Significance
Gain-of-function screens using overexpression genomic libraries are powerful tools for discovering drug target/resistance genes, but several limitations make this technique less amenable to high-throughput screening. Using cosmid-based functional screening coupled to next-generation sequencing, an approach that we term Cosmid Sequencing (or “Cos-Seq”), we followed the dynamics of cosmid enrichment during drug pressure in Leishmania, the parasite responsible for leishmaniasis, a neglected tropical disease. This improved and sensitive method has led to the identification and functional characterization of an unprecedented number of drug target/resistance genes against all drugs currently used to treat leishmaniasis.
Keywords: next-generation sequencing, Leishmania, functional cloning, Cos-Seq, resistance
Abstract
Innovative strategies are needed to accelerate the identification of antimicrobial drug targets and resistance mechanisms. Here we develop a sensitive method, which we term Cosmid Sequencing (or “Cos-Seq”), based on functional cloning coupled to next-generation sequencing. Cos-Seq identified >60 loci in the Leishmania genome that were enriched via drug selection with methotrexate and five major antileishmanials (antimony, miltefosine, paromomycin, amphotericin B, and pentamidine). Functional validation highlighted both known and previously unidentified drug targets and resistance genes, including novel roles for phosphatases in resistance to methotrexate and antimony, for ergosterol and phospholipid metabolism genes in resistance to miltefosine, and for hypothetical proteins in resistance to paromomycin, amphothericin B, and pentamidine. Several genes/loci were also found to confer resistance to two or more antileishmanials. This screening method will expedite the discovery of drug targets and resistance mechanisms and is easily adaptable to other microorganisms.
Leishmaniasis is a neglected tropical disease causing significant morbidity and mortality worldwide (1). It is caused by parasites of the genus Leishmania that cycle between the flagellar promastigote form in the gut of the insect vector and the nonmotile amastigote stage in the vertebrate host. Given the lack of an effective antileishmanial vaccine, control of leishmaniasis relies mainly on chemotherapy. Only a few antileishmanial drugs are available, and their efficacy is severely limited by toxicity, cost, and drug resistance (2, 3). New methods for expediting the discovery of drug targets and resistance mechanisms in Leishmania would aid the reassessment of current therapies and the development of new, effective drugs.
Next-generation sequencing (NGS) technologies have enabled the high-throughput and genome-scale screening of eukaryotic pathogens, and have been useful in identifying drug targets and elucidating drug resistance mechanisms (4, 5). The development of RNA interference (RNAi) target sequencing (RIT-Seq) in kinetoplastid parasites, such as Trypanosoma brucei (6), has revealed numerous genes associated with drug action (7); however, RNAi-based screening is not applicable to Leishmania, because most species lack functional RNAi machinery (8). In Leishmania spp., copy number variation and single-nucleotide polymorphism were detected in drug-resistant parasites using NGS (9, 10; reviewed in ref 11). Gain-of-function screening using genomic cosmid libraries is also a proven approach to studying drug resistance in Leishmania (12). Cosmid-based functional cloning was first implemented for studying lipophosphoglycan biosynthesis (13) and later successfully applied to study nucleoside transport (14, 15) and drug resistance (16–21). The technique has proven powerful, albeit with limitations; for instance, it is not easily amenable to high-throughput screening, because clones require individual characterization, and is biased toward the selection of cosmids conferring dominant phenotypes, leaving out less conspicuous candidates.
In this study, such limitations were alleviated by combining genome-wide cosmid-based functional screening with NGS, a strategy that we term Cosmid Sequencing (or “Cos-Seq”). This method allows us to study the dynamics of cosmid enrichment under selective drug pressure and to isolate an unprecedented number of both known and previously unidentified antileishmanial targets and resistance genes.
Results
The Cos-Seq Approach.
A library of partially digested Leishmania infantum genomic fragments (22) cloned into the cLHYG vector (23) was introduced into WT L. infantum, resulting in ∼12,000 clones that were pooled and grown for one passage under hygromycin B pressure to select transfectants. The mean size of the cosmid inserts was ∼40 kb, and the estimated coverage for the 33-Mb L. infantum genome was 15-fold. To ensure that the library was representative of the L. infantum genome (8,239 genes distributed over 36 chromosomes), we first sequenced an unselected population of L. infantum transfectants. Cosmids were extracted from early-stationary phase parasites. Libraries were prepared and sequenced using an Illumina HiSeq1000 platform. In all experiments (unselected or drug-based selections), two independent biological replicates were subjected to Cos-Seq. Analysis of fragments per kilobase per million mapped reads (FPKM) for each gene showed excellent coverage, with 8,081 genes (98% of the genome) and few underrepresented loci (SI Appendix, Fig. S1).
For Cos-Seq selection, the pooled population was subjected to incremental concentrations of antileishmanials, starting at the EC50, and then doubling the concentration at each consecutive passage (Fig. 1A). The proportion of parasites that harbor cosmids providing a selective advantage is expected to rise at each drug increment, thereby enriching with specific subpopulations of parasites. We also assessed a plateau selection scheme in which parasites were maintained for several passages at intermediate selection steps (Fig. 1A), which might favor the propagation of parasites bearing cosmids providing a more modest selective advantage. Parasites were likewise passaged in the absence of drug to control for the selection of cosmids conferring selectable growth fitness traits unrelated to drug pressure (12).
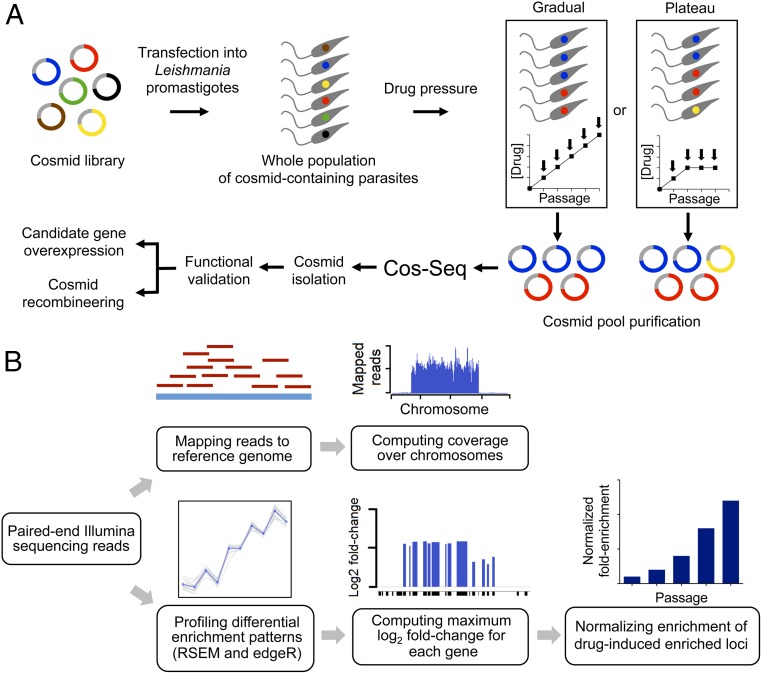
Fig. 1.
Overview of the Cos-Seq approach. (A) A WT L. infantum cosmid library cloned into the cLHYG vector (22, 23) is introduced into drug-susceptible L. infantum parasites. Pooled transfectants are submitted to incremental drug pressure starting at 1× EC50 and then increasing the drug concentration by twofold at each consecutive passage (from 1× EC50 to 16× EC50 depending on the drug; gradual selection strategy). Alternatively, a plateau selection scheme was used that allows parasites to adapt to a fixed drug concentration for two or three passages (plateau selection strategy). Cosmids are extracted from each selection step and purified for subsequent Illumina sequencing. The composition of cosmid pools at each drug/passage increment is determined from the NGS data, and cosmids of interest are isolated for functional validation. The relevant resistance genes are identified by gene overexpression studies and/or cosmid recombineering (29). (B) Sequencing reads are mapped to the reference genome, and gene coverage is inferred from the mapping data. Gene abundance and differential enrichment profiles are generated using RSEM (26) and edgeR (27), respectively. Genes are clustered according to their enrichment profiles. Genes located on the same cosmid are expected to have similar enrichment profiles. Gene abundance ratios are computed on a per-gene basis and normalized to the drug-free control.
For the genome-wide profiling of drug-enriched loci, paired-end sequencing reads of cosmid-derived libraries from each selection step were independently aligned with the L. infantum JPCM5 reference genome (tritrypdb.org/tritrypdb/) (24) using BWA software (25), which allowed assessment of the genome-wide reads coverage and the location of enriched genomic regions at single-nucleotide resolution (Fig. 1B). Genes enriched by incremental drug pressure were further analyzed using RSEM (26) and edgeR (27), and then clustered according to their enrichment pattern using R scripts packaged with the Trinity software (28). Clusters of genes whose abundance consistently increased with drug pressure were pooled, and for each gene, the variation of abundance at each drug increment (or passage for the plateau selection scheme) relative to baseline levels was established. This generated a map of the genomic loci enriched by drug selection. Gene enrichment was normalized to drug-free control levels (Materials and Methods and Fig. 1B). Cosmids carrying the enriched loci were isolated from the cosmid pool in bacteria and transfected in L. infantum to confirm their role in resistance. Finally, the relevant resistance genes were validated using single gene overexpression in WT parasites and/or cosmid recombineering (29), which allowed the generation of partially deleted cosmids by homologous recombination in bacteria (Fig. 1A).
Proof of Concept for Cos-Seq Using Methotrexate Selection.
With the possible exception of amphotericin B (AMB), which targets ergosterol-containing membranes, the mode of action of current antileishmanials remains unknown. Although methotrexate (MTX) is not used against Leishmania, it is nonetheless very active and has been extensively studied. Leishmania cells selected for MTX resistance undergo amplification of the dihydrofolate reductase-thymidylate synthase (DHFR-TS) and pteridine reductase 1 (PTR1) genes, which encode primary (30, 31) and secondary targets of MTX (32, 33), respectively. Because of this unique combination of drug target relationship with MTX, we first validated Cos-Seq with parasites selected under MTX pressure.
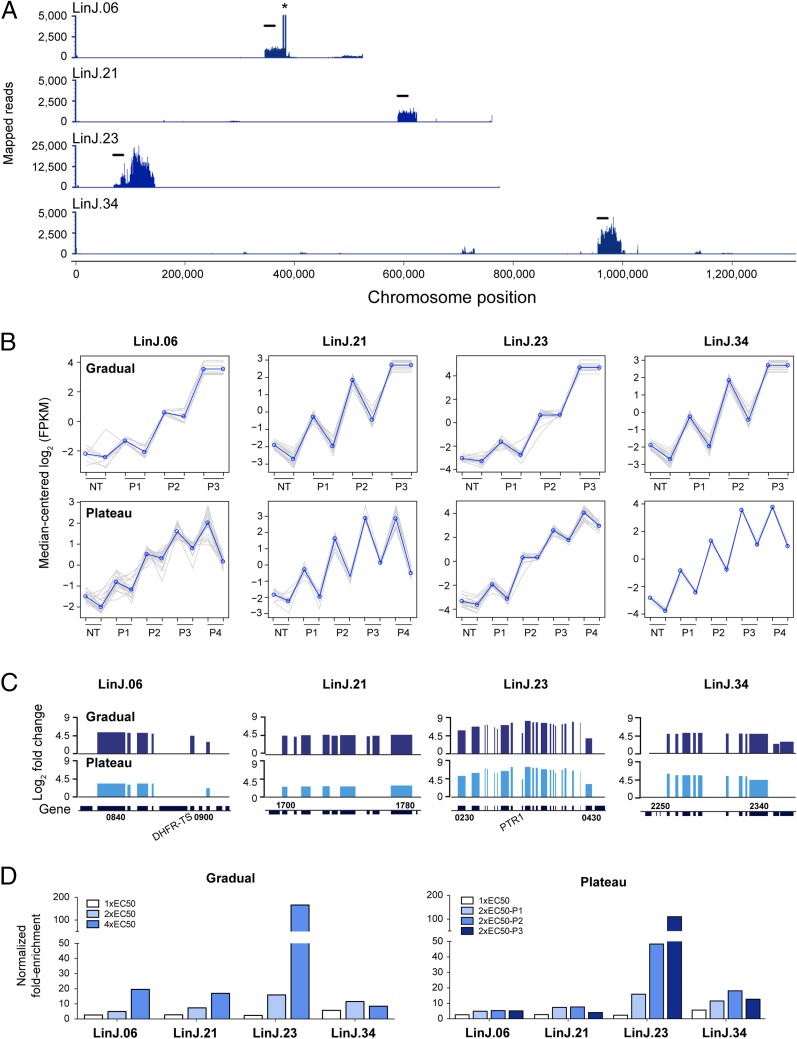
L. infantum transfectants were selected with MTX using the gradual (1×, 2×, 4×, and 8× EC50) and plateau (1× EC50, followed by three passages at 2× EC50) selection schemes (Fig. 1A). In both cases, parasites entered the stationary phase within 8 d (SI Appendix, Fig. S2). Cosmids isolated from parasites selected at 8× EC50 were not sequenced, because at this stage, parasites were well adapted and likely to be highly enriched in a low-diversity cosmid population (SI Appendix, Fig. S2). Using 16-fold enrichment (or log2-fold change ≥4) from baseline to the highest drug concentration (or from baseline to the last passage for the plateau scheme) as the minimum selection criterion, four loci with enriched consecutive genes on chromosomes 6, 21, 23, and 34 were identified (Fig. 2). Although these four loci were highlighted by gradual selection, only two (chromosomes 23 and 34) were also detected by plateau selection (SI Appendix, Fig. S3). Visualization of mapped reads from the 4× EC50 sample revealed regions of higher read density ranging from 34 kb to 75 kb, i.e., corresponding to the insert of one and two overlapping cosmids, respectively (Fig. 3A). Indeed, several loci were covered by overlapping cosmids (Table 1 and SI Appendix, Table S1).
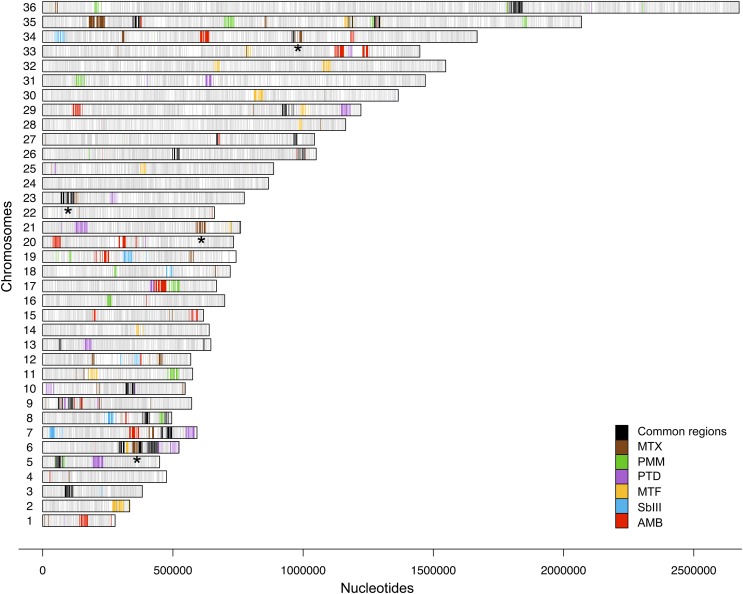
Fig. 2.
Genome-wide distribution of drug-enriched loci. Genes significantly enriched by antileishmanials as revealed by Cos-Seq are shown on one of the 36 Leishmania chromosomes. Genes enriched by both gradual and plateau selection schemes are included. Gray bars represent the gene positions on each chromosome. Colored bars represent genes enriched by the antileishmanials tested according to the color code shown at right. Only genes enriched with a mean log2-fold change of ≥4 are depicted. Black bars highlight loci enriched by at least two drugs and represent possible cross-resistance genes. Asterisks denote the genomic loci enriched by MTX, which are characterized in more detail in Fig. 3. The drug-specific Cos-Seq profiles for gradual and/or plateau selections are provided in SI Appendix, Figs. S3, S6–S9, and S11 for MTX, SbIII, MTF, AMB, PTD, and PMM, respectively. Details on the gene content of Cos-Seq enriched loci are presented in SI Appendix, Table S3.
Fig. 3.
Cos-Seq identification of loci implicated in MTX resistance. (A) Visualization of four representative MTX-enriched loci on chromosomes 6, 21, 23, and 34 as delimited by regions of higher read density. For LinJ.06, the asterisk denotes a bias in read counts coming from the DHFR-TS flanking regions originating from the cLHYG backbone (23). (Scale bars: 20 kb.) (B) Plots of gene clusters sharing similar Cos-Seq profiles from which the four representative MTX-enriched loci were recovered by gradual and plateau selections. Gray lines represent individual genes, and blue lines denote the average profile per cluster. Gene abundance is expressed on the y-axis as log2-transformed FPKM values centered to the median FPKM. Samples are ordered on the abscissa according to the selection procedure, from nontreated (NT) samples to the third drug increment (P3) for gradual selection or the fourth passage (P4) for plateau selection. Gene abundance for the two biological replicates is also shown. “Staircase” patterns are due to differences in gene abundance at baseline between the replicates. For gradual selection, sequencing of the second 4× EC50 replicate (P3) failed, leaving only a single 4× EC50 sample, which was used twice for the analysis. (C) Gene enrichment scores for the four representative MTX-enriched loci. Log2-transformed variation of gene abundance is determined between the last drug concentration (P3 for gradual selection) or passage (P4 for plateau selection) and the nontreated baseline (NT in B). The positions of DHFR-TS and PTR1 on chromosomes 6 and 23, respectively, are shown. Gene numbers indicate the first and last genes found on each enriched locus. (D) Fold enrichment of the four representative cosmids at each increment in MTX concentration (Left) or passage (Right), as normalized to the drug-free control.
Table 1.
Genomic loci enriched in the Cos-Seq screens and isolated for functional validation
| Drug | Cosmid/locus (no. of cosmids)† | Gene start‡ | Gene stop‡ | Genomic position | Cosmid fold resistance§ | Resistance gene | Gene product | Gene fold resistance¶ | Functional validation |
| MTX | LinJ.06 (1) | 0840 | 0900 | 344645–384085 | 3.3 ± 0.7* | LinJ.06.0890 | DHFR-TS | ND | |
| LinJ.21 (1) | 1700 | 1780 | 587933–622138 | 1.9 ± 0.4* | ND | ||||
| LinJ.23 (3) | 0240 | 0350 | 81869–117583 | >10*** | LinJ.23.0310 | PTR1 | Fig. 4C | Recomb | |
| LinJ.34 (4) | 2250 | 2340 | 953269–996693 | 2.2 ± 0.5* | LinJ.34.2310 | Phosphatase 2C-like | 3.5 ± 0.3** | Overexp | |
| LinJ.34.2320 | Phosphatase 2C-like | 3.7 ± 0.3** | Overexp | ||||||
| SbIII | LinJ.06a (1) | 0760 | 0810 | 289892–321083 | 1.2 ± 0.4 | ND | |||
| LinJ.07a (1) | 0070 | 0130 | 23843–57940 | 1.4 ± 0.1* | ND | ||||
| LinJ.07b (1) | 0170 | 0200 | 68913–76234 | 1.4 ± 0.2* | ND | ||||
| LinJ.08a (2) | 0610 | 0640 | 235475–268745 | 1.5 ± 0.3** | LinJ.08.0630 | P299 | ND | ||
| LinJ.12a (1) | 0470 | 0530 | 285805–320739 | 2.1 ± 0.4** | ND | ||||
| LinJ.12b (1) | 0580 | 0640 | 349890–374094 | 2.0 ± 0.2*** | LinJ.12.0610 | Ser/Thr phosphatase | 1.9 ± 0.1** | Overexp | |
| LinJ.18b (1) | 1150 | 1220 | 467206–504253 | 1.7 ± 0.2** | ND | ||||
| LinJ.19c (1) | 0690 | 0800 | 301964–345759 | 1.6 ± 0.4* | ND | ||||
| LinJ.23 (2) | 0240 | 0350 | 81869–117583 | 3.3 ± 0.6*** | LinJ.23.0290 | MRPA | ND | ||
| LinJ.34a (2) | 0130 | 0230 | 44686–78023 | 2.0 ± 0.4** | LinJ.34.0220 | ARM58 | ND | ||
| AMB | LinJ.06b (1) | 0940 | 1050 | 399192–437862 | 1.0 ± 0.1 | ND | |||
| LinJ.26b (1) | 2560 | 2670 | 975536–1010825 | 1.3 ± 0.1** | LinJ.26.2620 | Hypothetical | 1.3 ± 0.1 ** | Recomb and overexp | |
| LinJ.29b (2) | 2210 | 2270 | 921880–963007 | 1.1 ± 0.1 | ND | ||||
| LinJ.36b (1) | 4850 | 4950 | 1785378–1824575 | 1.1 ± 0.2 | ND | ||||
| MTF | LinJ.06a (1) | 0760 | 0810 | 289892–321083 | 1.2 ± 0.3 | ND | |||
| LinJ.26b (1) | 2560 | 2670 | 975536–1010825 | 1.7 ± 0.2* | ND | ||||
| LinJ.29b (2) | 2210 | 2270 | 921880–963007 | 2.6 ± 0.4** | LinJ.29.2250 | C-8 sterol isomerase | 2.7 ± 0.3*** | Overexp | |
| LinJ.29c (3) | 2320 | 2400 | 985978–1020668 | 2.5 ± 0.3** | ND | ||||
| LinJ.30 (2) | 2220 | 2300 | 816801–850574 | 3.2 ± 0.4*** | LinJ.30.2270 | Phospholipid-translocating ATPase | 2.0 ± 0.2*** | Overexp | |
| LinJ.33a (2) | 2100 | 2140 | 773766–799895 | 2.0 ± 0.3** | ND | ||||
| LinJ.35c (3) | 2870 | 2970 | 1157995–1190965 | 2.5 ± 0.3** | ND | ||||
| LinJ.36b (2) | 4850 | 4950 | 1785378–1824575 | 2.5 ± 0.3* | ND | ||||
| PTD | LinJ.06b (6) | 0940 | 1050 | 399192–437862 | 2.2 ± 0.2** | LinJ.06.1010 | Hypothetical | 1.8 ± 0.1*** | Overexp |
| LinJ.06c (6) | 0990 | 1100 | 414996–452236 | 2.7 ± 0.2*** | LinJ.06.1010 | Hypothetical | 1.8 ± 0.1*** | Overexp | |
| LinJ.26b (1) | 2560 | 2670 | 975536–1010825 | 1.2 ± 0.1 | ND | ||||
| LinJ.27b (1) | 2210 | 2280 | 961442–992831 | 2.3 ± 0.2*** | ND | ||||
| LinJ.29d (1) | 2760 | 2870 | 1146381–1181330 | 1.6 ± 0.1** | ND | ||||
| LinJ.31b (1) | 1440 | 1470 | 631444–656471 | 3.6 ± 0.3*** | LinJ.31.1460 | PRP1 | Fig. 5C | Recomb | |
| PMM | LinJ.06b (6) | 0940 | 1050 | 399192–437862 | 2.6 ± 0.2** | LinJ.06.1010 | Hypothetical | 2.1 ± 0.1*** | Overexp |
| LinJ.36b (2) | 4850 | 4950 | 1785378–1824575 | 1.4 ± 0.2* | ND |
ND, not determined; overexp, gene overexpression; recomb, cosmid recombineering. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
A single representative cosmid is reported for each enriched locus. Additional cosmids were often enriched, however, and the total number of distinct cosmids recovered per loci is indicated in parentheses. SI Appendix, Table S1 provides the genomic coordinates of the additional cosmids.
Genes found in the cosmid are indicated, including both partial and full ORFs. Previously unidentified resistance genes that were revealed in this study are highlighted in bold type.
The ratio of drug EC50 values for parasites transfected with isolated cosmids compared with cLHYG-transfected parasites. Data are the mean ± SD of three biological replicates unless otherwise indicated. Differences were statistically evaluated by unpaired two-tailed t test.
The ratio of drug EC50 for parasites transfected with the target gene cloned into pSP72αHYGα plasmid relative to empty vector-transfected parasites. Data are the mean ± SD of three biological replicates unless otherwise indicated. Differences were statistically evaluated using unpaired two-tailed t test.
The increase in gene abundance was similar in the two biological replicates (Fig. 3B). The loci enriched on chromosomes 6 and 23 covered the DHFR-TS (LinJ.06.0890) and PTR1 (LinJ.23.0310) genes, respectively, encoding the primary and secondary targets of MTX and representing two known mechanisms of MTX resistance, whereas enriched genomic regions on chromosomes 21 and 34 included genes not previously associated with MTX resistance (Fig. 3C). Additional loci, located on chromosomes 12, 13, 19, and 35, were also enriched by MTX selection (SI Appendix, Fig. S3) but were not characterized any further. Future functional work may clarify the role of the latter loci in MTX resistance.
Cosmids isolated from parasites growing at 2× EC50 and 4× EC50 were transformed into Escherichia coli. Cosmids corresponding to the regions of chromosomes 6, 21, 23, and 34 that were enriched as determined by NGS (Fig. 3A) were isolated by colony hybridization experiments using PCR-generated probes for single genes included in the enriched regions. Cosmids were purified and individually transfected in WT L. infantum. All four cosmids induced MTX resistance on parasite transfection, ranging from 1.9-fold for the LinJ.21-derived cosmid up to >10-fold for the PTR1-encoding cosmid (Table 1). MTX resistance levels correlated well with the fold enrichment during selection; that is, cosmids with a high enrichment profile (e.g., LinJ.23) provided stronger resistance than less abundant cosmids (e.g., LinJ.21 and LinJ.34) (Fig. 3D and Table 1). Interestingly, cosmid LinJ.34 was more abundant at the penultimate step of both selection schemes (Fig. 3D).
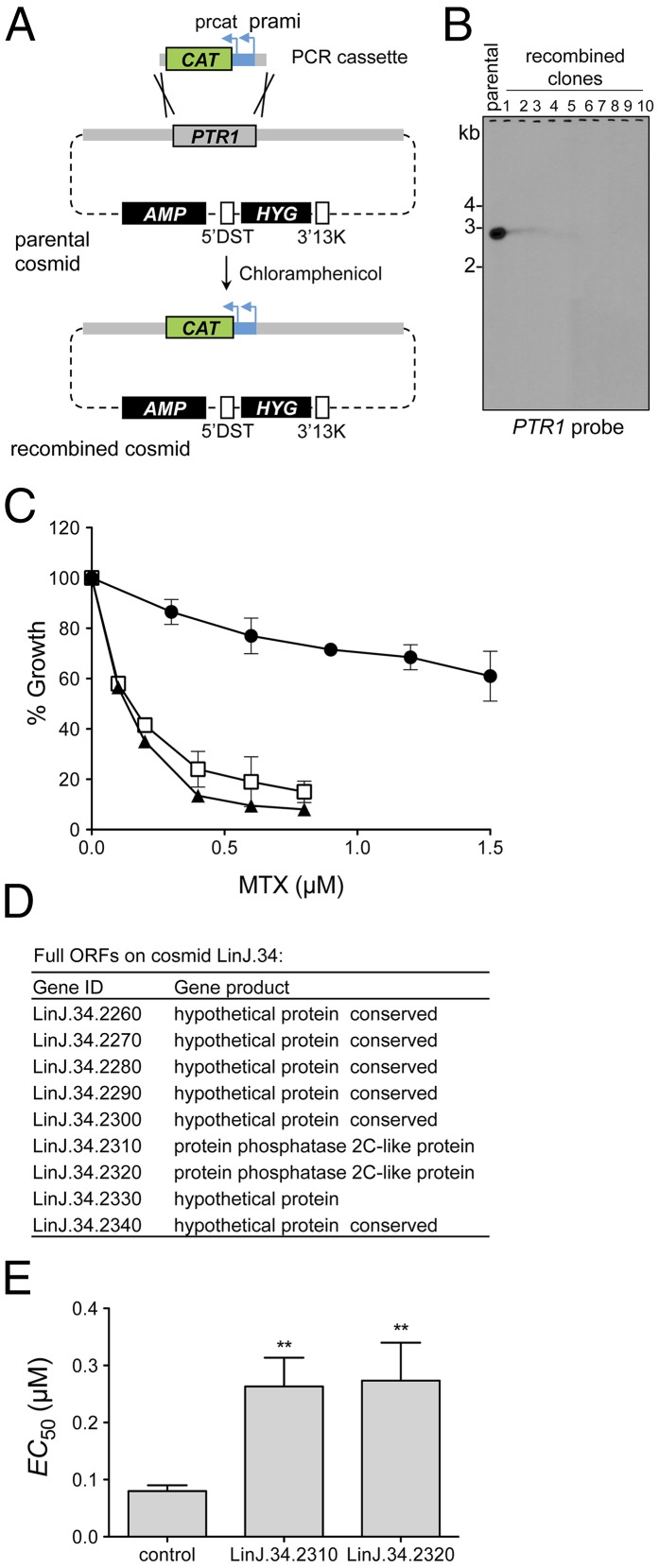
One approach that we used to identify unknown resistance genes within cosmids enriched via Cos-Seq was cosmid recombineering, which was initially developed for genetic studies in Toxoplasma gondii (29), and consists in homologous recombination-guided gene deletion within cosmids in E. coli. As a proof of principle for Leishmania, the PTR1 gene in cosmid LinJ.23 was replaced with the chloramphenicol acetyltransferase (CAT) gene, which confers chloramphenicol resistance in E. coli (Fig. 4A). After two selection rounds, PTR1 was replaced with the CAT gene in all recombined cosmid clones, as confirmed by Southern hybridization with a PTR1 probe (Fig. 4B). L. infantum transfected with a PTR1-null recombinant cosmid (clone 5) lost MTX resistance compared with parental cosmid LinJ.23 (Fig. 4C).
Fig. 4.
Identification of MTX resistance genes from Cos-Seq–enriched cosmids. (A) Schematic overview of the cosmid recombineering approach. The parental cosmid used as target for gene deletion is purified and transformed into E. coli EL250. A PCR cassette containing 50-bp PTR1 flanking sequences fused to a CAT selectable marker is introduced into the cosmid-transformed E. coli EL250 (Materials and Methods). The PTR1 gene on the parental cosmid is deleted by homologous recombination, and the recombined clones are recovered by selection on chloramphenicol agar plates. AMP and HYG are selectable markers for ampicillin and hygromycin B, respectively, and 5′DST and 3′13K are the 5′- and 3′-flanking regions of the DHFR-TS gene, respectively (23). (B) Southern blot analysis of EcoRI-digested cosmids recovered after two rounds of selection and analyzed with a PTR1 probe. Complete deletion of PTR1 was confirmed for each recombinant cosmid clone tested. (C) Effect of MTX on growth kinetics of L. infantum WT parasites transfected with parental cosmid LinJ.23 (circle) or with a representative PTR1-null LinJ.23 cosmid (clone 5 in B) (triangle). Parasites transfected with empty cLHYG vector served as controls (square). Data are the mean ± SD of three biological replicates. (D) Chromosome 34 genes harbored by cosmid LinJ.34 enriched in the Cos-Seq MTX screen. (E) Genes LinJ.34.2310 and LinJ.34.2320 encoding phosphatase 2C-like proteins were subcloned into the pSP72αHYGα plasmid and transfected in WT L. infantum for drug susceptibility determination. The control lane shows parasites transfected with the empty plasmid. The MTX EC50 values (mean ± SD) were determined from three biological replicates and statistically analyzed using an unpaired two-tailed t test. **P ≤ 0.01.
The ability of Cos-Seq to identify previously unidentified MTX resistance genes was assessed with the characterization of cosmid LinJ.34. This cosmid encodes seven hypothetical proteins in addition to two partly homologous (41% identity) phosphatase 2C-like proteins (Fig. 4D). A recent study has highlighted the role of protein phosphatase 2A in the mode of MTX action in mammalian cells (34). Thus, considering that MTX has similar mechanisms of action and entails comparable resistance mechanisms in Leishmania and mammalian cells (30, 31, 35), the two phosphatase-related genes emerged as reasonable candidates. Indeed, on the transfection of LinJ.34.2310 and LinJ.34.2320 in WT L. infantum, 3.5-fold and 3.7-fold increases in MTX resistance, respectively, were recorded (Fig. 4E and Table 1).
High-Throughput Screening of the Five Major Antileishmanial Drugs by Cos-Seq.
Cos-Seq was used to screen cosmid enrichment on selection of transfected L. infantum cells with the five major clinically relevant antileishmanials: antimony (SbIII), miltefosine (MTF), paromomycin (PMM), AMB, and pentamidine (PTD). The experimental protocol described above for MTX was applied for screening with these five drugs (SI Appendix, Fig. S4 and Table S2). Using this strategy, 76 genomic loci enriched by at least one of the five antileishmanials were identified (Fig. 2 and SI Appendix, Table S3). Of these, 12 were also enriched on subculturing in the absence of drug (SI Appendix, Fig. S5 and Table S3), thus narrowing the genuine candidates to 64 antileishmanial-enriched loci (Fig. 2). Interestingly, 12 of these drug-enriched loci were common to at least two distinct drug screens (Fig. 2 and SI Appendix, Tables S3 and S4). The high number of drug-enriched loci suggests that the five antileishmanials studied might act via different targets and/or trigger distinct resistance mechanisms, whereas the 12 shared loci may correspond to more universal mechanisms as part of a global stress response.
SbIII Resistance Genes.
Although the mode of SbIII’s action against Leishmania is poorly understood, mechanisms of resistance have been extensively described and involve the modulation of SbIII detoxifying pathways (36, 37), altered uptake (19, 38), increased efflux (39), and intracellular sequestration mediated by the ABC-thiol transporter multidrug resistance protein A (MRPA) (37, 40–42). The Cos-Seq screen for SbIII revealed 11 enriched cosmids (Fig. 2 and SI Appendix, Fig. S6 and Table S3), all of which except LinJ.03b were isolated for further characterization. On individual transfection into WT L. infantum, all cosmids but one (LinJ.06a) induced a significant upshift in SbIII EC50, ranging from 1.4-fold to 3.3-fold relative to control (Table 1). There was good agreement between cosmid fold enrichment and drug resistance level (Table 1 and SI Appendix, Fig. S6B). The cosmid conferring the highest resistance level was derived from chromosome 23 and contains the well-known resistance gene MRPA (40) (Table 1). Two other cosmids enriched through SbIII selection were derived from chromosomes 8 and 34 and contain the LinJ.08.0630 (P299) and LinJ.34.0220 (ARM58) genes, respectively, which have been implicated in SbIII resistance (43, 44) (Table 1).
As proof of principle for Cos-Seq, we sought to isolate at least one new resistance gene for each drug tested. LinJ.12b was among the most enriched cosmids in both gradual and plateau SbIII selections (SI Appendix, Fig. S6B), and thus was selected for further analysis. This cosmid encodes five full ORFs (LinJ.12.0590 to LinJ.12.0630) (Table 1). On the independent overexpression of the five ORFs in WT L. infantum, only LinJ.12.0610 induced levels of SbIII resistance similar to those observed with the LinJ.12b cosmid itself (Table 1). LinJ.12.0610 is a serine/threonine phosphatase protein with a conserved protein phosphatase-2A (PP2A) domain and two EF-hand motifs within a fused C-terminal domain (45), which may relate to the known role of SbIII as a protein phosphatase inhibitor (46). Reactive oxygen species (ROS) induced on cancer drug treatment have been shown to inactivate PP2A in mammalian cells (47). Because antimonials are potent ROS inducers in Leishmania (48), LinJ.12.0610 overexpression might allow the parasite to tolerate ROS produced on exposure to SbIII.
MTF Resistance Genes.
The MTF Cos-Seq screen revealed 23 enriched loci (Fig. 2 and SI Appendix, Fig. S7 and Table S3), five of which were common to selection with other antileishmanials (Fig. 2 and SI Appendix, Tables S3 and S4). Eight cosmids were individually isolated for characterization, seven of which imparted MTF resistance between 1.7- and 3.2-fold (Table 1). MTF resistance is an easily selected phenotype as a result of single-point mutations in the MTF transporter (MT) (10, 21). The latter is a loss-of-function event that would not be selected in the current screen. MTF is an alkylphosphocholine that mainly interferes with alkyl-lipid metabolism, phospholipid biosynthesis, and membrane composition and fluidity in Leishmania (49–51). Moreover, modification of lipid membrane content has been described in MTF-resistant lines (52). We found two genes, on cosmids LinJ.29b (LinJ.29.2250) and LinJ.30 (LinJ.30.2270), that are associated with ergosterol biosynthesis and phospholipid translocation and might be candidates for MTF resistance. Indeed, the independent overexpression of these genes in WT L. infantum induced 2.7-fold and 2-fold resistance to MTF, respectively (Table 1).
AMB Resistance Genes.
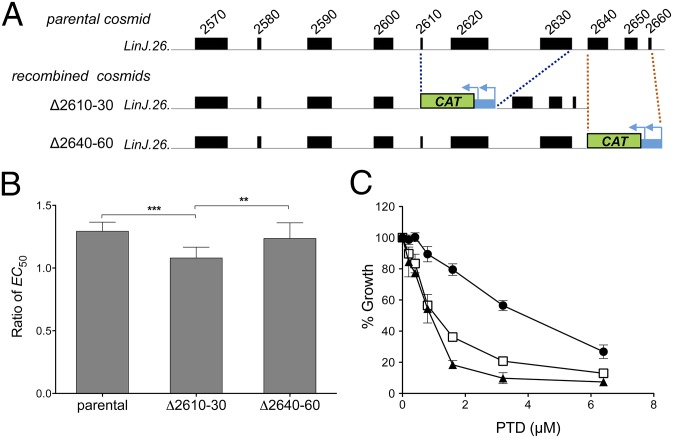
The AMB Cos-Seq selection yielded 31 enriched loci (Fig. 2 and SI Appendix, Fig. S8 and Table S3); however, 11 of these loci underwent enrichment without applying drug selection (SI Appendix, Fig. S5 and Table S3). Among the four most enriched loci (from 16- to >600-fold) (SI Appendix, Fig. S8B), only cosmid LinJ.26b conferred significant (albeit low-level) resistance (Table 1). We combined cosmid recombineering and gene overexpression to pinpoint the gene responsible for AMB resistance. From the parental cosmid, we first generated two different recombined cosmids, Δ2610–30 and Δ2640–60, each bearing three contiguous gene deletions (Fig. 5A). On transfection into WT L. infantum, only Δ2610–30 lost the AMB resistance phenotype (Fig. 5B). We then individually overexpressed LinJ.26.2610, LinJ.26.2620, and LinJ.26.2630, among which LinJ.26.2620 was found to be solely responsible for AMB resistance conferred by the cosmid LinJ.26b (Table 1). The hypothetical LinJ.26.2620 protein is of unknown function and contains several transmembrane domains and a secretory signal peptide, suggesting a plasma membrane location, where it might interact with AMB (53).
Fig. 5.
From cosmids to antileishmanial drug resistance genes. (A) Cosmid recombineering applied to cosmid LinJ.26b, which confers AMB resistance. Two segments of three consecutive genes at the 3′ end of cosmid LinJ.26b were independently replaced with a CAT marker, yielding recombined cosmids Δ2610–30 and Δ2640–60. Cosmid Δ2610–30 is deleted for the LinJ.26.2610, LinJ.26.2620, and LinJ.26.2630 genes, and cosmid Δ2640–60 is deleted for the LinJ.26.2640, LinJ.26.2650, and LinJ.26.2660 genes. (B) AMB EC50 ratio (candidate cosmid/empty vector) for L. infantum WT parasites transfected with the parental cosmid LinJ.26b or with its recombined derivative cosmids Δ2610–30 and Δ2640–60. Parasites transfected with empty cLHYG served as controls. Data are the mean ± SD of two independent experiments, each performed with five biological replicates and statistically analyzed using an unpaired two-tailed t test. **P ≤ 0.01; ***P ≤ 0.001. (C) Effect of PTD on the growth kinetics of L. infantum WT parasites transfected with cosmid LinJ.31b (circle) and its PRP1-null derivative generated by cosmid recombineering (triangle). The same strategy used for PTR1 deletion (Fig. 4A) was used here for PRP1. Parasites transfected with empty cLHYG served as controls (square). Data are the mean ± SD of three biological replicates.
PTD Resistance Genes.
The PTD screen identified 21 enriched loci, one of which was also enriched in the absence of PTD (Fig. 2 and SI Appendix, Fig. S9 and Table S3). The gene encoding the ABC transporter pentamidine resistance protein 1 (PRP1), which is known to confer PTD resistance (18), was enriched as part of cosmid LinJ.31b, and its role in resistance was confirmed by cosmid recombineering (Fig. 5C). Cosmid LinJ.31b was seen in the plateau selection scheme but not in gradual selection (SI Appendix, Fig. S9B), suggesting that PRP1 is involved mostly in low-level PTD resistance. LinJ.06b was the most enriched cosmid in gradual selection (SI Appendix, Fig. S9B), and on retransfection in WT L. infantum, it induced a 2.2-fold resistance to PTD (Table 1). This cosmid was also enriched in the PMM and AMB screens (SI Appendix, Tables S3 and S4). The region covered by LinJ.06b contains five genes overlapping with cosmid LinJ.06c (SI Appendix, Table S3), which is also implicated in PTD resistance (SI Appendix, Fig. S10). The independent overexpression of these five genes identified LinJ.06.1010 as the gene responsible for resistance to both PTD and PMM (Table 1). LinJ.06.1010 encodes a protein of unknown function that contains leucine-rich repeat domains. Further investigation should help to elucidate the common involvement of LinJ.06.1010 in resistance to these two drugs.
PMM Resistance Genes.
A total of 21 enriched loci were obtained via PMM selection, three of which were also selected in the drug-free control (Fig. 2 and SI Appendix, Fig. S11 and Table S3). Six of the 18 loci specifically enriched by PMM selection were isolated on chromosomes 6, 11, 17, 35, and 36 for further validation (SI Appendix, Fig. S11A). Of these, four conferred very high levels (≥20-fold) of PMM resistance. We found that these cosmids encode the neomycin phophotransferase marker NEO, a well-known marker of PMM resistance (54). These NEO-encoding cosmids might have arisen from the rare, incorrect ligation of NEO gene fragments from the cLHYG backbone (23) while generating the cosmid library. The two remaining cosmids were derived from chromosomes 6 (LinJ.06b) and 36 (LinJ.36b), did not include the NEO marker, and conferred bona fide PMM resistance (2.6-fold and 1.4-fold, respectively) (Table 1). The LinJ.06.1010 gene derived from LinJ.06b, with its leucine-rich repeat domain, was responsible for resistance to both PMM and PTD (Table 1 and SI Appendix, Table S4).
Cos-Seq Detects Genomic Loci Associated with Cross-Resistance.
Cos-Seq highlighted 12 cosmids enriched by selection with at least two antileishmanials (Fig. 2 and SI Appendix, Tables S3 and S4). A subset of three of such shared cosmids (LinJ.06b, LinJ.26b, and LinJ.36b) were isolated and individually transfected in WT L. infantum for phenotype confirmation. As indicated above, LinJ.06b contains a single gene (LinJ.06.1010), which is responsible for both PTD and PMM resistance (Table 1 and SI Appendix, Table S4). LinJ.26b was enriched in the AMB, PTD, and MTF screens and conferred resistance to AMB and MTF on retransfection (Table 1 and SI Appendix, Table S4). We found that the gene LinJ.26.2620, which encodes a putative membrane protein, is the determinant of AMB (Table 1) but not of MTF resistance (SI Appendix, Table S4), and thus cosmid LinJ.26b may contain at least two resistance genes. Finally, LinJ.36b was enriched by AMB, PMM, and MTF selection, and resistance to PMM and MTF was confirmed on LinJ.36b retransfection in WT L. infantum (Table 1 and SI Appendix, Table S4). Additional work is needed to pinpoint the gene(s) responsible for the latter resistance phenotypes.
Genes Isolated by Cos-Seq Confer Resistance in Intracellular Amastigotes.
In this Cos-Seq proof of concept, we used the more tractable promastigote stage of the parasite. We tested the capacity of a subset of genes isolated by Cos-Seq to produce resistance in intracellular amastigote parasites. The MRPA gene is known to confer resistance to Pentostam, the SbV-containing drug (a prodrug that is reduced to SbIII) in intracellular parasites (41), and this was further confirmed here (SI Appendix, Fig. S12A). We also observed the ability of LinJ34.0220, LinJ.29.2250, and LinJ.06.1010 to confer resistance to SbV, MTF, and PMM, respectively, in intracellular parasites (SI Appendix, Fig. S12 B–D) at levels similar to those observed with the promastigote stage (Table 1).
Discussion
Our Cos-Seq method has led to identification of an unprecedented number of resistance/target genes in the parasite Leishmania. Using the model drug MTX, we enriched for a DHFR-TS–containing cosmid and ongoing Cos-Seq screens using drugs with known targets (i.e., terbinafine, 5-fluorouracil, and ketoconazole) confirmed the recovery of the relevant target genes by Cos-Seq. For the five antileishmanials tested, we isolated many previously unidentified genes. Additional work is needed to determine whether some of these genes are genuine drug targets. It is conceivable that these antiquated drugs act on multiple minor targets, which may correspond to some of the cosmids identified via Cos-Seq. More specific drugs are needed, and several active lead compounds are now publicly available (55). The Cos-Seq technique could be used with these new molecules to pinpoint their putative targets, which could be helpful for further drug improvements. Some of the genes identified by Cos-Seq are clearly drug-resistance genes, however, such as the ABC transporters MRPA and PRP1, whose implications in SbIII and PTD resistance, respectively, are well established (18, 40).
Although overexpression of these proteins leads to a gain-of-function resistance phenotype, Cos-Seq does not lend itself to isolation of loss-of-function transporter mutations, such as for the MTF transporter MT (10). Additional technical limitations associated with Cos-Seq include the failure to detect nonprotein drug targets or selection against specific regions whose overexpression would be toxic. Some of the loci enriched by Cos-Seq were represented by single cosmids (Table 1), and having higher coverage libraries also may increase the likelihood of isolating all of the genes associated with a phenotype. To complement Cos-Seq, an alternative strategy for detecting loss-of-function mutants such as RNAi-based RIT-Seq developed for T. brucei (6) is likely needed.
Although Leishmania is RNAi-negative, this system is active in its Viannia subgenus (8), and combining loss-of-function and gain-of-function strategies might provide a major advancement toward understanding drug/microorganism interactions. Cos-Seq nonetheless allows detection of indirect resistance mechanisms resulting from drug transport alterations. For example, modification of membrane lipid content has been described in MTF-resistant lines (52), and the two genes characterized in the MTF Cos-Seq screen, which pertain to ergosterol biosynthesis and phospholipid translocation, might well trigger MTF resistance via membrane fluidity/permeability changes.
Our screens highlighted many candidate targets for SbIII, MTF, and PTD action and/or resistance, but were less useful for PMM selection because Cos-Seq spuriously captured the accidental contamination of the cosmid library by NEO markers originating from cLHYG vector fragments. The AMB screen revealed several cosmids, but only one conferred a low resistance level. So far, only one case of AMB resistance has been reported in clinical isolates (56), and we found no evidence of AMB resistance in L. infantum parasites derived from HIV patients after multiple courses of AMB (57). Thus, AMB resistance indeed may represent a rare, unlikely event in Leishmania, which is consistent with our difficulty in finding AMB resistance genes by Cos-Seq. Interestingly, we have identified several cosmids conferring resistance to at least two antileishmanials. This could have resulted from the linkage of two distinct genes on a single cosmid or from a single gene causing resistance to multiple drugs, such as LinJ.06.1010. Although different antileishmanials might act via independent targets, common effectors, such as ROS production, are clearly shared by various drugs (48), and the genes isolated in this work indeed might include resistance factors able to counteract generic cell death effectors.
Clinical drug resistance in Leishmania is restricted mostly to antimonials, for which a number of genes were found to correlate with resistance (2, 3). The Cos-Seq screen using SbIII identified MRPA, a gene found to be amplified in clinical isolates (37). The HSP83-1 protein was previously implicated in clinical resistance to SbIII (58), and its gene was significantly enriched in the SbIII screen, but only at early time points in the plateau selection and with a 3.1-fold enrichment (i.e., below our cutoff of ≥16-fold), and thus was not reported. This highlights the sensitivity of Cos-Seq and suggests that more thorough analyses using less stringent exclusion criteria also may yield relevant data. It is salient to point out that one of the main mechanisms of antimonial resistance in clinical isolates is a loss of function of AQP1 (59), which would not be captured by Cos-Seq.
In conclusion, using cosmid-based functional screening coupled to next-generation sequencing, we have identified an unprecedented number of drug resistance/target genes against all drugs currently used to treat Leishmania. We also have provided a complete list of genomic loci that are directly or indirectly linked to drug resistance, pinpointing numerous putative drug resistance/target genes. We have obtained a proof of concept for the Cos-Seq method using promastigote parasites, whose handling is more experimentally tractable, but have shown for a selection of genes that resistance is also observed in the intracellular amastigotes. Cos-Seq is currently adapted with intracellular parasites infecting macrophages, and could even be applied in in vivo animal models. The Cos-Seq method might apply not only to drug resistance, but also to the screening of any selectable phenotype, and to organisms other than Leishmania as long as they can replicate extrachromosomal elements. The screening of overexpression libraries has been described for a plethora of organisms, and it would be relatively easy to revisit such attempts with NGS, possibly expanding their outcome. As an example, we have implemented a similar approach for identifying genes that enable resistance to antibiotics in Escherichia coli and found more genes than by conventional screening on agar plates, including genes not known previously to be associated with antibiotics. Indeed, our experience has been for associating specific genes to a given phenotype, NGS surpasses any previously used technique.
Materials and Methods
Experimental procedures are described in detail in SI Appendix.
Parasite Culture and Transfection.
WT L. infantum (MHOM/MA/67/ITMAP-263) parasites were maintained as promastigotes at 25 °C in SDM-79 or M199 medium supplemented with 10% (vol/vol) heat-inactivated FBS and 5 µg/mL hemin. Leishmania promastigotes were transfected by electroporation as described previously (32).
Cos-Seq Selection.
Two biological replicates were included for each drug screen, as well as for the control in the absence of drug. For each drug screen, cosmid-harboring L. infantum parasites were thawed in 10 mL of the appropriate culture medium and then incubated at 25 °C for 24 h. The culture was diluted into 50 mL of the same medium supplemented with 600 µg/mL HYG and then incubated at 25 °C until reaching late-log phase (3–4 d). Parasites were further diluted 1:50 in 50 mL of fresh medium containing 600 µg/mL HYG and MTX, SbIII, MTF, AMB, PMM, or PTD at a concentration equal to the EC50 value of the respective drug, followed by incubation at 25 °C. Parasite growth was monitored daily by measuring A600 until late-log phase was reached (SI Appendix, Fig. S4), whereupon a 1-mL aliquot was transferred to 50 mL of fresh culture medium containing 600 µg/mL HYG as well as the appropriate antileishmanial at 2× EC50. Another 10-mL aliquot of the same culture at 1× EC50 was pelleted and stored at −80 °C, and the remaining volume was used to extract the cosmid pool for Illumina sequencing (see below).
The same procedure was repeated by using a twofold increment of drug concentration at each consecutive passage (gradual selection) or by passaging the parasites at the same intermediary concentration (plateau selection) for two or three additional rounds (SI Appendix, Table S2). In parallel, parasites were grown in the sole presence of 600 µg/mL HYG (i.e., without any additional drug) for the same number of passages to monitor basal fluctuations in cosmid abundance in the absence of antileishmanial-related selection.
Cosmid Extraction and Purification for Illumina Sequencing.
Cosmids were extracted from parasites by SDS/alkali lysis and phenol/CHCl3 extraction, followed by RNase treatment. Leishmania has an unusual single mitochondrion containing a compact DNA network (kinetoplastid DNA; kDNA) composed of minicircles and maxicircles (60). These circular DNA molecules are copurified with cosmids during SDS/alkali lysis and substantially contaminate the extracted cosmid pools. These contaminating fragments were separated from the pool of cosmids by electrophoresis on 1% low-melting point agarose gel, from which high-molecular weight DNA cosmid bands of ∼50 kb were excised and purified.
Paired-End Sequencing Library Preparation.
Here 50 ng of purified cosmid DNA was used for paired-end library preparation using the Nextera DNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions, and sequenced using an Illumina HiSeq system at a final concentration of 8 pM.
Genome Coverage and Quality Control.
Reads from each sample were independently aligned to the L. infantum JPCM5 genome (tritrypdb.org/tritrypdb/) (24) using BWA software (25). BAM files were processed with SAMStat (61) for quality and mapping statistics. BEDTools (62) was used to convert from the BAM format to the BED format.
Gene Enrichment Analysis.
Genes enriched by Cos-Seq were detected using Trinity software (28), which uses Bowtie (63) and RSEM (26) for aligning reads and computing gene abundance, respectively. Enriched genes were clustered using edgeR (27) (false discovery rate ≤0.001) and plotted using R scripts included in the Trinity package. Only genes with a log2-fold change of ≥4 were retained.
Cosmid Recombineering.
Cosmid recombineering, previously developed for T. gondii (29), allows targeted gene deletions on cosmids guided by homologous recombination in E. coli EL250. Cosmids were introduced into E. coli EL250 (a kind gift from Boris Striepen, University of Georgia, Athens, GA) by electroporation as described previously (29). A PCR cassette covering the CAT gene and pcat and pami promoters was amplified from the pEVP3 plasmid (64) for chloramphenicol selection. The PCR primers (SI Appendix, Table S5) included ∼50-bp of flanking sequences derived from the targeted locus. The PCR cassettes were electroporated into the appropriate cosmid-containing E. coli EL250 and then spread onto plates containing 12.5 µg/mL chloramphenicol. Gene deletions on recombined cosmids were confirmed by PCR and/or Southern blot analysis.
Supplementary Material
Acknowledgments
We thank Steve Beverley (Washington University) for the cLHYG shuttle vector, Boris Striepen (University of Georgia) for the EL250 bacteria and advice on the cosmid recombineering method, Frédéric Raymond (Laval University) for help with the R graphing, and Richard Poulin for a critical reading of the manuscript. This work was supported by the Canadian Institutes of Health Research Grant 1323 (to M.O.). M.O. holds a Canada Research Chair in Antimicrobial Resistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E. Ghedin is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena (accession no. PRJEB9592).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520693113/-/DCSupplemental.
References
- 1.World, Health Organization . Sustaining the Drive to Overcome the Global Impact of Neglected Tropical Diseases: Second WHO Report on Neglected Tropical Diseases. World Health Organization; Geneva, Switzerland: 2013. [Google Scholar]
- 2.Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clin Microbiol Rev. 2006;19(1):111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouellette M, Drummelsmith J, Papadopoulou B. Leishmaniasis: Drugs in the clinic, resistance and new developments. Drug Resist Update. 2004;7(4–5):257–266. doi: 10.1016/j.drup.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Kotze AC, et al. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Int J Parasitol Drugs Drug Resist. 2014;4(3):164–184. doi: 10.1016/j.ijpddr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn D, Duraisingh MT. Antiparasitic chemotherapy: From genomes to mechanisms. Annu Rev Pharmacol Toxicol. 2014;54:71–94. doi: 10.1146/annurev-pharmtox-011613-135915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alsford S, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21(6):915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alsford S, et al. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482(7384):232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lye LF, et al. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6(10):e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downing T, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21(12):2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coelho AC, et al. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl Trop Dis. 2012;6(2):e1512. doi: 10.1371/journal.pntd.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leprohon P, Fernandez-Prada C, Gazanion É, Monte-Neto R, Ouellette M. Drug resistance analysis by next-generation sequencing in Leishmania. Int J Parasitol Drugs Drug Resist. 2014;5(1):26–35. doi: 10.1016/j.ijpddr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clos J, Choudhury K. Functional cloning as a means to identify Leishmania genes involved in drug resistance. Mini Rev Med Chem. 2006;6(2):123–129. doi: 10.2174/138955706775476028. [DOI] [PubMed] [Google Scholar]
- 13.Ryan KA, Garraway LA, Descoteaux A, Turco SJ, Beverley SM. Isolation of virulence genes directing surface glycosyl-phosphatidylinositol synthesis by functional complementation of Leishmania. Proc Natl Acad Sci USA. 1993;90(18):8609–8613. doi: 10.1073/pnas.90.18.8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasudevan G, et al. Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc Natl Acad Sci USA. 1998;95(17):9873–9878. doi: 10.1073/pnas.95.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter NS, et al. Cloning of a novel inosine-guanosine transporter gene from Leishmania donovani by functional rescue of a transport-deficient mutant. J Biol Chem. 2000;275(27):20935–20941. doi: 10.1074/jbc.M002418200. [DOI] [PubMed] [Google Scholar]
- 16.Kündig C, Haimeur A, Légaré D, Papadopoulou B, Ouellette M. Increased transport of pteridines compensates for mutations in the high-affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite Leishmania tarentolae. EMBO J. 1999;18(9):2342–2351. doi: 10.1093/emboj/18.9.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotrim PC, Garrity LK, Beverley SM. Isolation of genes mediating resistance to inhibitors of nucleoside and ergosterol metabolism in Leishmania by overexpression/selection. J Biol Chem. 1999;274(53):37723–37730. doi: 10.1074/jbc.274.53.37723. [DOI] [PubMed] [Google Scholar]
- 18.Coelho AC, Beverley SM, Cotrim PC. Functional genetic identification of PRP1, an ABC transporter superfamily member conferring pentamidine resistance in Leishmania major. Mol Biochem Parasitol. 2003;130(2):83–90. doi: 10.1016/s0166-6851(03)00162-2. [DOI] [PubMed] [Google Scholar]
- 19.Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol. 2005;57(6):1690–1699. doi: 10.1111/j.1365-2958.2005.04782.x. [DOI] [PubMed] [Google Scholar]
- 20.Genest PA, et al. A protein of the leucine-rich repeats (LRRs) superfamily is implicated in antimony resistance in Leishmania infantum amastigotes. Mol Biochem Parasitol. 2008;158(1):95–99. doi: 10.1016/j.molbiopara.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Victoria FJ, Gamarro F, Ouellette M, Castanys S. Functional cloning of the miltefosine transporter: A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J Biol Chem. 2003;278(50):49965–49971. doi: 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- 22.Lamontagne J, Papadopoulou B. Developmental regulation of spliced leader RNA gene in Leishmania donovani amastigotes is mediated by specific polyadenylation. J Biol Chem. 1999;274(10):6602–6609. doi: 10.1074/jbc.274.10.6602. [DOI] [PubMed] [Google Scholar]
- 23.Ryan KA, Dasgupta S, Beverley SM. Shuttle cosmid vectors for the trypanosomatid parasite Leishmania. Gene. 1993;131(1):145–150. doi: 10.1016/0378-1119(93)90684-u. [DOI] [PubMed] [Google Scholar]
- 24.Aslett M, et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(Database issue):D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks CF, et al. The Toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe. 2010;7(1):62–73. doi: 10.1016/j.chom.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coderre JA, Beverley SM, Schimke RT, Santi DV. Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica. Proc Natl Acad Sci USA. 1983;80(8):2132–2136. doi: 10.1073/pnas.80.8.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubeda JM, et al. Modulation of gene expression in drug-resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9(7):R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11(10):3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callahan HL, Beverley SM. A member of the aldoketo reductase family confers methotrexate resistance in Leishmania. J Biol Chem. 1992;267(34):24165–24168. [PubMed] [Google Scholar]
- 34.Fernández-Pérez MP, et al. Suppression of antifolate resistance by targeting the myosin Va trafficking pathway in melanoma. Neoplasia. 2013;15(7):826–839. doi: 10.1593/neo.13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fotoohi AK, Albertioni F. Mechanisms of antifolate resistance and methotrexate efficacy in leukemia cells. Leuk Lymphoma. 2008;49(3):410–426. doi: 10.1080/10428190701824569. [DOI] [PubMed] [Google Scholar]
- 36.Wyllie S, Cunningham ML, Fairlamb AH. Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J Biol Chem. 2004;279(38):39925–39932. doi: 10.1074/jbc.M405635200. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee A, et al. Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase, and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J Antimicrob Chemother. 2007;59(2):204–211. doi: 10.1093/jac/dkl494. [DOI] [PubMed] [Google Scholar]
- 38.Monte-Neto R, et al. Intrachromosomal amplification, locus deletion and point mutation in the aquaglyceroporin AQP1 gene in antimony-resistant Leishmania (Viannia) guyanensis. PLoS Negl Trop Dis. 2015;9(2):e0003476. doi: 10.1371/journal.pntd.0003476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay R, et al. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci USA. 1996;93(19):10383–10387. doi: 10.1073/pnas.93.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Légaré D, et al. The Leishmania ATP-binding cassette protein PGPA is an intracellular metal-thiol transporter ATPase. J Biol Chem. 2001;276(28):26301–26307. doi: 10.1074/jbc.M102351200. [DOI] [PubMed] [Google Scholar]
- 41.El Fadili K, et al. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob Agents Chemother. 2005;49(5):1988–1993. doi: 10.1128/AAC.49.5.1988-1993.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leprohon P, et al. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37(5):1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhury K, Zander D, Kube M, Reinhardt R, Clos J. Identification of a Leishmania infantum gene mediating resistance to miltefosine and SbIII. Int J Parasitol. 2008;38(12):1411–1423. doi: 10.1016/j.ijpara.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Nühs A, et al. A novel marker, ARM58, confers antimony resistance to Leishmania spp. Int J Parasitol Drugs Drug Resist. 2013;4(1):37–47. doi: 10.1016/j.ijpddr.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mills E, Price HP, Johner A, Emerson JE, Smith DF. Kinetoplastid PPEF phosphatases: Dual acylated proteins expressed in the endomembrane system of Leishmania. Mol Biochem Parasitol. 2007;152(1):22–34. doi: 10.1016/j.molbiopara.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pathak MK, Yi T. Sodium stibogluconate is a potent inhibitor of protein tyrosine phosphatases and augments cytokine responses in hemopoietic cell lines. J Immunol. 2001;167(6):3391–3397. doi: 10.4049/jimmunol.167.6.3391. [DOI] [PubMed] [Google Scholar]
- 47.Nakahata S, Morishita K. PP2A inactivation by ROS accumulation. Blood. 2014;124(14):2163–2165. doi: 10.1182/blood-2014-08-594093. [DOI] [PubMed] [Google Scholar]
- 48.Moreira W, Leprohon P, Ouellette M. Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis. 2011;2:e201. doi: 10.1038/cddis.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rakotomanga M, Blanc S, Gaudin K, Chaminade P, Loiseau PM. Miltefosine affects lipid metabolism in Leishmania donovani promastigotes. Antimicrob Agents Chemother. 2007;51(4):1425–1430. doi: 10.1128/AAC.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent IM, et al. Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. Int J Parasitol Drugs Drug Resist. 2013;4(1):20–27. doi: 10.1016/j.ijpddr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imbert L, et al. Identification of phospholipid species affected by miltefosine action in Leishmania donovani cultures using LC-ELSD, LC-ESI/MS, and multivariate data analysis. Anal Bioanal Chem. 2012;402(3):1169–1182. doi: 10.1007/s00216-011-5520-3. [DOI] [PubMed] [Google Scholar]
- 52.Rakotomanga M, Saint-Pierre-Chazalet M, Loiseau PM. Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug–membrane interactions. Antimicrob Agents Chemother. 2005;49(7):2677–2686. doi: 10.1128/AAC.49.7.2677-2686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mbongo N, Loiseau PM, Billion MA, Robert-Gero M. Mechanism of amphotericin B resistance in Leishmania donovani promastigotes. Antimicrob Agents Chemother. 1998;42(2):352–357. doi: 10.1128/aac.42.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gueiros-Filho FJ, Beverley SM. On the introduction of genetically modified Leishmania outside the laboratory. Exp Parasitol. 1994;78(4):425–428. doi: 10.1006/expr.1994.1048. [DOI] [PubMed] [Google Scholar]
- 55.Peña I, et al. New compound sets identified from high-throughput phenotypic screening against three kinetoplastid parasites: An open resource. Sci Rep. 2015;5:8771. doi: 10.1038/srep08771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purkait B, et al. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob Agents Chemother. 2012;56(2):1031–1041. doi: 10.1128/AAC.00030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lachaud L, et al. Parasite susceptibility to amphotericin B in failures of treatment for visceral leishmaniasis in patients coinfected with HIV type 1 and Leishmania infantum. Clin Infect Dis. 2009;48(2):e16–e22. doi: 10.1086/595710. [DOI] [PubMed] [Google Scholar]
- 58.Vergnes B, et al. A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol Cell Proteomics. 2007;6(1):88–101. doi: 10.1074/mcp.M600319-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Mandal S, Maharjan M, Singh S, Chatterjee M, Madhubala R. Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and -resistant clinical isolates of Leishmania donovani from India. J Antimicrob Chemother. 2010;65(3):496–507. doi: 10.1093/jac/dkp468. [DOI] [PubMed] [Google Scholar]
- 60.de Souza W, Attias M, Rodrigues JC. Particularities of mitochondrial structure in parasitic protists (Apicomplexa and Kinetoplastida) Int J Biochem Cell Biol. 2009;41(10):2069–2080. doi: 10.1016/j.biocel.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 61.Lassmann T, Hayashizaki Y, Daub CO. SAMStat: Monitoring biases in next-generation sequencing data. Bioinformatics. 2011;27(1):130–131. doi: 10.1093/bioinformatics/btq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164(1):123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.