Significance
The conservation and cross-reactivity between species of the T-cell receptor (TR)-V regions and restricting major histocompatibility (MH) molecules characterizing innate-like T cells, natural killer T (NKT) and mucosal-associated invariant T (MAIT), indicate important functions for these cells. Yet, we show that the two MAIT-specific genes, TRAV1 and MR1, have been lost at least three times during the evolution of mammals. In the rabbit, which has few NKT cells and no MR1, we found a candidate invariant TR-α (iTRA) chain and another mammalian MH1Like molecule that seem to coevolve in mammals. Thus, at least three iTRA/MH-like systems were selected during mammalian evolution. The new MH1Like molecule may present a distinct set of antigens to a new innate-like T-cell subset. This study emphasizes the coevolution of TR and MH molecules.
Keywords: MHC, TCR, MAIT, evolution, mammals
Abstract
Whereas major histocompatibility class-1 (MH1) proteins present peptides to T cells displaying a large T-cell receptor (TR) repertoire, MH1Like proteins, such as CD1D and MR1, present glycolipids and microbial riboflavin precursor derivatives, respectively, to T cells expressing invariant TR-α (iTRA) chains. The groove of such MH1Like, as well as iTRA chains used by mucosal-associated invariant T (MAIT) and natural killer T (NKT) cells, respectively, may result from a coevolution under particular selection pressures. Herein, we investigated the evolutionary patterns of the iTRA of MAIT and NKT cells and restricting MH1Like proteins: MR1 appeared 170 Mya and is highly conserved across mammals, evolving more slowly than other MH1Like. It has been pseudogenized or independently lost three times in carnivores, the armadillo, and lagomorphs. The corresponding TRAV1 gene also evolved slowly and harbors highly conserved complementarity determining regions 1 and 2. TRAV1 is absent exclusively from species in which MR1 is lacking, suggesting that its loss released the purifying selection on MR1. In the rabbit, which has very few NKT and no MAIT cells, a previously unrecognized iTRA was identified by sequencing leukocyte RNA. This iTRA uses TRAV41, which is highly conserved across several groups of mammals. A rabbit MH1Like gene was found that appeared with mammals and is highly conserved. It was independently lost in a few groups in which MR1 is present, like primates and Muridae, illustrating compensatory emergences of new MH1Like/Invariant T-cell combinations during evolution. Deciphering their role is warranted to search similar effector functions in humans.
During infections, pathogens give rise to a variety of compounds able to stimulate the adaptive immune response. The compounds activating T cells can be peptides, glycolipids, and also unconventional small molecules, such as derivatives of the riboflavin (vitamin B2) biosynthesis pathway. Most conventional T cells recognize peptide–major histocompatibility complexes in which the peptide is embedded in the groove of the major histocompatibility (MH) proteins. MH class I (MH1) and MH class II (MH2) proteins present the peptide to CD8+ and CD4+ T cells, respectively. There are three classical MH1 molecules (also known as MH1a) in humans (HLA-A, HLA-B, and HLA-C) and in mice [MH1-K (H2-K), MH1-D (H2-D), and MH1-L (H2-L)]. Classic MH1 molecules are highly polymorphic with numerous alleles. The high variability of the amino acids constituting the groove leads to the presentation of distinct sets of peptides according to the alleles. There are three types of nonclassical MH1 molecules (also known as MH1b) characterized by limited polymorphism and presenting specific peptides (1). In humans, these molecules are HLA-E, HLA-F, and HLA-G, and in mice, they are MH1-M (H2-M), MH1-Q (H2-Q), and MH1-T (H2-T). A number of proteins with structural similarity to MH1 but which do not present peptides are known as MH1Like molecules (2). Examples of MH1Like proteins are CD1 (CD1A, CD1B, CD1C, CD1D, and CD1E) and MR1 in humans and CD1D and MR1 in mice. MH1Like proteins are not polymorphic. However, the groove of each kind of MH1Like protein displays specific biophysical features that lead to the presentation of different classes of compounds, such as glycolipids for CD1 (3, 4) or derivatives of the ubiquitous 5-amino-ribityl-uracil (5-A-RU), a microbial precursor of riboflavin (vitamin B2), for MR1 (5).
The high polymorphism of the classical MH1 reflects the rapid ability of the pathogens (especially viruses) to harbor mutations in their sequences that may prevent binding to the groove, and thereby their recognition by T cells. The lack of polymorphism of the MH1Like proteins is probably related to the difficulty for the pathogens to lose whole metabolic pathways; hence, these proteins are likely more focused on the presentation of antigens (Ags) originating from bacteria or yeasts, rather than from viruses. Indeed, the ligands presented by the MH1Like proteins are structural glycolipids in the case of CD1 or derivatives of chemical intermediates necessary for the synthesis of important coenzymes for MR1. For CD1D, the ligands can also be endogenous molecules, such as alpha-glycosyl-ceramides up-regulated in case of stress (6), which would represent a “danger” signal (7). No endogenous ligands are yet known for MR1.
The way in which T cells recognize Ags presented by classical and MH1Like molecules differs: T-cell receptor (TR) of conventional CD8+ T cells is made of TR alpha (TRA) and TR beta (TRB) chains that are encoded by rearrangements of any TRAV and TRAJ and any TRBV, TRBD, and TRBJ genes, respectively, with large variations in the complementarity determining region 3 (CDR3) length and amino acid composition. In contrast, T cells that are restricted by MH1Like molecules use semi-invariant TR usually made of an invariant TR-α (iTRA) chain coming from one particular TRAV/TRAJ rearrangement with a given CDR3 length, paired with a TRB chain using a limited number of TRBV genes combined with any TRBJ and without apparent restriction of the CDR3 sequence (8). The three types of semiinvariant αβ T cells described so far in humans are (i) CD1B-restricted T cells that use a TRAV1-2 (previously named Vα7.2)–TRAJ9 chain with a germline-encoded CDR3 lacking N diversity to recognize glucose monomycolate found in Mycobacterium tuberculosis, leading to their denomination as germline-encoded, mycolic acid-reactive (GEM) T cells (9); (ii) natural killer T (NKT) cells that use an invariant TRAV10 (previously named Vα24)-TRAJ18 chain to recognize alpha-glycosyl-ceramides presented by CD1D (3, 10); and (iii) mucosal-associated invariant T (MAIT) cells that use an invariant TRAV1-2 rearrangement to TRAJ33, TRAJ12, or TRAJ20 to recognize unstable pyrimidine adducts derived from 5-A-RU that are presented by MR1 (5, 11, 12). Other semiinvariant T cells also have been suspected in humans through the detection of other invariant TR chains (13).
From an evolutionary standpoint, classical MH1 proteins are subjected to strong evolutionary pressures that maintain a high level of polymorphism, with coevolution of the MH1 alleles and the pathogens. This finding is reflected by the excess of amino acid changes (nonsynonymous substitutions) between sequences of the MH1 binding groove of different species (14, 15). Notably, whereas the number of classical MH1 genes is usually two to three per species in mammals, the number of nonclassical MH1 and MH1Like genes is highly variable from one species to another (16). For instance, the MH1b genes are highly duplicated in mice and the MH1-M3 (H2-M3) that presents N-formyl-methionine peptides in mice is absent in humans (17). Five different CD1 (CD1A–CD1E) are present in humans, whereas only one, CD1D, is found in mice. In the absence of CD1B in mice, there is no homolog of GEM T cells in this species.
In fact, only two MH1Like proteins, CD1D and MR1, and their corresponding semiinvariant T cells, NKT cells and MAIT cells, are highly conserved in mammals (3, 10, 15, 18). Human and mouse NKT cells recognize alpha-galactosyl-ceramide (α-GC) presented by either human or mouse CD1D (19) despite only ≈70% identity between sequences of their α1 and α2 domains constituting the lipid-binding groove (20). Similarly, human and mouse MAIT cells recognize bacterially derived compounds presented by human and mouse MR1 proteins that are highly similar, with ≈90% identity (5, 15, 21, 22). Signs of purifying selection are apparent for the amino acids constituting the binding groove of MR1, indicating a strong selection pressure against diversification (15, 22). MR1 has not been as extensively duplicated as CD1 and remains the unique member of its type. Thus, its sequence appears to be more stable than other MH1Like proteins during evolution.
As for the TR, evolutionarily conserved features of the CDR1 and CDR2 of several TRBV genes have been proposed to enable a conserved docking mode of the TR onto the classical MH (23), although this remains controversial (24, 25). For TRA, the analysis is complex because the number of TRAV genes may be very large, with multiple subgroups and wide variation across species (≈50 to >350 within mammals). Typically, there are only one or two genes for TRAV1 and TRAV10 (or TRAV11 in mice) that characterize MAIT and NKT cells, respectively. TRAV1 interacts mainly with MR1, which is highly conserved. This finding is in stark contrast to most other TRAV subgroups, which are often much larger, interact with polymorphic classical MH1, and diversify during evolution.
This result leads to the question of whether the conservation of MR1 sequence would impart specific evolutionary pressures to the corresponding TRAV1 genes in comparison to TRAV genes used in conventional T cells. Based on the genomic data that are now available in many species, we studied the coevolution of both TRAV1 and MR1. In comparison to other TRAV genes, the CDR1 and CDR2 of TRAV1 are more constrained and the TRAJ33 used by MAIT cells is highly conserved during evolution. This finding is consistent with the idea that both MR1 and TRAV1 are evolutionarily locked by the interaction with a nonvariable compound. Unexpectedly, we observed that functional TRAV1 and MR1 are missing in three distinct groups of mammals, raising the question of the presence of compensatory mechanisms in species lacking MAIT cells. We examined the diversity of the MH1Like proteins across mammals, and found evidence of other MH1Like and TRAV proteins that display common features with MR1/TRAV1 and are present in MAIT-less species.
Results
MH1Like Proteins MR1 and CD1 Show Different Evolution Dynamics.
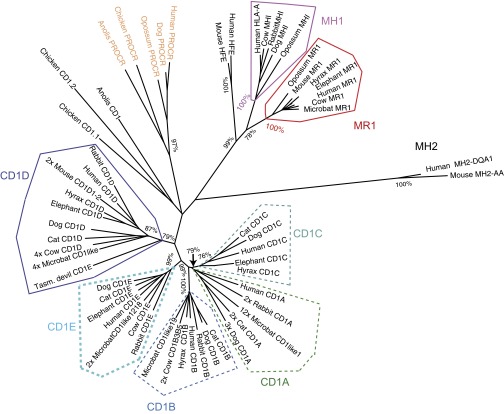
We first retrieved the available MR1 and CD1 sequences from public databases and compared the evolution of their α1 and α2 domains in mammals using mammalian and reptilian MH1 and MH1Like sequences for comparison and MH2 sequence as an outgroup (Fig. 1). MR1 is closely related to other MH1Like genes, such as HFE, PROCR (previously EPCR), and CD1 (Fig. 1 and Dataset S1). HFE and MR1 orthologs were found only in mammals, as reported by Tsukamoto et al. (26). Several molecules related to MH1 in turtles and crocodiles display some similarities with MR1, and might be related to the MR1 ancestor (Fig. S1A). A number of amino acids contacting the T-cell receptor (TR) are shared between mammalian MR1 and reptile sequences [including R43, R68, G69, and W70 in α1 and L64, Y66, E73, and W78 in α2 (using IMGT unique numbering for G domains (20); for example, K45 corresponds to the residue classically numbered K43 that is critical for binding 5-A-RU derivatives)], whereas they are lacking in CD1D or class II sequences. However, these reptile sequences lack critical motifs conserved in all MR1 (Fig. S1A); in particular, the amino acids involved in recognition of 5-A-RU derivatives (including R9, S13, K45, H59, and Y63 in α1 and R7 and I9 in α2) are found only in mammalian MR1, and not in related sequences from reptiles. A direct orthology relationship between these genes is also not supported by conserved synteny data. Because MR1 is present in marsupials but was not found in the monotreme platypus, this gene, with all its hallmarks, was probably selected during the evolution of the ancestors of living marsupials and eutherians. Strikingly, the neighbor-joining (NJ) tree and similarity scores of the α1 and α2 domains of MH1, CD1, and MR1 show that MR1 sequences have evolved less rapidly compared with MH1 or CD1 (Fig. 1 and Dataset S2 A and B), likely reflecting the conserved structure of the Ag presented.
Fig. 1.
NJ distance tree of MH1a, MR1, CD1, and related sequences. Protein sequences of α1-α2 domains were aligned using ClustalW, and a NJ tree was computed using Molecular Evolutionary Genetics Analysis 6 (MEGA6) (pairwise deletion, bootstrap: n = 1,000). Significant bootstrap values are indicated for critical nodes. Species: Reptilia: Anolis carolinensis, lizard; Gallus gallus, chicken; primates: Homo sapiens, human; rodents, etc.: Mus musculus, mouse; Oryctolagus cuniculus, rabbit; Laurasiatheria: Canis familiaris, dog; Felis catus, cat; Bos taurus, cow; Myotis lucifugus, microbat; Afrotheria: Procavia capensis, hyrax; Loxodonta africanus, elephant; marsupials: Monodelphis domesticus, opossum; Sarcophilus harrisii, Tasmanian devil.
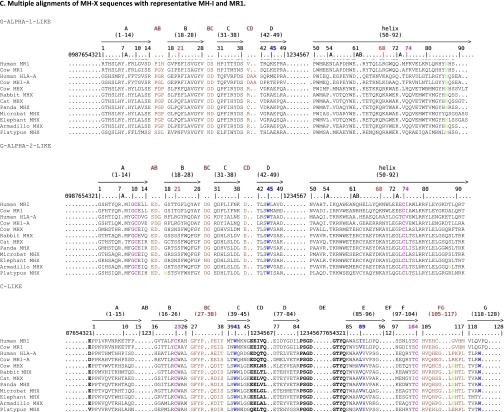
Fig. S1.
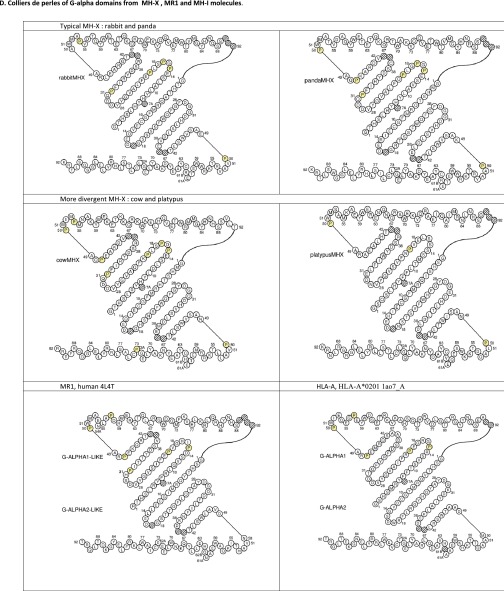
Multiple alignments and collier de perles of MR1, MR1-related, and MHX sequences. (A) Multiple alignments of MR1-related sequences found in reptiles with MR1 and other MH1 or MH1LIKE (α1 and α2 domains). MR1-related sequences from the tortoise (Chrysemys picta: XP_008175672) and alligators [Alligator mississippiensis (mis.): MR1-rel1:XP_014459775, MR1-rel2:XP_014462490; Alligator sinensis (sin.): XP_006032342] were aligned with human MR1 (NP_001522) and CD1D (P15813), opossum MR1 (XP_007480975), and mouse MH2 (H2-D1b; NP_034510). Positions of amino acids involved in MR1 interaction with the TR are highlighted in light blue, whereas positions of amino acids involved in MR1 interaction with the ligand are highlighted in yellow. Lysine 45 (43 in ref. 5) in the first domain is particularly crucial for 5-OP-RU recognition. The cysteines C11 and C74 of the conserved G-ALPHA2-LIKE/G-ALPHA2 disulfide bridge are highlighted in pink. Other colors follow the IMGT chart (www.imgt.org/IMGTScientificChart/RepresentationRules/colormenu.php#h1_36). Sequence numbering follows the IMGT unique numbering for G domains (20). (B) Multiple alignment of translation of mr1 pseudogene sequences found in carnivores with human MR1. Conserved stop codons are boxed. The black/blue color corresponds to exons in the human MR1, and red residues denote splicing sites. Conserved mutations are shown (stop codons or deletions are boxed). Species are as follows: human, Homo sapiens; dog, Canis familiaris; ferret, Mustela putorius; cat, Felix catus; and panda, Ailuropoda melanoleuca. (C) Multiple alignments of MHX sequences with representative MH-1 and MR1. The cysteines C11 and C74 of the conserved G-ALPHA2-LIKE/G-ALPHA2 disulfide bridge are highlighted in pink. The asparagines of N-glycosylation sites are highlighted in green, and other colors follow the IMGT chart (www.imgt.org/IMGTScientificChart/RepresentationRules/colormenu.php#h1_3). Sequence numbering follows the IMGT unique numbering for G domains (20). (D) Colliers de perles of G alpha domains from MHX, MR1, and MH-1 molecules. Collier de perle representations provide graphical 2D representations of the Gα domain of MH. Proline AAs are highlighted. Hatched circles correspond to missing positions according to the IMGT unique numbering.
In contrast to MR1, genes related to CD1 are found in birds and other reptiles, although no CD1 was found in fish, Xenopus, and other amphibians, or even in the platypus (Fig. 1). Reptilian CD1 genes do not seem to be specific orthologs of any of the human CD1 isotypes, because their extracellular part appears as an outgroup of the PROCR + CD1 branch of phylogenetic trees (e.g., they do not cluster with any of mammalian CD1 isotypes). However, they share a significant level of similarity with mammalian CD1 sequences, and likely are related to them.
There is apparently only one complete CD1 gene available from marsupials: The Tasmanian devil (Sarcophilus harrisii) genome contains a gene annotated as “CD1E” but more related to the CD1D group; a partial sequence is also present in the opossum. In contrast, eutherian CD1 sequences cluster in five main groups corresponding to human CD1A, CD1B, CD1C, CD1D, and CD1E. These groups have representatives in all of the main branches of eutherians. Hence, the diversification of CD1 likely occurred early in Placentalia evolution, after the separation with marsupials. The duplicates were mostly conserved in most lineages, and sometimes greatly amplified. The case of the microbat (Myotis lucifugus), but not the megabat (Pteropus vampyrus), is remarkable, with a massive diversification, especially for CD1A (Fig. 1). Apparently, only one or two CD1 genes, which are similar to CD1D, remain in the rat and mouse; however, this is not the case in the squirrel, indicating that the loss of CD1A–CD1C genes is not a general feature of the whole rodent group.
Thus, although CD1-like genes are older than MR1, these observations indicate that CD1 diversification happened (long) after MR1 appearance. However, CD1 sequences from different isotypes are much more divergent that MR1 in distant species.
MR1 Was Pseudogenized in Several Groups of Mammals.
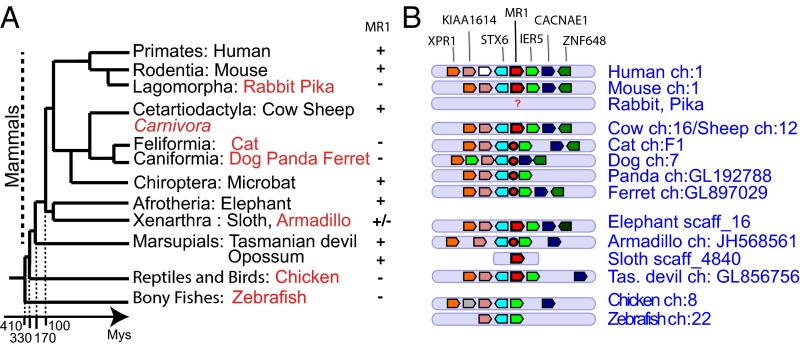
Surprisingly, a systematic survey of mammalian genomes showed that MR1 is not present in all eutherian groups (Fig. 2A), because it was not found in any carnivore genome, in the rabbit and pika genomes, or in the armadillo (Xenarthra). A comparison of the MR1 neighborhood across mammals shows that this region is, in fact, well conserved but contains a mutated MR1 sequence in a number of cases.
Fig. 2.
Absence of coding MR1 gene in carnivores, lagomorphs, and the armadillo. (A) Schematic representation of phylogenetic relationships between the main groups of mammals. Species in which no functional MR1 was found are shown in red. Note that the time scale is not linear. (B) Conserved synteny of the MR1 region across vertebrates, adapted from the Genomicus representation; MR1 genes are represented by a red polygon framed in black, and MR1 pseudogenes are represented by a red sphere framed in black. The color code corresponds to the different genes of the region.
Four genomes of carnivores are available: cat (Felix catus), dog (Canis lupus), panda (Ailuropoda melanoleuca), and ferret (Mustela putorius furo). The MR1 genomic region is well conserved in these species, with a translocation or perhaps misassembly in the dog genome, but contains a MR1 pseudogene in the expected position close to the marker stx6 (Fig. 2B). These pseudogenes are heavily mutated with a number of gaps or stop codons inside exons, of which distribution may reflect the phylogenetic relationships inside carnivores. For example, the cysteine 11 of the CELLE motif in the α2 domain [ImMunoGeneTics (IMGT) positions 10–15] is mutated in all four species, but is substituted by S in the cat and by a STOP codon in sequences from the dog, ferret, and panda, which are more closely related to each other (Fig. S1B).
No counterpart of MR1 could be identified in the rabbit (Oryctolagus cuniculus) and pika (Ochotona princeps), suggesting that it may also have been lost in lagomorphs. Best BLAST hits using human MR1 bait share about 40% identity with it, and were, in fact, more similar to human MH1 sequences. No conserved synteny could be found in genomic data in these two species, because markers (or pairs of markers) of the regions are found on different short scaffolds that are not connected in the current assemblies. Hence, it is not clear if the region has been extensively rearranged during evolution or if the available genomic data do not allow the reconstruction. A functional MR1 is also missing in the armadillo genome (Dasypus novemcinctus), in which a mutated MR1 sequence was found at the expected location between ier5 and stx6 markers as described above for carnivores (Fig. 2B). However, a functional gene was found in a short scaffold of the sloth (Choloepus hoffmanni), another representative of Xenarthra, suggesting that the pseudogenization observed in the armadillo is either restricted to this group or might be an artifact.
Taken together, these observations indicate that MR1 is highly conserved across most mammals, including marsupials, but has been lost or inactivated in several clades of eutherians (Table 1).
Table 1.
Correlation between the presence of functional MH1Like genes restricting invariant TR, and selected TRAV across mammals
| Mammal | MR1* | CD1B | TRAV1 | TRAV5 | CD1D | TRAV10 | TRAV22 | MHX | TRAV41 |
| Human | + | + | + | + | + | + | + | − | + (like) |
| Mouse | + | − | + | + | + | + | + | − | − |
| Rabbit | − | + (?) | − | + | + | + | + | + | + |
| Pika | − | + (?) | − | + | + | − | + | + | − (?) |
| Cow | + | + | + | + | + | + | + | + | + |
| Cat | − | + | − | + | + | + | + | + | + |
| Dog | − | + | − | + | + | + | + | ? | − |
| Panda | − | + | − | + | + | + | + | + | + |
| Ferret | − | − | − | + | + | + | + | + | + |
| Microbat | + | + | + | + | + | − | + | + | + |
| Elephant | + | − (?) | + | + | + | + | − | + | + |
| Sloth | + | − (?) | + | − (?) | + | − | + | + | + |
| Armadillo | − | − | − | + | + | + | + | + | + |
| Opossum | + | − | + | + | cd1† | − (?) | − (?) | − | − |
| Tasmanian devil | + | − | + | + | cd1† | − (?) | − (?) | − | − |
Gene names (human IMGT nomenclature).
Indicates that the best BLAST hit for a human sequence shows some significant similarity to it but is not clearly its closest counterpart.
Consistent Loss of TRAV1 Gene in Species in Which MR1 Is Mutated.
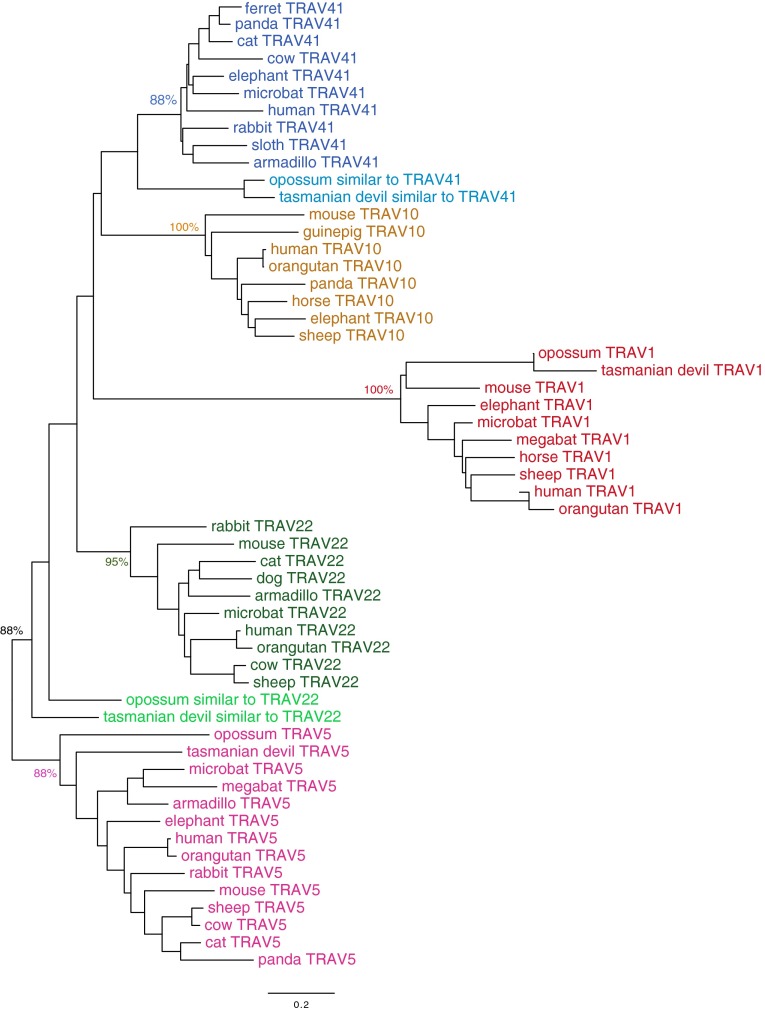
MR1-restricted MAIT cells use an iTRA chain that is produced by a rearrangement of TRAV1-2 to the TRAJ33 gene. This rearrangement and the MAIT specificity are conserved between species, at least between humans, cattle, and mice. Because MR1 is mostly recognized by TR comprising TRAV1-2, we investigated whether the lack of functional MR1 in some mammalian species would be correlated to a lack of TRAV1. The closest relatives of human TRAV1-2 were identified by a reciprocal BLAST approach in 15 representative species of mammals: two marsupials (opossum and Tasmanian devil), two xenarthrans (armadillo and sloth), an afrotherian (elephant, Loxodonta africana), two lagomorphs (rabbit and pika), a rodent (mouse), a primate (human), a bat (microbat), four carnivores (cat, dog, panda, and ferret), and a cetartiodactyl (cow). Putative counterparts of other human TRAV genes, including TRAV5, TRAV10, and TRAV22, were also identified across the same list of species. A NJ analysis was then performed to identify the orthology groups across these TRAV sequences (Fig. S2).
Fig. S2.
Distance (NJ) tree of TRAV1, TRAV5, TRAV10, TRAV22, and TRAV41 across mammals. TRAV sequences (Dataset S1) were aligned using ClustalW, and a NJ tree was computed using MEGA6 (NJ, bootstrap: n = 1,000; pairwise deletion). Key bootstrap values validating the TRAV groups (depicted in different colors) are shown.
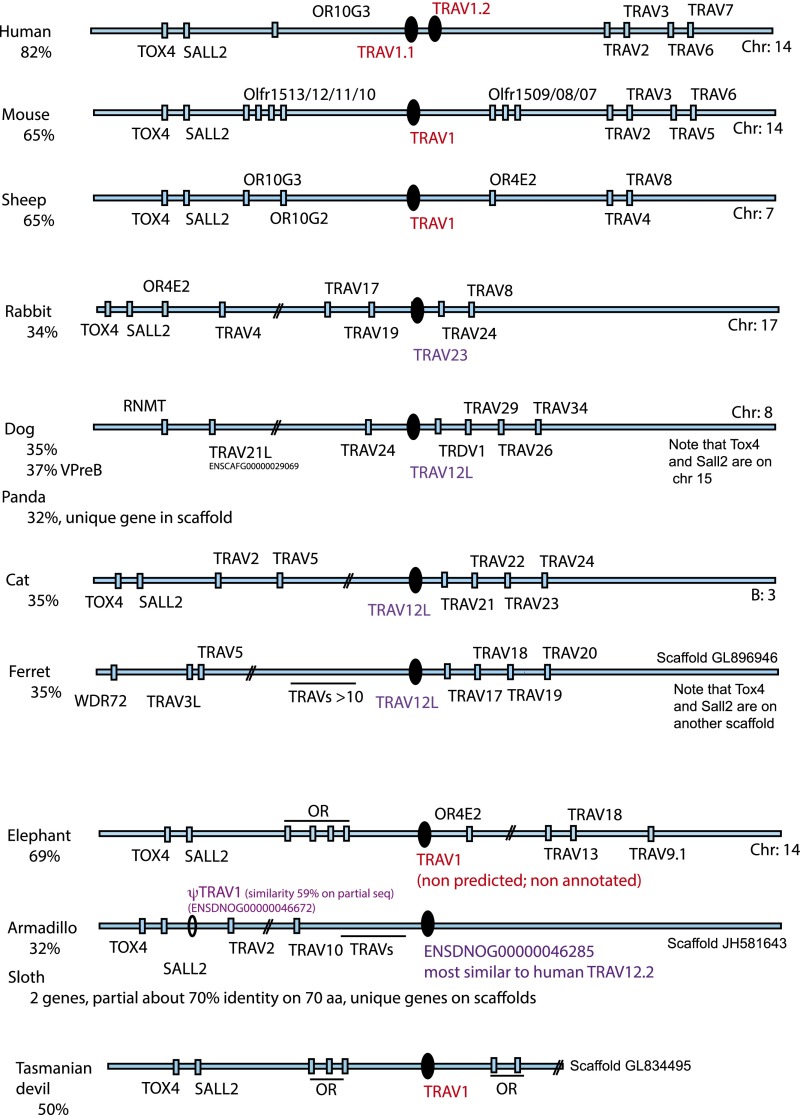
Human and mouse TRAV1 genes are located at the 5′ end of the TRAV locus (IMGT locus representation, www.imgt.org) (27), close to several conserved markers, such as tox4, sall2, and a number of olfactory receptor genes. The localization of the TRAV1 best BLAST hits within the locus was analyzed in each species and is represented in Fig. S3. All TRAV1 orthologous sequences (Fig. S2) were located at the beginning of the TRAV locus, close to tox4 and sall2, confirming the phylogenetic predictions and the validity of our approach.
Fig. S3.
Location of relevant TRAV in the TRA locus of different mammalian species. Locations of most similar counterparts to human TRAV1 in the TRA loci of different mammals are shown. TRAV loci are represented with conserved markers found on the 5' side of the locus (TOX4, SALL2, ORs) when they were present in the available contigs (Ensembl release 77). TRAV1 genes are represented in red; when TRAV1 was absent, the TRAV gene most similar to human TRAV1 is represented in purple. The percentage of sequence identity between human TRAV1 and the best match in the targeted species is given on the left.
Strikingly, the absence of a TRAV1-like gene at the beginning of the locus was systematically observed in all species lacking a functional MR1 gene (Table 1): in all four carnivores, in the rabbit and pika, and even in the armadillo. Within Xenarthra, it is remarkable that the armadillo lacks both functional MR1 and TRAV1, in contrast to the related sloth, in which a functional MR1 and a typical TRAV1 were found. Because sequences related to TRAV2 and TRAV5 (Table 1) were identified in lagomorphs, carnivores, and the armadillo, the lack of TRAV1 in these clades was not due to a global change of the 5′ side of the TRAV locus, but rather to a targeted loss of the TRAV1 gene.
Taken together, these results indicate that MR1 and TRAV1 are locked in an evolutionary unit, and MR1 remnants found in different species lacking TRAV1 suggest that the loss of TRAV1 likely preceded MR1 inactivation. Notably, TRAV1 is also used in humans by GEM T cells that are restricted by CD1B (9). No correlation between the presence/absence of TRAV1 and CD1B was found (Table 1), suggesting that other TRAV genes are used by CD1B-restricted T cells in species lacking TRAV1.
The TRA chain expressed by human CD1D-restricted NKT cells results from the rearrangement of TRAV10 to the TRAJ18 gene. In contrast to MR1, CD1D is found in lagomorphs and carnivores, as well as in all of the main groups of eutherians we investigated (Table 1). Accordingly, TRAV10 is widely distributed across eutherians: This gene is apparently missing only in the microbat, which has CD1D, as well as in the sloth (but not the armadillo) and pika (but not the rabbit). In the rabbit, a typical CD1D was found (Fig. 1), which is effectively expressed, as shown by the presence of multiple perfect matches in the Transcriptome Shotgun Assembly (TSA) database (e.g., GBCT01064921.1); a gene encoding a protein 74% similar to the human TRAV10 was also identified on chromosome 17, but was absent from all EST databases or the TSA database. These observations, as well as our own sequencing experiments (discussed below), suggested that the TRAV10/TRAJ18 combination with the canonical length characterizing NKT cells is expressed at low levels, suggesting, at best, a very low frequency of NKT cells in the rabbit. Moreover, the frequency of NKT cells was below the detection level when staining blood lymphocytes with α-GC–loaded murine CD1D tetramers.
Altogether, these observations suggest that CD1D was not lost in large groups of eutherians and indicate that the presence/absence of a TRAV10 gene used by the corresponding invariant TR is not strictly correlated to the occurrence of a functional CD1D.
TRAV1 CDR1 and CDR2 Are Highly Conserved Across Mammals.
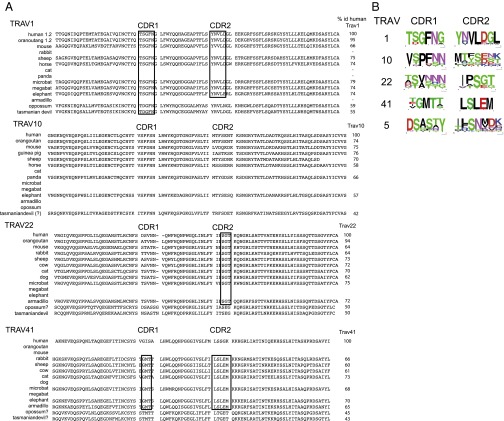
To test the hypothesis of a particular evolutionary pressure exerted on TRAV1 in comparison to TRAV expressed by conventional TR, we performed multiple alignments of sequences from various mammals and analyzed the variability of CDR1 and CDR2 loops (Fig. 3A).
Fig. 3.
Comparison of CDR1 and CDR2 of selected TRAV sequences across mammals. (A) Multiple alignments of TRAV1, TRAV10, TRAV22, and TRAV41 V exons from selected representative species of mammals are shown. CDR1 and CDR2 follow the IMGT definition, and highly conserved motifs in CDR are boxed. The percentage of identity of each sequence to the corresponding human TRAV is given to the right of the alignment. (B) CDR1 and CDR2 were extracted from multiple alignments, and their sequence variation was represented by frequency sequence logos. Note that the TRAV41 logo is based on conserved typical TRAV41 sequences, thus not including human and marsupials, which lack MHX. The Seqlogo of TRAV5 is provided as a control of TRAV not involved in the invariant TR chain.
Overall, the level of similarity of complete TRAV sequences between humans and other species is relatively constant, and TRAV1 does not appear particularly conserved. However, SeqLogo representations show that TRAV1 CDR1 and CDR2 are both particularly well conserved, whereas TRAV5 CDR2 and TRAV22 CDR1 are highly variable. Regarding TRAV10, which encodes the iTRA chain of NKT cells, the CDR2 that does not interact with the CD1D/Ag complex is also highly variable; in contrast, TRAV10 CDR1, which contacts the glycoside head of the Ag presented by CD1D, shows an intermediate level of conservation (Fig. 3). Thus, conservation of CDR loops reveals situations in which a TRAV and the corresponding MH1LikeAg complex are evolutionarily locked by specific interactions. TRAV1 and MAIT cells constitute a perfect example. Notably, it has already been observed that the most conserved amino acids in TRAV1 CDR make contact with MR1 (28).
A Well-Conserved TRAJ33 Is Present in All of the Main Groups of Mammals.
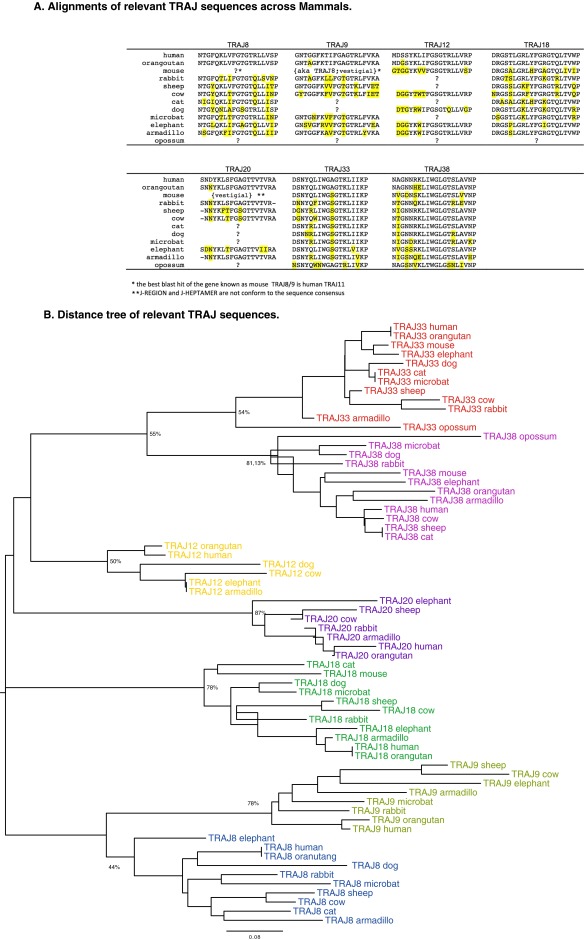
The orthologs of selected TRAJ genes were also identified in representative species of the main groups of mammals, and NJ analysis was performed to confirm that they constitute consistent evolutionary subsets (Fig. S4). Counterparts of the human TRAJ33, which is used by the iTRA chain of MAIT cells, were found in all species investigated as for TRAJ38. Thus, in contrast to TRAV1, this gene has not been lost in species in which there is no functional MR1. There was no correlation between the occurrence of TRAJ12 and TRAJ20, which are also used by human MAIT cells, and the presence/absence of MR1. With regard to human TRAJ18, which is part of the iTRA chain of NKT cells, counterparts were found in all eutherian groups but not in any marsupial species, which parallels the absence/presence of the CD1D gene.
Fig. S4.
Characteristics of relevant TRAJ sequences across mammals. (A) Alignments of relevant TRAJ sequences across mammals. Multiple alignments of the best match for TRAJ8, TRAJ9, TRAJ12, TRAJ18, TRAJ20, TRAJ33, and TRAJ38 from selected representative species of mammals are shown. Differences from human sequences are highlighted. (B) Distance tree of relevant TRAJ sequences. TRAJ sequences were aligned using Clustal, and a NJ tree was computed using MEGA6 (NJ, bootstrap: n = 1,000, pairwise deletion). Key bootstrap values corresponding to the TRAV groups (depicted in different colors) are shown.
Notably TRAJ9, which, together with TRAV1, is part of the semiinvariant TR of human GEM T cells, was not found in mice and the opossum (Fig. S4), which could fit with the absence of the CD1B gene in these species (Table 1). TRAJ9 was not retrieved in the dog, ferret, panda, or cat despite the presence of CD1B; this is, in fact, consistent with the lack of TRAV1 in carnivores, and suggests that GEM-like T cells restricted by CD1, if they exist, would use a different TRAV/TRAJ rearrangement to produce their iTRA chain.
ITRA Chains Expressing TRAV41 Are Found in the Rabbit in the Absence of MAIT Cells.
Our findings indicate that several groups of mammals lack typical MAIT cells. The case of the rabbit is particular, because this species lacks MR1 and TRAV1 and does not harbor detectable NKT cells in the blood. In the hypothesis of a compensatory mechanism, we searched for other innate T cells and MH1Like/iTRA systems in this species. For this purpose, we performed a 5′ RACE analysis of TRA transcripts, using a TRAC-specific primer, in three independent individuals and subjected the RACE products to deep sequencing. Reads were aligned from the 3′ end, and TRAJ segments were recognized through the detection of the F/YGXG (canonical or related) motif. TRAV segments were detected through the occurrence of the Y/FC (canonical or related) motif, upstream of the TRAJ sequence. Because the reads were performed from the 3′ end, only partial sequences of TRAV were obtained, but they were long enough to identify V genes.
We aggregated sequences sharing the same TRAV and TRAJ and encoding a CDR3 of a given length, and computed the frequency of such sequence sets, which we named “VJL combinations” (for V/J/length), within the total number of reads. We then compared the contribution of each rabbit to this frequency (Fig. 4A). Consistent with the CD1D tetramer data, we only detected very few occurrences of the iTRA chain corresponding to NKT cells: The corresponding TRAV10/TRAJ18 VJL represented 0.02%, 0.003%, and 0.01% of all functional VJLs in the blood of the three rabbits studied. Because the CDR3 sequence of this VJL was heterogeneous, it might just correspond to conventional T cells.
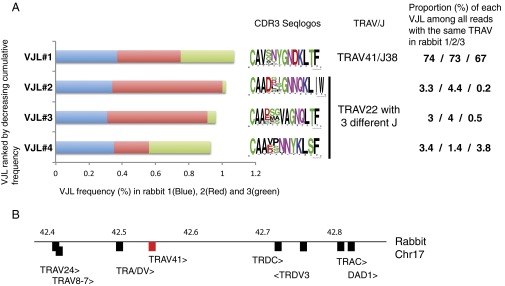
Fig. 4.
Rabbit TRAV41 shows features typical of iTRA chains. (A) Frequency of the VJL (TRAV/TRAJ/CDR3 length) combinations in repertoires sequenced from the blood cells of three independent rabbits. The relative frequency of VJL in each animal is denoted by a given color, and the frequency sequence logo of the CDR3 sequences encoded in the VJL is represented on the right. The unique VJL in which TRAV41 is implicated is ranked first according to the cumulative frequency over the three rabbits, and is well distributed between the individuals; the following less abundant VJL combinations use TRAV22 associated with different TRAJ segments, and encode CDR3 of variable sequence. They represent a minor fraction (0.2–4.5%) of transcripts comprising TRAV22, whereas VJL#1 represents a major fraction (≈70%) of transcripts comprising TRAV41. (B) Location of the TRAV41 within the TRA/D locus is on rabbit chromosome 17 (Chr17).
However, among the most frequent VJLs, two were found at significant frequencies in each individual (VJL#1 and VJL#4) (Fig. 4A). VJL#1 was particularly interesting because it showed several features suggestive of a TR invariant alpha chain (1): This VJL was consistently the most represented VJL in all three rabbits, accounting for 0.37%, 0.38%, and 0.32% of all reads with a functional VJL (2). VJL#1 expresses the rabbit counterpart of human TRAV41 (Fig. 4B), which is found in combination with 13 TRAJ, for a total number of VJLs of only 17 (3). Remarkably, TRAV41 CDR1 is highly conserved with a GMT motif and CDR2 is strictly identical (LSLEM) across many mammal species as different as the rabbit, cat, elephant, and armadillo (Fig. 3), and CDR3 within VJL#1 shows a limited variability (Seqlogo in Fig. 4A). VJL#4 was also well represented in all three individuals (Fig. 4A). It comprised TRAV22, but this gene is found in as many as 238 VJL, with 50 different TRAJ and highly diverse CDR3 length and composition (e.g., Seqlogo of VJL#2, VJL#3, and VJL#4); additionally, TRAV22 CDR1 and CDR2 are not as highly conserved as in TRAV41 or TRAV1; Fig. 3). Notably, VJL#4 represented only 3.4%, 1.4%, and 3.8% of the TRAV22-like sequences, whereas VJL#1 represented 74%, 74%, and 67% of the TRAV41-like sequences in the three rabbits. Altogether, these data suggest that VJL#4 does not correspond to an iTRA chain, whereas VJL#1 does.
These results strongly suggest the presence of T cells with iTRA chains in the rabbit that would not correspond to MAIT or NKT cells.
An Alternative Highly Conserved MH1Like Sequence Is Found Across Eutherian Mammals, Including Species in Which MR1 Has Been Inactivated.
The independent pseudogenization of MR1 in several groups of mammals raises the question of how its functions would be fulfilled in the species that have lost it, because this sequence is highly conserved and subjected to strong purifying selection pressure in other mammals (15). The presence of an iTRA chain in the rabbit further supports the idea that the function of MR1 may be performed by other MH1Like proteins, restricting another specific T-cell subset.
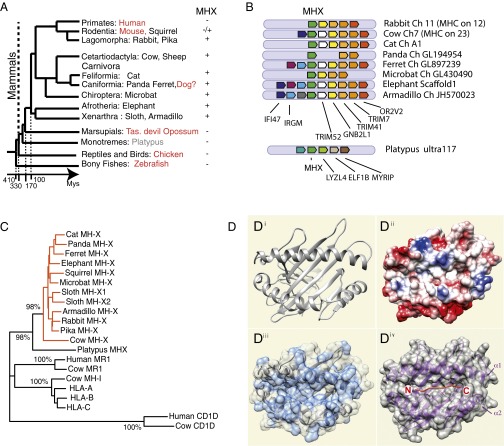
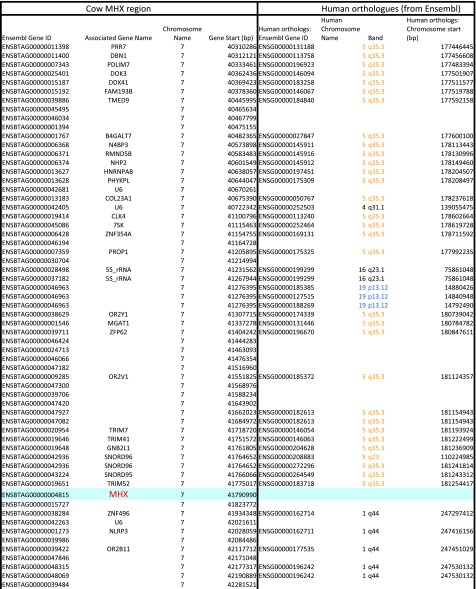
We therefore looked for putative MH1Like sequences in the rabbit and other mammalian species lacking MR1 (Dataset S2C). In addition to the “old” MH1Like molecules [e.g., HFE, FCGRT (previously FcRn), PROCR] (1), we found a gene, which we named MHX, encoding an MH1-related molecule. Although MHX is absent from the mouse, the rat, and primates, it is present in most groups of mammals, including Xenarthra, Afrotheria, Chiroptera, Artiodactyla, carnivores, and Lagomorpha, as well as in a rodent, the squirrel (Fig. 5A). The MHX gene is part of a conserved group of synteny comprising trim7, gnb2l2, trim 41, and trim52 (Fig. 5B). The MHX region is not in the MHC itself, but is linked to one of its paralogons (29) (Fig. S5). Thus, the MHX gene is located on cow chromosome 7 and rabbit chromosome 11, in a region with many genes of which human orthologs are located in the human 5q11-q35 or 19p13 region (Fig. S5), whereas the cow MHC locus is on chromosome 23 and the rabbit MHC locus is on chromosome 12. It could be noted that MR1 is also on a MHC paralogous region, located on the human genome 1q25.3. Such a location of MHX in paralogs of the bona fide MHC might suggest that MR1 and MHX could derive from ancient MH1Like genes from the original paralogons produced by Ohno’s whole-genome duplications. However, many cis-duplications and translocations occurred during evolution, and alternative explanations cannot be excluded.
Fig. 5.
MHX is a previously unidentified MH1Like gene found across mammals. (A) Schematic representation of phylogenetic relationships between the main groups of mammals. Species in which no functional MHX was found are shown in red. Note that the time scale is not linear. (B) MHX gene is located in a conserved genomic context across eutherians, whereas the related sequence from the platypus is not in the same microsynteny gene set. (C) Phylogenetic analysis (NJ, pairwise deletion, bootstrap: n = 1,000) shows that MHX genes constitute a distinct branch of MH1Like sequences. (D) Molecular modeling of rabbit MHX α1-α2 region. (D, i) Ribbon diagram of rabbit MHX based on the structure of H-2Kb. By convention, the α1 helix is at the top of the top view, placing a potential peptide N terminus to the left. (D, ii) Coulombic surface coloring (red is positive, and blue is negative); the groove is open on the side of the peptide N terminus as for a MH2 molecule and closed on the side of the peptide C terminus as for an MH1 molecule, but it is neutral. (D, iii) Surface hydrophobicity of the α1 and α2 domains (blue) showing that the C-terminal part of the groove is hydrophobic. (D, iv) For comparison, ribbon and surface representations of the α1-α2 domains of H-2Kb showing how the peptide is accommodated in a groove that is closed at the peptide N and C termini.
Fig. S5.
MHX neighborhood on cow chromosome 7 and rabbit chromosome 11 are counterparts of MHC human paralogous regions. Synteny analysis was extended to 50 flanking genes on each side of rabbit and cow MHX. Many of the closest human orthologs of these genes, as given by Ensembl, are located on MHC paralogous regions, especially 5q11Eq26 (in red) and 19p13 (in blue), or in linked regions like 5q35 (in orange).
No MHX gene could be identified in the dog, perhaps due to misassembly or incomplete genomic data. However, this gene is present in caniformes, because it is found in the ferret, the panda, the black bear (Ursus americanus), and the Weddel seal (Leptonychotes weddellii). No MHX sequence was found in marsupials (opossum and Tasmanian devil), but a related sequence was identified in the platypus, which, however, appears as an outgroup of all MHX sequences in the phylogenetic tree (Fig. 5C). Moreover, it lacks many conserved amino acids across MHX sequences (Fig. S1C) and is not located in the same genomic context as the eutherian MHX genes, suggesting it may not be a true ortholog. Interestingly, the α1 and α2 domains of cow MHX show particular features and appear to be more similar to MH1, compared with MHX from other species. Overall, MHX and MR1 distributions in mammalian genomes are rather complementary.
MHX sequences constitute a well-supported branch in the phylogenetic trees (Fig. 5C). Sequence analysis suggests that MHX encodes an MH1Like molecule, like MR1 and CD1. As in other MH1Like G domains, there is only one amino acid (IMGT position 39) in the MHX α1 CD turn (in contrast to three amino acids in the MH1 α1 CD turn) and two amino acids (IMGT positions 15 and 16) in the α2 AB turn (instead of three amino acids in the MH1 α2 AB turn) (Fig. S1 C and D). Although MHX lacks an alanine in position 54 in α1, a typical feature of most MH1Like sequences, this feature has also been observed in a few MH1Like α1 domains (IMGT protein display, www.imgt.org) (20). Molecular modeling of rabbit MHX along the H-2Kb sequence (Fig. 5D) showed that MHX displays a typical MH groove. This groove is open at the “N terminus” but closed at the “C terminus” (Fig. 5 D, i). The C-terminus pocket is dominated by hydrophobic residues (Fig. 5 D, ii and iii), but many positively charged residues are found in the floor of the groove. Altogether, the features of the groove look rather similar to those of an MH1 molecule without any indication for a particular type of ligand.
We then analyzed MHX expression and polymorphism in the rabbit, because this species does not have MAIT or NTK but likely expresses invariant TR; hence, MHX may be more important in this context. The rabbit MHX is expressed in a number of tissues as shown by the 45 ESTs from the lungs, liver, skin, brain, testis, kidney, eye, and muscle [tblastn Expect (E) value < 9e-90 of the genome-predicted MHX onto TSA]. Importantly, only two positions were found to be polymorphic among the available rabbit ESTs: position 47 (L/P) and position 89 (S/T) in the α1 and α2 D strand and helix, respectively (alignments in Fig. S1C illustrate numbering). Furthermore, deep sequencing data from the spleen of three individuals and from the mammary glands of nine individuals were analyzed. The diversity of MHX sequences was very low, and the distribution of substitutions between ESTs and the reference MHX sequence was similar within the α1-α2 region and in the rest of the molecule, suggesting that MHX α1-α2 domains are not polymorphic. In contrast, a significant diversity was found in MH1 sequences, and this diversity was higher in the α1-α2 region (Dataset S2D).
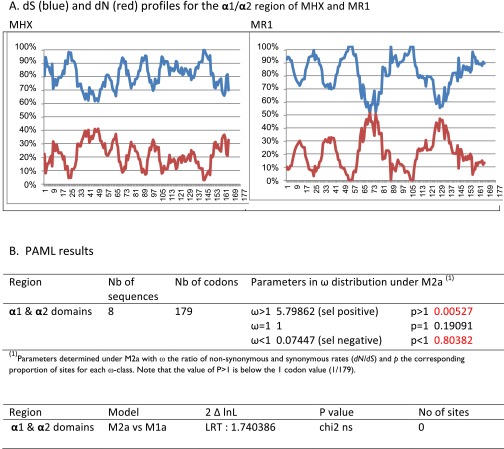
As in MR1, the α1 and α2 domains of MHX are highly conserved across mammals (Dataset S2E). As previously reported for MR1 (15), amino acid changes [nonsynonymous (dN) substitutions] were minimal and silent [synonymous (dS)] substitutions were predominant (Fig. S6A). The ratio ω = dN/dS is an indication for negative selection (ω < 1), neutral evolution (ω = 1), or positive selection (ω >1), and we used Phylogeny Analysis by Maximum Likelihood (PAML) to test whether specific sites were under positive selection. The model of substitution distribution M2a was used to test this hypothesis; a value of ω >1 was detected for only 0.05% of sites and the likelihood-ratio test result was not significant, rejecting the positive selection hypothesis and more in favor of a purifying selection process (Fig. S6B).
Fig. S6.
Nonsynonymous and synonymous substitutions in MHX sequences. (A) Synonymous substitution is predominant in MHX as in MR1. The MHX (Left) and MR1 (Right) sequences from eight representative mammalian species were analyzed for the proportion of synonymous (dS) and nonsynonymous (dN) substitutions, which were plotted to corresponding codons using a sliding-window model for each continuous 30-codon set. MHX sequences analyzed are from the rabbit, microbat, cat, panda, ferret, squirrel, elephant, and armadillo. The dS (blue) and dN (red) profiles for the α1-α2 region of MHX and MR1 are shown. (B) PAML results and test based on likelihood-ratio test (LRT_ calculations). The ratio ω = dN/dS is an indication for negative selection (ω < 1), neutral evolution (ω = 1), or positive selection (ω >1). The site-specific model analysis using the PAML software package allows testing of whether specific sites are under positive selection. The model of substitution distribution M2a was used to test the positive selection hypothesis against the nested null model M1a. A value of ω >1 was detected for 0.05% of sites under M2a and the LRT was not significant, rejecting the positive selection hypothesis. Δ InL, difference between log likelihood values (lnL1 for the alternative and lnL0 for the null models); Nb, number; ns, not significant; sel, selection.
Altogether, the similarities between the phylogenetic features of MR1 and MHX suggest that the MHX molecule may present an evolutionarily conserved, hence likely, nonpeptidic Ag. Although our data do not provide any direct evidence that the iTRA found in the rabbit is expressed by T cells restricted by MHX, a comparative analysis of the TRAV41 gene in mammals reveals an interesting pattern. The distribution of TRAV41 across mammals is consistent with the distribution of MHX, except that it is present in humans, which lack MHX (Fig. 5 and Table 1). Strikingly, CDR1 is highly conserved and CDR2 is strictly identical (LSLEM) within mammals that possess a MHX gene (Fig. 3); in contrast, the human TRAV41 sequence, although similar overall to the sequence of TRAV41 from other species, has divergent CDR1 and CDR2 (Fig. 3). Hence, the canonical CDR1 and CDR2 of TRAV41 correlate with the presence of MHX.
Discussion
The high conservation in mammals of both MR1 and the iTRA chain of MAIT cells indicates important functions for these cells. We confirm that MR1 stands apart in comparison to classical MH1 and to other MH1Like proteins restricting conventional or invariant T cells, with a much slower rate of evolution/genetic changes. The appearance of MR1 in early mammals indicates that it does not represent an ancestral molecule fulfilling primordial functions in vertebrates. The independent loss or inactivation of both the MR1 and TRAV1 genes in three instances suggests that the functions of MAIT cells is either dispensable or that another T-cell subset restricted by another MH1Like molecule has taken over their role. The repeated loss/inactivation of TRAV1/MR1 might also be related to a deleterious role of MAIT cells in some environments. Moreover, the MR1 remnants found in species lacking TRAV1 suggest that the loss of TRAV1 preceded MR1 inactivation. This finding is also consistent with the idea that because TRAV1 is also used by conventional T cells, the disruption of MR1 should not have led to the loss of this TRAV. Hence, our results suggest that the only function of MR1 is to present Ags to MAIT cells and that the evolutionary pressure operates mainly on the MR1-restricted T cells that use TRAV1. The way in which MHC restriction is imposed to TR is still debated, with variable importance given to two different mechanisms: (i) a selected, germline-encoded physical interaction between TR V domains and MH molecules leading to intrinsic “rules of engagement” and (ii) a restriction imposed by the CD4/CD8 coreceptor environment that restricts TR-induced signaling and T-cell thymic selection (25, 30). Our data strongly argue for a coevolution model in the case of MR1 and TRAV1 with structural properties engrafted in the germ line. Because the recognition mode of MR1 by the invariant MAIT TR is highly reminiscent of the classical TR/MH recognition (31), our data argue for the coevolution model in the general debate on the origin of TR/MH interaction. Our findings raise two main questions regarding (i) the function of the MR1/MAIT cell system in mammals and (ii) how this function would be fulfilled in the absence of the MR1/MAIT cell system.
The evolutionary conservation of MR1 is greater than the evolutionary conservation of other MH1Like molecules, such as HFE, AZGP1 (previously ZAG), or FCGRT (32) (Fig. 1), which are not subjected to pathogen-induced selection because they are not involved in Ag presentation. The excess of synonymous substitutions in the sequences of the two MR1 groove domains indicated a purifying selection process (15), further emphasizing the selection pressure on this molecule. Because 65–70% conservation of primary amino acid sequences between species is sufficient to preserve specific features (e.g., binding of specific classes of ligands) for other MH superfamily proteins, such as classical MH1, MH2, or MH1Like proteins (e.g., CD1D) (32, 33), this stronger conservation of MR1 sequence may reflect additional constraints on the external sides of the groove. One hypothesis could be that MR1 is part of a multiprotein complex with strong interactions with chaperon proteins. This hypothesis fits with the particular trafficking of MR1 proteins, which, in the absence of ligand, remain mostly intracellular in a poorly characterized compartment (15, 34).
Despite these evolutionarily conserved features, MR1 is not an ancient MH1Like molecule because its specific features were probably selected in the lineage leading to marsupials and placental mammals about 170 Mya (26, 35). Because MR1 presents microbe-derived pyrimidine adducts to MAIT cells, the function of these cells could be related to regulation of microbe/host interactions specifically in marsupials and eutherians. Although marsupials lack a fully developed placenta and the fetus is nourished by a mother-derived yolk-like structure (36), the fetus is transiently attached to the uterine wall through a primitive placenta in some species (36). Because recent data suggest that bacteria can often be found in the placenta (37), MAIT cells could be involved in placenta/microbiota interactions, although no data are available on MR1 expression or MAIT cell occurrence in the placenta. However, the independent loss of functional MR1 in several mammalian groups indicates that MAIT cells are not required in mammals. It should be noted that species lacking MR1 do not share common features with regard to placental structure. Lagomorphs and the armadillo (MR1−) and primates (MR1+) have a hemochorial placenta, whereas the placenta of both Carnivora (MR1−) and Proboscidea [elephant (MR1+)] is endotheliochorial and cattle (MR1+) placenta is epitheliochorial (38).
MAIT cells are abundant in the liver, which drains all of the digestive tract blood (39, 40). The liver is in direct contact with gut microbiota-derived compounds and translocated bacteria. MAIT cells could be involved in the antibacterial firewall function that has been proposed for the liver (41). However, from an evolutionary perspective, the gut-liver portal vascular system is also present in reptiles and birds, which argues against the hypothesis that the MR1/MAIT cell system emerged in parallel with the intestinal liver portal system to control translocated bacteria. Still, MAIT cells could be involved in this function for specific commensals or pathogens that would only be present in mammals, or may represent a mechanism specific to this group of vertebrates. Moreover, one hypothesis to explain the disappearance of MR1 and MAIT cells in some species could have been specific gut microbiota related to specific diets. However, dogs and cats are carnivorous, whereas bears and armadillos are omnivorous and rabbits and related species (and even, secondarily, the panda) are strictly vegetarian, indicating that there is no direct correlation between diet and MR1/MAIT cell system occurrence.
We looked for MH1Like molecules that would be present in the species lacking MR1 (Dataset S2C). In addition to the “old” MH1Like molecules (e.g., HFE, FCGRT, PROCR) (1) that are well conserved in mammals, numerous more recent MH1Like molecules are found across species. One of those MH1Like molecules, which we called MHX, was present in groups lacking MR1 (Table 1). This sequence is typical of an MH1Like molecule with the α1 and α2 domains and the constant domain harboring canonical conserved amino acids. Molecular modeling of the rabbit sequence revealed a groove presenting hydrophobic residues and a closed end on the C-terminus side but more charged and open-ended on the N-terminus side, without any clear indication with regard to the nature of the ligand.
It is not clear whether the absence of this sequence in the dog and sloth is real (and recent) or more likely due to incomplete genomic data or misassembly. Indeed, this gene is clearly present in caniformes because it is present in the ferret, the panda, the black bear, and the Weddel seal. Notably, this MHX gene is not found in humans and mice, although this gene can also be found in species harboring MR1, such as cattle, elephants, and bats. MHX shares many features with MR1, including restriction to mammals (Fig. 5A), lack of polymorphism and unusual conservation between species (Dataset S2 D and E), signs of purifying selection (Fig. S6), and independent loss in several species.
With regard to MH1 genes that are better known, the number of CD1 genes varies from one to two (mouse) to >10. In the microbat, numerous CD1 genes are found, whereas (classic?) MH1 genes (e.g., XP_014304883) contain insertions within their peptide-binding grooves, which would affect the nature and diversity of peptides presented to T cells (42). Notably, the number of CD1 genes and other “recent” MH1Like genes seems to be higher in species lacking MR1 [following Rodgers and Cook (1)]: For instance, the cat harbors five CD1 and 10 other “recent” MH1Like genes, whereas the armadillo has only one CD1 but 49 MH1Like genes. Overall, our data suggest some kind of compensation between the different MH1 and MH1Like proteins that would allow presentation of Ags of different natures. According to the species and the type of presenting genes (MH1 or MH1Like), the conventional or semiinvariant T-cell responses would focus on peptides or on compounds of various chemical natures, respectively. Such a compensation could be reminiscent of the high number of MH1 genes found in species in which MH2 genes have been lost, such as cod (43), or are nonpolymorphic, such as the axolotl (44).
Although MHX is not present in humans and mice, it is important to determine the nature of the Ags presented by this molecule and the type of T cells that respond to it. Indeed, the functions of MHX is probably fulfilled by another MH1Like molecule in humans and mice. Knowledge of the specific selective pressure being exerted on MHX may provide insights on the type of pathogens and epitopes targeted by the human T cells fulfilling a similar function and allow their identification.
Materials and Methods
Identification and Analysis of MH1Like and TRAV Sequences.
Genes of interest were extracted from the Ensembl database (releases 78–82); known sequences from human, mouse, or other species were used in BLAST EST, TSA, and genome databases to look for the most similar sequences in species in which genes of interest were not annotated. Best BLAST hits were subjected to reverse BLAST analysis to reference species, and were used for phylogenetic analysis to test orthology relationships. Multiple alignments and NJ distance trees were produced using MEGA6 (45). Synteny analyses were performed using Genomicus (46). Regarding TRAV sequences, the topology of phylogenetic trees, rather than absolute thresholds of similarity/identity, was used to assign nomenclature; IMGT numbering and alignments were performed according to Lefranc (20).
Molecular Modeling of MHX.
After sequence alignment, rabbit MHX was modeled using SWISS-MODEL software to generate MHX.pdb. The viewed Protein Data Bank file was modeled using H-2Kb as a template and was viewed using Chimera for analysis and figures.
Analysis of Synonymous and Nonsynonymous Mutations in MHX Sequences.
Coding sequences of MHX from the rabbit, microbat, cat, panda, ferret, squirrel, elephant, and armadillo were aligned using MEGA6, and the proportion of synonymous (dS) and nonsymonymous (dN) substitutions was determined. PAML was used to test positive selection as described in Fig. S6.
Samples and RNA Extraction for Sequencing of Rabbit TRA Transcripts.
Ten milliliters of blood was collected from three rabbits, and mononuclear cells were isolated using Ficoll reagent (GE Healthcare) as recommended by the manufacturer. After isolation, cells were resuspended in 500 μL of RNA lysing guanidine-thiocyanate containing buffer (RLT) + β-mercaptoethanol buffer, and RNA extraction, together with DNA digestion, was performed using an RNeasy Mini kit (Qiagen).
TRA Repertoire Sequencing.
TRA repertoire sequencing was performed using a 5′ RACE SMARTer PCR technique utilizing a gene-specific reverse primer and a MiSeq apparatus (Illumina), followed by deconvolution of the reads as described in SI Materials and Methods.
SI Materials and Methods
TRA Repertoire Sequencing.
Reverse transcription (RT) was carried out using the SMARTScribe Reverse Transcriptase kit (Clontech). Briefly, RT was performed as follow: 1 μg of RNA was mixed with 2 μL of Oc_GSP1 primer (12 μM) and 3.5 μL of Rnase-free/DNase-free H2O and then incubated for 3 min at 72 °C, followed by 2 min at 42 °C. At room temperature, 4 μL of 5× First Strand Buffer, 2 μL of DTT, 2 μL of SMARTScribe Reverse Transcriptase, 2 μL of dNTP (10 mM), 2 μL of SMARTer IIA primer (12 μM), and 0.5 μL of RNase inhibitor were added to the mix and incubated at 42 °C for 90 min and at 70 °C for 10 min.
Then, double-stranded cDNA was obtained with PCR assay carried out using 5 μL of RT product, 32 μL of Rnase-free/DNase-free H2O, 2 μL of CS1-Oc_GSP2 primer (10 μM), 2 μL of S1/L1 primer mix (0.4 μM/2 μM), 5 μL of 10× PCR buffer, 2 μL of MgCl2 (20 mM), 1 μL of dNTP (10 mM), and 5 μL of Platinum Taq DNA Polymerase (Invitrogen). PCR cycling conditions were performed as follows: 94 °C for 3 min, followed by four cycles at 94 °C for 30 s, 70 °C for 30 s, and 72 °C for 30 s; four cycles at 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 30 s; 24 cycles at 94 °C for 30 s, 66 °C for 30 s, and 72 °C for 30 s; and a final step at 72 °C for 10 min. Primers are available upon request.
Fluidigm Access Array primers (Fluidigm) containing barcodes and MiSeq adapators (Illumina) were incorporated on amplicons by PCR. PCR cycling conditions were 95 °C for 2 min, followed by 10 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and a final step at 72 °C for 5 min. The PCR mix was assembled using 1/100 dilution of cDNA, primer mix, and High Fidelity Taq.
After PCR, amplicons were purified using Agencourt AMPure XP (Beckman Coulter) and pooled using a SequalPrep Normalization Kit (Life Technologies) as recommended by the manufacturer.
Finally, a normalized amplicon library was sequenced using MiSeq sequencing technologies [version 3 kit, 2 × 300-bp end reads, Institut Curie Next Generation Sequencing (ICGEX) platform; Illumina].
Data Analysis.
Reads were checked for quality using sickle (se -t sanger -q 20 -l 252 -g -f) (47) and corrected for errors with spades (–only-error-correction –careful) (48). Then, reads were full-length dereplicated using vsearch (49) and clustered using swarm (-d 1 -f -t 20) (50). Abundance of each cluster per sample was also recorded for downstream analysis. A representative sequence from each cluster was then screened for Y[FHILY]C and [FW][AG][ADEGHIKLPQRST]G pattern detection and V, CDR3, and J sequence segment extraction. Sequences sharing the same TRAV and TRAJ and encoding a CDR3 of a given length were then aggregated into VJL, and the frequency of such VJL sequence sets was computed within the total number of reads in which a TRAV, TRAJ, and CDR3 have been identified over the three rabbits. Sequences occurring less than three times were eliminated. The contribution of each rabbit to the frequency of each VJL was then computed and analyzed as shown in Fig. 4.
Supplementary Material
Acknowledgments
We thank Véronique Duranthon for discussion and for sharing rabbit EST data; and Pierre Pontarotti for advice about positive selection analysis. We also thank the Broad Institute Genomics Platform for giving us access to additional EST databases from rabbit tissues (sequencing was funded by NIH Grant NS083660). O.L.’s group is the “Equipe Labellisée de la Ligue Contre le Cancer.” This work was supported by the Institut National de la Recherche Agronomique, Institut National de la Santé et de la Recherche Médicale, Institut Curie, Agence Nationale de la Recherche [Blanc and Labex DCBIOL (Dendritic Cell Biology)], Mérieux Fondation, and the Association de la Recherche sur la Sclérose en Plaque.
Footnotes
Conflict of interest statement. O.L. received grants from Agence Nationale de la Recherche (ANR) during the study, and received personal fees from NESTEC outside the time of the submitted work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600674113/-/DCSupplemental.
References
- 1.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5(6):459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 2.Duprat E, Lefranc MP, Gascuel O. A simple method to predict protein-binding from aligned sequences—Application to MHC superfamily and beta2-microglobulin. Bioinformatics. 2006;22(4):453–459. doi: 10.1093/bioinformatics/bti826. [DOI] [PubMed] [Google Scholar]
- 3.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 4.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15(10):643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509(7500):361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 6.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian α-linked glycosylceramides. Immunity. 2014;41(4):543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 8.Treiner E, Lantz O. CD1d- and MR1-restricted invariant T cells: Of mice and men. Curr Opin Immunol. 2006;18(5):519–526. doi: 10.1016/j.coi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14(7):706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180(3):1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Bourhis L, Mburu YK, Lantz O. MAIT cells, surveyors of a new class of antigen: Development and functions. Curr Opin Immunol. 2013;25(2):174–180. doi: 10.1016/j.coi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Reantragoon R, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210(11):2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Schaik B, et al. Discovery of invariant T cells by next-generation sequencing of the human TCR α-chain repertoire. J Immunol. 2014;193(10):5338–5344. doi: 10.4049/jimmunol.1401380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci USA. 2009;106(20):8290–8295. doi: 10.1073/pnas.0903196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parham P, et al. Diversity of class I HLA molecules: Functional and evolutionary interactions with T cells. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):529–543. doi: 10.1101/sqb.1989.054.01.063. [DOI] [PubMed] [Google Scholar]
- 17.Takada T, et al. Species-specific class I gene expansions formed the telomeric 1 mb of the mouse major histocompatibility complex. Genome Res. 2003;13(4):589–600. doi: 10.1101/gr.975303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tilloy F, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189(12):1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brossay L, et al. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188(8):1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefranc MP, et al. IMGT unique numbering for MHC groove G-DOMAIN and MHC superfamily (MhcSF) G-LIKE-DOMAIN. Dev Comp Immunol. 2005;29(11):917–938. doi: 10.1016/j.dci.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11(8):701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 22.Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat Immunol. 2007;8(6):563–568. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- 23.Yin L, Scott-Browne J, Kappler JW, Gapin L, Marrack P. T cells and their eons-old obsession with MHC. Immunol Rev. 2012;250(1):49–60. doi: 10.1111/imr.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland SJ, et al. The T-cell receptor is not hardwired to engage MHC ligands. Proc Natl Acad Sci USA. 2012;109(45):E3111–E3118. doi: 10.1073/pnas.1210882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangarajan S, Mariuzza RA. T cell receptor bias for MHC: Co-evolution or co-receptors? Cell Mol Life Sci. 2014;71(16):3059–3068. doi: 10.1007/s00018-014-1600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukamoto K, Deakin JE, Graves JA, Hashimoto K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 2013;65(2):115–124. doi: 10.1007/s00251-012-0666-5. [DOI] [PubMed] [Google Scholar]
- 27.Lefranc MP, et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43(Database issue):D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel O, et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat Commun. 2013;4:2142. doi: 10.1038/ncomms3142. [DOI] [PubMed] [Google Scholar]
- 29.Flajnik MF, Kasahara M. Comparative genomics of the MHC: Glimpses into the evolution of the adaptive immune system. Immunity. 2001;15(3):351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 30.Garcia KC. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 2012;33(9):429–436. doi: 10.1016/j.it.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Sagaseta J, et al. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci USA. 2013;110(19):E1771–E1778. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol. 1998;161(8):4066–4077. [PubMed] [Google Scholar]
- 33.Martínez-Naves E, Lafuente EM, Reche PA. Recognition of the ligand-type specificity of classical and non-classical MHC I proteins. FEBS Lett. 2011;585(21):3478–3484. doi: 10.1016/j.febslet.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, et al. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med. 2008;205(5):1201–1211. doi: 10.1084/jem.20072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.dos Reis M, et al. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc Biol Sci. 2012;279(1742):3491–3500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freyer C, Zeller U, Renfree MB. The marsupial placenta: A phylogenetic analysis. J Exp Zoolog A Comp Exp Biol. 2003;299(1):59–77. doi: 10.1002/jez.a.10291. [DOI] [PubMed] [Google Scholar]
- 37.Aagaard K, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enders AC, Carter AM. What can comparative studies of placental structure tell us?—A review. Placenta. 2004;25(Suppl A):S3–S9. doi: 10.1016/j.placenta.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117(4):1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 40.Tang XZ, et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol. 2013;190(7):3142–3152. doi: 10.4049/jimmunol.1203218. [DOI] [PubMed] [Google Scholar]
- 41.Balmer ML, et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6(237):237ra66. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- 42.Ng JH, et al. Evolution and comparative analysis of the bat MHC-I region. Sci Rep. 2016;6:21256. doi: 10.1038/srep21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson AC, Stet RJ, Pilström L. Characterization of MHC class I and beta(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics. 1999;50(1-2):49–59. doi: 10.1007/s002510050685. [DOI] [PubMed] [Google Scholar]
- 44.Tournefier A, et al. Structure of MHC class I and class II cDNAs and possible immunodeficiency linked to class II expression in the Mexican axolotl. Immunol Rev. 1998;166:259–277. doi: 10.1111/j.1600-065x.1998.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louis A, Murat F, Salse J, Crollius HR. GenomicusPlants: A web resource to study genome evolution in flowering plants. Plant Cell Physiol. 2015;56(1):e4. doi: 10.1093/pcp/pcu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi NA, Fass JN. 2011. sickel—A Windowed Adaptive Trimming Tool for FASTQ Files Using Quality (version 1.33). Available at https://github.com/najoshi/sickle. Accessed April 27, 2016.
- 48.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rognes T, Mahé F, Flouri T. 2015. VSEARCH. Available at https://github.com/torognes/vsearch. Accessed April 27, 2016.
- 50.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.