Significance
Copper-dependent lytic polysaccharide monooxygenases (LPMOs) are key players in the enzymatic conversion of biomass. LPMOs catalyze oxidative cleavage of glycosidic bonds in a process involving molecular oxygen and an electron donor, such as cellobiose dehydrogenase (CDH). Using protein NMR and isothermal titration calorimetry we have studied the interactions between a fungal LPMO and three soluble substrates and CDH. The results reveal which areas on the LPMO surface interact with the varying substrates and unambiguously show that both the substrate and CDH bind to a region that is centered around the copper site. The data presented here suggest that electron transfer occurs before substrate binding, providing important new leads for understanding the reaction mechanism of LPMOs.
Keywords: lytic polysaccharide monooxygenase, LPMO, cellulose, xyloglucan, cellobiose dehydrogenase
Abstract
Lytic polysaccharide monooxygenases (LPMOs) are copper-dependent enzymes that catalyze oxidative cleavage of glycosidic bonds using molecular oxygen and an external electron donor. We have used NMR and isothermal titration calorimetry (ITC) to study the interactions of a broad-specificity fungal LPMO, NcLPMO9C, with various substrates and with cellobiose dehydrogenase (CDH), a known natural supplier of electrons. The NMR studies revealed interactions with cellohexaose that center around the copper site. NMR studies with xyloglucans, i.e., branched β-glucans, showed an extended binding surface compared with cellohexaose, whereas ITC experiments showed slightly higher affinity and a different thermodynamic signature of binding. The ITC data also showed that although the copper ion alone hardly contributes to affinity, substrate binding is enhanced for metal-loaded enzymes that are supplied with cyanide, a mimic of O2−. Studies with CDH and its isolated heme b cytochrome domain unambiguously showed that the cytochrome domain of CDH interacts with the copper site of the LPMO and that substrate binding precludes interaction with CDH. Apart from providing insights into enzyme–substrate interactions in LPMOs, the present observations shed new light on possible mechanisms for electron supply during LPMO action.
The polysaccharides in chitinous and lignocellulosic biomass compose large sources of organic carbon and are attractive substrates in biorefineries for the production of biofuels and value-added products. However, the exploitation of these resources is hindered by polysaccharide recalcitrance, which hampers enzymatic depolymerization.
Traditionally, it was thought that hydrolytic enzymes were solely responsible for the degradation of chitin and cellulose. A fundamental change in this model was triggered by the discovery of copper-dependent redox enzymes today known as lytic polysaccharide monooxygenases (LPMOs) (1–8). LPMOs are abundantly present in biomass-degrading microbes and make use of molecular oxygen and an external electron donor to cleave polysaccharides through hydroxylation of one of the carbons in the scissile glycosidic bond (4, 5, 9–13). LPMOs can accept electrons from cellobiose dehydrogenase (CDH) (3, 14, 15) or a variety of small molecule reducing agents such as ascorbate and gallic acid (4, 5) as well as lignin-derived redox mediators (16). Each LPMO reaction cycle is postulated to consume two electrons (3, 5, 6).
Due to the potentially major role of LPMOs in enzymatic biomass conversion and the uniqueness of their catalytic power, there is great interest in unraveling the molecular basis of LPMO activity. This interest has resulted in the discovery and characterization of several LPMOs, currently classified in auxiliary activity (AA) families 9, 10, 11, and 13 in the carbohydrate-active enzymes (CAZy) database (17–19). Bacterial LPMOs occur in family AA10, whereas LPMOs of fungal origin belong to families AA9, AA11, and AA13. LPMOs show large diversity, in terms of domain and sequence composition, as well as in terms of substrate specificity, oxidative regioselectivity, and product profiles. To date, LPMO activity has been demonstrated for β-1,4 glycosidic bonds in chitin (5), cellulose (2), soluble cellulose oligosaccharides (20), hemicelluloses (21), and xylan (22), as well as α-1,4 glycosidic bonds in starch (23). LPMOs acting on β-1,4-linked glucans oxidize either C1 or C4 or show mixed oxidative regioselectivity leading to the formation of both C1 and C4 oxidized products.
NcLPMO9C (also known as NCU02916 or NcGH61-3) is a two-domain C4-oxidizing AA9 LPMO from Neurospora crassa that is active on β-1,4 glycosidic bonds in cellulose, cellodextrins, and β-glucan hemicelluloses (20, 21, 24, 25). In addition to its catalytic domain, NcLPMO9C contains a carbohydrate-binding module belonging to the CBM1 family. The recently published crystal structure of the catalytic domain of NcLPMO9C (26) displays a typical core LPMO structure: two β-sheets forming a β-sandwich fold with several loops protruding from the β-sandwich. The enzyme has a flat surface, which contains the copper coordination site [a common feature for all LPMOs (4, 9, 27)]. The copper ion is coordinated by a histidine brace composed by the N-terminal amino group (His1), the side chain of His-1 (Nδ1), and the side chain of His83 (Nε2). The hydroxyl group of a characteristic tyrosine residue, Tyr166, further shapes the copper site by occupying one of the axial coordination positions.
NMR spectroscopy studies have previously shown that the surface surrounding the conserved copper-binding site is responsible for the interaction of the chitin-active AA10 LPMO CBP21 from Serratia marcescens to crystalline chitin (9), and a similar substrate binding surface has been suggested for family AA9 LPMOs on the basis of docking and crystallography studies (13, 28). An intriguing question is how the LPMO interacts with CDH during electron transfer, especially when the LPMO is bound to the substrate, which would make the copper site inaccessible to CDH. According to the postulated LPMO mechanisms, CDH has to deliver two electrons to the LPMO during each reaction cycle (6, 10, 29). Docking studies have suggested that surface residues close to the copper site interact with the cytochrome domain of CDH during electron transfer (ET) (14). An alternative CDH docking site that would not be blocked by substrate binding has also been suggested (13). Experimental data that could shed light on the interactions of AA9 LPMOs with their substrates and CDH are lacking.
In the present study, we have used isothermal titration calorimetry (ITC) and NMR spectroscopy techniques to analyze the structure and dynamic features of NcLPMO9C in solution. We have mapped the residues involved in the interaction between the catalytic domain of NcLPMO9C and three soluble substrates: cellulose hexasaccharide (Glc6), xyloglucan 14-mer (XG14 = a cellooctaose backbone with substitutions; SI Materials and Methods), and polymeric xyloglucan from tamarind seeds (polyXG). Furthermore, we have used the unique possibilities offered by the NMR assignment to map the interaction of NcLPMO9C with full-length CDH from N. crassa and its isolated heme b-type cytochrome domain (CYT). Thus, we obtained insights into how fungal LPMOs interact with their substrates and CDH.
Results
Secondary Structure and Mobility in Solution.
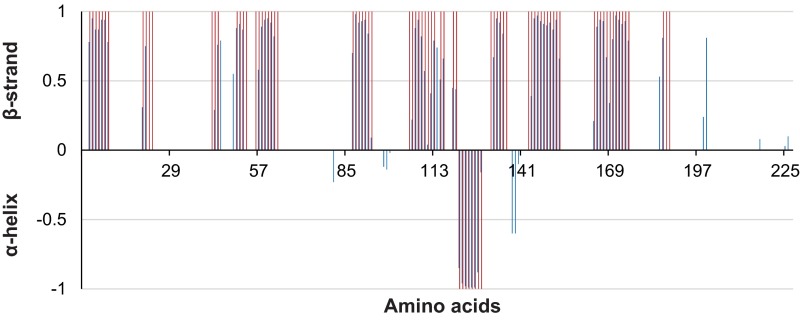
To verify that the overall NMR structure of NcLPMO9C in solution corresponds to the X-ray crystal structure, the presence of secondary structure elements was analyzed using TALOS-N. This analysis demonstrated that the solution structure and the X-ray crystal structure comprise the same secondary structure elements (Fig. S1), indicating that the two structures are very similar.
Fig. S1.
Secondary structure of NcLPMO9C in solution, as derived from secondary chemical shifts. Secondary chemical shifts allow estimation of dihedral angles, which in turn can be used to predict secondary structure propensities, which are indicated by the blue bars. The pink bars indicate secondary structure assignments based on the crystal structure, and comparison shows that there is excellent agreement between the secondary structure elements observed in solution and those seen in the X-ray crystallographic structure (PDB ID: 4D7U) (26). Such high similarity is not unexpected and has previously been shown to be valid for another LPMO, CBP21, for which both the NMR (PDB ID: 2LHS) (9) and the X-ray (PDB ID: 2BEM) (52) structures are available.
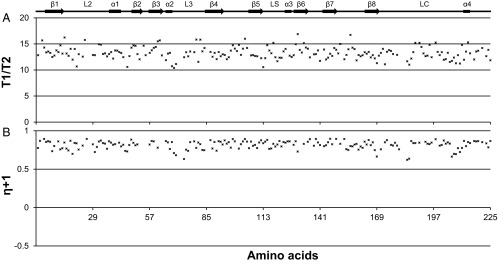
NcLPMO9C is able to bind a variety of substrates, which could indicate a flexible binding surface. To gain insight into this issue, 15N-{1H} NOEs as well as T1 and T2 relaxation times (picosecond and nanosecond timescales) were measured. Both the 15N-{1H} NOEs and the relaxation data are relatively featureless and show the characteristics of a rigid protein (Fig. S2), as also observed previously for an AA10 LPMO (9).
Fig. S2.
The 15N relaxation data and 15N-{1H} heteronuclear NOEs for apo-NcLPMO9C. (A) Longitudinal relaxation time/transverse relaxation time (T1/T2). The T1:T2 ratio is a direct measure of the correlation time for overall rotational tumbling of the protein. The average T1:T2 ratio for NcLPMO9C was calculated to be 13.8 ± 0.7 (SD), which, assuming a spherical particle, corresponds to an overall rotational correlation time, τc, of 11.8 ± 0.5 ns (SD) (44), indicating a tightly packed structure. The Northeast Structural Genomics Consortium has a database with NMR-determined correlation times for globular proteins with different molecular weights. The protein WR73 (which is a 21.9-kDa globular protein) has a longer τc = 13.0 ns. The database is available online at www.nmr2.buffalo.edu/nesg.wiki/NMR_determined_Rotational_correlation_time (accessed March 22, 2016). (B) Steady-state 15N-{1H} NOEs measured for the backbone amide nitrogen atoms.
Substrate Binding.
The interaction of apo-NcLPMO9C with different ligands was probed by measuring changes in chemical shifts in 15N-HSQC and 13C-aromatic-HSQC spectra upon titration with three substrates (Glc6, XG14, and polyXG) and GlcNAc6. The larger chemical shift changes are likely to occur at the ligand binding interface. Additional chemical shift changes, which are normally smaller, may be observed for nuclei that are near but not directly involved in the interface or as the result of propagating conformational changes upon binding (30).
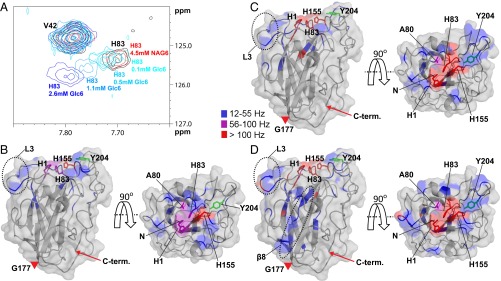
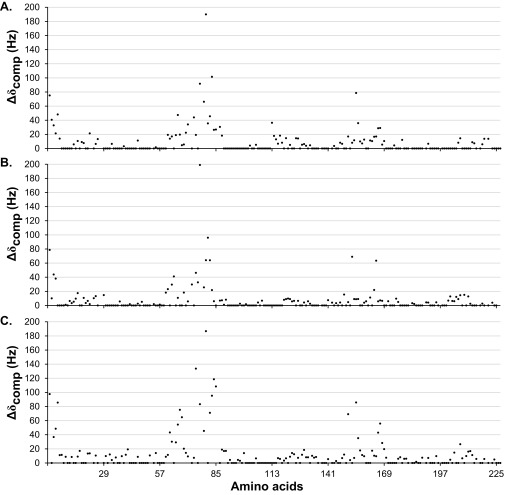
All ligands except chitin-derived GlcNAc6 showed binding to an area on the surface of NcLPMO9C clustered around the histidine brace (His1 and His83) (Fig. 1 and Fig. S3). All substrates had a substantial effect on the chemical shifts of residues His1, Ala80, His83 and His155, whereas the effect on the chemical shifts of other amino acids varied according to the substrates used. A large surface loop [also known as the LC loop (31)] showing considerable variation among LPMOs but also containing a highly conserved tyrosine, Tyr204, which has been suggested to contribute to cellulose binding (13, 31), was generally little affected by substrate binding and was more affected by the binding of XG14 and polyXG than by the binding of Glc6 (Fig. 1). Generally, the longer XG substrates (Fig. 1 C and D) had more extended effects on chemical shifts than Glc6 (Fig. 1B). Differences were observed for a surface loop containing His64, which is part of an insertion [also known as L3 (26)] that only occurs in a subgroup of AA9 LPMOs and that is present in both xyloglucan active LPMOs that have been described so far (26, 32). This region was more affected by the binding of XG14 and, particularly, polyXG compared with Glc6. Compared with other substrates, binding of polyXG had more predominant effects on residues located further away (>15 Å) from the active site surface, such as Tyr166–Cys169 in the β8-strand (Fig. 1D and Fig. S3).
Fig. 1.
Interaction of apo-NcLPMO9C with substrates. (A) Overlay of an area of interest from the 15N-HSQC spectrum for NcLPMO9C (black) in the presence of 4.5 mM GlcNAc6 (labeled as NAG6; red) or increasing concentrations of Glc6 (from lighter to darker blue). The HN/N chemical shift of Val42 is not affected by the interaction, and therefore, the peak is shown as a reference. (B–D) Compound change in chemical shifts larger than 12 Hz (Fig. S3) upon substrate binding mapped on the structure of NcLPMO9C. The backbone of NcLPMO9C (shown in cartoon and surface representation) is colored according to the compound change in chemical shift (15N-HSQC) upon adding 2.6 mM Glc6 (B), 1.3 mM XG14 (C), or 4.2 µM polyXG (D) using the indicated coloring scheme (gray coloring represents no change). The NcLPMO9C structure is shown by a side view (Left) and a top view (Right). The side chains of residues His1, Ala80, His83, and His155 are shown in stick representation. In addition, the side chain of Tyr204 is shown in green. The positions of the L3 loop and the β8-strand are marked on the structures. The LC loop spans the stretch from Gly177 (marked with a red triangle) to the C terminus (marked with a red arrow). The N-terminal amino group (His1) is not observed in 15N-HSQC spectra because of its fast exchange. The 13C-aromatic HSQC spectra showed clear changes in chemical shift for this residue, with all three substrates, with the strongest effects (a vanished signal) being observed with XG14 and polyXG. Based on these observations, for illustrative purposes, His-1 is colored purple (B) or red (C and D) in the figures.
Fig. S3.
Compound change in chemical shift (15N-HSQC) upon adding (A) 2.6 mM Glc6, (B) 1.3 mM XG14, or (C) 4.2 µM polyXG to apo-NcLPMO9C. No change in the chemical shifts was observed upon adding 4.5 mM GlcNAc6. For His1, clear effects on the side chain chemical shifts were observed in a 13C-aromatic HSQC spectrum.
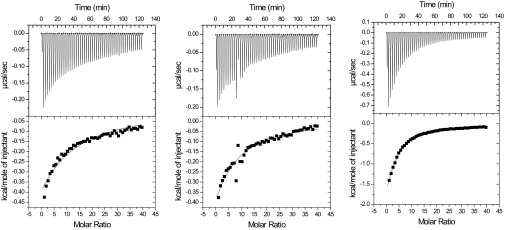
Initially, NMR studies were done using the apo-enzyme, one reason being the paramagnetic relaxation enhancement effect of Cu2+ that would make a larger portion of the substrate binding surface invisible in the NMR experiments (9, 33). To gain further insight into substrate binding, the effect of bivalent metal ions and the effect of CN−, an analog of superoxide known to inhibit LPMO activity (5), we then carried out a series of ITC experiments (Fig. S4), the results of which are summarized in Table 1. Loading the apo-enzyme with copper had little effect on substrate affinity, and both enzyme forms bound XG14 slightly better than Glc6. A clear increase in affinity was observed in the presence of cyanide, for both substrates but more so for Glc6. This result suggests that the enzyme–substrate interaction is strengthened during the first steps of the LPMO reaction, where a Cu2+–superoxide complex is likely to emerge (29).
Fig. S4.
Thermograms (Upper) and binding isotherms with theoretical fits (Lower) for the binding of 4.0 mM XG14 to 15 μM of NcLPMO9C apo (Left), in the presence of Cu2+ (Middle), and in the presence of Cu2+/CN− (Right) at t = 25 °C.
Table 1.
Thermodynamic parameters for binding of XG14 and Glc6 to various forms of NcLPMO9C at t = 25 °C in 20 mM MES (pH 5.5)
| Active site ligand | Kd* | ΔG°† | ΔH°† | −TΔS°† |
| XG14 | ||||
| apo | 0.42 ± 0.02 | −4.6 ± 0.1 | −10.8 ± 1.0 | 6.2 ± 1.0 |
| Cu2+ | 0.33 ± 0.04 | −4.7 ± 0.1 | −10.5 ± 0.5 | 5.8 ± 0.5 |
| Cu2+/CN−‡ | 0.14 ± 0.01 | −5.3 ± 0.1 | −16.7 ± 0.4 | 11.4 ± 0.4 |
| Zn2+/CN− | 0.22 ± 0.02 | −5.0 ± 0.1 | −9.1 ± 1.0 | 4.1 ± 1.0 |
| Glc6 | ||||
| apo | 1.1 ± 0.1 | −4.0 ± 0.1 | −4.5 ± 0.5 | 0.5 ± 0.5 |
| Cu2+ | 0.81 ± 0.08§ | −4.3 ± 0.2§ | −2.5 ± 0.5§ | −1.8 ± 0.5§ |
| Cu2+/CN−‡ | 0.13 ± 0.01 | −5.3 ± 0.1 | −9.2 ± 0.2 | 3.9 ± 0.2 |
Metal-loaded enzymes were obtained by adding solutions of CuCl2 or ZnSO4 to the apo-enzyme, to reach a final concentration of 30 µM. Cyanide was added to a final concentration of 1 mM. For practical reasons these experiments were done with the full-length enzyme. Thermograms are shown in Fig. S4.
In mM.
In kcal⋅mol−1.
Analogous experiments with only the catalytic domain were done, showing essentially similar results.
Data from ref. 26.
The ITC data (Table 1) confirm that Glc6 and XG14 have different binding modes. First, the affinity for XG14 is higher compared with Glc6, except for the Cu2+/CN− situation, where the two ligands bind with similar affinities. Second, for all LPMO forms, the thermodynamic signatures of Glc6 and XG14 binding differed considerably; the enthalpic effects of binding of XG14 were much larger compared with Glc6. As a prelude to further NMR experiments (where use of Cu2+ was to be avoided), an experiment with a Zn2+-loaded enzyme was also conducted, showing slightly weaker binding compared with Cu2+.
Based on the results from the ITC experiments, additional NMR experiments were carried out, using Zn2+-loaded enzyme, in the presence of cyanide, without revealing other or additional substrate interactions compared with the data discussed above.
Docking of Cellulose Hexamer.
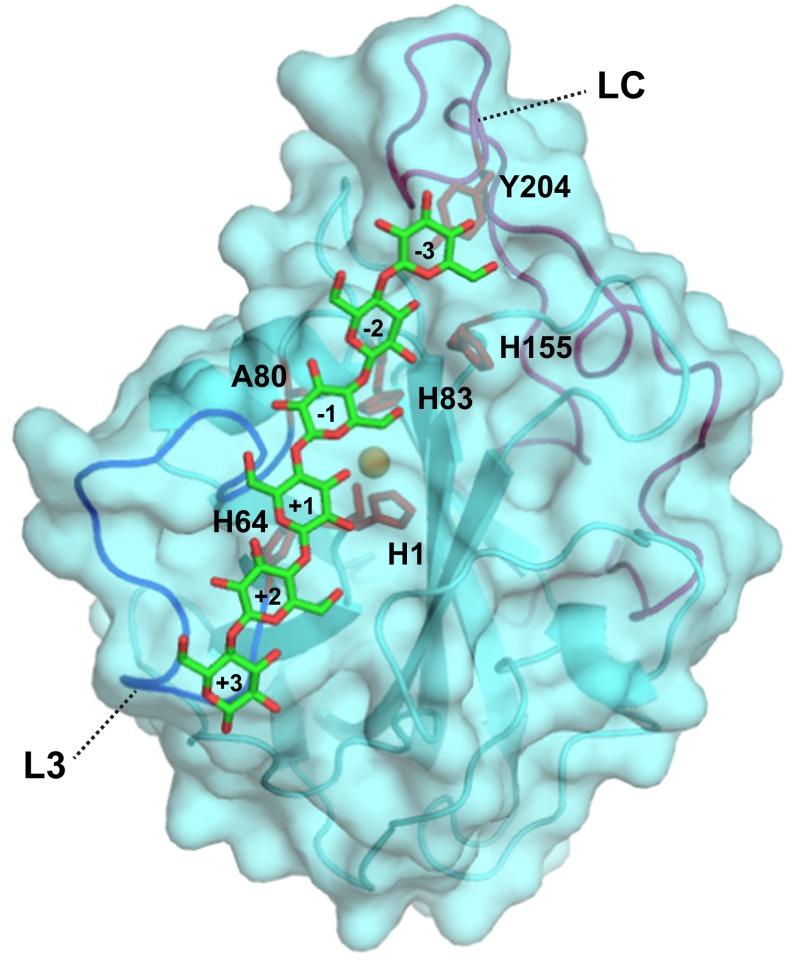
Docking was used to obtain a conceptual understanding of how binding might occur, using a simple docking algorithm combined with restraints obtained from the NMR data. The results showed that the surface patch surrounding the copper site appears to be a preferred interaction surface for Glc6 (Fig. 2) and that the interaction energies are favorable. The results suggest that Tyr204 is not involved in binding of this short substrate, whereas residues in the L3 loop (His64), which is characteristic for this group of LPMOs, are more prominent.
Fig. 2.
Interaction model of Glc6 and NcLPMO9C produced by HADDOCK (39). The backbone is shown as a cartoon and surface, and the side chains of residues known from the NMR experiments to be strongly affected by substrate binding (His1, His64, Ala80, His83, and His155) are shown as sticks. In addition, the picture shows the side chain of a selected surface residue (Tyr204) that shows a high degree of sequence conservation and that may be involved in substrate binding, possibly without an effect of substrate binding on the compound change in chemical shift (15N-HSQC). In the shown complex, the scissile glycosidic bond is located at 5 Å from the copper atom (orange sphere). The L3 loop (containing His64) and the long LC loop (containing Tyr204) are displayed in blue and magenta, respectively. The sugar residues are numbered by subsite, where the sugar that is closest to His1 is sugar +1, in accordance with recent crystallographic data (28) (Discussion). The HADDOCK energies were (with SD) as follows: van der Waals energy = −39.6 ± 2.7 kcal⋅mol−1, electrostatic energy = −27.8 ± 7.7 kcal⋅mol−1, and desolvation energy = −8.5 ± 3.2 kcal⋅mol−1.
Interaction with CDH and CYT.
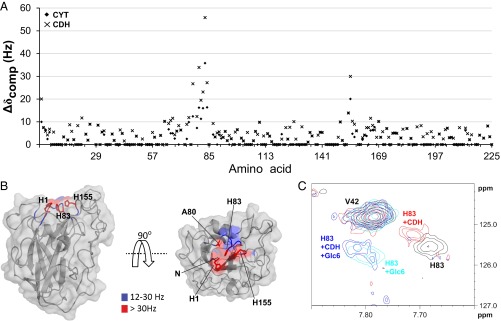
To identify amino acids involved in the interaction of the LPMO with CDH and CYT, changes in chemical shifts in the 15N-HSQC and 13C-aromatic-HSQC spectra were measured upon addition of either protein to NcLPMO9C. Both CDH and CYT showed binding to an area on the surface of NcLPMO9C clustered around the copper coordination site, the effects being somewhat more pronounced for CDH than for CYT (Fig. 3). Thus, the amino acid clusters that were affected by the binding of these proteins were similar to the regions that were most affected in the substrate titration experiments. However, compared with the substrates, and especially the xyloglucan substrates, the interaction areas on the LPMO are more focused on the catalytic center, i.e., His1, Ala80, His83, and His155, with minimal effects on the LC and L3 loops.
Fig. 3.
Interaction of apo-NcLPMO9C with CDH and CYT. (A) Compound change in chemical shifts (15N-HSQC) for each amino acid in NcLPMO9C upon addition of CYT (rhombi) or CDH (crosses). (B) Compound change in chemical shifts larger than 12 Hz mapped on the NcLPMO9C structure. The backbone of NcLPMO9C (shown in cartoon and surface representation) is colored using the indicated coloring scheme. The side chains of residues His1, Ala80, His83, and His155 are shown in stick representation. The 13C-aromatic HSQC spectra showed clear changes in chemical shift for His1, and this residue was treated as described in the legend of Fig. 1. (C) Overlay of an area of interest from the 15N-HSQC spectrum for 0.10 mM apo-NcLPMO9C in the absence of an interaction partner (black) and in the presence of 0.12 mM CDH (red), 2.6 mM Glc6 (cyan), or both (blue).
A competition experiment was carried out to verify that the same residues were involved in binding substrates and CYT/CDH. Indeed, upon addition of excess Glc6 to a mixture of NcLPMO9C and CDH, the chemical shifts of the HN/N signals in the 15N-HSQC spectrum returned to values similar to those observed for the titration end-point of NcLPMO9C with Glc6 (Fig. 3C). This result unambiguously shows that CDH and Glc6 bind to the same area on the LPMO.
Despite considerable efforts, we have so far not been able to establish conditions that allow monitoring of the CDH–LPMO interaction by ITC, an interaction that likely is transient in nature.
Discussion
The complex mechanism that LPMOs use to oxidize their substrates involves the interplay between the LPMO, its substrate, the copper ion, molecular oxygen, and an electron donor. Thus, a fundamental understanding of the interaction between the LPMO and each of these factors is necessary for unraveling the molecular basis of the enzymatic mechanism. In the current study, we have used NMR and ITC techniques to provide unprecedented insight into key interactions between NcLPMO9C, its substrates, and a likely natural electron donor, CDH, as they would occur in solution.
It is challenging to obtain information about the interaction of LPMOs with polymeric substrates because these are mostly large, insoluble, and even partially crystalline, which complicates or precludes most experimental approaches. The ability of NcLPMO9C to act on soluble substrates allowed us to probe these interactions by titrating the protein with the substrates and measuring perturbations in the chemical shifts observed by NMR. The results show that the substrate binding surface is centered around the copper site and extends over the relatively flat surface of the LPMO, involving residues in surface loops that are remote from the catalytic center and that show considerable sequence variation. The NMR data show that the L3 and LC loops interact more strongly with XG14 and polyXG than with smaller substrates such as Glc6. The L3 loop only occurs in a subgroup of LPMOs that includes the two LPMOs for which xyloglucan activity has been described so far [NcLPMO9C (26) and PaLPMO9H (32)]. The ITC data showed differences in binding affinity between Glc6 and XG14 and revealed considerable differences in the thermodynamic signatures of binding. Binding of XG14 was associated with considerably larger beneficial enthalpic effects, suggesting more extended binding interactions, thus confirming the NMR data.
During the course of this study, Frandsen et al. (28) reported the crystal structure of an AA9 LPMO, LsLPMO9A, in complex with Glc6 (PDB ID: 5ACI). The docking model of Fig. 2 and the crystal structure show similar binding in the area near the copper-site, spanning subsites −2 to +2. Frandsen et al. showed that the +1 sugar docks onto His1. The NMR data do not provide the same level of atomic accuracy, meaning that defining the +1 sugar in the docking model of Fig. 2 is somewhat arbitrary. Fig. 2 suggests that the binding mode is −3 to +3; it could also be −2 to +4 but certainly not −4 to +2. The main difference between the studies is that the crystal structure shows binding from −4 to +2, whereas the NMR data suggest binding from −3 to +3 or −2 to +4. This difference is accompanied by interesting structural variation. The L3 loop, which is important for interactions in the +3/+4 subsites (Figs. 1 and 2 and Fig. S3), is only 7 residues long in LsLPMO9A, compared with 14 in NcLPMO9C, leading to a quite different interaction surface, albeit with conservation of His64 (His66 in LsLPMO9A). Furthermore, relative to LsLPMO9A, NcLPMO9C carries a six-residue insertion right next to Tyr204 (Tyr203 in LsLPMO9A), which changes the binding surface near subsite −3/−4.
Notably, binding of polyXG to NcLPMO9C perturbed chemical shifts on amino acids that are far from the interaction surface (Fig. 1D and Fig. S3). This might be due to indirect effects that result in minor conformational rearrangements in the protein, rather than direct surface binding interactions. Some of these conformational rearrangements occurred on the β8-strand containing the copper site tyrosine Tyr166.
The ITC data show that the presence of Cu2+ hardly affects the strength of ligand binding. This observation shows that copper binding does not affect the conformation of the substrate-binding surface, which coincides with the observed rigidity of the protein. Using NMR, it has been shown previously (9) that the only structural effect of metal ion binding is a tightening of the N terminus of the protein (His1). This observation also suggests that the copper ion is too recessed to interact directly with the substrate, as one would indeed conclude from enzyme–substrate complexes that are available in the literature (6, 28, 29, 31). We observed stronger ligand binding in the presence of cyanide. Cyanide is a known inhibitor of LPMOs (5) and a known copper-binding analog of the superoxide ion (34). Cu2+–CN− complexation is analogous to the Cu2+–O2− complex emerging in the first step of LPMO catalysis initiated by a reduced LPMO (29). Our data thus suggest that ligand binding is enhanced upon single electron transfer from Cu1+ in the reduced LPMO to molecular oxygen.
Because of the shared timescales of molecular motions and spin-precession frequencies, we were able to perform NMR measurements that provided dynamic information about the protein. The results show that the backbone of NcLPMO9C is inflexible (Fig. S2). Furthermore, the data were used to calculate the rotational correlation time (τc) for NcLPMO9C, which was found to be smaller (τc = 11.8 ± 0.5 ns for 23.3 kDa NcLPMO9C) than what would be expected for a globular protein of similar molecular weight (τc = 13.0 ns for a 21.9 kDa globular protein; Fig. S2). The same is true for CBP21 (9), confirming that LPMOs are compact, rigid proteins, which is an advantageous feature for proteins involved in ET reactions (35).
Significant progress has been made in understanding the role of CDH in the oxidative degradation of cellulose (3, 14, 15). The CYT domain of CDH contains a heme b prosthetic group that, after being reduced by the catalytic dehydrogenase domain of CDH, proceeds to reduce the LPMO through an intermolecular electron transfer (ET) event (3, 14). In early work, CDH was proposed to interact with a conserved region centered around a Pro–Gly–Pro motif situated on a side of the LPMO that is opposite of the copper-binding surface, which implies that long-range electron transfer through the LPMO would take place. Upon the determination of the crystal structure of full-length CDH, Tan et al. (14) concluded from docking studies that CDH is likely to interact directly with the copper. The present NMR data show that CDH indeed interacts directly with the copper site and that the interaction involves a relatively narrow surface patch near that site (Fig. 3). Thus, it would seem that the ET event occurs directly between the heme b of CYT and the copper ion in NcLPMO9C. This patch contains amino acids that are also involved in the interaction with substrates, and we show that NcLPMO9C will not bind Glc6 and CDH simultaneously. The changes in chemical shift upon interaction of NcLPMO9C with CYT or CDH were small compared with changes in the chemical shifts caused by the binding of substrates, which is indicative of weak protein–protein interactions (36).
Knowing that CDH by itself is capable of supplying the LPMO with the two electrons it needs per catalytic cycle and the fact that CDH cannot bind to the LPMO in the presence of substrate would imply that these electrons are present in the LPMO before substrate binding. This is a puzzling thought because reduction of the copper transfers only one electron to the LPMO. The presence of two electrons before substrate binding could be taken to imply that LPMOs have the ability to somehow store a second electron. Alternatively, the formation of the copper–superoxo adduct initiated by the first transferred electron could lead to subsequent oxidation of a tyrosine or a tryptophan residue. To complete the reaction cycle, CDH would donate two electrons: one to reduce Cu2+ to Cu1+ and another one to return the tyrosine or tryptophan radical to its normal form (37). LPMOs are exceptionally rich in aromatic residues and contain a conserved cluster of aromatic residues in vicinity of the copper site, whose π-electron clouds may facilitate intraprotein ET.
Altogether, this study provides insight into LPMO–substrate interactions as well as the first experimental evidence to our knowledge demonstrating that the electron transfer mechanism between CYT and LPMO is based on the direct interaction of the heme b and LPMO copper sites. The location of the binding site for CDH, the binding competition between CDH and substrate (Fig. 3), and the increased binding strength observed in the LPMO–Cu2+–CN−–ligand complex suggest that all electrons necessary for the reaction are present in the LPMO before substrate binding.
Materials and Methods
Detailed information for all experimental procedures is provided in SI Materials and Methods.
Sample Preparation.
The isotope-labeled catalytic domain of NcLPMO9C used in NMR experiments was recombinantly produced in Pichia pastoris cultivated in isotope-enriched (13C, 15N) minimal medium and purified by multiple chromatographic steps as described previously (26). Nonlabeled full-length NcLPMO9C and its catalytic domain were produced and purified as previously described. Because the LPMO was produced in P. pastoris, His1 was not methylated (26).
Full-length CDH and its isolated CYT, obtained as a proteolytic degradation product, were produced in P. pastoris and purified by subsequent hydrophobic interaction and ion exchange chromatography steps, followed by deglycosylation. Homogeneous CDH and CYT preparations were obtained by size exclusion chromatography.
NMR Spectroscopy.
NMR data for apo-NcLPMO9C in 25 mM sodium phosphate buffer (pH 5.5) and 10 mM NaCl were obtained using a Bruker Ascend 800-MHz spectrometer and a Bruker Avance III 600-MHz spectrometer. The NMR assignment of NcLPMO9C has been published elsewhere (38).
Detection of residues involved in the interactions between NcLPMO9C and different ligands (Glc6, XG14, polyXG, CYT, and CDH) was accomplished by measuring chemical shift perturbations in 15N-HSQC spectra upon adding a ligand to 80–100 µM NcLPMO9C.
ITC.
Dissociation constants and thermodynamics data for binding of Glc6 and XG14 to various forms of NcLPMO9C were determined with a VP-ITC system from Microcal, Inc., essentially as described previously (26), using 20 mM MES buffer (pH 5.5) at t = 25 °C.
SI Materials and Methods
Sample Preparation.
Cloning, protein expression production, and purification of the uniformly isotope-labeled (15N and 13C) catalytic domain of NcLPMO9C (26) and of unlabeled full-length NcLPMO9C (24), as well as conditions for NMR measurements (30), have been described previously. The catalytic domain of NcLPMO9C is not glycosylated (26), whereas full-length NcLPMO9C carries O-glycosylation in the additional linker and/or carbohydrate-binding module (24). For some experiments (where Zn2+ was used), the buffer was exchanged using protein spin concentrators (Vivaspin 6 PES; 5 kDa MWCO; Sartorius) to 20 mM sodium-acetate (pH 5.5), 10 mM NaCl, and 0.5 mM ZnCl2 in 90% (vol/vol) H2O/10% (vol/vol) D2O.
Recombinant production of Neurospora crassa CDH IIA in Pichia pastoris was carried out as previously described (40). Culture supernatant was harvested from the bioreactor 72 h after methanol induction and contained CDH as well as its proteolytic degradation product CYT. After diafiltration with a Microza UF module (SLP-1053; 10 kDa MWCO; Pall Corporation) the proteins, buffered in 50 mM sodium acetate (pH 5.5) containing 20% (wt/vol) (saturated) (NH4)2SO4, were loaded onto a Phenyl-Sepharose-FF column (GE Healthcare) and eluted by applying a linear gradient to 50 mM sodium acetate buffer (pH 5.5). Fractions containing CDH and CYT were separately pooled, changed to a 100 mM sodium acetate buffer (pH 5.5), loaded onto a DEAE-Sepharose column (GE Healthcare), and eluted with the same buffer containing 1 M NaCl. Fractions containing CDH or CYT were pooled and deglycosylated with 5,000 U endoglycosidase Hf and 300 U α-1,2/3-mannosidase (New England Biolabs) per mg protein in a 100 mM sodium acetate buffer (pH 5.5) containing 5 mM ZnCl2 for 48 h at 8 °C (14). Size exclusion chromatography with a Superdex 75 column (GE Healthcare) was used to remove glycosidases and low-molecular weight compounds and resulted in homogeneous CDH (6.0 mg⋅mL−1) and CYT (10 mg⋅mL−1) preparations in 100 mM sodium acetate buffer (pH 5.5).
Polymeric xyloglucan from tamarind seeds (polyXG, average Mw = 225 kDa), cellulose hexasaccharide (Glc6), chitin hexasaccharide (GlcNAc6), and the sample referred to as XG14 were obtained from Megazyme. XG14 is a mixture of longer xyloglucan oligomers that is dominated by species with a cellooctaose backbone with sequence XXXGXXXG (where X stands for glucose with a xylose substitution), where up to three (most commonly one) of the xyloses carry an additional galactose substitution (Megazyme, product number O-XGHDP).
Protein concentration was determined by measuring the A280 of the protein solution and deducing the protein concentration based on the theoretical extinction coefficient [calculated using the ProtParam tool; web.expasy.org/tools/protparam/ (41)].
NMR Spectroscopy.
NMR spectra of 80–200 µM NcLPMO9C samples were recorded at 25 °C on a Bruker Ascend 800 MHz spectrometer Avance III HD equipped with a 5-mm Z-gradient CP-TCI (H/C/N) cryogenic probe at the NT-NMR-Center/Norwegian NMR Platform (NNP). NMR relaxation measurements were recorded on a Bruker Avance III 600 MHz spectrometer equipped with a 5-mm Z-gradient CPP-TCI (H/C/N) cryogenic probe at the Department of Chemistry and Bioscience, Aalborg University. NMR data were processed using Bruker TopSpin version 3.5. NMR spectral analysis was performed using CARA version 1.5.5 (42). The NMR assignment of the catalytic domain of NcLPMO9C has been published elsewhere (30). The 15N-{1H} heteronuclear NOEs were derived with Protein Dynamic Center software version 2.3.1 from Bruker BioSpin using two independently measured and integrated 15N-{1H} heteronuclear correlated spectra with and without 1H saturation (43). Nuclear magnetic relaxation time measurements of 15N nuclei (T1 and T2) were analyzed with Protein Dynamic Center software version 2.3.1 from Bruker BioSpin, using exponential fitting of data from 15N heteronuclear single quantum coherence (HSQC)-type spectra that had been acquired with different relaxation delays (43, 44). Secondary structure elements were analyzed using the web-based version of the TALOS-N software (spin.niddk.nih.gov/bax/nmrserver/talosn/) (45) using the backbone chemical shifts (N, HN, C’, Cα, and Hα) and some side chain chemical shifts (Cβ and Hβ).
Interaction Studies with NMR.
The Kd values for binding to copper-saturated NcLPMO9C at pH 5.5, determined by Borisova et al. (26) using ITC, were used to estimate the amounts of Glc6 and polyXG added to apo-NcLPMO9C. For Glc6, the titration points were 0.1, 0.5, 1.1, and 2.6 mM. For polyXG, the titration points were 0.1 µM (20 µg/mL), 0.2 µM (41 µg/mL), 0.7 µM (146 µg/mL), 2.4 µM (538 µg/mL), and 4.2 µM (944 µg/mL). For XG14, the titration points were 0.05 mM (133 µg/mL), 0.1 mM (267 µg/mL), 0.2 mM (400 µg/mL), and 1.3 mM (2822 µg/mL). As a negative control, two titration points were recorded for GlcNAc6 at 0.9 mM and 4.4 mM. The 1D-proton, 15N-HSQC, and 13C-aromatic-HSQC spectra were recorded for each titration point.
To map the interaction of CYT and CDH with NcLPMO9C, the proteins were added to apo-NcLPMO9C in a 1.1:1 ratio. The 1D-proton, 15N-HSQC, and 13C-aromatic-HSQC spectra were recorded before and after the addition of the proteins.
Mapping of the residues involved in the interaction between NcLPMO9C and each of the three substrates, CYT, or CDH was accomplished by measuring chemical shift changes in the N and HN atoms of the backbone of NcLPMO9C. A compound change in chemical shift was calculated using the following formula:. Δδcomp is the absolute change in chemical shift (Hz). ΔδH is the change in chemical shift of the amide proton (Hz). ΔδN is the change in chemical shift of the amide nitrogen atom (Hz). x is the constant used to achieve equal contributions from changes in N and HN shifts (9, 46). Significant compound changes in chemical shifts were defined as those larger than twice the spectral resolution (Δδcomp > 12 Hz).
Affinity Experiments: ITC.
Isothermal titration calorimetry (ITC) experiments were performed with a VP-ITC system from Microcal, Inc. Solutions were thoroughly degassed before experiments to avoid air bubbles in the calorimeter. Chelex-treated 20 mM MES buffer (pH 5.5) at t = 25 °C was used for all binding studies. For binding, a 15 µM enzyme solution was placed in the reaction cell with a volume of 1.42 mL, and 4.0 mM of XG14 or Glc6 was placed in the ITC syringe. Aliquots of 4 µL were injected into the reaction cell at 180-s intervals with a stirring speed of 260 rpm. The titrations were complete after 50 injections.
ITC data were collected automatically using the Microcal Origin v.7.0 software accompanying the VP-ITC system. Before further analysis, all data were corrected for heat of dilution by subtracting the heat produced by titrating ligand into buffer alone. The data were fitted using a nonlinear least-squares algorithm using a single-site binding model used by the Origin software that accompanies the VP-ITC system, yielding the stoichiometry (n), the equilibrium binding association constant (Ka), and the enthalpy change (ΔHr°) of the reaction. Errors in ΔHr°, Kd, and ΔGr° were obtained as SDs of at least three experiments. Errors in ΔSr° and −TΔSr° were obtained as propagation of errors.
The shape of the ITC binding curve may be described by the so-called Wiseman c value, which can be expressed as follows: c = nKa[M]t, where n is the stoichiometry of the reaction, Ka is the equilibrium binding association constant, and [M]t is the protein concentration (47). The c values preferably need to be in the range of 10 < c < 1,000 to obtain meaningful calculations of Ka. For the titration of XG14 or Glc6 in all forms of NcLPMO9C, binding isotherms were hyperbolic, indicating weak binding and a c value below 10 (47, 48). In such cases, binding thermodynamics can be obtained even if c is in the range of 0.01 < c < 10 if a sufficient portion of the binding isotherm is used for analysis (48). This is achieved by ensuring a high molar ratio of ligand versus protein at the end of the titration, accurate knowledge of the concentrations of both ligand and receptor, an adequate level of signal-to-noise in the data, and known stoichiometry. These conditions were all met in the experimental setup used here.
Docking.
To gain further insight into the interaction between Glc6 and NcLPMO9C, high ambiguity-driven biomolecular docking was carried out using the Easy Interface of HADDOCK 2.2 (haddock.science.uu.nl/services/HADDOCK2.2/haddockserver-easy.html) (39). A PDB file containing the X-ray crystal structure of the catalytic domain of NcLPMO9C (26) was obtained from the Protein Data Bank (PDB ID: 4D7U), and protons were added to the residues using PyMol (49). Active residues were defined as those with a compound chemical shift larger than twice the spectral resolution (Δδcomp > 12 Hz) and a relative solvent accessibility of either main chain or side chain above 15%, as determined by MolMol (50). Passive residues were determined automatically by HADDOCK as those within a radius of 6.5 Å of any active residue and a relative solvent accessibility above 15% (39). The structure of a fully extended conformation of Glc6 was constructed using GLYCAM Carbohydrate Builder (glycam.org), and the PDB file was prepared as an input for HADDOCK using PRODRG (davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg) (51).
Acknowledgments
This work was financed by strategic funds from the Norwegian University of Science and Technology and the MARPOL project, a FRINAT project, and the Norwegian NMR Platform (Grants 221576, 214613, and 226244 from the Research Council of Norway, respectively). The NMR laboratory at Aalborg University is supported by the Obel, SparNord, and Carlsberg Foundations. Part of the work was supported by the European Commission through the INDOX project (FP7-KBBE-2013-7-613549).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Assigned chemical shifts have been deposited in BioMagResBank (www.bmrb.wisc.edu/) (entry no. 26717).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602566113/-/DCSupplemental.
References
- 1.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VGH. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45–57. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forsberg Z, et al. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011;20(9):1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 4.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 6.Kim S, Ståhlberg J, Sandgren M, Paton RS, Beckham GT. Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc Natl Acad Sci USA. 2014;111(1):149–154. doi: 10.1073/pnas.1316609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beeson WT, Vu VV, Span EA, Phillips CM, Marletta MA. Cellulose degradation by polysaccharide monooxygenases. Annu Rev Biochem. 2015;84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 8.Hemsworth GR, Johnston EM, Davies GJ, Walton PH. Lytic polysaccharide monooxygenases in biomass conversion. Trends Biotechnol. 2015;33(12):747–761. doi: 10.1016/j.tibtech.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Aachmann FL, Sørlie M, Skjåk-Bræk G, Eijsink VGH, Vaaje-Kolstad G. NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc Natl Acad Sci USA. 2012;109(46):18779–18784. doi: 10.1073/pnas.1208822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beeson WT, Phillips CM, Cate JHD, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134(2):890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 11.Hemsworth GR, et al. The copper active site of CBM33 polysaccharide oxygenases. J Am Chem Soc. 2013;135(16):6069–6077. doi: 10.1021/ja402106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsberg Z, et al. Comparative study of two chitin-active and two cellulose-active AA10-type lytic polysaccharide monooxygenases. Biochemistry. 2014;53(10):1647–1656. doi: 10.1021/bi5000433. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Beeson WT, 4th, Phillips CM, Marletta MA, Cate JHD. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure. 2012;20(6):1051–1061. doi: 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan T-C, et al. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat Commun. 2015;6(May):7542–7553. doi: 10.1038/ncomms8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langston JA, et al. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol. 2011;77(19):7007–7015. doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westereng B, et al. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep. 2015;5:18561–18670. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41–55. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo Leggio L, et al. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun. 2015;6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10(2):122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaksen T, et al. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289(5):2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agger JW, et al. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci USA. 2014;111(17):6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frommhagen M, et al. Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol Biofuels. 2015;8(1):101–113. doi: 10.1186/s13068-015-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vu VV, Beeson WT, Span EA, Farquhar ER, Marletta MA. A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111(38):13822–13827. doi: 10.1073/pnas.1408090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kittl R, Kracher D, Burgstaller D, Haltrich D, Ludwig R. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol Biofuels. 2012;5(1):79–92. doi: 10.1186/1754-6834-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vu VV, Beeson WT, Phillips CM, Cate JHD, Marletta MA. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc. 2014;136(2):562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 26.Borisova AS, et al. Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J Biol Chem. 2015;290(38):22955–22969. doi: 10.1074/jbc.M115.660183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Frandsen KEH, et al. The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat Chem Biol. 2016;12(4):298–303. doi: 10.1038/nchembio.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjaergaard CH, et al. Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111(24):8797–8802. doi: 10.1073/pnas.1408115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd Ed. Elsevier Academic Press; Amsterdam: 2007. Protein-ligand binding interfaces; pp. 755–760. [Google Scholar]
- 31.Wu M, et al. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the Basidiomycota fungus Phanerochaete chrysosporium. J Biol Chem. 2013;288(18):12828–12839. doi: 10.1074/jbc.M113.459396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennati-Granier C, et al. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels. 2015;8:90–104. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertini I, Pierattelli R. Copper(II) proteins are amenable for NMR investigations. Pure Appl Chem. 2004;76(2):321–333. [Google Scholar]
- 34.Paci M, Desideri A, Rotilio G. Cyanide binding to Cu, Zn superoxide dismutase. An NMR study of the Cu(II), Co(II) derivative. J Biol Chem. 1988;263(1):162–166. [PubMed] [Google Scholar]
- 35.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402(6757):47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 36.Ubbink M. The courtship of proteins: understanding the encounter complex. FEBS Lett. 2009;583(7):1060–1066. doi: 10.1016/j.febslet.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 37.Warren JJ, Winkler JR, Gray HB. Redox properties of tyrosine and related molecules. FEBS Lett. 2012;586(5):596–602. doi: 10.1016/j.febslet.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courtade G, et al. Backbone and side-chain 1H, 13C, and 15N chemical shift assignments for the apo-form of the lytic polysaccharide monooxygenase NcLPMO9C. Biomol NMR Assign. 2016 doi: 10.1007/s12104-016-9683-x. [DOI] [PubMed] [Google Scholar]
- 39.de Vries SJ, van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5(5):883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 40.Sygmund C, et al. Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation. Appl Environ Microbiol. 2012;78(17):6161–6171. doi: 10.1128/AEM.01503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handook. Springer; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 42.Keller R. The Computer Aided Resonance Assignment Tutorial. CANTINA Verlag; Goldau, Switzerland: 2004. [Google Scholar]
- 43.Farrow NA, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 44.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 45.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56(3):227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulder FAA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J Mol Biol. 1999;292(1):111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 47.Wiseman T, Williston S, Brandts JF, Lin L-N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179(1):131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 48.Turnbull WB, Daranas AH. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125(48):14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 49.DeLano WL, Lam J. PyMOL: A communications tool for computational models. Abstr Pap Am Chem Soc. 2005;230:1371–1372. [Google Scholar]
- 50.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 51.Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 52.Vaaje-Kolstad G, Houston DR, Riemen AHK, Eijsink VGH, van Aalten DMF. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J Biol Chem. 2005;280(12):11313–11319. doi: 10.1074/jbc.M407175200. [DOI] [PubMed] [Google Scholar]