Significance
Ecological scaling laws are intensively studied for their predictive power and universal nature but often fail to unify biodiversity across domains of life. Using a global-scale compilation of microbial and macrobial data, we uncover relationships of commonness and rarity that scale with abundance at similar rates for microorganisms and macroscopic plants and animals. We then show a unified scaling law that predicts the abundance of dominant species across 30 orders of magnitude to the scale of all microorganisms on Earth. Using this scaling law combined with the lognormal model of biodiversity, we predict that Earth is home to as many as 1 trillion (1012) microbial species.
Keywords: biodiversity, microbiology, macroecology, microbiome, rare biosphere
Abstract
Scaling laws underpin unifying theories of biodiversity and are among the most predictively powerful relationships in biology. However, scaling laws developed for plants and animals often go untested or fail to hold for microorganisms. As a result, it is unclear whether scaling laws of biodiversity will span evolutionarily distant domains of life that encompass all modes of metabolism and scales of abundance. Using a global-scale compilation of ∼35,000 sites and ∼5.6⋅106 species, including the largest ever inventory of high-throughput molecular data and one of the largest compilations of plant and animal community data, we show similar rates of scaling in commonness and rarity across microorganisms and macroscopic plants and animals. We document a universal dominance scaling law that holds across 30 orders of magnitude, an unprecedented expanse that predicts the abundance of dominant ocean bacteria. In combining this scaling law with the lognormal model of biodiversity, we predict that Earth is home to upward of 1 trillion (1012) microbial species. Microbial biodiversity seems greater than ever anticipated yet predictable from the smallest to the largest microbiome.
The understanding of microbial biodiversity has rapidly transformed over the past decade. High-throughput sequencing and bioinformatics have expanded the catalog of microbial taxa by orders of magnitude, whereas the unearthing of new phyla is reshaping the tree of life (1–3). At the same time, discoveries of novel forms of metabolism have provided insight into how microbes persist in virtually all aquatic, terrestrial, engineered, and host-associated ecosystems (4, 5). However, this period of discovery has uncovered few, if any, general rules for predicting microbial biodiversity at scales of abundance that characterize, for example, the ∼1014 cells of bacteria that inhabit a single human or the ∼1030 cells of bacteria and archaea estimated to inhabit Earth (6, 7). Such findings would aid the estimation of global species richness and reveal whether theories of biodiversity hold across all scales of abundance and whether so-called law-like patterns of biodiversity span the tree of life.
A primary goal of ecology and biodiversity theory is to predict diversity, commonness, and rarity across evolutionarily distant taxa and scales of space, time, and abundance (8–10). This goal can hardly be achieved without accounting for the most abundant, widespread, and metabolically, taxonomically, and functionally diverse organisms on Earth (i.e., microorganisms). However, tests of biodiversity theory rarely include both microbial and macrobial datasets. At the same time, the study of microbial ecology has yet to uncover quantitative relationships that predict diversity, commonness, and rarity at the scale of host microbiomes and beyond. These unexplored opportunities leave the understanding of biodiversity limited to the most conspicuous species of plants and animals. This lack of synthesis has also resulted in the independent study of two phenomena that likely represent a single universal pattern. Specifically, these phenomena are the highly uneven distributions of abundance that underpin biodiversity theory (11) and the universal pattern of microbial commonness and rarity known as the microbial “rare biosphere” (12).
Scaling laws provide a promising path to the unified understanding and prediction of biodiversity. Also referred to as power laws, the forms of these relationships, y ∼ xz, predict linear rates of change under logarithmic transformation [i.e., log(y) ∼ zlog(x)] and hence, proportional changes across orders of magnitude. Scaling laws reveal how physiological, ecological, and evolutionary constraints hold across genomes, cells, organisms, and communities of greatly varying size (13–15). Among the most widely known are the scaling of metabolic rate (B) with body size [M; B = BoM3/4 (13)] and the rate at which species richness (i.e., number of species; S) scale with area [A; S = cAz (16)]. These scaling laws are predicted by powerful ecological theories, although evidence suggests that they fail for microorganisms (17–19). Beyond area and body size, there is an equally general constraint on biodiversity, that is, the number of individuals in an assemblage (N). Often referred to as total abundance, N can range from less than 10 individuals in a given area to the nearly 1030 cells of bacteria and archaea on Earth (6, 7). This expanse outstrips the 22 orders of magnitude that separate the mass of a Prochlorococcus cell (3⋅1−16 kg) from a blue whale (1.9·105 kg) and the 26 orders of magnitude that result from measuring Earth’s surface area at a spatial grain equivalent to bacteria (5.1⋅1026 μm2).
Here, we consider whether N may be one of the most powerful constraints on commonness and rarity and one of the most expansive variables across which aspects of biodiversity could scale. Although N imposes an obvious constraint on the number of species (i.e., S ≤ N), empirical and theoretical studies suggest that S scales with N at a rate of 0.25–0.5 (i.e., S ∼ Nz and 0.25 ≤ z ≤ 0.5) (20–22). Importantly, this relationship applies to samples from different systems and does not pertain to cumulative patterns (e.g., collector’s curves), which are based on resampling (20–22). Recent studies have also shown that N constrains universal patterns of commonness and rarity by imposing a numerical constraint on how abundance varies among species, across space, and through time (23, 24). Most notably, greater N leads to increasingly uneven distributions and greater rarity. Hence, we expect greater N to correspond to an increasingly uneven distribution among a greater number of species, an increasing portion of which should be rare. However, the strength of the relationships, whether they differ between microbes and macrobes, and whether they conform to scaling laws across orders of magnitude are virtually unknown.
If aspects of diversity, commonness, and rarity scale with N, then local- to global-scale predictions of microbial biodiversity could be within reach. Likewise, if these relationships are similar for microbes and macrobes, then we may be closer to a unified understanding of biodiversity than previously thought. To answer these questions, we compiled the largest publicly available microbial and macrobial datasets to date. These data include 20,376 sites of bacterial, archaeal, and microscopic fungal communities and 14,862 sites of tree, bird, and mammal communities. We focused on taxonomic aspects of biodiversity, including species richness (S), similarity in abundance among species (evenness), concentration of N among relatively low-abundance species (rarity), and number of individuals belonging to the most abundant species (absolute dominance, Nmax). We use the resulting relationships to predict Nmax and S in large microbiomes and make empirically supported and theoretically underpinned estimates for the number of microbial species on Earth.
Results and Discussion
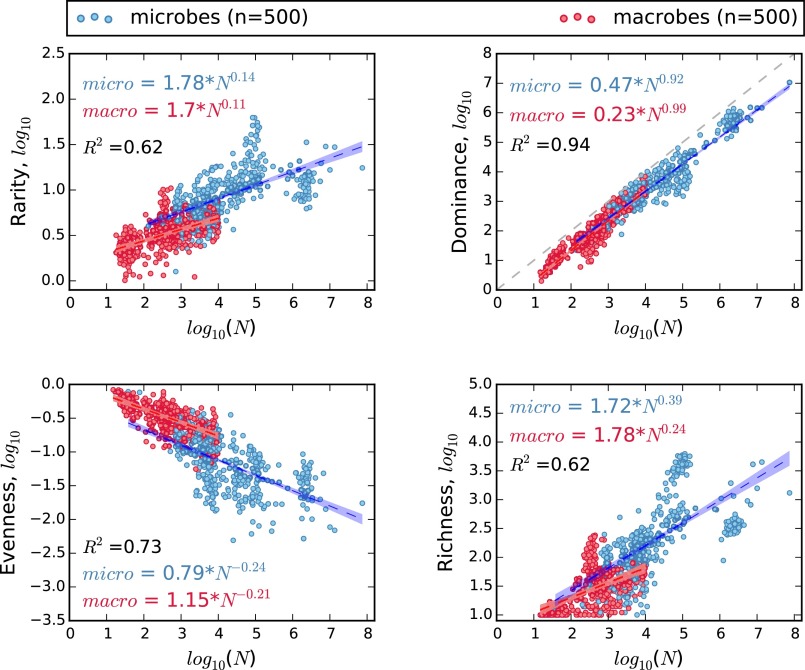
As predicted, greater N led to an increase in species richness, dominance, and rarity and a decrease in species evenness. Rarity, evenness, and dominance scaled across seven orders of magnitude in N at rates that differed little, if at all, between microbes and macrobes (Fig. 1). We found that richness (S) scaled at a greater rate for microbes (z = 0.38) than for macrobes (z = 0.24), but still, it was near the expected range of 0.25 ≤ z ≤ 0.5 (Fig. 1). However, for a given N, microbes had greater rarity, less evenness, and more species than macrobes (Fig. 1). As a result, microbes and macrobes are similar in how commonness and rarity scale with N but differ in ways that support the exceptional nature of the microbial rare biosphere. The most unifying relationship that we observed was a nearly isometric (i.e., 0.9 < z < 1.0) scaling of dominance (Nmax). When extended to global scales, this dominance scaling law closely predicts the abundance of dominant ocean bacteria. Using the lognormal model of biodiversity, published estimates of global microbial N, and published and predicted values of Nmax, we predict that Earth is occupied by 1011–1012 microbial species. This estimate is also supported by the scaling of S with N.
Fig. 1.
Microbial communities (blue dots) and communities of macroscopic plants and animals (red dots) are similar in the rates at which rarity, absolute dominance, and species evenness scale with the number of individuals or genes reads (N). However, for a given N, microbial communities have greater rarity, less evenness, and greater richness than those of macroorganisms. Coefficients and exponents of scaling equations are mean values from 10,000 bootstrapped multiple regressions, with each regression based on 500 microbial and 500 macrobial communities chosen by stratified random sampling. Each scatterplot represents a single random sample; hulls are 95% confidence intervals.
Scaling Relationships Point to an Exceptional Rare Biosphere.
Across microbial and macrobial communities, increasing N led to greater rarity, greater absolute dominance, less evenness, and greater species richness (Fig. 1 and SI Appendix, Figs. S5–S9). Bootstrapped multiple regressions revealed that the significance of differences between microbes and macrobes with regard to rarity and evenness was dependent on sample size. Larger samples suggested significant differences but were less likely to pass the assumptions of multiple regression (Materials and Methods and SI Appendix, Fig. S5). Although based on disparate types of data (i.e., counts of individual organisms vs. environmental molecular surveys), absolute dominance scaled at similar rates for microbes and macrobes (Fig. 1). Each relationship was best fit by a power law as opposed to linear, exponential, or semilog relationships (SI Appendix, Table S1).
Since being first described nearly a decade ago (25), the rare biosphere has become an intensively studied pattern of microbial commonness and rarity (12). Although its general form reiterates the ubiquitously uneven nature of ecological communities, our results suggest that microbial communities are exceptional in degrees of rarity and unevenness. Although artifacts sometimes associated with molecular surveys may inflate disparities in abundance or generate false singletons, our findings suggest that relationships of rarity, dominance, evenness, and richness were robust to the inclusion or exclusion of singletons and different percentage cutoffs in sequence similarity (SI Appendix, Figs. S8 and S9). Naturally, the inclusion of unclassified sequences led to higher taxonomic richness. As a result of this large-scale comparison, we suggest that the rare biosphere is driven by the unique biology and ecology of microorganisms. Examples are the ability of small populations to persist in suboptimal environments through resilient life stages, the ability of microbes to disperse long distances and colonize new habitats, the capacity of microbes to finely partition niche axes, and the greater ability of asexual organisms to maintain small population sizes (12).
Predicted Scaling of Species Richness (S).
Scaling exponents (z) for the relationship of species richness (S) to N fell near or within the predicted range (i.e., 0.25 < z < 0.5) (20–22) (Fig. 1 and Table 1). Despite variation in the relationship among datasets (SI Appendix, Fig. S7), the error structure across datasets was largely symmetrical (SI Appendix, Fig. S5). Across datasets, z varied more greatly for macrobes (0.07–1.23) than for microbes (0.20–0.46), which more closely resembled the expected relationship (Table 1 and SI Appendix, Fig. S7). However, pooling all data to make use of the full range of N and average out idiosyncrasies across datasets provided a stronger overall relationship and produced an exponent (z = 0.51) nearly identical to that observed in other empirical studies (20–22).
Table 1.
Scaling relationship for abundance (N) and different measures of diversity for microbial and macrobial datasets
| Dataset | Rarity | Dominance | Evenness | Richness |
| EMP (n = 14,615) | 0.2 (0.30) | 1.01 (0.67) | −0.44 (0.42) | 0.46 (0.42) |
| MG-RAST (n = 1,283) | 0.06 (0.20) | 0.98 (0.97) | −0.17 (0.32) | 0.20 (0.45) |
| HMP (n = 4,303) | 0.14 (0.14) | 1.02 (0.70) | −0.33 (0.18) | 0.29 (0.13) |
| TARA (n = 139) | −0.26 (0.02) | 1.02 (0.13) | 0.06 (0.00) | 0.29 (0.13) |
| BBS (n = 2,769) | 0.16 (0.086) | 1.0 (0.54) | −0.32 (0.22) | 0.32 (0.19) |
| CBC (n = 1,412) | 0.16 (0.39) | 1.07 (0.90) | −0.35 (0.44) | 0.22 (0.48) |
| FIA (n = 10,355) | 0.07 (0.01) | 1.34 (0.68) | −0.45 (0.27) | 0.07 (0.02) |
| GENTRY (n = 222) | 0.46 (0.27) | 0.29 (0.038) | −0.19 (0.05) | 1.24 (0.46) |
| MCDB (n = 103) | 0.07 (0.07) | 1.07 (0.91) | −0.16 (0.20) | 0.09 (0.19) |
Values are scaling exponents; coefficients of determination (r2) are in parentheses. Datasets are the Earth Microbiome Project (EMP), the Argonne National Laboratory metagenomic server (MG-RAST) rRNA amplicon projects, the Human Microbiome Project (HMP), the Tara Oceans Expedition (TARA), the North American Breeding Bird Survey (BBS), the Christmas Bird Count (CBC), the Forest Inventory and Analysis (FIA), the Gentry tree transects (GENTRY), and the Mammal Community Database (MCDB). TARA was the only dataset where N ranged over less than an order of magnitude, leading results for the TARA to be inconclusive. For most datasets, Nmax scaled almost isometrically with N. For all datasets except TARA, evenness decreased with N, while rarity increased. For birds and all microbe datasets, S scaled near the predicted range of 0.25–0.5.
Expansive Dominance Scaling Law.
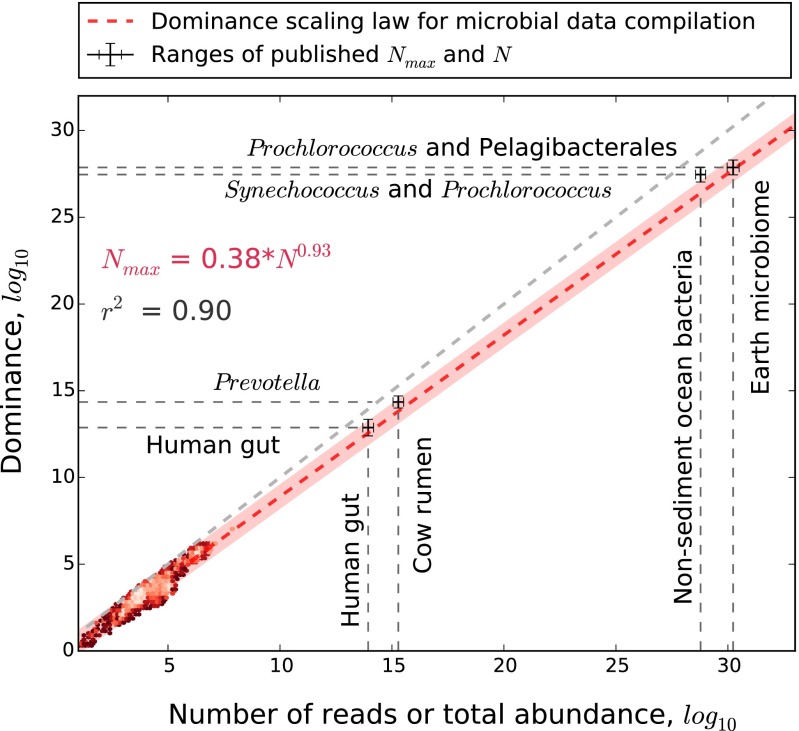
Although greater N naturally leads to greater absolute dominance (Nmax) (26), this relationship is rarely explored and to our knowledge, has not been studied as a scaling law. We found that Nmax scaled with N at similar and nearly isometric rates (i.e., 0.9 < z < 1.0) for microbes and macrobes across seven orders of magnitude (Fig. 1) (R2 = 0.94). Based on the strength of this result, we tested whether this scaling law holds at greater scales of N. We used published estimates for N and Nmax from the human gut (27, 28), the cow rumen (29, 30), the global ocean (nonsediment), and Earth (6, 7, 31, 32). In each case, we found that Nmax fell within the 95% prediction intervals of the dominance scaling law (Fig. 2). Although derived from datasets where n < 108, the dominance scaling law predicted the global abundance of some of the most abundant bacteria on Earth [Pelagibacterales (SAR11); Prochlorococcus marinus] within an order of magnitude of prior estimates (31, 32). As a result, this dominance scaling law seems to span an unprecedented 30 orders of magnitude in N, extending to the upper limits of abundance in nature. The only other biological scaling law that approaches this expanse is the 3/4 power scaling of metabolic power to mass, which holds across 27 orders of magnitude (33).
Fig. 2.
The dominance-abundance scaling law (dashed red line) predicts the abundance of the most abundant microbial taxa (Nmax) up to global scales. The pink hull is the 95% prediction interval for the regression based on 3,000 sites chosen by stratified random sampling (red heat map) from our microbial data compilation. Gray cross-hairs are ranges of published estimates of N and Nmax for large microbiomes, including Earth (6, 7, 31, 32) (Materials and Methods, Approximating Ranges of Nmax for Large Microbiomes). The light-gray dashed line is the 1:1 relationship. The scaling equation and r2 only pertain to the scatterplot data.
Predicting Global Microbial S Using N and Nmax.
Knowing the number of species on Earth is among the greatest challenges in biology (34–37). Historically, scientists have estimated global richness (S) by extrapolating rarefaction curves and rates of accumulation, often without including microorganisms (36–38). Although estimates of global microbial S exist, they range from 104 to 109, rely on cultured organisms, precede large-scale sequencing projects, and are often based on the extrapolation of statistical estimators (e.g., rarefaction and Chao). These approaches also lack the theoretical underpinnings that distinguish extrapolations of statistical estimates from predictions of biodiversity theory. As an alternative approach to estimating S, we leveraged our scaling relationships with a well-established model of biodiversity.
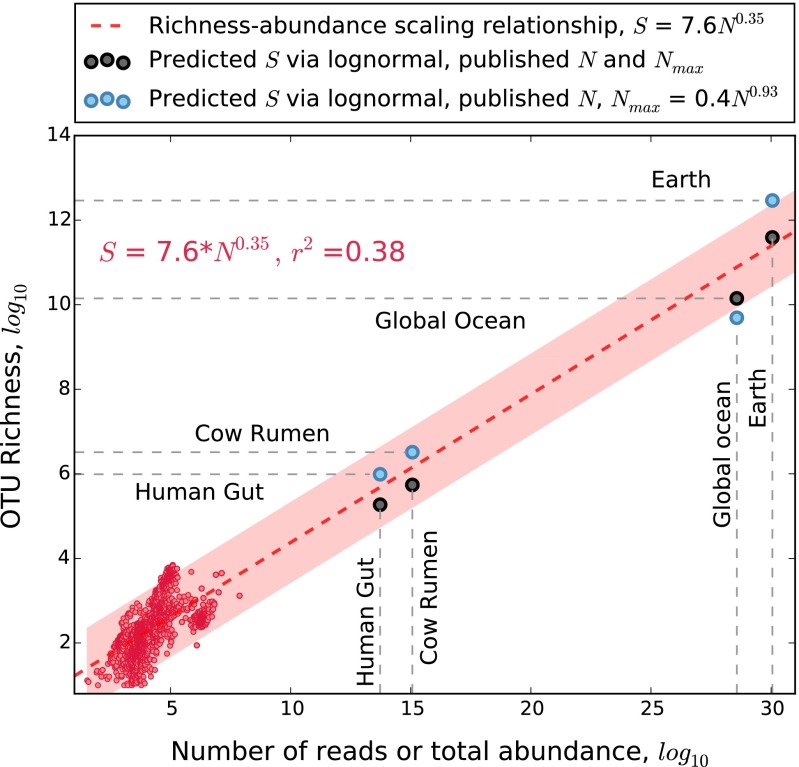
Based on the scaling of S with N (Fig. 1), we would expect a global microbial S of 2.1 ± 0.14⋅1011 species. However, this risky type of exercise would extrapolate 26 orders of magnitude beyond the available data (Fig. 2). Instead, we used the dominance scaling law and one of the most successful models of biodiversity (i.e., the lognormal distribution) to make a theoretically underpinned prediction of global microbial S (35, 39). The lognormal predicts that the distribution of abundance among species is approximately normal when species abundances are log-transformed (20). An extension of the central limit theorem, the lognormal arises from the multiplicative interactions of many random variables (20, 39). Although historically used to predict patterns of commonness and rarity, the lognormal was later derived to predict S using N and Nmax (35). This derivation of the lognormal led to predictions of S for habitats ranging in size from a milliliter of water to an entire lake and speculations of S for the entire ocean.
To our knowledge, the lognormal is the only general biodiversity model that has been derived to predict S using only N and Nmax as inputs. We used the lognormal to predict microbial S in two ways. First, we used published estimates of N and predicted the values of Nmax using our dominance scaling law (Fig. 2). Second, we made predictions of S using published estimates of both N and Nmax (6, 7, 31, 32). Assuming that global microbial N ranges from 9.2⋅1029 to 3.2⋅1030 (6, 7), the lognormal predicts 3.2 ± 0.23⋅1012 species when Nmax is predicted from the dominance scaling law (Materials and Methods). However, using published estimates for Nmax ranging from 2.9⋅1027 to 2.4⋅1028 (31, 32), the lognormal model predicts a value of global microbial S that is on the same order of magnitude as the richness-abundance scaling relationship (i.e., 3.9 ± 0.05⋅1011 species) (Fig. 3). The general agreement between the lognormal model and the richness scaling relationship is encouraging given the magnitude of these predictions.
Fig. 3.
The microbial richness-abundance scaling relationship (dashed red line) supports values of S predicted from the lognormal model using the published ranges of N and Nmax (gray dots) as well as ranges of Nmax predicted from the dominance scaling law (blue dots). The pink hull is the 95% prediction interval for the regression based on 3,000 sites chosen by stratified random sampling (red scatterplot). The scaling equation and r2 value are based solely on the red scatterplot data. SEs around predicted S are too small to illustrate.
Our predictions of S for large microbiomes are among the most rigorous to date, resulting from intersections of empirical scaling, ecological theory, and the largest ever molecular surveys of microbial communities. However, several caveats should be considered. First, observed S for the Earth Microbiome Project (EMP) differed greatly depending on whether we used closed or open reference data (Materials and Methods), where S was ∼6.9⋅104 and 5.6⋅106, respectively. In our main study, we used the closed reference data owing to the greater accuracy of that approach and because 42% of all taxa in the open reference EMP dataset were only detected twice or less. Consequently, choices, such as how to assign operational taxonomic units (OTUs) and which primers or gene regions to use, need to be made cautiously and deliberately. Second, estimates of S will be much greater than observed when many species are detected only once or twice, such as with the EMP. Statistical estimators of S, such as rarefaction, jackknife, Chao, etc., are driven by singletons and doubletons (26). Third, it is difficult to estimate the portion of species missed when only a miniscule fraction of all individuals is sampled. For example, the intersection of the lognormal model and the richness scaling relationship suggests that S for an individual human gut could range from 105 to 106 species (Fig. 3). However, S of the human gut samples is often on the order of 103, whereas N is often less than 106. These sample sizes are vanishingly small fractions of the gut microbiome, even when many samples are compiled together. For example, compiling all 4,303 samples from the Human Microbiome Project (HMP) dataset yields only 2.2⋅107 reads, hardly sufficient for detecting 105–106 species among 1014 cells. Consequently, detecting the true S of microbiomes with large N is a profound challenge that requires many large samples.
Conclusion
We estimate that Earth is inhabited by 1011–1012 microbial species. This prediction is based on ecological theory reformulated for large-scale predictions, an expansive dominance scaling law, a richness scaling relationship with empirical and theoretical support, and the largest molecular surveys compiled to date. The profound magnitude of our prediction for Earth’s microbial diversity stresses the need for continued investigation. We expect the dominance scaling law that we uncovered to be valuable in predicting richness, commonness, and rarity across all scales of abundance. To move forward, biologists will need to push beyond current computational limits and increase their investment in collaborative sampling efforts to catalog Earth’s microbial diversity. For context, ∼104 species have been cultured, less than 105 species are represented by classified sequences, and the entirety of the EMP has cataloged less than 107 species, 29% of which were only detected twice. Powerful relationships like those documented here and a greater unified study of commonness and rarity will greatly contribute to finding the potentially 99.999% of microbial taxa that remain undiscovered.
Materials and Methods
Data.
Our macrobial datasets comprised 14,862 different sites of mammal, tree, and bird communities. We used a compilation of data that included species abundance data for communities distributed across all continents, except Antarctica (40). This compilation is based, in part, on five continental- to global-scale surveys: United States Geological Survey (USGS) North American Breeding Bird Survey (41) (2,769 sites), citizen science Christmas Bird Count (42) (1,412 sites), Forest Inventory Analysis (43) (10,356 sites), Alwyn Gentry’s Forest Transect Data Set (44) (222 sites), and one global-scale data compilation: the Mammal Community Database (45) (103 sites). We limited our Christmas Bird Count dataset to sites where N was no greater than 104 (i.e., the reported maximum N for the North American Breeding Bird Survey). Above that, estimates of N are not likely based on counts of individuals. No site is represented more than once in our data. Greater detail can be found elsewhere (appendix in ref. 40).
We used 20,376 sites of communities of bacteria, archaea, and microscopic fungi; 14,615 of these were from the EMP (1) obtained on August 22, 2014. Sample processing, sequencing, and amplicon data are standardized and performed by the EMP, and all are publicly available at qiita.microbio.me/. The EMP data consist of open and closed reference datasets. The Quantitative Insights Into Microbial Ecology (QIIME) tutorial (qiime.org/tutorials/otu_picking.html) defines closed reference as a classification scheme where any rRNA reads that do not hit a sequence in a reference collection are excluded from analysis. In contrast, open reference refers to a scheme where reads that do not hit a reference collection are subsequently clustered de novo and represent unique but unclassified taxonomic units. Our main results are based on closed reference data because of the greater accuracy of that approach and because 13% of all taxa in the open reference EMP dataset were only detected once, whereas 29% were only detected twice.
We also used 4,303 sites from the Data Analysis and Coordination Center for the NIH Common Fund-supported HMP (46). These data consisted of samples taken from 15 or 18 locations (including the skin, gut, vagina, and oral cavity) on each of 300 healthy individuals. The V3–V5 region of the 16S rRNA gene was sequenced for each sample. We excluded sites from pilot phases of the HMP as well as time series data. More detail on HMP sequencing and sampling protocols can be found at hmpdacc.org/micro_analysis/microbiome_analyses.php.
We included 1,319 nonexperimental PCR-targeted rRNA amplicon sequencing projects from the Argonne National Laboratory metagenomics server MG-RAST (47). Represented in this compilation were samples from arctic aquatic systems (130 sites; MG-RAST ID: mgp138), hydrothermal vents (123 sites; MG-RAST ID: mgp327) (48), freshwater lakes in China (187 sites; MG-RAST ID: mgp2758) (49), arctic soils (44 sites; MG-RAST ID: mgp69) (50), temperate soils (84 sites; MG-RAST ID: mgp68) (51), bovine fecal samples (16 sites; MG-RAST ID: mgp14132), human gut microbiome samples not part of the HMP (529 sites; MG-RAST ID: mgp401) (52), a global-scale dataset of indoor fungal systems (128 sites) (53), and freshwater, marine, and intertidal river sediments (34 sites; MG-RAST ID: mgp1829). Using MG-RAST allowed us to choose common parameter values for sequence similarity (i.e., 97% for species level) and taxa assignment, including a maximum e-value (probability of observing an equal or better match in a database of a given size) of 10−5, a minimum alignment length of 50 bp, and minimum percentage sequence similarities of 95%, 97%, and 99% to the closest reference sequence in the MG-RAST’s M5 rRNA database (47–54). Below, we analyze the MG-RAST datasets with respect to these cutoffs and reveal no significant effect on scaling relationships. Last, we included 139 “prokaryote-enriched” samples from 68 pelagic and mesopelagic locations, representing all major oceanic regions (except the Arctic) gathered by the Tara Oceans expedition (55). Among the taxa not included in our analyses are reptiles, amphibians, fish, large mammals, invertebrates, and protists. These taxa were absent, because large datasets to do not exist for their communities or because redistribution rights could not be gained for publication.
Quantifying Dominance, Evenness, Rarity, and Richness.
We calculated or estimated aspects of diversity (dominance, evenness, rarity, and richness) for each site in our data compilation. All analyses can be reproduced or modified for additional exploration by using the code, data, and directions provided at https://github.com/LennonLab/ScalingMicroBiodiversity.
Rarity.
Here, rarity quantifies the concentration of species at low abundance (26). Our primary rarity metric was the skewness of the frequency distribution of arithmetic abundance classes (Rskew), which are almost always right-skewed distributions (26). Because of the inability to take the logarithm of a negative skew, Rskew was given a modulo transformation. The log-modulo transformation adds a value of one to each measure of skewness and converts negative values to positive values, making them all positive and able to be log-transformed. We also quantified rarity using log-transformed abundances (Rlog-skew) (26). We present results for Rlog-skew in SI Appendix, Fig. S4.
Dominance.
Dominance refers to the abundance of the most abundant species, the simplest measure of which is the abundance of the most abundant species (absolute dominance; Nmax) (26). Relative dominance is also a common measure, and it is known as the Berger–Parker index (Nmax/N = DBP). We focus on Nmax in the main body because of the previously undocumented scaling with N and the ability to predict S using N, Nmax, and the lognormal model. We also calculated dominance as the sum of the relative abundance of the two most abundant taxa (i.e., McNaughton’s dominance) and Simpson’s diversity, which is more accurately interpreted as an index of dominance (26). We present results for dominance metrics other than Nmax in SI Appendix, Fig. S3.
Evenness.
Species evenness captures similarity in abundance among species (26, 56). We used five evenness metrics that perform well according to a series of statistical requirements (56), including lacking a strong bias toward very large or very small abundances, independence of richness (S), and scaling between zero (no evenness) and one (perfect evenness). These metrics included Smith and Wilson’s indices (Evar and EQ), Simpson’s evenness (E1/D), Bulla’s index (O), and Camargo’s index (E′) (26, 56). We present results for E1/D in Results and Discussion and results for the other four metrics in SI Appendix, Fig. S2.
Richness.
Richness (S) is the number of species observed or estimated from a sample. Estimates of S are designed to account for rare species that go undetected in unbiased surveys (26). We present results for observed S in the text along and results for six estimators of S (Chao1, ACE, jackknife, rarefaction, Margelef, and McHennick) in SI Appendix, Fig. S1.
Approximating Ranges of Nmax for Large Microbiomes.
Cow rumen.
The most dominant taxonomic unit (based on 97% sequence similarity in 16S rRNA reads) in the cow rumen is typically a member of the Provotella genus and has been reported to account for about 1.5–2.0% of 16S rRNA gene reads in a sample (29, 30). Assuming there are about 1015 microbial cells in the cow rumen (29, 30) and if these percentages are reflective of community-wide relative dominance (DBP), then Nmax of the cow rumen would be in the range of 1.5⋅1014 to 2⋅1014.
Human gut.
Deep sequencing of the human gut reveals that the most dominant taxon (based on 97% 16S rRNA sequence similarity) accounts for 10.6–12.2% of 16S rRNA gene reads in a sample (6, 28). Assuming these percentages are reflective of the microbiome at large and that there are about 1014 microbial cells in the human gut (5, 28, 46), then Nmax would be in the range from 1.06⋅1013 to 1.22⋅1013.
Global ocean (nonsediment) and Earth.
The most abundant microbial species on Earth has yet been determined. Perhaps, the best genus-level candidates (based on 97% 16S rRNA sequence similarity) are the marine picocyanobacteria Synechococcus and Prochlorococcus, with estimated global abundances of 7.0 ± 0.3⋅1026 and 2.9 ± 0.1⋅1027, respectively (32). Members of the SAR11 clade (i.e., Pelagibacterales) have an estimated global abundance of 2.0⋅1028 and may also be candidates for the most abundant microorganisms on Earth (31). We used SAR11 as the upper limit for the most dominant microbial species on Earth (i.e., the most abundant species cannot be more abundant than the most dominant order-level clade). We used 6.7⋅1026–3.0⋅1027 as the range for Nmax of the nonsediment global ocean and 2.9⋅1027–2.0⋅1028 as the range for Nmax of Earth. We used the range from 3.6⋅1028 to 1.2⋅1029 as the lower to upper range for the number of microbial cells in the open ocean (7) and from 9.2⋅1029 to 3.2⋅1030 (6) as the lower to upper range for the number of microbial cells on Earth.
Predictions of S for Large Microbiomes and Earth.
We used the methods described in Curtis et al. (35) to predict global microbial richness (S) using the lognormal species abundance model in ref. 39. Curtis et al. (35) used the lognormal to estimate microbial S in 1 g soil, 1 mL water, and an entire lake, and then, they speculate on what S may be for a ton of soil (many small ecosystems) and the entire ocean (many large ecosystems). The lognormal prediction of S is based on the ratio of total abundance (N) to the abundance of the most abundant species (Nmax) and the assumption that the rarest species is a singleton, Nmin = 1. In equation 1 from the work by Curtis et al. (35), according to the lognormal model, in communities with S(N) species, the number of taxa that contain N individuals is
where a is an inverse measure of the width of the distribution, with SD that is σ2 (a = [2ln2σ2]−1/2) and NO that is the most common (i.e., modal) abundance class. In equation 3 from the work by Curtis et al. (35), if it is assumed that the lognormal species abundance curve is not truncated and therefore, is symmetric about NO, then it can be shown that
The second method for estimating the spread of the lognormal distribution, a, is by knowing or assuming Nmin. By using equations 1 and 3 from the work by Curtis et al. (35) and the assumption that S(Nmin) = 1, S can be expressed in terms of a, Nmin, and Nmax. Curtis et al. (35) reason that S will not be sensitive to small deviations from the Nmin = 1 assumption and hence, that knowledge of Nmin, Nmax, and N allows equation 11 from the work by Curtis et al. (35) to be solved numerically for Preston’s a parameter and subsequently, S to be predicted using equation 10 from the work by Curtis et al. (35):
Curtis et al. (35) show that the above equation can be used to rewrite equation 5 in ref. 35 as equation 11 in ref. 35:
Equation 11 from the work by Curtis et al. (35) can be numerically solved for a, which is then used in their equation 10 to solve for S. We coded equations 1, 3, 10, and 11 from the work by Curtis et al. (35) into a Python script that can be used to recreate the results in the work by Curtis et al. (35) under the functions “alpha2” to derive a and “s2” to estimate S. In predicting S, we accounted for variability in N and Nmax by randomly sampling within their published ranges (SI Appendix, Fig. S14). This sampling strategy allowed us to generate means and SEs, which are often lacking from large-scale predictions of S.
Resampling and Dependence on Sample Size and Sequence Similarity.
We examined relationships of rarity, evenness, dominance, and richness to the number of individual organisms or gene reads (N) using 10,000 bootstrapped multiple regressions based on stratified random sampling of microbial and macrobial datasets. We examined the sensitivity of our results to sampling strategy, sample size, particular datasets, and the microbe/macrobe dummy variable, results of which can be found in SI Appendix, Figs. S5–S13. To use equal numbers of sites for macrobes and microbes in each multiple regression analysis, we used 100 sites from each macrobial dataset for a total of 500 randomly chosen sites. To obtain 500 sites from our microbial data, we used 50 randomly chosen sites from each microbial dataset having more than 100 sites and 20 randomly chosen sites from smaller datasets. We used the mean values of coefficients and intercepts (accounting for whether differences between microbes and macrobes were significant at P < 0.05; α = 0.05) from multiple regressions to estimate the relationships of rarity, evenness, dominance, and richness to N. We examined whether scaling relationships for microbial data were sensitive to the percentage cutoff in rRNA sequence similarity, which is used for binning sequences into OTUs. These analyses were restricted to datasets obtained from the MG-RAST but reveal no statistical differences caused by whether sequences were binned based on 95%, 97%, and 99% similarity.
Power Law Behavior vs. Other Functional Forms.
We tested whether relationships of richness, evenness, rarity, and dominance were better fit by a power law (log–log) than by linear, exponential, and semilog relationships (SI Appendix, Table S1). The power law model explained substantially greater variance or in the one case where it was nearly tied in explanatory power, had substantially lower Akaike information criterion (AIC) and Bayesian information criterion (BIC) values than other models.
Available Code and Data.
We used freely available open-source computing and version control tools. Analyses and figures can be automatically regenerated using Python scripts and data files in the public GitHub repository https://github.com/LennonLab/ScalingMicroBiodiversity. Analyses can be recreated step by step using the directions given in the repository.
Supplementary Material
Acknowledgments
We thank M. Muscarella, W. Shoemaker, N. Wisnoski, X. Xiao, S. Gibbons, E. Hall, and E. P. White for critical reviews. We thank the individuals involved in collecting and providing the data used in this paper, including the essential citizen scientists who collect the Breeding Bird Survey and Christmas Bird Count data; the researchers who collected, sequenced, and provided metagenomic data on the MG-RAST as well as the individuals who maintain and provide the MG-RAST service; the Audubon Society; the US Forest Service; the Missouri Botanical Garden; and Alwyn H. Gentry. This work was supported by National Science Foundation Dimensions of Biodiversity Grant 1442246 and US Army Research Office Grant W911NF-14-1-0411.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521291113/-/DCSupplemental.
References
- 1.Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: Successes and aspirations. BMC Biol. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523(7559):208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 3.Hug LA, et al. A new view of the tree of life. Nat Microbiol. 2016 doi: 10.1038/nmicrobiol.2016.48. [DOI] [Google Scholar]
- 4.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 5.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109(40):16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton Univ Press; Princeton: 2001. [DOI] [PubMed] [Google Scholar]
- 9.McGill BJ. Towards a unification of unified theories of biodiversity. Ecol Lett. 2010;13(5):627–642. doi: 10.1111/j.1461-0248.2010.01449.x. [DOI] [PubMed] [Google Scholar]
- 10.Harte J. Maximum Entropy and Ecology. Oxford Univ Press; New York: 2011. [Google Scholar]
- 11.McGill BJ, et al. Species abundance distributions: Moving beyond single prediction theories to integration within an ecological framework. Ecol Lett. 2007;10(10):995–1015. doi: 10.1111/j.1461-0248.2007.01094.x. [DOI] [PubMed] [Google Scholar]
- 12.Reid A, Buckley M. The Rare Biosphere: A Report from the American Academy of Microbiology. American Academy of Microbiology; Washington, DC: 2011. [Google Scholar]
- 13.Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. Toward a metabolic theory of ecology. Ecology. 2004;85(7):1771–1789. [Google Scholar]
- 14.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302(5649):1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie ME, Olff H. Spatial scaling laws yield a synthetic theory of biodiversity. Nature. 1999;400(6744):557–560. doi: 10.1038/23010. [DOI] [PubMed] [Google Scholar]
- 16.Lomolino MV. Ecology’s most general, yet protean pattern: The species‐area relationship. J Biogeogr. 2000;27(1):17–26. [Google Scholar]
- 17.DeLong JP, Okie JG, Moses ME, Sibly RM, Brown JH. Shifts in metabolic scaling, production, and efficiency across major evolutionary transitions of life. Proc Natl Acad Sci USA. 2010;107(29):12941–12945. doi: 10.1073/pnas.1007783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM. A taxa-area relationship for bacteria. Nature. 2004;432(7018):750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- 19.Green JL, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432(7018):747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- 20.May RM. Patterns of species abundance and diversity. In: Cody ML, Diamond JM, editors. Ecology and Evolution of Communities. Harvard Univ Press; Cambridge, MA: 1975. pp. 81–120. [Google Scholar]
- 21.May RM. The dynamics and diversity of insect faunas. In: Mount LA, Waloff N, editors. Diversity of Insect Faunas. Blackwell; Oxford: 1978. pp. 188–204. [Google Scholar]
- 22.Siemann E, Tilman D, Haarstad J. Insect species diversity, abundance and body size relationships. Nature. 1996;380:704–706. [Google Scholar]
- 23.Locey KJ, White EP. How species richness and total abundance constrain the distribution of abundance. Ecol Lett. 2013;16(9):1177–1185. doi: 10.1111/ele.12154. [DOI] [PubMed] [Google Scholar]
- 24.Xiao X, Locey KJ, White EP. A process-independent explanation for the general form of Taylor’s Law. Am Nat. 2015;186(2):E51–E60. doi: 10.1086/682050. [DOI] [PubMed] [Google Scholar]
- 25.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc Natl Acad Sci USA. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magurran AE, McGill BJ, editors. Biological Diversity: Frontiers in Measurement and Assessment. Vol 12 Oxford Univ Press; Oxford: 2011. [Google Scholar]
- 27.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4(11):430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 28.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jami E, Mizrahi I. Composition and similarity of bovine rumen microbiota across individual animals. PLoS One. 2012;7(3):e33306. doi: 10.1371/journal.pone.0033306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75(1):165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 31.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420(6917):806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 32.Flombaum P, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110(24):9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West GB, Woodruff WH, Brown JH. Allometric scaling of metabolic rate from molecules and mitochondria to cells and mammals. Proc Natl Acad Sci USA. 2002;99(Suppl 1):2473–2478. doi: 10.1073/pnas.012579799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.May RM. How many species are there on Earth? Science. 1988;241(4872):1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- 35.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99(16):10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on Earth and in the ocean? PLoS Biol. 2011;9(8):e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stork NE, McBroom J, Gely C, Hamilton AJ. New approaches narrow global species estimates for beetles, insects, and terrestrial arthropods. Proc Natl Acad Sci USA. 2015;112(24):7519–7523. doi: 10.1073/pnas.1502408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol Rev. 2004;68(4):686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston FW. The commonness, and rarity, of species. Ecology. 1948;29(3):254–283. [Google Scholar]
- 40.White EP, Thibault KM, Xiao X. Characterizing species abundance distributions across taxa and ecosystems using a simple maximum entropy model. Ecology. 2012;93(8):1772–1778. doi: 10.1890/11-2177.1. [DOI] [PubMed] [Google Scholar]
- 41.Sauer JR, et al. The North American Breeding Bird Survey 1966–2009, Version 3.23.2011. USGS Patuxent Wildlife Research Center; Laurel, MD: 2011. [Google Scholar]
- 42.National Audubon Society 2002 The Christmas Bird Count Historical Results. Available at netapp.audubon.org/cbcobservation/. Accessed April 14, 2016.
- 43.US Department of Agriculture . Forest Inventory and Analysis: National Core Field Guide (Phase 2 and 3), Version 4.0. US Department of Agriculture Forest Service, Forest Inventory and Analysis; Washington, DC: 2010. [Google Scholar]
- 44.Phillips O, Miller JS. Global Patterns of Plant Diversity: Alwyn H. Gentry’s Forest Transect Data Set. Missouri Botanical Garden Press; St. Louis: 2002. [Google Scholar]
- 45.Thibault KM, Supp SR, Giffin M, White EP, Ernest SKM. Species composition and abundance of mammalian communities. Ecology. 2011;92(12):2316. [Google Scholar]
- 46.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flores GE, et al. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ Microbiol. 2011;13(8):2158–2171. doi: 10.1111/j.1462-2920.2011.02463.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, et al. Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME J. 2013;7(7):1310–1321. doi: 10.1038/ismej.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu H, et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12(11):2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- 51.Fierer N, et al. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6(5):1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amend AS, Seifert KA, Samson R, Bruns TD. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc Natl Acad Sci USA. 2010;107(31):13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 55.Sunagawa S, et al. Tara Oceans coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348(6237):1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 56.Smith B, Wilson JB. A consumer’s guide to evenness indices. Oikos. 1996;76(1):70–82. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.