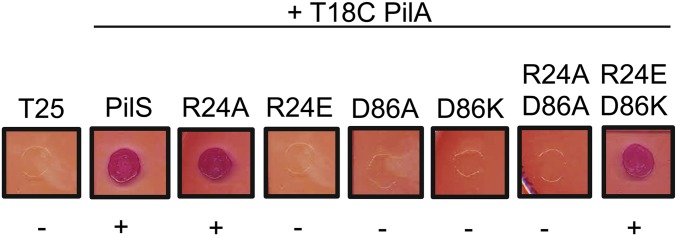
PilS and its homologs in other species contain a conserved, positively charged arginine residue in the first predicted TM (PilS R24 in P. aeruginosa) that we hypothesized might interact with PilA E5. R24 is not required for PilS activity, but an R24E substitution leads to loss of pilA autoregulation (Fig. 4A). As predicted, PilS R24E failed to interact with PilA (Fig. 4B). Interestingly, charge-swapped PilA E5K and PilS R24E variants also failed to interact (Fig. 4B), suggesting that the R24 has another role: potentially formation of a salt bridge with a second conserved charged residue in TM3 of PilS, D86. Charge alteration at either R24 or D86 disrupted the PilA–PilS interaction, but PilA interaction was restored with a PilS R24E D86K double charge-swapped mutant, suggesting that the charged TM residues control PilS conformation and thus its ability to interact with pilins (Fig. S4).
Fig. 4.
PilS R24E neither interacts with PilA E5K nor autoregulates pilA transcription in vivo. (A) The conserved R24 residue of PilS was substituted with a glutamic acid. When expressed from the chromosome, PilS R24E activated pilA transcription in the lux-pilA assay, but was no longer responsive to PilA overexpression. (B) In the T25-PilS fusion, the R24E PilS variant failed to interact with T18-WT PilA or T18-PilA E5K (gray bars).
Fig. S4.
PilS residues R24 and D86 are important for the PilA–PilS interaction. Individual charge swaps at positions R24 and D86 of PilS both disrupted the PilA–PilS interaction, which was restored in the BACTH plate assay when the mutations were combined.
A PilS Phosphatase Motif Is Required for PilA Autoregulation.
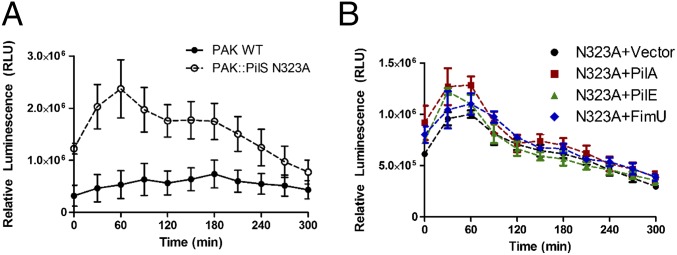
In earlier studies (21), PilS was predicted to have kinase and potentially phosphatase activities. We identified a canonical ExxN phosphatase motif (23) adjacent to the H319 residue that is the site of PilS phosphorylation and engineered a PAK chromosomal point mutant expressing PilS N323A, a substitution shown previously to disrupt SK phosphatase, but not kinase, activity (23). WT PAK and the PilS N323A mutant were each transformed with the pMS402-ppilA lux reporter to test the effect of this mutation on pilA transcription. The PilS N323A mutant had an approximately threefold increase in basal pilA transcription compared with WT (Fig. 5A). These data suggest that PilS N323A could lack phosphatase activity or have increased kinase activity.
Fig. 5.
A conserved PilS phosphatase motif is required for PilA autoregulation. A chromosomal PilS N323A substitution was made to disrupt the putative PilS phosphatase motif. (A) The PilS N323A mutant has significantly higher than WT levels of pilA transcription. (B) When PilA, PilE, or FimU are overexpressed in the PilS N3232A background, pilA promoter activity is comparable to the empty vector control. Error bars represent the SE calculated from three independent experiments.
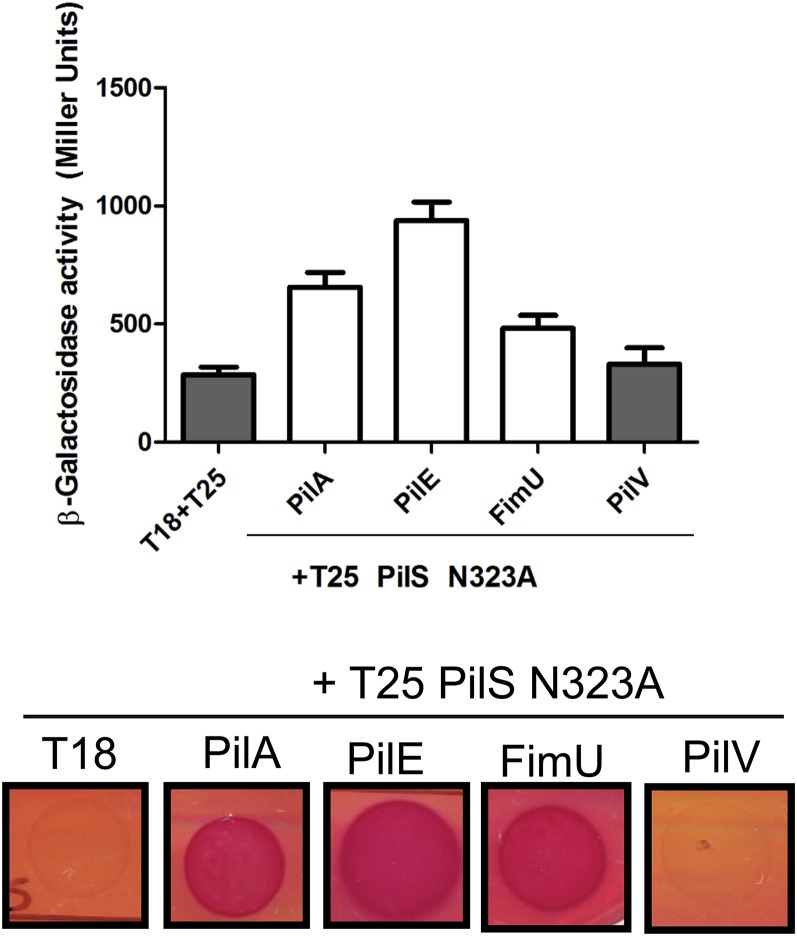
To distinguish between these possibilities, the PilS N323A mutant was cotransformed with pMS402-ppilA and empty pBADGr vector, or pBADGr-pilA, -pilE, or -fimU, and the ability of the pilins to modulate pilA transcription was tested. Unlike in the WT background (Fig. 2C), overexpression of PilA or PilE failed to reduce pilA transcription in the PilS N323A point mutant (Fig. 5B), even though the pilins interact with PilS N323A (Fig. S5). Together, these data suggest that PilA–PilS interactions likely down-regulate pilA transcription by promoting PilS phosphatase activity on PilR.
Fig. S5.
PilS N323A interacts with PilA, PilE, and FimU in the BACTH assay. Phosphatase-deficient PilS N323A was N-terminally tagged with T25 and assessed in the BACTH assay for its ability to interact with T18 major and minor pilin fusions. PilS N323A interacts with PilA, PilE, and FimU (white bars; P < 0.05), but not PilV, comparable to their interactions with WT PilS in Fig. 2.
Discussion
TCSs are widely used by bacteria for rapid adaptation to changes in their intra- and extracellular environments. However, the signals to which most SKs respond remain unknown (17). In this work, we provide evidence that PilA is a protein ligand for the SK, PilS, which controls pilin expression. We propose an autoregulatory model for the control of pilA transcription that is dependent on intrinsic PilS phosphatase activity (Fig. 6).
Fig. 6.
Model of pilA transcriptional autoregulation. (Left) When PilA levels in the inner membrane are high, PilA-PilS interactions occur more frequently and PilS dimers bound to PilA adopt a phosphatase-active conformation, deactivating PilR and thus reducing pilA transcription. (Right) When inner membrane pools of PilA are low, PilA–PilS interactions are less frequent. PilS remains in a kinase state and continues to up-regulate pilA transcription until PilA levels rise.
When levels of intracellular PilA are low, pilA transcription is significantly increased (11). In our model, depletion of PilA in the inner membrane leads to fewer PilA–PilS interactions. In the absence of such interactions, PilS adopts a kinase conformation and phosphorylates PilR, activating pilA transcription until intracellular PilA inventory increases. Conversely, when PilA levels are high, pilA transcription is dramatically reduced (11). Changes in PilA protein levels under the same conditions are more modest due to inherent pilin stability (11). It is also possible that PilS-PilR could impact PilA levels by regulating the expression of additional factors that indirectly impact pilA expression, contributing to the difference in magnitude between transcription and protein levels.
We propose a model where PilA–PilS interaction promotes a PilS phosphatase conformation, preventing further pilA transcription until intracellular pilin inventories become depleted. Although PilA interacts with PilS N323A (Fig. S5), the SK presumably no longer dephosphorylates PilR, and pilA transcription is elevated even when intracellular PilA levels are high. Similarly, our model explains why overexpression of only the cytoplasmic portion of PilS does not impair pilA transcription: lack of inhibitory signal input via interaction of PilA with the TM segments of PilS likely prevents propagation of the conformational changes that favor phosphatase activity. This hypothesis is supported by lack of reduction in native pilin levels when heterologous pilins are overexpressed in a strain that expresses a TM-less form of PilS, indicating that PilA autoregulation specifically requires the TM segments of PilS (Fig. 1B).
Although an activating signal for pilA transcription has not been identified, this work suggests that PilA is an inhibitory signal for its own transcription. The opposite paradigm was identified in Streptococcus bovis, where the kinase activity of the eight-TM SK BovK is partially controlled by its product, the lantibiotic HJ50, via interactions at the periplasm–inner membrane interface (37). Unlike PilA, HJ50 acts as an inducer of its own expression when it interacts with BovK, which, like PilS, lacks an extracytoplasmic signal detection domain (37).
Autoregulation in TCSs has also been demonstrated in some gram-negative systems. The most relevant example is the PhoPQ TCS of E. coli, which controls the transcription of a diverse set of genes in response to low magnesium (38–40), low pH, or exposure to antimicrobial peptides (38–40). MgrB is a small membrane peptide whose expression is positively regulated by PhoPQ. When MgrB is highly expressed, transcription of the entire PhoPQ regulon is decreased, whereas the regulon is up-regulated in mgrB mutants (41). MgrB interacts directly with PhoQ (41). Interestingly, autoregulation appears to be the only function of MgrB, whereas PilA has both autoregulatory and structural roles.
Intramembrane-signal sensing SKs have been most widely studied in gram-positive bacteria, where they often sense perturbations in the membrane itself (42, 43). Only a few gram-negative SKs that rely on intramembrane interactions for signal transduction have been identified and most require accessory proteins for signal transduction. For example, E. coli UhpB is a predicted eight-TM SK that modulates expression of a sugar phosphate transport protein, in response to extracellular glucose-6-phosphate (44). Unlike PilS, UhpB cannot detect its signal directly. Instead, UhpC, a single TM inner membrane protein, binds glucose-6-phosphate and interacts with UhpB in the inner membrane to drive downstream transcriptional activation (44). Thus, PilS represents one of the first gram-negative SKs that directly detect its ligand, PilA, in the inner membrane without the use of accessory proteins.
The N-terminal amino acid sequence of PilA is highly conserved, even among distantly related species (35, 45, 46), whereas the C-terminal domains of pilins, both major and minor, can be extremely divergent (24). Heterologous P. aeruginosa pilins, plus the naturally truncated pilin from G. sulfurreducens, interacted with PilS (Fig. 2) and reduced chromosomal PilA expression in P. aeruginosa (Fig. 1A), suggesting that the N terminus of PilA mediates both the interactions and their regulatory consequences. However, the pilin–PilS interaction alone is not sufficient for regulation, as specific point mutants of PilA (E5A or E5F) or minor pilins (FimU) interacted with PilS (Figs. 2 and 3) but failed to decrease PilA expression (Figs. 1C and 3). It is possible, although less likely, that these pilins interact with PilS at a different site than the WT pilin, leading to their inability to reduce pilA transcription. Although PilS–minor pilin interactions are probably physiologically irrelevant due to the low abundance of minor pilins in vivo (6, 47), the effects of their overexpression on pilA transcription gave insight into sequence specificity of PilS interaction and regulation. PilE and FimU each share 14 of 24 N-terminal residues with PilA, but the pattern of conservation is different (Fig. S1), implicating both overall similarity and the presence of specific residues in regulation. For example, P22 creates a kink in the N terminus and controls pilin angle in the membrane (10). The P22A mutation abolished PilA–PilS interactions and PilA autoregulation, and PilV, which lacks a proline at position 22, failed to interact stably with PilS. Very low levels of β-galactosidase activity in this sample could be indicative of a weak or transient interaction, although the plate-based assay supports lack of interaction (Fig. 2).
Also important for the PilA–PilS interaction and autoregulation of pilA transcription was the highly conserved E5 residue, present in PilA, PilE, and FimU—all of which interact with PilS—but also in PilV, which did not (Fig. 2). Thus, multiple contacts are important for the pilin–PilS interaction. Rare charged residues in TM segments are typically buried within multi-TM proteins or involved in protein–protein interactions (48, 49). PilA E5K or PilS R24E substitutions disrupted PilA–PilS interactions and dysregulated PilA expression, but the PilA E5K and PilS R24E charged-swapped pair failed to interact (Fig. 4B). Instead, interaction with WT PilA was restored when PilS R24E and D86K substitutions were combined (Fig. 4B), suggesting that these residues form a salt bridge that stabilizes a PilS conformation amenable to PilA binding.
Identification of the signals detected by SKs remains a significant challenge. Although we show here that PilA is an inhibitory signal for PilS, there may also be an activating signal, as hypothesized previously (15, 21). Recent studies showed that T4P and a number of other P. aeruginosa virulence factors are significantly up-regulated on surface contact, through increases in intracellular cAMP production (50, 51). Increased cAMP production leads to activation of pilSR expression by the cAMP-binding regulatory protein Vfr (52), which could account for increased pilA transcription on surface interaction. Alternatively, cAMP itself, or other molecules associated with the surface-responsive Chp chemotaxis system (50), could activate PilS and thus increase PilA levels.
Each pilus contains thousands of subunits whose synthesis consumes cellular resources, but the subunits can be recycled back to the inner membrane on disassembly to mitigate demand (53). We suggest that the inventory control mechanism identified here is used by T4P-expressing bacteria to regulate expression of major pilins in response to their levels in the inner membrane. The ability of the naturally truncated GS pilin to impair expression of the heterologous P. aeruginosa pilin gene suggests that it may be possible to design PilA N-terminal–mimetic peptides with the potential to block T4P expression and function.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains and plasmids used in this study are summarized in Tables S1 and S2. All vectors were constructed using standard cloning techniques and introduced into E. coli and P. aeruginosa using heat shock and electroporation, respectively. All E. coli and P. aeruginosa strains were grown in Lennox broth (LB) media (Bioshop) or LB 1.5% (wt/vol) agar plates in the presence of appropriate antibiotics. Antibiotic concentrations for E. coli strains were as follows: gentamycin, 15 µg/mL; ampicillin, 100 µg/mL; or kanamycin, 50 µg/mL. For the comparable P. aeruginosa strains, the following were used: gentamycin, 30 µg/mL; or kanamycin, 150 µg/mL. Deletion and Flp recombinase target (FRT) sequence insertion mutants were made as described in ref. 54.
Table S1.
Primers used in this study
| Primer name | Oligonucleotide sequence (5′→3′) |
| F1A_For | ATG AAA GCT CAA AAA GGC GCT ACC TTG ATC GAA CTG ATG |
| F1A_Rev | CAT CAG TTC GAT CAA GGT AGC GCC TTT TTG AGC TTT CAT |
| E5A_For | GGC TTT ACC TTG ATC GCC CTG ATG ATC GTG GTT G |
| E5A_Rev | CAA CCA CGA TCA TCA GGC CGA TCA AGG TAA AGC C |
| E5D_For | GGC TTT ACC TTG ATC GAC CTG ATG ATC GTG GTT G |
| E5D_Rev | CAA CCA CGA TCA TCA GGT CGA TCA AGG TAA AGC C |
| E5F_For | GGC TTT ACC TTG ATC TTT CTG ATG ATC GTG GTT |
| E5F_Rev | ACC CAC GAT CAT CAG AAA GAT CAA GGT AAA GCC |
| E5K_For | GGC TTT ACC TTG ATC AAA CTG ATG ATC GTG GTT G |
| E5K_Rev | CAA CCA CGA TCA TCA GTT TGA TCA AGG TAA AGC C |
| M7A_For | ACC TTG ATC GAA CTG GCG ATC GTG GTT GCG ATC |
| M7A_Rev | GAT CGC AAC CAC GAT CGC CAG TTC GAT CAA GGT |
| M7F_For | ACC TTG ATC GAA CTG TTT ATC GTG GTT GCG ATC |
| M7F_Rev | GAT CGC AAC CAC GAT AAA CAG TTC GAT CAA GGT |
| V9A_For | ATC GAA CTG ATG ATC GCG GTT GCG ATC ATC GGT |
| V9A_Rev | ACC GAT GAT CGC AAC CGC GAT CAT CAG TTC GAT |
| V9F_For | ATC GAA CTG ATG ATC TTT GTT GCG ATC ATC GGT |
| V9F_Rev | ACC GAT GAT CGC AAC AAA GAT CAT CAG TTC GAT |
| A11I_For | CTG ATG ATC GTG GTG ATC ATC ATC GGT ATC CTG |
| A11I_Rev | CAG GAT ACC GAT GAT GAT CAC CAC GAT CAT CAG |
| G14A_For | GTG GTG GCG ATC ATC GCT ATC CTG GCG GCA ATT |
| G14A_Rev | AAT TGC CGC CAG GAT AGC GAT GAT CGCCAC CAC |
| G14F_For | GTG GTT GCG ATC ATC TTT ATC TTG GCT GCA ATT |
| G14F_Rev | AAT TGC AGC CAA GAT AAA GAT GAT CGC AAC CAC |
| M16A_For | GCG ATC ATC GGT ATC GCG GCG GCA ATT GCC ATT |
| M16A_Rev | AAT GGC AAT TGC CGC CGC GAT ACC GAT GAT CGC |
| A18I_For | ATC GGT ATC CTG GCG ATA ATT GCC ATT CCC CAG |
| A18I_Rev | CTG GGG AAT GGC AAT TAT CGC CAG GAT ACC GAT |
| I19A_For | GGT ATC CTG GCG GCA GCT GCC ATT CCC CAG TAT |
| I19A_Rev | ATA CTG GGG AAT GGC AGC TGC CGC CAG GAT ACC |
| A20G_For | ATC CTG GCG GCA ATT GGC ATT CCC CAG TATA CAG |
| A20G_Rev | CTG ATA CTG GGG AAT GCC AAT TGC CGC CAG GAT |
| P22A_For | GCG GCA ATT GCC ATT GCC CAG TAT CAG AAC TAT G |
| P22A_Rev | CAT AGT TCT GAT ACT GGG CAA TGG CAA TTG CCG C |
| Q23A_For | GCA ATT GCC ATT CCC GCG TAT CAG AAC TAT GTT G |
| Q23A_Rev | CAA CAT AGT TCT GAT ACG CGG GAA TGG CAA TTG C |
| PilS N323A For | GCC CAT GAG ATC CGC GCC CGC CTG GGC GCG ATC |
| PilS N323A Rev | GAT CGC GCC CAG CGG GGC GCG GAT CTC ATG GGC |
| Lux-pPilA For | GTC GTG GGA TCC ACA GTC GAA TAT CTCCAT TGA TAT GTA TAG G |
| Lux-pPilA Rev | AGT CGT GGA TCC CTG AGA GGA GAA GGA AAT CGC AGA G |
| BACTH PilA For | GCG TCT AGA ACT GAT GAA AGC TCA AAA AGG CTT TAC |
| BACTH PilA Rev | CAT GAG CTC TCT GTT ATC ACA ACC TTT CGG AGT G |
| BACTH PilS For | GCA ACT GGA TCC GTG CGC AGT GCG CGC TGA ACG GC |
| BACTH PilS Rev | ACT TCT GAA TTC TCA GCC GGG TGG GTG CGT TTG AGT CG |
| BACTH FimU For | GCA TCT AGA CTT CAC CCT GAT CGA GTT GCT GAT |
| BACTH FimU Rev | CAT GAA TTC TCA ATA GCA TGA CTG GGG CGC CT |
| BACTH PilV For | GCA TCT AGA CTT CAG CAT GAT CGA AGT GCT GGT CG |
| BACTH PilV Rev | CAT GGT ACC TCA TGG CTC GAC CCT GAG G |
| BACTH PilE For | GCA TCT AGA CTT CAC GTT GCT GGA AAT GGT GGT GGT |
| BACTH PilE Rev | CAT GGT ACC TCA GCG CCA GCA GTC GTT GAC |
| PilScyt F1 | GTT GAA TTC GCC GGA AAA CCA GGA TC |
| PilScyt R1 | TCA GGA TCC GCG CAC GGT CGC CCT GGT CCG |
| PilScyt F2 | TCA GGA TCC CAG GAG CAG ACC GAA ACG CTG |
| PilScyt R2 | TCA AAG CTT GGC ATC CAG TTC CTC TGA CTC |
Restriction sites are underlined.
Table S2.
Strains and plasmids used in this study
| Strain or construct | Characteristics | Source |
| Strain | ||
| E. coli DH5α | F- φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk-, mk+) phoA supE44 thi-1 gyrA96 relA1 λ- | Invitrogen |
| E. coli BTH 101 | F-, cya-99, araD139, galE15, galK16, rpsL1 (Str r), hsdR2, mcrA1, mcrB1. | Euromedex |
| PAK WT | WT, group II T4P | (J. Boyd) |
| PA5196 | WT, group IV T4P, rectal clinical isolate | (24) |
| PA5196 pilA::FRT | PA5196 with an FRT scar in pilA | (24) |
| PAK pilA::FRT (NP) | FRT scar in pilA | This study |
| PAK pilT::FRT | FRT scar at position 540 pilT, T4P retraction-deficient strain | (56) |
| PAK pilA::FRT/pilT::FRT | FRT scar in pilA and FRT scar at position 540 in pilT | This study |
| Vector | ||
| pEX18Ap | Suicide vector used for gene replacement | (54) |
| pPS856 | Source of FRT-flanked gentamicin cassette | (54) |
| pFLP2 | Suicide vector encoding flp recombinase | (54) |
| pEX18Gm | Suicide vector used for gene replacement | (54) |
| pBADGr | Broad host range arabinose inducible vector used for complementation; ori araC-PBAD Gmr mob+ | (9) |
| pMS402 | Expression reporter plasmid carrying the promoterless luxCDABE gene; ori of pRO1614 | (55) |
| pUT18C | Derived from pUC19, plac, Ampr, Contains B. pertussis CyaA (225-399) (T18 domain) for N-terminal tagging | (32) |
| pKT25 | Derived from pSU40, plac, Kanr, Contains B. pertussis CyaA (1-224) (T25 domain) for N-terminal tagging | (32) |
| pMS402::PpilA | pMS402 vector with lux genes under control of the pilA promoter, cloned into the BamHI site | This study |
| pBADGr+ PA1244 pilA-V5 | pBADGr expressing PA1244 (group I) PilA with a C-terminal V5 epitope tag | This study |
| pBADGr+ PAK pilA | pBADGr expressing PAK (group II) PilA | (25) |
| pBADGr+PA14 pilA-V5 | pBADGr expressing PA14 (group III) PilA with a C-terminal V5 epitope tag | This study |
| pBADGr+PA5196 pilA-V5 | pBADGr expressing PA5196 (group IV) PilA with a C-terminal V5 epitope tag | This study |
| pBADGr+ PA1457 pilA-V5 | pBADGr expressing PA1457 (group V) PilA with a C-terminal V5 epitope tag | This study |
| pBADGr+fimU-V5 | pBADGr expressing the mature PAK minor pilin FimU with a C-terminal V5 epitope tag | This study |
| pBADGr+pilV-V5 | pBADGr expressing the mature PAK minor pilin PilV with a C-terminal V5 epitope tag | This study |
| pBADGr+pilE-V5 | pBADGr expressing the mature PAK minor pilin PilE with a C-terminal V5 epitope tag | This study |
| pKT25+pilS | BACTH plasmid expressing PAK PilS with an N-terminal T25 tag | This study |
| pKT25+pilS R24E | BACTH plasmid expressing PAK PilS R24E with an N-terminal T25 tag | This study |
| pKT25+pilS D86A | BACTH plasmid expressing PAK PilS D86A with an N-terminal T25 tag | This study |
| pKT25+pilS D86K | BACTH plasmid expressing PAK PilS D86K with an N-terminal T25 tag | This study |
| pKT25+pilS N323A | BACTH plasmid expressing phosphatase-deficient PAK PilS N323A with an N-terminal T25 tag | This study |
| pUT18C+pilA | BACTH plasmid expressing the full-length PAK PilA with an N-terminal T18 tag | This study |
| pUT18C+pilA F1A | pUT18C+pilA with a PilA F1A mutation (numbered according to mature pilin) | This study |
| pUT18C+pilA E5A | pUT18C+pilA with a PilA E5A mutation | This study |
| pUT18C+pilA E5D | pUT18C+pilA with a PilA E5D mutation | This study |
| pUT18C+pilA E5F | pUT18C+pilA with a PilA E5F mutation | This study |
| pUT18C+pilA E5K | pUT18C+pilA with a PilA E5K mutation | This study |
| pUT18C+pilA M7A | pUT18C+pilA with a PilA M7A mutation | This study |
| pUT18C+pilA M7F | pUT18C+pilA with a PilA M7F mutation | This study |
| pUT18C+pilA V9A | pUT18C+pilA with a PilA V9A mutation | This study |
| pUT18C+pilA A11I | pUT18C+pilA with a PilA A11I mutation | This study |
| pUT18C+pilA G14A | pUT18C+pilA with a PilA G14A mutation | This study |
| pUT18C+pilA G14F | pUT18C+pilA with a PilA G14F mutation | This study |
| pUT18C+pilA M16A | pUT18C+pilA with a PilA M16A mutation | This study |
| pUT18C+pilA A18I | pUT18C+pilA with a PilA A18I mutation | This study |
| pUT18C+pilA I19A | pUT18C+pilA with a PilA I19A mutation | This study |
| pUT18C+pilA A20G | pUT18C+pilA with a PilA A20G mutation | This study |
| pUT18C+pilA P22A | pUT18C+pilA with a PilA P22A mutation | This study |
| pUT18C+pilA Q23A | pUT18C+pilA with a PilA Q23A mutation | This study |
| pUT18C+ GS pilA | pUT18C carrying the G. sulfurreducens pilA gene | This study |
| pUT18C+ fimS | pUT18C carrying the PAK fimS gene | This study |
| pBADGr+pilA F1A | pBADGr+ PAK pilA with a PilA F1A mutation (numbered according to mature pilin) | This study |
| pBADGr+pilA E5A | pBADGr+ PAK pilA with a PilA E5A mutation | This study |
| pBADGr+pilA E5D | pBADGr+ PAK pilA with a PilA E5D mutation | This study |
| pBADGr+pilA E5F | pBADGr+ PAK pilA with a PilA E5F mutation | This study |
| pBADGr+pilA E5K | pBADGr+ PAK pilA with a PilA E5K mutation | This study |
| pBADGr+pilA M7A | pBADGr+ PAK pilA with a PilA M7A mutation | This study |
| pBADGr+pilA V9A | pBADGr+ PAK pilA with a PilA V9A mutation | This study |
| pBADGr+pilA A11I | pBADGr+ PAK pilA with a PilA A11I mutation | This study |
| pBADGr+pilA G14A | pBADGr+ PAK pilA with a PilA G14A mutation | This study |
| pBADGr+pilA M16A | pBADGr+ PAK pilA with a PilA M16A mutation | This study |
| pBADGr+pilA A18I | pBADGr+ PAK pilA with a PilA A18I mutation | This study |
| pBADGr+pilA I19A | pBADGr+ PAK pilA with a PilA I19A mutation | This study |
| pBADGr+pilA A20G | pBADGr+ PAK pilA with a PilA A20G mutation | This study |
| pBADGr+pilA P22A | pBADGr+ PAK pilA with a PilA P22A mutation | This study |
| pBADGr+pilA Q23A | pBADGr+ PAK pilA with a PilA Q23A mutation | This study |
| pBADGr+ GS pilA | pBADGr carrying the G. sulfurreducens pilA gene | This study |
| pEX18Ap+pilA::Gm FRT | Gent-FRT insertion in pilA | (25) |
| pEX18Gm::pilS N323A | pEX18Gm plasmid carrying PAK pilS with an N323A point mutation | This study |
| pEX18Gm::pilS R24E | pEX18Gm mating plasmid containing 500bp up and downstream of PAK pilS at position R24 (72 bp), where an R→E substitution was made | This study |
Site-Directed Mutagenesis.
Site-directed mutagenesis of PAK pilA was performed using the QuikChange protocol (Agilent Technologies) Following PCR amplification, reactions were then treated with 1 µL 10 U/µL FastDigest DpnI (Fermentas) for 2 h in a 37 °C water bath and transformed into chemically competent DH5α cells. Nucleotide substitutions were verified by DNA sequencing [McMaster Institute for Molecular Biology and Biotechnology (MOBIX)].
Lux-pilA Luminescent Reporter Assay.
pilA transcription in various P. aeruginosa strains was measured in 96-well plate liquid cultures similar to the method described in ref. 55. Briefly, bacterial strains carrying the pMS402+ppilA plasmid were grown for 16 h at 37 °C in 5 mL LB-Kan, 150 µg/mL Cultures were then diluted 1:20 in fresh media and grown to a standardized OD600 = 0.15, and the OD600 and relative luminescence of each culture were measured at 15-min intervals using a Synergy 4 microtiter plate reader (BioTek) for 5h. See SI Materials and Methods for additional details.
Twitching Motility Assay.
Twitching motility assays were performed as described previously (24). Briefly, 1% LB agar plates were stab inoculated to the plate–agar interface with a single P. aeruginosa colony and incubated at 37 °C for 24 h. The agar was then removed, and plates were stained with 1% (wt/vol) crystal violet for 20 min and washed with water to remove excess dye. Twitching zone diameter was measured in ImageJ (imagej.nih.gov/ij/; National Institutes of Health), and statistical significance was determined using a one-way ANOVA test.
Sheared Surface Protein Analysis.
Sheared surface protein preparations were conducted as described in ref. 56. Additional details are available in SI Materials and Methods.
Western Blot Analysis.
See SI Materials and Methods for details on protein sample preparation. Protein samples were separated on 15% SDS/PAGE and transferred to nitrocellulose membrane. Membranes were then blocked in 5% (wt/vol) milk solution in PBS. α-5196 PilA and α-PAK PilA rabbit polyclonal antibodies (Cedarlane Laboratories) were used at 1:7,500 dilutions. α-V5 monoclonal primary antibody and alkaline phosphatase-conjugated goat α-rabbit secondary antibody (both Sigma-Aldrich) were used at 1:3,000 dilutions. Blots were developed using manufacturers’ instructions. Blots were scanned and densitometry was performed using ImageJ and data from at least three independent experiments. One-way ANOVA analysis was used to determine significance of native pilin decreases in Graphpad Prism 5.01.
Bacterial Two-Hybrid β-Galactosidase Activity Assay.
Chemically competent E. coli BTH 101 cells were cotransformed with derivatives of the pUT18C and pKT25 plasmids—expressing the T18 and T25 domains of adenylate cyclase, respectively—and interactions were determined using a 96-well β-galactosidase assay as described in ref. 57 or on MacConkey agar supplemented with 1% maltose. A one-way ANOVA statistical analysis was performed on β-galactosidase assay results of four independent experiments with a Dunnett posttest to determine significance. Additional details can be found in SI Materials and Methods.
SI Materials and Methods
Pilin Overexpression Experiments.
Heterologous pilin genes of interest were cloned into the pBADGr vector at the EcoRI and HindIII sites, in most cases with a C-terminal V5 epitope tag (Table S2). Sequence-verified plasmids were transformed by electroporation into either PAK or PA5196 WT Pseudomonas strains and selected for on LB 1.5% agar plates supplemented with 30 µg/mL gentamicin. Single colonies were selected and grown overnight with shaking at 37 °C in liquid LB with gentamicin. The following day, samples were diluted 1:20 in fresh LB containing 30 µg/mL gentamicin and 0.1% arabinose to induce protein expression from pBADGr. Cultures were grown at 37 °C with shaking to a standardized OD600 = 0.6 (∼3 h). At this time, cells from 1 mL standardized cultures were collected by centrifugation, and the supernatants were discarded. Cell pellets were resuspended in 100 µL 1× SDS sample buffer [80 mM Tris (pH 6.8), 5.3% (vol/vol) β-mercaptoethanol, 10% (vol/vol) glycerol, 0.02% (wt/vol) bromophenol blue, and 2% (wt/vol) SDS] and boiled for 10 min in preparation for Western blot analysis.
Lux-pilA Luminescent Reporter Assay.
pilA transcription in various P. aeruginosa strains was measured in 96-well plate liquid cultures as described in ref. 55 with some modifications. Bacterial strains carrying the pMS402+ppilA plasmid, which has the luciferase (lux) genes under control of the pilA promoter, were grown with shaking for 16 h at 37 °C in 5 mL LB-Kan 150 µg/mL media. Gent 30 µg/mL was also added to those cultures containing pBADGr constructs. Following incubation, cultures were diluted 1:20 in 5 mL fresh media containing Kan 150 µg/mL and as needed, Gent 30 µg/mL cultures were grown to a standardized OD600 = 0.15. Standardized cultures (100 µL) were added in triplicate to a white wall, clear-bottom 96-well plate (3632 Costar; Corning). At t = 0 min, 0.1% arabinose was added to each well to induce protein expression from the pBADGr vector where applicable, and the plate was sealed. Plates were incubated in a Synergy 4 microtiter plate reader (BioTek) with moderate agitation at 37 °C, and OD600 and luminescence were measured every 15 min for 5 h, with the first read occurring immediately after arabinose was added and the plate was sealed. Relative luminescence was normalized to OD600 measured at the same time points. Data presented represent the mean ± SE of at least four independent experiments.
Sheared Surface Protein Analysis.
Sheared surface protein preparations were conducted as described in ref. 56. Briefly, strains of interest were streaked in a grid-like pattern on LB 1.5% agar plates (150 × 15 mm) containing antibiotic and 0.1% (wt/vol) L-arabinose where appropriate and grown overnight at 37 °C. Cells were gently scraped using sterile glass coverslips and resuspended in 4.5 mL PBS. Cell suspensions were vortexed vigorously for 30 s to shear off surface appendages, and cells were pelleted by centrifugation at 16,000 × g for 5 min. Supernatants were transferred to new 1.5-mL Eppendorf tubes and recentrifuged at 16,000 × g for 20 min to remove any remaining cellular debris. Supernatants containing sheared surface proteins were transferred to new Eppendorf tubes, and 5 M NaCl and 30% polyethylene glycol were added, to final concentrations of 0.5 M and 3%, respectively. Solutions were incubated on ice for 90 min, inverting tubes occasionally to precipitate proteins. Following incubation, precipitated proteins were collected by centrifugation. Pellets were resuspended in 150 µL 1× SDS sample buffer and boiled for 10 min in preparation for 15% SDS/PAGE analysis (3).
Bacterial Two-Hybrid β-Galactosidase Activity Assay.
To test for interactions between pilins and the sensor kinase PilS, chemically competent E. coli BTH 101 cells were cotransformed with derivatives of the pUT18C and pKT25 plasmid-expressing protein fusions with the T18 and T25 domains of adenylate cyclase, respectively. Interactions between pilins and PilS were determined using a 96-well β-galactosidase assay as described in ref. 57 or on MacConkey agar indicator media supplemented with 1% maltose. For the β-galactosidase assay, 5 mL liquid LB cultures supplemented with kanamycin and ampicillin were inoculated with the cotransformed BTH 101 cells and grown overnight with shaking at 37 °C. Samples were diluted 1:20 in 5 mL fresh media and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma-Aldrich) and grown to a standardized OD600 = 0.6 at 30 °C for optimal protein expression. Cells from 1 mL standardized culture were pelleted by centrifugation, resuspended in 400 µL PBS, and lysed with silica bead matrix by FastPrep (MP Biomedicals). Following lysis, tubes were centrifuged for 1 min at 16,000 × g to pellet the beads and cellular debris. Sample supernatants (50 µL, or an appropriate dilution in PBS totaling 50 µL) were pipetted into a 96-well plate, and 50 µL 2× reaction buffer [200 mM sodium phosphate buffer, pH 7.0, 2 mM MgCl2, 100 mM β-mercaptoethanol, and 14.4 nM o-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma-Aldrich)] was added to each well. Plates were incubated at 37 °C with rocking for 30 min. The reaction was stopped with 150 microlitres 1 M sodium carbonate, and A420 and A550 were measured using a Multiscan GO microplate reader (Thermo Scientific). β-Galactosidase activity was calculated in Miller Units (MU), using the following equation: MU = 1,000·[(A420 − (1.75A550)]/(OD600tv), where A is absorbance, t is time in minutes, and v is the lysate volume (mL). A one-way ANOVA statistical analysis was performed with a Dunnett posttest to determine significance, where the T18C-T25 empty vector control was used as the control value. Reported data represent the mean of at least four independent experiments ±SE.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512947113/-/DCSupplemental.
References
- 1.Hahn HP. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—A review. Gene. 1997;192(1):99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 2.Merz AJ, So M. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu Rev Cell Dev Biol. 2000;16:423–457. doi: 10.1146/annurev.cellbio.16.1.423. [DOI] [PubMed] [Google Scholar]
- 3.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradley DE. A function of Pseudomonas aeruginosa PAO polar pili: Twitching motility. Can J Microbiol. 1980;26(2):146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 5.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2(5):363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 6.Giltner CL, Habash M, Burrows LL. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol. 2010;398(3):444–461. doi: 10.1016/j.jmb.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen Y, et al. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem. 2015;290(1):601–611. doi: 10.1074/jbc.M114.616904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig L, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11(5):1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 9.Harvey H, Habash M, Aidoo F, Burrows LL. Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J Bacteriol. 2009;191(21):6513–6524. doi: 10.1128/JB.00943-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemkul JA, Bevan DR. Characterization of interactions between PilA from Pseudomonas aeruginosa strain K and a model membrane. J Phys Chem B. 2011;115(24):8004–8008. doi: 10.1021/jp202217f. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand JJ, West JT, Engel JN. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J Bacteriol. 2010;192(4):994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowles KN, et al. The putative Poc complex controls two distinct Pseudomonas aeruginosa polar motility mechanisms. Mol Microbiol. 2013;90(5):923–938. doi: 10.1111/mmi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd JM, Koga T, Lory S. Identification and characterization of PilS, an essential regulator of pilin expression in Pseudomonas aeruginosa. Mol Gen Genet. 1994;243(5):565–574. doi: 10.1007/BF00284205. [DOI] [PubMed] [Google Scholar]
- 14.Ishimoto KS, Lory S. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J Bacteriol. 1992;174(11):3514–3521. doi: 10.1128/jb.174.11.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu SS, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179(24):7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehl-Fie TE, Porsch EA, Miller SE, St Geme JW., 3rd Expression of Kingella kingae type IV pili is regulated by sigma54, PilS, and PilR. J Bacteriol. 2009;191(15):4976–4986. doi: 10.1128/JB.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 18.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 19.Jin S, Ishimoto KS, Lory S. PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis-acting sequence upstream of the pilin gene promoter. Mol Microbiol. 1994;14(5):1049–1057. doi: 10.1111/j.1365-2958.1994.tb01338.x. [DOI] [PubMed] [Google Scholar]
- 20.Ethier J, Boyd JM. Topological analysis and role of the transmembrane domain in polar targeting of PilS, a Pseudomonas aeruginosa sensor kinase. Mol Microbiol. 2000;38(4):891–903. doi: 10.1046/j.1365-2958.2000.02189.x. [DOI] [PubMed] [Google Scholar]
- 21.Boyd JM, Lory S. Dual function of PilS during transcriptional activation of the Pseudomonas aeruginosa pilin subunit gene. J Bacteriol. 1996;178(3):831–839. doi: 10.1128/jb.178.3.831-839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igo MM, Silhavy TJ. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170(12):5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh TN, Noriega CE, Stewart V. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc Natl Acad Sci USA. 2010;107(49):21140–21145. doi: 10.1073/pnas.1013081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kus JV, Tullis E, Cvitkovitch DG, Burrows LL. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology. 2004;150(Pt 5):1315–1326. doi: 10.1099/mic.0.26822-0. [DOI] [PubMed] [Google Scholar]
- 25.Kus JV, et al. Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with D-Araf by a novel GT-C family Arabinosyltransferase, TfpW. J Bacteriol. 2008;190(22):7464–7478. doi: 10.1128/JB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voisin S, et al. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J Bacteriol. 2007;189(1):151–159. doi: 10.1128/JB.01224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castric P. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 1995;141(Pt 5):1247–1254. doi: 10.1099/13500872-141-5-1247. [DOI] [PubMed] [Google Scholar]
- 28.Winsor GL, et al. Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39(Database issue):D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krushkal J, et al. Genome-wide survey for PilR recognition sites of the metal-reducing prokaryote Geobacter sulfurreducens. Gene. 2010;469(1-2):31–44. doi: 10.1016/j.gene.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Reardon PN, Mueller KT. Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J Biol Chem. 2013;288(41):29260–29266. doi: 10.1074/jbc.M113.498527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, et al. A Geobacter sulfurreducens strain expressing pseudomonas aeruginosa type IV pili localizes OmcS on pili but is deficient in Fe(III) oxide reduction and current production. Appl Environ Microbiol. 2014;80(3):1219–1224. doi: 10.1128/AEM.02938-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95(10):5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom MS, Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem. 1991;266(3):1656–1664. [PubMed] [Google Scholar]
- 34.Strom MS, Lory S. Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J Bacteriol. 1992;174(22):7345–7351. doi: 10.1128/jb.174.22.7345-7351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, et al. Alanine 32 in PilA is important for PilA stability and type IV pili function in Myxococcus xanthus. Microbiology. 2011;157(Pt 7):1920–1928. doi: 10.1099/mic.0.049684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng K, et al. Identification of ligand specificity determinants in lantibiotic bovicin HJ50 and the receptor BovK, a multitransmembrane histidine kinase. J Biol Chem. 2014;289(14):9823–9832. doi: 10.1074/jbc.M113.513150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prost LR, et al. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26(2):165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 39.García Véscovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: Environmental regulation of Salmonella virulence. Cell. 1996;84(1):165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 40.Bader MW, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122(3):461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 41.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5(12):e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mascher T. Bacterial (intramembrane-sensing) histidine kinases: Signal transfer rather than stimulus perception. Trends Microbiol. 2014;22(10):559–565. doi: 10.1016/j.tim.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70(4):910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Island MD, Kadner RJ. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175(16):5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imam S, Chen Z, Roos DS, Pohlschröder M. Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS One. 2011;6(12):e28919. doi: 10.1371/journal.pone.0028919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 47.Mattick JS, Whitchurch CB, Alm RA. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa--a review. Gene. 1996;179(1):147–155. doi: 10.1016/s0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- 48.Bañó-Polo M, et al. Polar/Ionizable residues in transmembrane segments: Effects on helix-helix packing. PLoS One. 2012;7(9):e44263. doi: 10.1371/journal.pone.0044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Illergård K, Kauko A, Elofsson A. Why are polar residues within the membrane core evolutionary conserved? Proteins. 2011;79(1):79–91. doi: 10.1002/prot.22859. [DOI] [PubMed] [Google Scholar]
- 50.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci USA. 2014;111(47):16860–16865. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Y, et al. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. MBio. 2015;6(1):26. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4(2):253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 53.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98(12):6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212(1):77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 55.Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4(11):e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. The platform protein is essential for type IV pilus biogenesis. J Biol Chem. 2013;288(14):9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battesti A, Bouveret E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods. 2012;58(4):325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]