Significance
The goal of precision medicine is to identify selective drugs that modulate disease-causing biomolecules. This slow process often involves developing a high-throughput screen to test millions of potential drugs to find a few that affect the biomolecule. Here, we describe a facile approach using a disease-causing biomolecule’s sequence to enable design of specific drugs, eliminating arduous and time-consuming screens. By using the sequence of a non–protein-coding, oncogenic RNA, we designed a drug specifically targeting the RNA’s folded structure. In cells and animals, the drug inhibits its target, killing cancer cells while leaving healthy cells unaffected. Thus, a preclinical anticancer drug candidate can be quickly designed from sequence.
Keywords: RNA, noncoding RNA, drug design, chemistry, nucleic acids
Abstract
The design of precision, preclinical therapeutics from sequence is difficult, but advances in this area, particularly those focused on rational design, could quickly transform the sequence of disease-causing gene products into lead modalities. Herein, we describe the use of Inforna, a computational approach that enables the rational design of small molecules targeting RNA to quickly provide a potent modulator of oncogenic microRNA-96 (miR-96). We mined the secondary structure of primary microRNA-96 (pri-miR-96) hairpin precursor against a database of RNA motif–small molecule interactions, which identified modules that bound RNA motifs nearby and in the Drosha processing site. Precise linking of these modules together provided Targaprimir-96 (3), which selectively modulates miR-96 production in cancer cells and triggers apoptosis. Importantly, the compound is ineffective on healthy breast cells, and exogenous overexpression of pri-miR-96 reduced compound potency in breast cancer cells. Chemical Cross-Linking and Isolation by Pull-Down (Chem-CLIP), a small-molecule RNA target validation approach, shows that 3 directly engages pri-miR-96 in breast cancer cells. In vivo, 3 has a favorable pharmacokinetic profile and decreases tumor burden in a mouse model of triple-negative breast cancer. Thus, rational design can quickly produce precision, in vivo bioactive lead small molecules against hard-to-treat cancers by targeting oncogenic noncoding RNAs, advancing a disease-to-gene-to-drug paradigm.
Many disease association studies have identified biomolecules that, if targeted selectively, could be exploited to provide precision lead therapeutics. Generally, drug-like compounds are screened to discover modulators of the protein products of genes, particularly enzymes (1). However, evidence is emerging that RNA is an attractive and unexploited target for precise medicine strategies. One major implication of the Encyclopedia Of DNA Elements (ENCODE) project was that noncoding RNAs play major roles in human biology, including disease biology (2). Furthermore, sequencing techniques that directly report the identity and abundance of RNAs (3, 4) have revealed many potentially druggable RNAs that are differentially expressed in healthy and disease-affected tissue, for which a precision medicine approach could be ideal.
Outside of the bacterial ribosome, however, RNA is considered undruggable with small molecules because of the difficulty in identifying high-affinity, selective compounds. Despite this challenge, there are many advantages of targeting RNA with small molecules. For example, RNA secondary structure can be rapidly annotated from sequence (5–9), thereby identifying functional regions that could be inactivated by small molecule binding. We reasoned that gene expression data could be quickly converted into precise, lead therapeutic modalities by combining these methods with rational design strategies.
As part of a program to rationally drug RNAs from sequence, we established a database of RNA motif–small molecule binding partners (10). The database is comprised of privileged interactions identified by a library vs. library screen named 2D combinatorial screening (2DCS) (11, 12). In 2DCS, a library of array-immobilized small molecules is probed for binding to a library of small RNA motifs that are likely to be present in biological RNAs (11, 12). These interactions are then mined against genomic RNAs to provide lead therapeutics in a target agnostic manner by Inforna (10), a computational approach developed in our laboratory. That is, Inforna parses RNA secondary structures, regardless of whether they are determined by phylogeny, structural methods, or computation, and compares them with the RNA motif–small molecule database. Overlap provides lead compounds. Previously, Inforna identified the lead benzimidazole 1, which avidly bound to the RNA motif present in a Drosha nuclease-processing site in the microRNA-96 (miR-96) hairpin precursor (10), an oncogenic microRNA (miRNA) implicated in breast cancer (13). This miRNA suppresses the production of proapoptotic forkhead box O1 (FOXO1) protein in cancer cells. Targeting the miRNA with an oligonucleotide (an antagomir) or compound 1 selectively inhibits production of mature miR-96 and triggers apoptosis in breast cancer cells at micromolar concentrations (10, 13).
Herein, we used Inforna for the facile lead optimization of 1, providing highly potent compounds that impede tumorigenesis in vivo. Inforna identified that a bis-benzimidazole (2) bound to a 1 nt × 1 nt internal loop adjacent to the Drosha site bound by 1 (Fig. 1). The optimal dimeric compound that targets both sites, Targaprimir-96 (3), binds primary microRNA-96 (pri-miR-96) with low nanomolar affinity (>40-fold more avidly than 1) and is >400-fold more potent than 1 for inhibiting miR-96 biogenesis in breast cancer cells. The effect of 3 is specific to breast cancer cells, because nontumorigenic mammary epithelial cells are unaffected. Furthermore, target validation studies using Chemical Cross-Linking and Isolation by Pull-Down (Chem-CLIP) (14, 15) demonstrate that 3 engages pri-miR-96 in cells, and rescue experiments are consistent with the proposed mechanism of action of 3. Compound 3 has a favorable pharmacokinetic profile in mice, and thus we studied the effect of the compound on tumor growth in an in vivo preclinical model of triple-negative breast cancer (TNBC). Indeed, 3 inhibited tumor growth, showing that it is possible to target oncogenic noncoding RNAs through small molecules as a potential treatment for breast cancer.
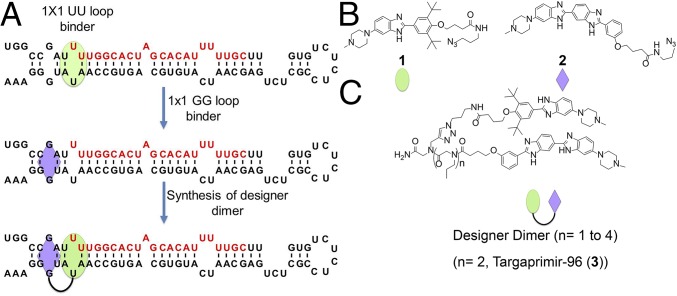
Fig. 1.
Inforna enabled a structure-based approach to rationally design a high-affinity, dimeric small molecule that targets the oncogenic miR-96 hairpin precursor. (A) Structure of the miR-96 hairpin precursor. The mature miRNA is written in red letters, and binding sites for 1 and 2 are indicated with a green circle and purple diamond, respectively. (B) Structures of RNA-binding modules 1 (10) and 2. (C) Structures of the designer dimeric compounds, in which RNA-binding modules 1 and 2 are displayed on a peptoid backbone. The number of propylamine spacing modules (n) controls the distance between 1 and 2. The compound with two spacing modules (n = 2) is named Targaprimir-96 (3), the most potent inhibitor of miR-96 processing.
Results and Discussion
Rational Design of a Dimeric Compound That Targets pri-miR-96.
Previously, 1 was found to precisely target the Drosha site in the miR-96 hairpin precursor (Fig. 1 A and B), inhibiting its biogenesis, derepressing downstream targets, and triggering apoptosis in breast cancer cells (10). However, this compound had micromolar activity, limiting its applicability in vivo. To optimize 1 to provide a more potent modulator of miRNA dysfunction, we used Inforna to mine the pri-miRNA’s secondary structure in an attempt to identify small molecule “modules” that are predicted to bind motifs nearby the Drosha processing site where 1 binds (Fig. 1A). Indeed, Inforna predicted that compound 2, a bis-benzimidazole, binds to the 1 × 1 GG internal loop adjacent to the Drosha site (Fig. 1 A and B). Thus, we used 1 and 2 as modules to design a dimeric compound that would enhance the avidity of the compounds for pri-miR-96 and hence, its potency (Fig. 1C).
Previously, we have shown that dimeric compounds can be designed to bind adjacent motifs simultaneously in an RNA by using spacing rules to span different distances (16). Such a multivalent approach has been shown to improve avidity and potency to study and manipulate biology in many systems (17–23). For example, a modular approach has been transformationally applied to target the DNA minor groove with polyamides by using eponymous Dervan rules (24). An analogous modular approach for small molecules targeting RNA is emerging and has provided a rapid enhancement of affinity and selectivity, because molecular recognition depends on the identity of the RNA-binding modules, their RNA-binding preferences, and the spacing between the modules (20).
We, therefore, designed a small library of dimers in which the distance between 1 and 2 was varied. Previous studies on spacing rules for targeting RNA internal loops separated by different distances showed that two to four propylamine spacing modules spanned the distance between two internal loops separated by 2 bp (16, 20), the same distance separating the 1 × 1 GG internal loop targeted by 2 and the Drosha site targeted by 1. We screened the library of dimers for their ability to inhibit biogenesis of miR-96 in MDA-MB-231 cells, a model of TNBC (Fig. 2). As anticipated, these studies showed that two propylamine spacers between the RNA-binding modules provided the most potent compound, which we named Targaprimir-96 (3; indicating that the compound targets a pri-miRNA and pri-miR-96 in particular).
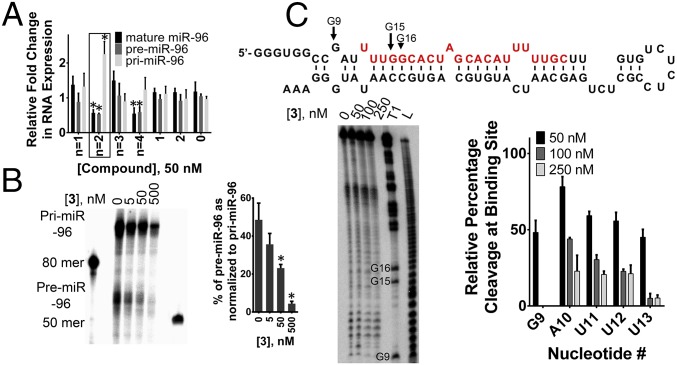
Fig. 2.
Studying the effect of designer dimeric compounds on miR-96 biogenesis. (A) Effects of designer dimeric compounds (50 nM) with varied spacing between the RNA-binding modules (n = 1 to n = 4) on pri-, pre-, and mature miR-96 levels in MDA-MB-231 TNBC cells. The compound with n = 2, as indicated in the box, is the most potent inhibitor of miR-96 processing and named 3. (B) Effect of 3 on the in vitro processing of pri-miR-96 by Drosha, indicating that the compound binds the desired site. Error bars are the SDs in the measurements. *P < 0.05 as determined by a two-tailed Student t test (n ≥ 3). (C) Compound 3 binds the predicted site as evidenced by protection from S1 nuclease cleavage. Lane T1 indicates cleavage by T1 nuclease under denaturing conditions (cleaves Gs). Lane L indicates a hydrolysis ladder.
We next performed a dose–response of 3 by assessing the reduction of mature miR-96 levels and determined an IC50 of ∼50 nM (SI Appendix, Fig. S1). Because 3 was designed to inhibit Drosha processing, we also studied the effect of the compound on pri- and precursor microRNA-96 (pre-miR-96) levels. As expected, treatment of TNBC with 3 boosted the amount of the pri-miRNA and decreased the levels of the pre-miRNA and mature miRNA in a dose-dependent manner (Fig. 2A and SI Appendix, Fig. S1). We further assessed inhibition of pri-miR-96 processing by 3 in vitro. Using a radioactively labeled model of pri-miR-96 and cell extracts in which Drosha was overexpressed, 3 inhibited cleavage of pri-miR-96 at low-nanomolar concentrations (Fig. 2B), thus validating that the compound binds the desired sites in the miRNA precursor. Furthermore, nanomolar concentrations of 3 protected the predicted binding site from cleavage by S1 nuclease, which supports the observed in vitro activity and establishes the binding site of 3 (Fig. 2C).
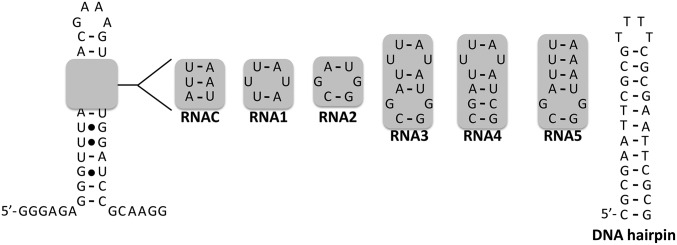
The binding affinities of 3 for various RNAs and DNAs were measured to assess the bonus in avidity that the dimeric compound provides over monomers 1 and 2 (Fig. 3 and Table 1). These studies showed that 1 bound to the motif in miR-96’s Drosha processing site (RNA1) (Fig. 3) with a Kd of 1,300 nM. Mutation of the UU loop to an AU pair (RNAC) eliminates binding. Likewise, our binding studies show that 2 binds the desired 1 × 1 GG loop (RNA2) with a Kd of 1,500 nM but does not bind to RNA1 or control RNAC. We also measured the affinities of 1 and 2 to a model RNA that contains both the Drosha target site for 1 and the adjacent target site for 2 (RNA3) (Fig. 3), affording Kd values of 3,400 and 2,700 nM, respectively.
Fig. 3.
Secondary structures of the nucleic acids to which the binding of monomeric small molecules 1 and 2 and dimeric compound 3 was assessed. RNAC is a control RNA that does not contain target sites for 1 or 2 and has a single-nucleotide change relative to RNA1. RNA1 contains the motif present in the Drosha processing site of pri-miR-96. RNA2 contains the 1 × 1 nt GG internal loop that is adjacent to the Drosha site. RNA3 contains both the Drosha site and the adjacent 1 × 1 nt GG internal loop. RNA4 contains the UU internal loop present in the Drosha site and a single-nucleotide change to mutate the 1 × 1 nt GG internal loop to a GC pair. RNA5 contains the GG internal loop, but the UU loop has been mutated to an AU pair.
Table 1.
Binding of various small molecules to RNAs and a DNA hairpin
| Small molecule | RNAC | RNA1 | RNA2 | RNA3 | RNA4 | RNA5 | DNA hairpin |
| 1 | >30 | 1.3 ± 0.1 | 9.4 ± 0.1 | 3.4 ± 0.1 | 1.3 ± 0.2 | 7.4 ± 0.7 | >30 |
| 2 | >2 | >20 | 1.5 ± 0.2 | 2.7 ± 0.3 | >20 | 1.0 ± 0.1 | 0.2 ± 0.02 |
| Targaprimir-96 (3) | >4 | 1.2 ± 0.3 | 0.9 ± 0.2 | 0.085 ± 0.01; 0.020 ± 0.007* | 1.2 ± 0.3 | 1.5 ± 0.2 | >50 |
Kd values are reported in micromolar.
This measurement was completed in the presence of PEG 8000, a molecular crowding agent that allows in vitro conditions to mimic the molecularly crowded environment in cells.
Dimeric 3 binds RNA3 with a Kd of 85 nM, an increase of ∼40-fold relative to monomer 1 and ∼30-fold relative to 2. As expected, 3 forms a 1:1 complex with RNA3 as assessed by a Jobs plot (25), indicating that the compound binds both internal loops simultaneously (SI Appendix, Fig. S2). Mutation of the GG or UU loop to a base pair reduces binding affinity to afford Kd values of ∼1,200 nM (GG to GC mutation; RNA4) and ∼1,500 nM (UU to UA mutation; RNA5), similar to the affinities of the respective monomers. The cellular environment is best mimicked by using molecular crowding agents, and addition of these agents can affect the binding of ligands to RNA (26). Thus, binding of 3 to RNA3 was measured in the presence of PEG 8000, which afforded a Kd of 20 nM or a fourfold enhancement.
Because selective binding of small molecules to RNA over DNA is an important consideration, we measured the affinities of 1, 2, and 3 for an AT-rich DNA hairpin (Fig. 3 and Table 1). Although 3 does not bind to the DNA hairpin (Kd > 50,000 nM), 2 binds to this DNA with a Kd of 200 nM. Thus, appendage of 2 to 1, a molecule that does not bind DNA because it has bulky t-butyl groups (12, 27), ablates the binding of 3 to DNA. Taken together, control of target binding in multivalent ligands depends on the identity of the modules individually and collectively and the distance between them, which has been previously observed (20, 28).
Collectively, these rigorous binding analyses show that 3 is highly RNA-selective and recognizes both the 1 × 1 nt GG and 1 × 1 nt UU loops to provide high affinity, effectively discriminating against a variety of related targets.
Targaprimir-96 Triggers Apoptosis in TNBC.
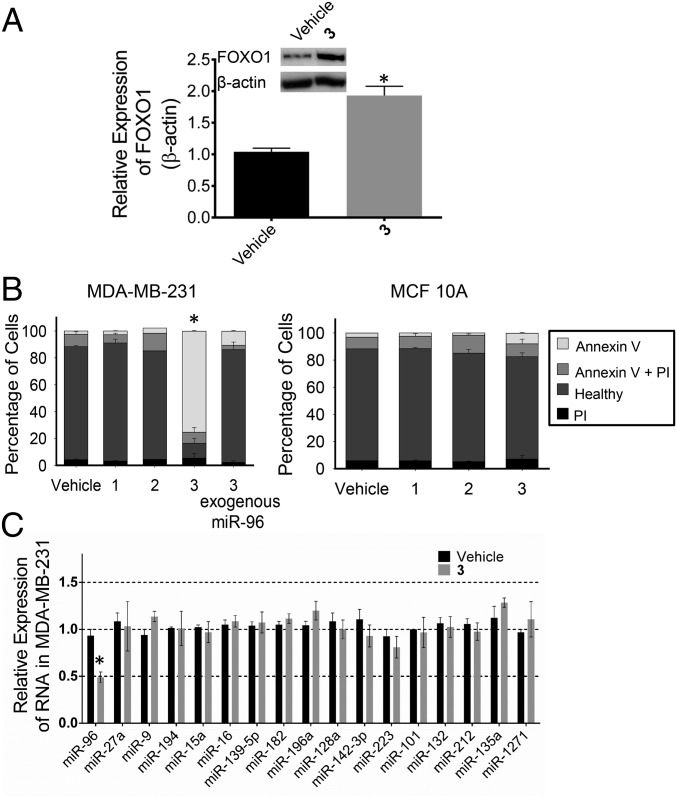
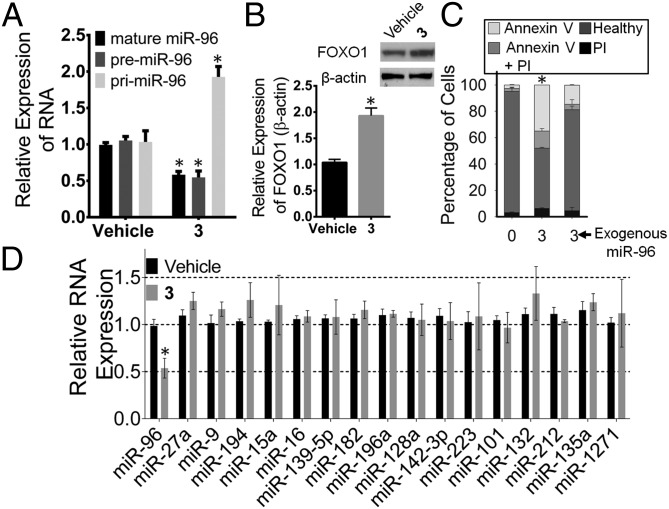
Various experiments were completed to assess the biological impact of 3. In MDA-MB-231 TNBC cells, 50 nM compound selectively inhibited biogenesis of the miR-96 hairpin precursor (Fig. 2A and SI Appendix, Fig. S1). Previously, it was shown that miR-96 contributes to cancer by silencing FOXO1 mRNA’s translation (13). Thus, the effect of the compound on FOXO1 protein expression was assessed. These studies showed that treatment with 50 nM 3 boosted expression of FOXO1 by approximately twofold (Fig. 4A). Additionally, 3 triggered early apoptosis in MDA-MB-231 cells as assessed by Annexin V staining but had no significant effect on a nontumorigenic mammary epithelial cell line (MCF 10A) (Fig. 4B).
Fig. 4.
Effect of 3 in TNBC and nontumorigenic mammary epithelial breast cells. (A) Effect of 3 on FOXO1 protein expression, because miR-96 silences FOXO1 protein to suppress apoptosis (13). (B) Effects of compounds 1, 2, and 3 (50 nM) on MDA-MB-231 (TNBC) and MCF 10A (nontumorigenic mammary epithelial breast) cells with or without forced overexpression of pri-miR-96. Compound 3 only triggers apoptosis in MDA-MB-231 cells, and this effect is ablated upon exogenous expression of pri-miR-96. (C) Profiling of all miRNAs predicted to target FOXO1 mRNA by using TargetScan (37) in 3-treated MDA-MB-231 cells. Error bars are the SDs in the measurements and in some cases, too small to be seen. *P < 0.05 as determined by a two-tailed Student t test (n ≥ 3). PI, propidium iodide.
The selectivity of 3 for triggering apoptosis through the miR-96 circuit was assessed in two ways: (i) by forced overexpression of the target followed by compound treatment and (ii) by profiling of other miRNAs predicted to regulate FOXO1 expression. The amount of apoptosis triggered by 3 in cells that overexpressed pri-miR-96 was significantly diminished (Fig. 4B), suggesting that the compound triggers apoptosis by inhibiting miR-96 biogenesis.
Using quantitative real-time PCR (qRT-PCR), we profiled the effect of 3 (50 nM) on the levels of miRNAs that have potential to modulate FOXO1 mRNA (n = 16), including miR-27a and miR-182 (transcribed as a cluster with miR-96), which were previously shown to silence FOXO1 expression in breast cancer cells (13). Among these miRNAs, only miR-96 expression was affected (Fig. 4C). These data indicate that down-regulation of miR-96 is sufficient to increase FOXO1 expression and induce apoptosis in MDA-MB-231 cells. Thus, 3 can counteract the effects of a single miRNA to promote cancer cell death.
We next studied the potency of 3 in breast cancer cell line 4175 derived from MDA-MB-231, which has been selected to metastasize to the lung (Fig. 5) (29). In agreement with our studies in MDA-MB-231, 3 (50 nM) inhibits biogenesis of miR-96 as determined by measurement of mature (decreased), pre-miR-96 (decreased), and pri-miR-96 (increased) levels (Fig. 5A). Furthermore, treatment with 3 increases FOXO1 levels (Fig. 5B) and triggers apoptosis (Fig. 5C), whereas forced expression of pri-miR-96 decreases the ability of 3 to induce apoptosis (Fig. 5C). As observed in MDA-MB-231 cells, among 16 miRNAs with potential to regulate FOXO1 levels, only miR-96 is affected (Fig. 5D).
Fig. 5.
Effect of 3 in 4175 lung metastatic breast cancer cells derived from MDA-MB-231. (A) Effect of 3 (50 nM) on pri-, pre-, and mature miR-96 levels. (B) Effect of 50 nM 3 on FOXO1 protein expression. (C) Ability of 3 (50 nM) to trigger apoptosis with or without forced overexpression of pri-miR-96. (D) Profiling of all miRNAs predicted to target FOXO1 mRNA by using TargetScan (37) in 3-treated 4175 cells. Error bars are the SDs in the measurements. *P < 0.05 as determined by a two-tailed Student t test (n ≥ 3). PI, propidium iodide.
Target Engagement in Cells by Chem-CLIP and Competitive Chem-CLIP.
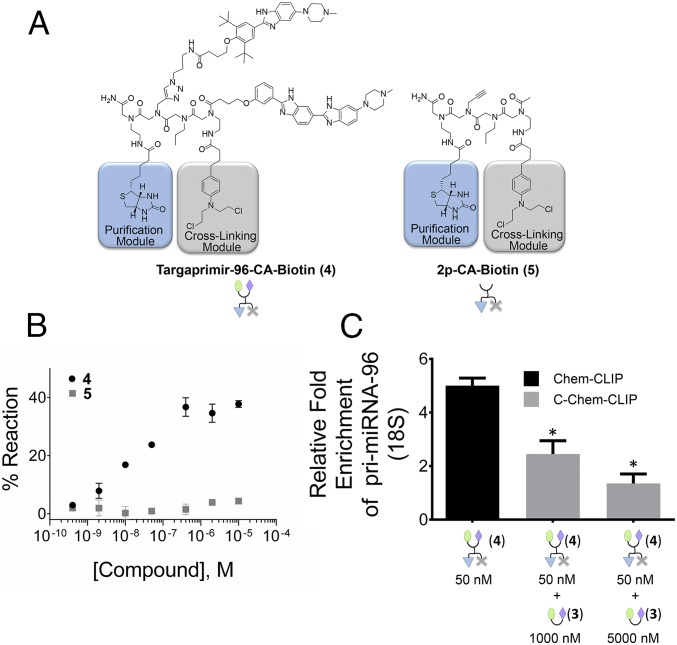
To determine if pri-miR-96 is a direct cellular target of 3, we used Chem-CLIP and Competitive Chem-CLIP (C-Chem-CLIP). In these approaches, small molecules that target RNA are appended with a nucleic acid reactive module that allows small molecules to be conjugated to their cellular RNA targets. Additionally, the compound is appended with a biotin module that allows for isolation of biotinylated products. Thus, Chem-CLIP is a steady-state experiment, whereby cross-linking could trap the intended and other transient interactions, because the products are stable once cross-linked. To enable these approaches, Targaprimir-96-CA-Biotin (4) was synthesized (Fig. 6A). Indeed, 4 selectively reacts with pri-miR-96 in vitro (Fig. 6B). A control compound, in which the RNA-binding modules are removed [2p-CA-Biotin (5)] (Fig. 6A) does not react with pri-miR-96 (Fig. 6B). Application of 50 nM 4 to MDA-MB-231 breast cancer cells and quantification of the isolated targets showed an approximately fivefold enrichment of pri-miR-96 (Fig. 6C).
Fig. 6.
Chem-CLIP and C-Chem-CLIP (14, 15) to study the engagement of pri-miR-96 by 3. (A) Structure of the Targaprimir-96-CA-Biotin (4) Chem-CLIP probe and the control compound that lacks RNA-binding modules [2p-CA-Biotin (5)]. (B) In vitro assessment of the compounds in A for reacting with pri-miR-96. (C) Chem-CLIP and C-Chem-CLIP profiling in MDA-MB-231 cells and quantification by qRT-PCR showing that 4 enriches the pri-miR-96 target in the pull-down. Coaddition of 3 (1,000 or 5,000 nM) and 4 (50 nM) depleted the amount of pri-miR-96 in the pull-down. Error bars are the SDs in the measurements. *P < 0.05 as determined by a two-tailed Student t test (n ≥ 3).
In C-Chem-CLIP, cells are cotreated with the Chem-CLIP probe and the unreactive parent compound. Cellular targets that are bound selectively by the parent compound are depleted in the pull-down. That is, C-Chem-CLIP accounts for nonselective effects due to the cross-linking and purification modules. As expected, 3 reduces the amount of pri-miR-96 pulled down by 4 in a dose-dependent manner (Fig. 6C).
Pharmacological Testing of 3.
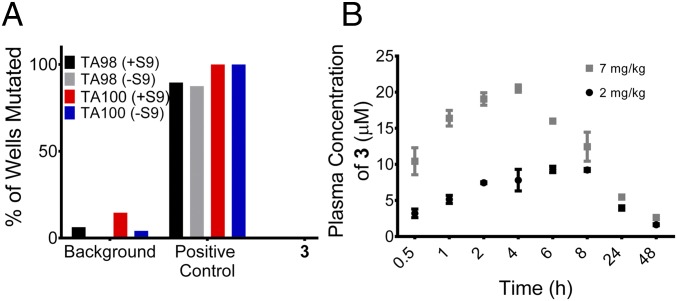
Because we were interested in the in vivo application of 3, its mutagenicity was measured by an Ames test (30, 31). There is potential for any compound that binds nucleic acids to be mutagenic, which would limit therapeutic potential. Fortuitously, 3 was negative throughout the Ames test and is thus not mutagenic under the conditions and strains evaluated (Fig. 7A).
Fig. 7.
Pharmacological profiling of 3 for mutational liability and stability. (A) Results of the Ames test, which measures the mutagenicity of a compound and thus, its potential to cause cancer. TA98 and TA100 are two strains of S. typhimurium; +S9 and −S9 indicate the presence and absence of rat liver extract S9, respectively. Nitrofluorene, amino anthracene, and sodium azide were used as positive controls. (B) Amount of 3 in mouse plasma as a function of time after i.p. injection of 2 and 7 mg/kg compound.
Drug Metabolism and Pharmacokinetics Analysis of 3.
Compound 3 is a dimeric compound that has a greater molecular weight than small molecules that obey the guidelines by Lipinski et al. (32) for small molecule drug development. Because of this potential liability, drug metabolism and pharmacokinetics (DMPK) studies were completed in vivo. Mice were treated with different concentrations of compound (2 or 7 mg/kg) by i.p. injection, and the amount of intact compound in blood plasma was measured as a function of time. For both doses, the amount of intact compound in plasma peaks at ∼4 h (Fig. 7B). Importantly, even 48 h postinjection, the concentration of 3 remaining in plasma is much greater than the 50 nM cellular concentration that triggered apoptosis: 1.6 μM for the 2 mg/kg dosage and 1.9 μM for the 7 mg/kg dosage (Figs. 4B and 7B). Thus, 3 has great potential to be bioactive in vivo.
Targaprimir-96 (3) Inhibits Tumor Growth in a Mouse Model of TNBC.
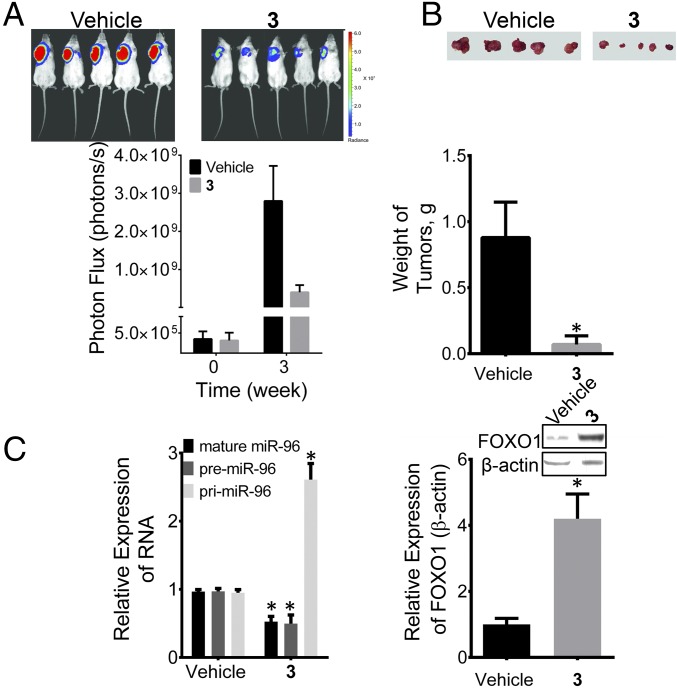
Collectively, our cellular and DMPK data indicate that 3 has potential to inhibit tumor growth in vivo. Nod/SCID mice were injected with a variant of MDA-MB-231 that expresses luciferase, or MDA-MB-231-luc, and allowed to develop tumors. After tumor development, mice were treated with 3 (10 mg/kg) every other day over the course of 21 d. Indeed, 3 significantly decreased tumor growth as assessed by both luciferase measurements and the size and weight of the excised tumor (Fig. 8 A and B). In agreement with cellular assays (Figs. 2A and 4A), profiling of the tumors showed that addition of 3 decreased levels of mature miR-96 by ∼50% and increased levels of pri-miR-96 (Fig. 8C), with a concomitant increase of FOXO1 (Fig. 8C). Furthermore, no toxicity was observed. These studies suggest that small molecules that target miRNAs in vivo have promise not only as chemical probes of RNA function, but also as future anticancer therapeutics.
Fig. 8.
In vivo studies of 3 on tumor growth in MDA-MB-231 TNBC. (A) Treatment of Nod/Scid mice xenografted with MDA-MB-231-luc cells with 3 inhibits tumor growth. (B) Images of the excised tumors showing decreased tumor size and mass. (C) Effect of 3 on miR-96 biogenesis (Left) and FOXO1 protein levels (Right) in tumors excised from untreated and treated mice. Error bars are the SDs in the measurements. *P < 0.05 as determined by a two-tailed Student t test (n = 5).
Conclusions and Outlook.
One goal of precision medicine is to assess the gene expression profile of disease-affected tissues and tailor patient treatment accordingly. Gene expression studies on tumor biopsies can quickly provide accurate information on genes that are up-regulated by measuring RNA levels. The most common way to directly target disease-associated RNAs is with oligonucleotide-based therapeutics that recognize RNAs by using Watson–Crick base pairing (33–35). Recognition of RNA targets by using sequence complementarity is a powerful technology, but one that still has obstacles to overcome. Herein, we present a complementary strategy, in which RNA structures are targeted with designer small molecules.
By using Inforna to identify multiple sites in a target that can be bound by small molecules and multivalency to design precision compounds that accommodate these sites simultaneously, an in vivo active compound targeting an oncogenic miRNA was identified. The observation that designer approaches can allow the rapid identification and subsequent lead optimization of bioactive compounds bodes well for the approach having broad utility for many other disease-associated RNAs that have a folded structure required for biological activity. Importantly, these studies also advance a paradigm, in which a disease-associated RNA is validated and its structure is used to design and optimize small molecules in rapid fashion. The applicability of this approach to other targets is likely to be enhanced as more information on the RNA motifs that bind small molecules is acquired, which could be enabled by 2DCS (11, 36).
Materials and Methods
General Methods.
General experimental procedures are given in the SI Appendix.
RNA Isolation and qRT-PCR of miRs.
Total RNA was extracted from cells that were cultured in either 6- or 12-well plates using a Quick-RNA Miniprep Kit (Zymo Research) per the manufacturer’s protocol. Approximately 200 ng of total RNA was used in reverse transcription (RT) reactions, which were completed using a miScript II RT Kit (Qiagen) per the manufacturer’s protocol. qRT-PCR was performed on a 7900HT Fast Real Time PCR System (Applied Biosystem) using Power SYBR Green Master Mix (Applied Biosystems). All primer sets were purchased from Integrated DNA Technologies (IDT) or Eurofins (SI Appendix, Table S1). The expression levels of mature miRNAs, pre-miR-96, and pri-miR-96 were normalized to U6 small nuclear RNA.
Chem-CLIP and C-Chem-CLIP.
Cells were cultured as described above in a 100-mm dish and treated with either 50 nM 4 (Chem-CLIP) or 50 nM 4 and 1 or 5 µM 3 (C-Chem-CLIP) for 24 h. Total RNA was extracted using TRIzol reagent (Ambion) per the manufacturer’s protocol. Approximately 30 µg of total RNA was used for pull-down by incubating with 50 µL of high-capacity streptavidin agarose beads (Thermo Scientific) in 1× phosphate-buffered saline (PBS) (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) for 30 min at room temperature. After centrifugation, the supernatant was removed, and the resin was washed with 1× PBS twice. RNA was eluted from the streptavidin beads by incubation with 100 µL of 1× Elution Buffer [10 mM EDTA, and 95% (vol/vol) formamide] at 65 °C for 20 min. The eluted RNA was cleaned up using the Quick-RNA Miniprep Kit per the manufacturer’s protocol. qRT-PCR was completed simultaneously for pri-miR-96 and 18S rRNA as described above using 50 ng of total RNA in the RT reaction. Expression levels of pri-miR-96 were normalized to 18S rRNA before and after pull-down by using the ΔΔCt (cycle threshold) method, and the ratio of their expression levels afforded enrichment.
Reaction of 4 and 5.
To determine if 4 or 5 reacts with the miR-96 hairpin precursor in vitro, 5 μL of 32P-labeled miR-96 hairpin precursor (∼50,000 cpm) was diluted in a total volume of 300 μL 1× PBS. The RNA was folded by heating at 60 °C for 5 min and slowly cooling to room temperature. Compound was then added at various concentrations, and the solutions were incubated for at least 4 h at room temperature. Next, 10 μL of streptavidin resin was added to the samples, which were incubated for an additional 30 min at room temperature. After centrifugation, the supernatant was removed, and the resin was washed with 1× PBS supplemented with 0.1% (vol/vol) Tween 20. The amount of radioactivity in the supernatant and wash and associated with the beads was measured using a Beckman Coulter LS6500 Liquid Scintillation Counter.
Ames Test.
Mutagenicity was evaluated using the MUTA-ChromoPlate Kit (Environmental Bio-Detection Products Inc.) according to manufacturer’s suggested protocol. Two strains of Salmonella typhimurium were used to detect frameshift mutations (TA98) and base pair substitutions (TA100). Mutagenic potential was studied in the presence (+S9) and absence (−S9) of rat liver extract S9. Nitrofluorene, amino anthracene, and sodium azide were used as positive controls. Mutagenicity was evaluated after 5 d of 3 exposure, which was tested at 10, 2, 0.4, 0.08, 0.02, 0.003, and 0.006 µM (n = 6 each).
DMPK Studies.
A six-mouse mini-pharmacokinetics (PK) assessment was used to determine the PK profile of 3. FVB/n mice were dosed i.p. with 3 at 2 or 7 mg/kg with 25-μL blood draws at indicated time points. Drug levels were determined by liquid chromotography/MS-MS using an ABSciex 5500.
Assessing Potency of 3 in Mouse Xenograft Model of TNBC.
NOD/Scid mice (B6.CB17-Prkdcscid/Sz; Jackson Labs) were housed in the Scripps Florida vivarium, and all experiments using live animals were approved by the Scripps Florida Institutional Animal Care and Use Committee. Female mice (n = 10; 5–7 wk old) were used for xenograft studies.
MDA-MB-231-luc cells (2 million) were suspended in 1× Dulbecco's phosphate-buffered saline (DPBS) supplemented with 0.9 mg/mL matrigel and injected s.c. into mice mammary fat pads. Tumor growth was monitored by injecting 100 µL of 30 mg/mL d-luciferin i.p. Imaging was completed over 2–5 min using a Xenogen IVIS 2000 Live Imaging System. Luminescence signals were quantified using IVIS analysis software by drawing a circular/oval region of interest around the luminescent signal. Mice were administered 10 mg/kg 3 by i.p. injection every 48 h after the tumor was detectable (at least 100,000 photons per second) by luminescence imaging. Tumors were monitored weekly by luminescent imaging. qRT-PCR and Western blot analyses were performed on mice 3 wk postvisualization of the tumor by luciferase imaging.
Compound Synthesis.
Details on synthesis and characterization of dimers (Fig. 1 and SI Appendix, Figs. S3–S6 and Scheme S1) as well as 4 and 5 (Fig. 6 and SI Appendix, Figs. S7 and S8 and Scheme S2) are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. John L. Cleveland for helpful advice, Prof. Joan Massagué for the 4175 cell line, Kimberley Lowe for assistance with flow cytometry, and Xiangming Kong for assistance with NMR spectroscopy. This work was funded by National Institutes of Health Grants R01-GM9455 and R01-GM9455-S1 (to M.D.D.) and The Nelson Fund for Therapeutic Development (M.D.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523975113/-/DCSupplemental.
References
- 1.Dews M, et al. The myc-miR-17∼92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res. 2010;70(20):8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 3.Farazi TA, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiang XF, et al. miR-20a promotes prostate cancer invasion and migration through targeting ABL2. J Cell Biochem. 2014;115(7):1269–1276. doi: 10.1002/jcb.24778. [DOI] [PubMed] [Google Scholar]
- 5.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101(19):7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 7.García-Sacristán A, et al. A magnesium-induced RNA conformational switch at the internal ribosome entry site of hepatitis C virus genome visualized by atomic force microscopy. Nucleic Acids Res. 2015;43(1):565–580. doi: 10.1093/nar/gku1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460(7256):711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 10.Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10(4):291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disney MD, et al. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130(33):11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 12.Velagapudi SP, Seedhouse SJ, French J, Disney MD. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J Am Chem Soc. 2011;133(26):10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284(35):23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan L, Disney MD. Covalent small-molecule-RNA complex formation enables cellular profiling of small-molecule-RNA interactions. Angew Chem Int Ed. 2013;52(38):10010–10013. doi: 10.1002/anie.201301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly E, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173(3):856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Childs-Disney JL, Tsitovich PB, Disney MD. Using modularly assembled ligands to bind RNA internal loops separated by different distances. ChemBioChem. 2011;12(14):2143–2146. doi: 10.1002/cbic.201100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. J Am Chem Soc. 2002;124(50):14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 18.Gordon EJ, Sanders WJ, Kiessling LL. Synthetic ligands point to cell surface strategies. Nature. 1998;392(6671):30–31. doi: 10.1038/32073. [DOI] [PubMed] [Google Scholar]
- 19.Kitov PI, et al. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403(6770):669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 20.Lee MM, et al. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: Targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009;131(47):17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37(20):2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Sucheck SJ, et al. Design of bifunctional antibiotics that target bacterial rRNA and inhibit resistance-causing enzymes. J Am Chem Soc. 2000;122(21):5230–5231. [Google Scholar]
- 23.Michael K, Wang H, Tor Y. Enhanced RNA binding of dimerized aminoglycosides. Bioorg Med Chem. 1999;7(7):1361–1371. doi: 10.1016/s0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 24.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg Med Chem. 2001;9(9):2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 25.Huang CY. Determination of binding stoichiometry by the continuous variation method: The Job plot. Methods Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- 26.Ohyashiki M, Ohyashiki JH, Hirota A, Kobayashi C, Ohyashiki K. Age-related decrease of miRNA-92a levels in human CD8+ T-cells correlates with a reduction of naïve T lymphocytes. Immun Ageing. 2011;8(1):11. doi: 10.1186/1742-4933-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sando S, Narita A, Aoyama Y. Light-up Hoechst-DNA aptamer pair: Generation of an aptamer-selective fluorophore from a conventional DNA-staining dye. ChemBioChem. 2007;8(15):1795–1803. doi: 10.1002/cbic.200700325. [DOI] [PubMed] [Google Scholar]
- 28.Pushechnikov A, et al. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: Application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131(28):9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ames BN, Durston WE, Yamasaki E, Lee FD. Carcinogens are mutagens: A simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci USA. 1973;70(8):2281–2285. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ames BN, Mccann J, Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975;31(6):347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 32.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 33.Stein CA, Cheng YC. Antisense oligonucleotides as therapeutic agents--is the bullet really magical? Science. 1993;261(5124):1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 34.Swayze EE, Bhat B. The medicinal chemistry of oligonucleotides. In: Crooke ST, editor. Antisense Drug Technology: Principles, Strategies, and Applications. 2nd Ed. CRC; Boca Raton, FL: 2008. pp. 143–182. [Google Scholar]
- 35.Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 36.Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem Biol. 2007;2(11):745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.