Significance
The HRD (HMG-CoA reductase degradation) pathway is a conserved eukaryotic route of protein destruction, removing misfolded proteins by ER-associated degradation (ERAD). Hrd1 and Hrd3 are each required for substrate degradation. Hrd1 is the core E3 ligase of the HRD pathway, but the function of Hrd3 has remained unclear. Loss of Hrd3 causes extreme destabilization and loss of Hrd1. Thus, it had not been possible to ascertain whether Hrd3 had ERAD functions distinct from Hrd1 stabilization. We have leveraged our understanding of Hrd1 degradation to uncouple Hrd1 stability from Hrd3 presence, allowing us to demonstrate unequivocally that Hrd3 has a direct and essential role in ER quality control. This work sets the stage for biologically relevant, translatable studies of this critical stress management pathway.
Keywords: HRD pathway, HRD3, ERAD, quality control, ubiquitin
Abstract
The HRD (HMG-CoA reductase degradation) pathway is a conserved route of endoplasmic reticulum-associated degradation (ERAD), by which misfolded ER proteins are ubiquitinated and degraded. ERAD substrates are ubiquitinated by the action of the Hrd1 RING-H2 E3 ligase. Hrd1 is always present in a stoichiometric complex with the ER membrane protein Hrd3, which is also required for HRD-dependent degradation. Despite its conserved presence, unequivocal study of Hrd3 function has been precluded by its central role in Hrd1 stability. Loss of Hrd3 causes unrestricted self-degradation of Hrd1, resulting in significant loss of the core ligase. Accordingly, the degree to which Hrd3 functions independently of Hrd1 stabilization has remained unresolved. By capitalizing on our studies of Usa1 in Hrd1 degradation, we have devised a new approach to evaluate Hrd3 functions in ERAD. We now show that Hrd3 has a direct and critical role in ERAD in addition to Hrd1 stabilization. This direct component of Hrd3 is phenotypically as important as Hrd1 in the native HRD complex. Hrd3 was required the E3 activity of Hrd1, rather than substrate or E2 recruitment to Hrd1. Although Hrd1 can function in some circumstances independent of Hrd3, these studies show an indispensable role for Hrd3 in living cells.
Endoplasmic reticulum-associated degradation (ERAD) is a highly conserved quality control pathway responsible for the degradation of both misfolded and normal proteins that limits cellular stress and cytotoxicity caused by an accumulation of misfolded ER proteins (1, 2). ERAD occurs through the ubiquitin proteasome pathway, wherein ER-localized ubiquitin ligases covalently modify substrates by attaching multiple copies of the protein ubiquitin, thus tagging them for degradation by cytosolic 26S proteasome (3–5).
In Saccharomyces cerevisiae, the highly conserved HRD (HMG-CoA† reductase degradation) pathway degrades a variety of ERAD substrates, including misfolded luminal proteins (ERAD-L substrates) such as CPY* and integral membrane proteins (ERAD-M substrates) such as Hmg2 (6, 7). Briefly, ERAD-L substrates are ubiquitinated by the HRD complex, which includes the ubiquitin RING-H2 ligase Hrd1, complex members Hrd3, Usa1, and Der1, and luminal factors implicated in substrate selection (3, 8–11). Conversely, ERAD-M substrates require only Hrd1 and Hrd3 for in vivo degradation (12, 13). Although Hrd1 functions as the core catalytic subunit of the HRD E3 ligase, Hrd3’s function has remained enigmatic. Both Hrd1 and Hrd3 are highly conserved, and Hrd1 is rate limiting for degradation of misfolded substrates (3, 12, 14, 15). In yeast, the steady-state levels of Hrd1 strongly depend on Hrd3. At native levels of the HRD components, Hrd3 is required for Hrd1 stability: In a hrd3Δ, Hrd1 half-life drops to less than 15 min, resulting in very low levels (12). This concomitant loss of Hrd1 is due to rapid autodegradation, catalyzed by the RING domain of Hrd1 (16).
Because of Hrd3’s strong role in maintaining Hrd1 stability, it has remained unclear whether Hrd3 has any other independent ERAD functions. A “stability only” model for Hrd3 function is supported by numerous instances in which overexpression of Hrd1 suppresses ERAD defects observed in a hrd3Δ (12, 14, 15). In contrast, a “direct effect” model for Hrd3 is implied by our early observation that a large deletion of Hrd3 (350 amino acids) stabilizes Hrd1 but does not allow Hmg2 degradation (12). Resolving these two models is important for understanding this highly conserved quality control pathway.
Until now, there has been no way to study any function of Hrd3 independent of Hrd1 stabilization. Our studies of Usa1 have now allowed us to address this problem. We had found that Hrd1 self-degradation observed in the hrd3Δ strongly depends on Usa1: Hrd1 destabilization is absent in the complete deletion of Usa1 or when the Usa1 protein has only its cytoplasmic ubiquitin-like (UBL) domain deleted (Usa1-ΔUBL) (16). However, unlike a simple usa1Δ, deletion of the UBL domain of Usa1 does not affect ERAD-L or ERAD-M (16). Thus, use of the USA1-ΔUBL allele in place of the native gene provides a powerful platform for addressing the two views of Hrd3 function in ERAD.
Using this and other approaches, we have observed that Hrd3 was vital for Hrd1-mediated substrate degradation. At endogenous levels of Hrd1, the absence of Hrd3 rendered stable Hrd1 inactive toward all substrates tested. The importance of Hrd3 was also clear at high levels of Hrd1 used to test Hrd1 autonomy in ERAD (17, 18). Hrd3 was not required for Hrd1–substrate interaction nor Hrd1 multimerization, but was required for Hrd1-mediated E2–substrate interactions. These results unequivocally demonstrate a direct role of Hrd3 in the activity of the HRD complex and show the importance of Hrd3 in the intrinsic functions of the HRD complex.
Results
Hrd3 Was Not Required for Hrd1 Stability in a USA1-ΔUBL Strain.
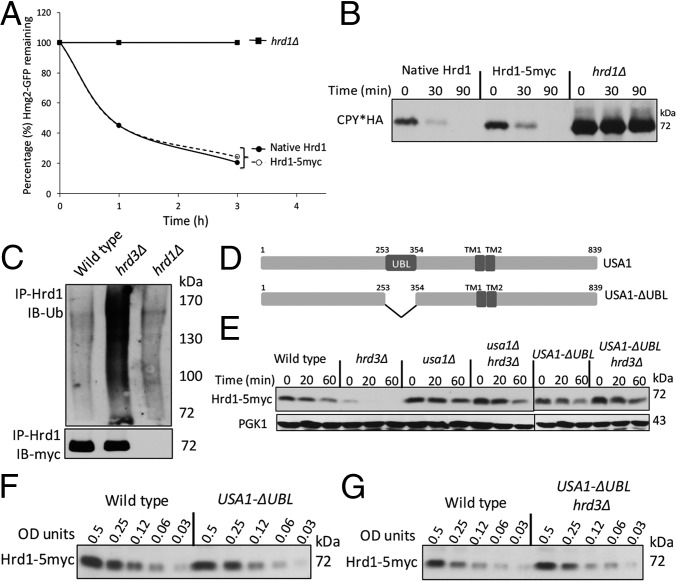
In this study, we used an epitope-tagged Hrd1 (Hrd1-5myc). To confirm its normalcy, we compared the degradation rates of ERAD-M substrate Hmg2-GFP in a strain expressing either untagged Hrd1 or Hrd1-5myc each from single integrants driven by the native promoter. We quantitated Hmg2-GFP degradation by using flow cytometry after cycloheximide addition (Chx chase). Untagged Hrd1 and Hrd1-5myc were identically efficient in Hmg2-GFP degradation (Fig. 1A). They were also identically efficient in degradation of ERAD-L substrate CPY*HA (Fig. 1B). Finally, Hrd1-5myc underwent the expected rapid degradation absence of Hrd3 (Fig. 1C) (16). These results showed that Hrd1-5myc behaved identically to native untagged Hrd1.
Fig. 1.
Hrd1-5myc was stable and functional. (A) Flow cytometry analysis of Hmg2-GFP degradation in hrd1∆ strains with indicated Hrd1 plasmids by cycloheximide (Chx) chase. (B) Chx chase for CPY*HA with anti-HA immunoblotting in strains with indicated Hrd1 plasmids. (C) Ubiquitination analysis of Hrd1-5myc in the indicated strains upon immunoprecipitation (IP) with anti-Hrd1 antibody. (D) Schematic of Usa1 and Usa1-∆UBL (an internal deletion of 100 amino acids). (E) Indicated strains were analyzed for Hrd1-5myc degradation by Chx chase. (F) Strains with Wt USA1 or USA1-∆UBL allele were lysed and analyzed by anti-myc IB for Hrd1. (G) Dilution IB as in Fig. 1F between USA1 (Wt) or USA1-ΔUBL/hrd3Δ strains.
Hrd1 self-ubiquitination in hrd3∆ depends on a region of Usa1 called the UBL domain (amino acids 254–353; Fig. 1D and ref. 16). A deletion of USA1 caused complete loss of ERAD-L and impacted degradation of ERAD-M substrates (ref. 16 and this work). However, USA1 with a deletion of only the UBL domain (called USA1-ΔUBL) fully supported both ERAD-L and ERAD-M but stabilized Hrd1: Hrd1-5myc was rapidly degraded in a hrd3∆ strain but was strongly stabilized in USA1-ΔUBL/hrd3∆ strains (Fig. 1E). To accurately assess Hrd1 steady-state levels in various strains, we used dilution immunoblotting (Fig. 1 F and G). The presence of USA1-ΔUBL allele had no effect on steady-state levels of Hrd1-5myc compared with isogenic wild-type strain. Additionally, Hrd1-5myc levels were also identical between wild-type and USA1-ΔUBL/hrd3∆ strain because of the lack of Hrd1 self-degradation. Therefore, the USA1-ΔUBL allele provided an ideal genetic platform to investigate the role of Hrd3 independent of effects on Hrd1 stability.
A Direct Role for Hrd3 in ERAD-L.
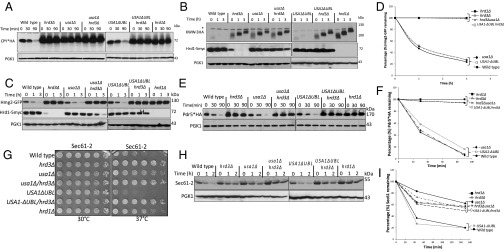
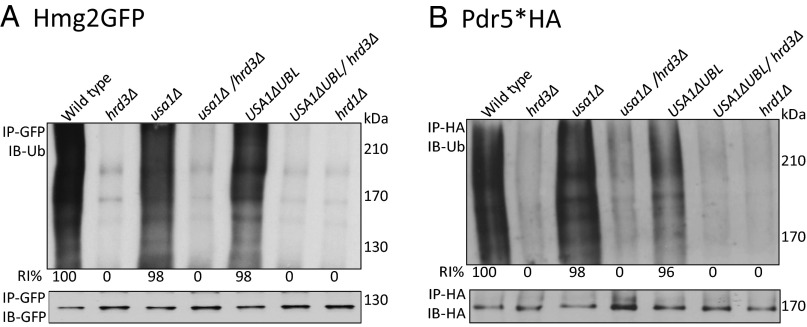
Hrd3 is thought to be directly involved in degradation of ERAD-L substrates because of its observed abilities to recruit luminal ERAD factors and to bind misfolded substrates (19–21). Nevertheless, in the hrd3Δ null, the resulting low levels of destabilized Hrd1 could explain the ERAD defect. To test whether Hrd3 was directly required for ERAD-L, we performed Chx chase for CPY*HA in hrd3Δ, usa1Δ, and the newly made USA1-ΔUBL/hrd3Δ strain where Hrd1 levels are identical to those in the corresponding HRD3 strain. CPY* was rapidly degraded in a wild-type and USA1-ΔUBL strain (Fig. 2A), but was stable in either hrd3∆ or USA1-ΔUBL/hrd3∆ strains, to the same extent as a hrd1Δ null. A second ERAD-L substrate KWW behaved identically in the same tests (Fig. 2B). Thus, both ERAD-L substrates showed complete HRD3 dependence in a USA1-ΔUBL/hrd3Δ strain where Hrd1 was stable and present at wild-type levels (Fig. 2B, Middle). Note, in striking contrast to the USA1-ΔUBL strains, the complete block in ERAD-L observed in the included usa1∆, as expected from our previous studies (16). This difference emphasizes the need for the USA1-∆UBL allele in studying Hrd3 function in ERAD.
Fig. 2.
Hrd3 dependence of ERAD substrate degradation. (A) Chx chase of CPY*HA in the indicated strains. Incubation time following Chx addition is indicated in minutes. Anti-PGK1 is included as a loading control in all experiments. (B) KWW-3HA degradation by Chx chase. (C) Hmg2-GFP degradation by Chx chase. (D) Hmg2-GFP degradation measured by flow cytometry. Mean fluorescence of 10,000 cells at the 0-h point was taken as 100%. Error bars represent ± SEM. (E) Pdr5*HA degradation by Chx chase. (F) Pdr5*HA blots from three different experiments as in Fig. 2E were quantified by Typhoon 9400 and ImageQuant 5.2 where t = 0 was taken as 100%. Error bars represent ± SEM. (G) Indicated strains were spotted at fivefold dilutions on plates and grown at 30 °C or 37 °C for 3 d. (H) Sec61-2 degradation as in A. Cultures were preincubated at 37 °C for 15 min before Chx addition. (I) Sec61 degradation blots from three different experiments were quantified and represented as described in Fig. 2F. Error bars represent ± SEM.
A Direct Role for Hrd3 in ERAD-M.
It was perhaps not surprising that Hrd3 is directly involved in ERAD-L, independent of Hrd1 stability, because of its proposed roles in luminal factor recruitment and substrate binding. However, ERAD-M does not appear to require any luminal factors. We next used the USA1-ΔUBL/hrd3Δ strain to study the role of Hrd3 in ERAD-M independent of effects on Hrd1 stability, starting with the eponymous substrate Hmg2-GFP. Hmg2-GFP undergoes Hrd1-dependent degradation in wild-type cells and is completely stable in either hrd1Δ or hrd3Δ strains (12, 13, 22). Hmg2-GFP was also completely stabilized by removal of HRD3 in USA1-ΔUBL/hrd3∆, although Hrd1 levels were unaffected, observed by Chx chase (Figs. 1G and 2C), or flow cytometry (Fig. 2D). Again, stabilization by removal of HRD3 in a USA1-ΔUBL/hrd3∆ background was identical to that observed in a hrd1∆ (Fig. 2 C and D), although Hrd1 levels were unaffected (Fig. 2C, Middle).
Two other ERAD-M substrates, Pdr5* and Sec61-2, showed the same direct Hrd3 dependence in the USA1-ΔUBL/hrd3∆ strain. Pdr5* is a point mutant of ABC transporter Pdr5, rendering it an ERAD-M substrate (7). Pdr5* was rapidly degraded both in the wild-type and USA1-ΔUBL strains (Fig. 2E), whereas there was a complete block in the degradation of Pdr5* when hrd3∆ was included in either of the backgrounds (Fig. 2E). Stabilization of Pdr5* in USA1-ΔUBL/hrd3∆ strain was as strong as that seen in a hrd1∆ strain (Fig. 2F). Strains carrying the SEC61-2ts allele, encoding the ERAD substrate Sec61-2, are temperature sensitive because of Hrd1-mediated degradation of essential ER membrane protein Sec61 (23, 24). When the HRD pathway is nonfunctional, SEC61-2ts strains can grow at the normally nonpermissive temperature (37 °C). We analyzed the growth of SEC61-2ts strains expressing various combinations of USA1 and HRD3 at 30 °C and 37 °C. All strains grew at similar rates at 30 °C (Fig. 2G), whereas at 37 °C, there were substantial differences in the growth. SEC61-2ts strains with wild-type USA1 or USA1-∆UBL were severely impaired for growth at 37 °C. Conversely, strains also harboring a hrd3Δ showed robust growth at 37 °C identical to that of the hrd1∆ (Fig. 2G). We also directly tested Sec61-2 stability in these strains by Chx chase and immunoblotting. The results were entirely in agreement with the growth assays: Sec61-2 was stable in all of the strains at 30 °C. When strains with wild-type USA1 or USA1-∆UBL were shifted to 37 °C, Sec61-2 was rapidly degraded, but was fully stable when hrd3∆ was included in either background (Fig. 2H). Again, Sec61 stability in the USA1-ΔUBL/hrd3Δ strain was comparable to that seen in a hrd1∆ (Fig. 2 H and I). As with the other two ERAD-M substrates tested, the USA1-ΔUBL/hrd3∆ strain exhibited strong stabilization of Sec61 despite wild-type levels of Hrd1. Surprisingly, the lone usa1Δ null mutant allowed the SEC61-2ts strain growth at nonpermissive temperature, indicating Sec61 stabilization. Conversely, the USA1-ΔUBL strain showed poor growth because of wild-type Hrd1 activity at nonpermissive temperatures. Chx chase confirmed that Sec61-2 was stabilized in usa1∆, but not USA1-ΔUBL. These results demonstrated that the usa1∆ can have significant effects on degradation of some ERAD-M substrates and emphasize the importance of using the USA1-ΔUBL allele.
Taken together, these results for the first time, to our knowledge, show a direct role for Hrd3 in both ERAD-L and ERAD-M, such that the Hrd1 protein is nonfunctional as an E3 when Hrd3 is absent and all other HRD complex members are at native levels.
Hrd1–Substrate Interactions Were Not Affected by Deletion of Hrd3.
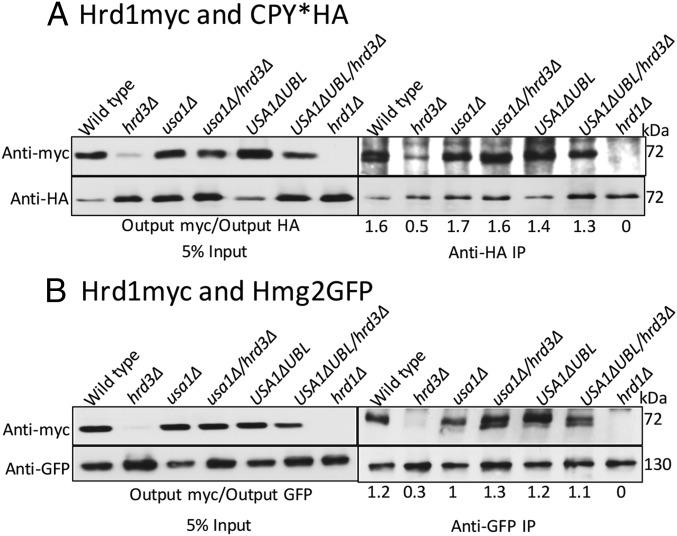
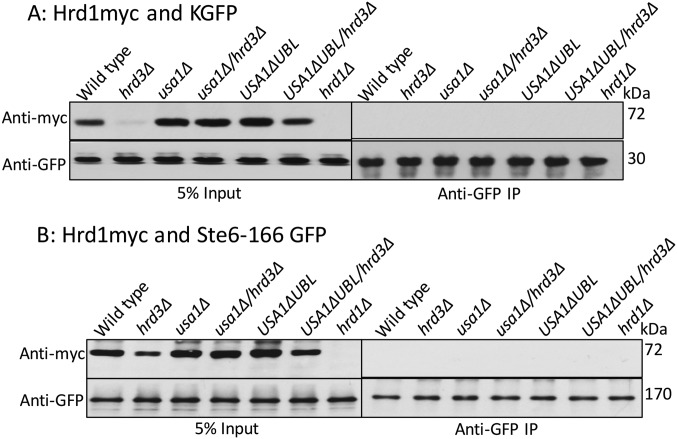
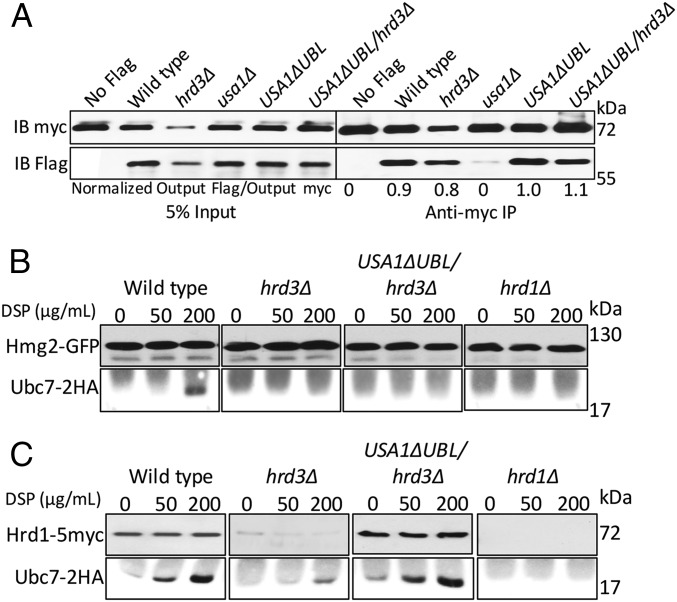
We next explored how Hrd3 affects the action of the HRD complex. We first tested the role of Hrd3 in Hrd1–substrate interaction, using the USA1-ΔUBL approach to investigate these Hrd3 effects. The classic ERAD-L substrate CPY* was immunoprecipitated from lysates of various strains coexpressing Hrd1-5myc in presence or absence of Hrd3, followed by SDS/PAGE and immunoblotting for Hrd1-5myc and CPY*HA (Fig. 3A). A control strain containing an empty vector (instead of Hrd1 plasmid) was included in each experiment. Binding of CPY* to Hrd1 was clear, and unaffected by the presence or absence of Hrd3 or Usa1, indicating that Hrd3 or Usa1 are not essential for binding of ERAD-L substrates to Hrd1. Even when Hrd1 protein levels were low in hrd3∆ the CPY*–Hrd1 interaction was detected (Fig. 3A). As a specificity control, we tested Hrd1 binding to ER luminal protein KGFP (25) that is not a HRD substrate and found no detectable association (Fig. S1A).
Fig. 3.
ERAD substrates interaction with Hrd1 in absence of Hrd3. (A) CPY*HA and Hrd1-5myc interaction was analyzed by co-IP. The IP was analyzed for presence of Hrd1-5myc. The output ratios were calculated by fluorChemE and alphaviewQ from three different experiments. (B) Interaction between Hmg2-GFP and Hrd1-5myc as in Fig. 3A.
Fig. S1.
ERAD substrates interaction with Hrd1 in absence of Hrd3. (A) As a negative control, luminal ER protein KGFP and Hrd1-5myc interaction was analyzed by co-IP. The IP was analyzed for presence of Hrd1-5myc. (B) As a negative control, ER membrane protein Ste6-166-GFP and Hrd1-5myc interaction was analyzed by co-IP. The IP was analyzed for presence of Hrd1-5myc as in A.
We similarly tested the role of Hrd3 in Hrd1 interaction with ERAD-M substrate Hmg2. Hmg2-GFP was immunoprecipitated with anti-GFP antibodies from lysates coexpressing Hrd1-5myc in presence or absence of Hrd3, followed by Hrd1-5myc or Hmg2-GFP immunoblotting (Fig. 3B). Again, an empty vector control was included in each experiment. Hrd1-5myc was similarly bound to Hmg2 in wild-type and hrd3∆ strains, indicating that Hrd3 is not required for ERAD-M substrate binding to Hrd1. The non-HRD ERAD-M substrate Ste6-166-GFP was included as a specificity control and was unable to bind Hrd1-5myc (Fig. S1B). These results indicated that Hrd3 does not alter Hrd1–ERAD substrate interactions. Thus, it appeared that the direct Hrd3 ERAD effects were not due to altering E3–substrate interactions.
Hrd3 Was Required for Hrd1 Ubiquitination Activity.
We next tested the effect of Hrd3 on Hrd1 E3 ligase activity, using two different ERAD-M substrates. We first carried out in vivo ubiquitination assays for Hmg2-GFP in the indicated strains by immunoprecipitation followed by ubiquitin or GFP blotting. In either a hrd1∆ or hrd3∆ strain, there was a severe block in Hmg2-GFP ubiquitination. We observed the same profound block in Hmg2-GFP ubiquitination in USA1-∆UBL/hrd3∆ with normal levels of Hrd1 (Fig. 4A). A similar effect was seen with ERAD-M substrate Pdr5* (Fig. 4B). These results indicated that Hrd3 is directly required for Hrd1 activity at native levels of the RING protein. Importantly, in both cases, the defect in substrate ubiquitination brought about by the hrd3∆ in the USA1-∆UBL background is as profound as a hrd1∆. It is interesting to note (and see below) that the activation of Hrd1 by Hrd3 was also observed in the simple usa1Δ strain for both of these ERAD substrates.
Fig. 4.
Hrd3 dependence of ERAD substrate ubiquitination. (A) Hmg2-GFP ubiquitination was assayed in the indicated strains by IP. The percentage intensities (RI%) were quantified by fluorChemE and alphaviewQ from three different experiments. (B) Prd5* ubiquitination was assayed as described in Fig. 4A by using anti-HA IP.
Hrd3 Strongly Activated Overexpressed Hrd1.
The above studies using USA1-ΔUBL showed that Hrd3 has a direct role in Hrd1 activity: Removal of HRD3 caused an ERAD deficiency as severe as a hrd1Δ null, and it appeared to operate on substrate ubiquitination rather than substrate recruitment. By all measures, USA1-ΔUBL supported normal ERAD-L and ERAD-M. Nevertheless, it was important to examine this direct role of Hrd3 independent of the USA1-ΔUBL allele to allay concerns that this USA1 variant had some spurious involvement.
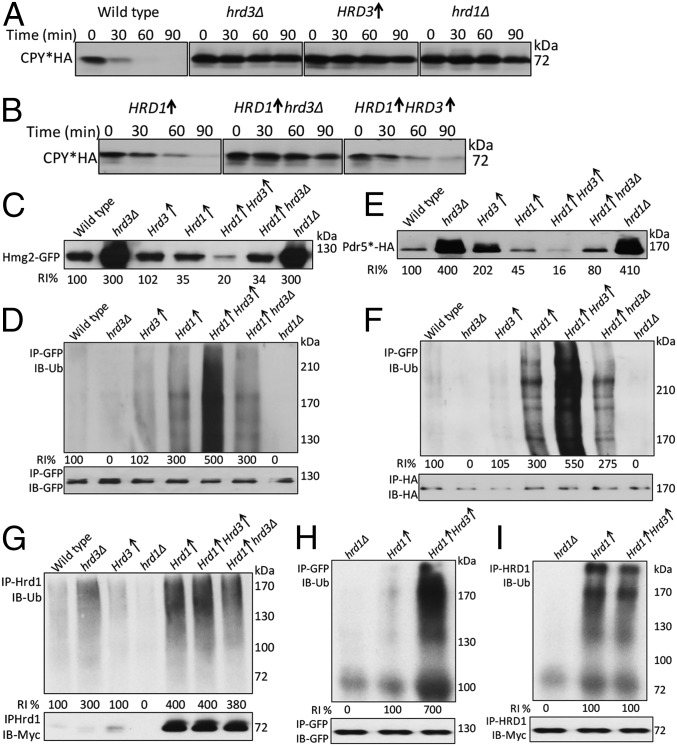
To independently test Hrd3 effects, we used strains overexpressing Hrd1. This approach is particularly relevant because Hrd1 overexpression has long been observed to suppress the hrd3Δ null, implying that Hrd1 might function independently of Hrd3 at high levels (12, 14, 15). If Hrd3 is a direct activator of Hrd1, we posited that addition of Hrd3 would increase HRD activity even at high levels of Hrd1. We first assessed this overexpression approach for ERAD-L because elevated Hrd1 is sufficient to drive this branch (15, 19). We directly compared various combinations of Hrd1 and Hrd3 expression for degradation of CPY*. In the wild-type strain, degradation of CPY* is very brisk and, as expected, loss of either Hrd3 or Hrd1 showed complete stabilization of the substrate. Additionally, overexpression of Hrd3 alone also causes stabilization of CPY* (Fig. 5A), perhaps because excess Hrd3 sequesters Hrd1 or other ERAD components (15). Simple overexpression of Hrd1 in a hrd3Δ partially restored degradation of CPY* with a half-life of approximately 45 min compared with a hrd3Δ alone (Fig. 5A vs. Fig. 5B) (19). However, addition of a genomic copy of Hrd3 significantly enhanced the effects of overexpressed Hrd1, but increasing Hrd3 above genomic levels had no additional effect (Fig. 5B, Left vs. Right). This effect may also be due to the more complex role of Hrd3 itself as an organizer of a variety of rate-limiting factors needed for ERAD-L, or perhaps to a different stoichiometry of Hrd1 used in ERAD-L and ERAD-M.
Fig. 5.
Hrd3 requirement of overexpressed Hrd1. (A) CPY*HA degradation was analyzed by Chx chase. Arrows indicate expression from strong TDH3 promoter for the respective genes. (B) CPY*HA degradation in the indicated strains as in Fig. 5A. (C) Steady-state levels of Hmg2-GFP in the indicated strains. The RI% were calculated as in Fig. 4A. (D) Hmg2-GFP ubiquitination was assayed as described in Fig. 4A. (E) Steady-state levels of Prd5*-HA in the indicated strains. (F) Ubiquitination of Prd5* was assayed as in Fig. 4B. (G) Ubiquitination of Hrd1-5myc was assayed as in Fig. 1C. (H) In vitro Hmg2-GFP ubiquitination was assayed. (I) In vitro ubiquitination of Hrd1-5myc was assayed.
We next tested effect of Hrd3 addition to overexpressed Hrd1 on ERAD-M. First, we compared the steady-state levels of ERAD-M substrate Hmg2-GFP in various strains (Fig. 5C). As expected, Hmg2 is significantly lower when Hrd1 is overexpressed in a wild-type strain or in a hrd3Δ (3, 12, 19) (Fig. 5 C and D). However, expressing Hrd3 from the same strong promoter as that driving Hrd1 caused ∼fivefold lower steady-state levels of Hmg2-GFP compared with Hrd1 alone (Fig. 5C, lane 4 vs. 5). We also saw ∼fivefold increased Hmg2-GFP ubiquitination when Hrd3 was cooverexpressed with Hrd1. The ERAD-M substrate Pdr5*HA behaved in a similar manner: Its steady-state levels were also substantially lowered by increasing Hrd3 along with Hrd1 (Fig. 5E), and Pdr5* ubiquitination was ∼fivefold greater when Hrd3 and Hrd1 were cooverexpressed (Fig. 5F). We confirmed that, as in the native expression case, Hmg2-Hrd1 was not enhanced by increasing Hrd3 (Fig. S2). These results indicated that Hrd3 directly and significantly stimulated the E3 activity of Hrd1 toward ERAD-M substrates. Furthermore, although Hrd1 is clearly rate limiting for substrate degradation and ubiquitination in both wild-type and hrd3Δ strains, the combination of the two proteins at similar levels caused a profound increase in Hmg2 ubiquitination.
Fig. S2.
Overexpression of Hrd3 did not alter Hrd1–substrate interaction. The interaction between Hmg2-GFP and TDH3-driven Hrd1-5myc was tested by co-IP. Microsomes were isolated from logarithmically growing cells and subjected to an anti-GFP IP. The immunoprecipitate was analyzed by SDS/PAGE and immunoblotting for presence of Hrd1 using myc antibodies. Arrows indicate expression from strong TDH3 promoter of the respective genes.
To test whether Hrd3-dependant activation of Hrd1 was due to a general activation of Hrd1 E3 activity, we examined Hrd1 self-ubiquitination in the overexpression strains. Despite the strong effects on ERAD-M substrate ubiquitination, addition of Hrd3 caused no change in Hrd1 self-ubiquitination (Fig. 5G). These results indicated that the Hrd3-dependent activation of Hrd1 was specific for ubiquitin transfer to ERAD substrates. We tested the effect of Hrd3 on Hrd1 E3 activity in vitro (13, 17). The assay involves a microsome donor strain with overexpressed Hmg2-GFP and Hrd1 in a ubc7Δ strain and a cytosol strain overexpressing Ubc7 in a hrd1Δ strain. As predicted from the in vivo studies, in vitro Hmg2-GFP ubiquitination was strikingly elevated (∼sevenfold) when Hrd3 was overexpressed along with Hrd1 (Fig. 5H). As in the in vivo studies, Hrd1 self-ubiquitination was unaffected by the presence or absence overexpressed Hrd3 (Fig. 5I).
Hrd3 Did Not Affect Hrd1 Multimerization.
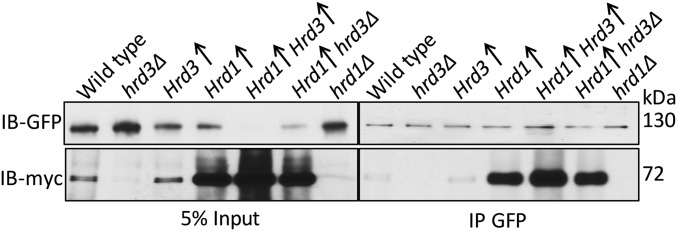
Hrd1 forms Usa1-dependent multimers, and this feature of the Hrd1 has been posited to be important in ERAD (20). We wondered whether the profound activation of Hrd1 by Hrd3 was due to effects on Hrd1 self-association. To address this concern, we expressed two differently tagged Hrd1s (Hrd1-5myc and Hrd1-flag) and examined multimerization by coimmunoprecipitation. Specifically, Hrd1-5myc was immunoprecipitated from the lysates and immunoblotted for flag and myc (Fig. 6A). For each experiment, an empty vector, no-Hrd1-flag control was included. We confirmed that Hrd1 multimerization, as indicated by coprecipitation, was completely absent in the usa1Δ background (Fig. 6A). However, Hrd1 multimerization was equal to wild type in the USA1-ΔUBL strain. Furthermore, hrd3Δ had no effect on Hrd1 multimerization in the USA1-ΔUBL where Hrd1 levels remain identical to wild type. These results indicated Hrd3 did not affect Hrd1 multimerization.
Fig. 6.
Hrd3 requirement in multimerization and E2 recruitment. (A) Hrd1-Flag and Hrd1-5myc interaction was tested by co-IP. The IP was analyzed for presence of Hrd1-Flag. The output ratios were calculated by fluorChemE and alphaviewQ from three different experiments. (B) Analysis of cross-linking between Hmg2-GFP and Ubc7-2HA. Spheroplasts were harvested from DSP-treated isogenic strains and subjected to an anti-GFP IP, followed by IB for GFP and HA. (C) The cross-linking between Hrd1-5myc and Ubc7-2HA was tested as in Fig. 6B.
In the course of these studies, we made a separate observation concerning the role of multimerization in the HRD pathway. In many of the in vivo experiments above, we included usa1Δ strain to contrast them to the more subtle USA1-ΔUBL allele extensively used above. Accordingly, the effect of presence or absence of Hrd3 in these simple usa1Δ nulls can be viewed as a test of Hrd3’s action when Hrd1 multimerization is not permitted. In the ubiquitination studies of ERAD-M substrates Hmg2 and Pdr5* (Fig. 4 A and B), loss of Hrd3 in the usa1Δ strain caused a loss of ubiquitination as strong as that seen in the hrd1Δ, indicating that the activating effects of Hrd3 do not require the presence of Usa1. Thus, Hrd3 can strongly activate Hrd1 to ubiquitinate several ERAD substrates in the apparent absence of Hrd1 multimerization.
Hrd3 Was Required for E2–Substrate Interaction.
The Hrd1 RING ligase catalyzes ubiquitin transfer from Ubc7 (the principal E2) to ERAD substrates. We used an in vivo cross-linking assay to directly study association of Ubc7 with ERAD-M substrate Hmg2 and Hrd1 (12). We expressed 2HA-tagged Ubc7 in a variety of strains expressing Hmg2-GFP. Briefly, cells were spheroplasted followed by treatment with the membrane-permeable bifunctional cross-linking reagent DSP. Hmg2 was immunoprecipitated from various strains and immunoblotted for the presence of cross-linked and, thus, coprecipitated Ubc7-2HA. As previously published, Ubc7 was cross-linked to Hmg2 in a Hrd1- and DSP-dependent manner (Fig. 6B). The lack of Hrd3 completely abrogated Hmg2-GFP/Ubc7 interaction, even in the USA1-ΔUBL/hrd3Δ strain where Hrd1 levels are identical to wild type (Fig. 6C). Consistent with the other assays, the presence of Hrd3 was as important as Hrd1 itself: The effect of hrd3Δ was as strong as that seen in the negative control hrd1Δ despite there being wild-type levels of Hrd1 in the USA1-ΔUBL/hrd3Δ strain. These results establish that Hrd3 is necessary for Hrd1-dependent interaction of E2 and Hmg2 at native levels of Hrd1. Ubc7-Hrd1 cross-linking (Fig. 6C) was unaffected by the presence or absence of Hrd3, as expected from the other results above.
Taken together, these studies indicated that Hrd3 activates Hrd1 for ERAD, causing ubiquitination and degradation rates far in excess of what is observed by elevated Hrd1 alone, and far in excess of the rates observed when all of the components are present at native levels. In fact, at genomic levels of Hrd1, Hrd3 is absolutely required for either branch of ERAD, in addition to its long-standing role in preserving Hrd1 stability.
Discussion
Although Hrd3 was one of the first discovered ERAD factors, the details of its function have remained cryptic because of its role in stabilizing Hrd1 (12, 16). Here, we showed a direct and strong role of Hrd3 in both branches of ERAD-independent Hrd1 stabilization. To uncouple the two functions, we used a strain harboring the USA1-ΔUBL allele; in the USA1-ΔUBL background, Hrd1 is stable in the presence of a hrd3Δ null and so has levels identical to those found in wild-type strains. Using this approach, we showed that loss of Hrd3, even when Hrd1 was stable (Fig. 1E), still drastically compromised both branches of HRD-dependent ERAD (Fig. 2). In fact, a hrd3Δ strain was as ERAD deficient as a hrd1Δ strain, indicating that Hrd3 has a role in the HRD complex as important as the core E3 subunit Hrd1. This unifying feature of Hrd3 being directly important in both branches of the HRD pathway is consistent with the broadly conserved presence of both Hrd1 and Hrd3 in all organisms with the HRD pathway.
Hrd3 has been proposed to serve as an “interaction scaffold” for mediating substrate–HRD complex association. Nevertheless, we could not discern an effect of Hrd3 on a specific association between either ERAD-M or ERAD-L substrates and Hrd1. This observation could mean that the primary direct role of Hrd3 is distinct from mediating Hrd1–substrate interactions. This idea is perhaps consistent with the observation that Hrd1 alone can specifically recognize CPY* with an apparent Km in the low nanomolar range (18). In ERAD-M as well, substrate ubiquitination was fully Hrd3-dependent at endogenous levels of Hrd1 despite any effects on Hrd1–substrate interaction (Fig. 4).
The USA1-ΔUBL allele of the USA1 gene appears to function entirely normally for each ERAD substrate tested. Nevertheless, we tested for a role of Hrd3 independent of the presence of this useful variant. We thus examined the effect of increasing Hrd3 when Hrd1 was overexpressed in otherwise normal strains. Indeed, an addition of sufficient Hrd3 to strains overexpressing Hrd1 had an impressive effect on Hrd1-dependent degradation and ubiquitination of several substrates (Fig. 5 C and D). This result was especially poignant because overexpression of Hrd1 has been one of the ways that we and others have surmised that Hrd3 might be dispensable for the HRD pathway. The presence of a genomic copy of Hrd3 enhanced the effects of overexpressed Hrd1 for ERAD-L, but increasing Hrd3 higher than genomic levels had no additional effect (Fig. 5B). Use of this same approach revealed a striking, direct effect of Hrd3 on Hrd1 ligase activity in ERAD-M (Fig. 5 D and F). Although overexpressed Hrd1 can enhance ERAD-M as expected, its efficiency in both degradation and ubiquitination of these substrates was increased ∼fivefold in the presence of similarly overexpressed Hrd3 both in vivo and in vitro (Fig. 5 D, F, and H). In contrast, Hrd1 self-ubiquitination was unaffected by overexpressing Hrd3 from a strong promoter (Fig. 5 G and I).
We further used the USA1-ΔUBL allele to explore the nature of Hrd3’s role in Hrd1 action. We found that Hrd3 had no effect on Hrd1 multimerization (Fig. 6A). As expected, we confirmed the absence of Hrd1 multimerization in a usa1Δ strain (Fig. 6A). Of note, Hrd3-depenedent activation of Hrd1 was still observable in the usa1Δ strain where multimerization does not occur, with or without Hrd3. Accordingly, at least by this criterion, multimerization cannot be a sine qua non for Hrd1 action or Hrd3 enhancement of Hrd1. Using a cross-linking assay to query E2–substrate interaction, we found that Ubc7 was not able to interact with an ERAD-M substrate in the absence of Hrd3 even when Hrd1 levels were identical to wild type (Fig. 6B). Here also, the effect of the hrd3Δ was identical to that of a hrd1Δ, again indicating the key role of Hrd3 in the HRD complex activity. In contrast, Ubc7 cross-linking to Hrd1 itself was independent of Hrd3 (Fig. 6C), consistent with the lack of effect of Hrd3 on Hrd1 self-ubiquitination both in vivo and in vitro (Fig. 5 G and I). These results highlight the selective and strong importance of Hrd3 for Hrd1-mediated E2/substrate interaction.
Taken together, the direct role of Hrd3 is clearly evident, and Hrd3 is indispensable at endogenous levels of Hrd1. At the same time, it is clear that sufficient Hrd1 will allow some degree of ERAD in the absence of Hrd3. What is not clear is the extent to which Hrd1 action is the same in the presence or absence of Hrd3. Is Hrd3 simply enhancing the same catalytic process, or is it altering the enzymology of Hrd1? This question is particularly relevant because many studies including our own have capitalized on the dispensability of Hrd3 for examination of Hrd1 in microsomes or reconstituted systems (15, 17). Certainly these approaches are useful in studying aspects of the HRD pathway although caution must be exerted in marginalization of Hrd3 in the light of its unveiled importance in Hrd1 action. Because the Hrd3 homolog SEL1-L similarly stabilizes Hrd1 in mammalian cells (26), these studies of the dual roles of Hrd3 as a stabilizer and an activator will have interesting connections to the larger eukaryotes; we anticipate SEL1-L will be similarly involved in both ensuring Hrd1 levels and activity in the mammalian context as well.
In conclusion, the present study adds a new dimension to our understanding of Hrd3 action in ERAD. We have demonstrated an indispensable role of Hrd3 in the E3 activity of HRD complex at endogenous levels. This activation occurred over a wide range of Hrd1 expression and can be observed both in vivo and in vitro. Hrd3 appeared to enhance the efficacy of ubiquitin transfer to substrates, but not the general activity of Hrd1 nor the association of substrates with Hrd1. Hrd3 thus should be considered an integral component in the normal function of this widely conserved quality control ERAD complex.
Materials and Methods
Plasmid Construction, Yeast, and Bacterial Strains.
A complete table of the plasmids and yeast strains used in this study is provided in Tables S1 and S2.
Degradation Assays.
Cycloheximide chase assays were performed by the addition of cycloheximide to log-phase cultures followed by lysis at the indicated times. A more detailed description is provided in SI Materials and Methods.
Ubiquitination Assay.
Ubiquitination of Hmg2p-GFP or Pdr5*HA was performed as described (13). A more detailed description is provided in SI Materials and Methods.
In Vivo Cross-Linking.
The in vivo cross-linking assay was essentially done as described by ref. 12. A more detailed description is provided in SI Materials and Methods.
SI Materials and Methods
Plasmid Construction, Yeast, and Bacterial Strains.
All plasmids were constructed with standard molecular biology techniques as described (13). PCR was carried out as described (13), and plasmids constructed by using PCR were sequence-verified (Eton Bioscience). A detailed description and complete plasmid table is provided in Tables S1 and S2. The KWW (pRH1960), KGFP (pRH1549), and CPY* (pRH1377) plasmids were a generous gift from Davis Ng, National University of Singapore, Singapore. The Pdr5* plasmid (pRH2312) was a generous gift from Dieter H. Wolf, University of Stuttgart, Stuttgart, Germany.
Escherichia coli DH5a were grown in LB media with ampicillin. Yeast strains were grown at 30 °C unless noted in minimal media supplemented with dextrose and amino acids. All strains were isogenic.
Degradation Assays.
Cycloheximide chase assays were performed by the addition of cycloheximide to log-phase cultures followed by glass bead lysis in 100 μL of SUME [SDS, UREA, MOPS, and EDTA (1% SDS, 8 M Urea, 10 mM MOPS, pH 6.8, 10 mM EDTA)] with protease inhibitors at the indicated times, as described (13). Equal loading of gels was confirmed by India ink staining of the respective nitrocellulose membranes. GFP levels were analyzed in living cells (10,000 cells per sample) by flow cytometry of log-phase cultures using an Accuri machine (BD Biosciences) and Flowjo software. All non-GFP strain quantitation was performed by using Typhoon 9400 and ImageQuant 5.2 software (GE Healthcare). In such experiments, a representative Western blot was quantified and the data points were graphed.
Dilution Assays.
Indicated yeast strains were grown to high density (A600 > 1.5) in YPD medium overnight at 30 °C. Cells were then pelleted, washed with water once, and beginning with an A600 of 0.35, fivefold serial dilutions were made and spotted onto the appropriate dropout plates followed by incubation at the indicated temperatures for 2 d (30 and 35 °C) or 3 d (37 °C).
Ubiquitination Assay.
Ubiquitination of Hmg2p-GFP or Pdr5* HA was examined in log-phase cells as described (13) by IP followed by ubiquitin or substrate immunoblotting. Briefly, 5 OD of log-phase cells were pelleted. Cells were resuspended in 100 μL of SUME with protease inhibitors and N-ethyl maleimide (NEM). Later, 100 μL of glass beads were added and the tubes were vortexed for 3–5 min with 1-min interval to lyse the cells. The lysates were clarified to remove unbroken cells. One milliliter of IP buffer [150 mM NaCl, 15 mM Na2HPO4, 2% (vol/vol) Triton X-100, 0.1% SDS, 0.5% DOC, 10 mM EDTA, pH 7.5] with protease inhibitors and NEM was added to the lysates, and the mix was centrifuged for 5 min at 14,000 × g. The supernatant was removed, and 15 μL of the respective polyclonal antibody was added. The mix was incubated overnight at 4 °C. One hundred microliters of Protein-A Sepharose (Amersham Biosciences) in IP buffer (50% wt/vol) was added and incubated for 2 h. Beads were washed once with IP buffer and once with IP wash buffer (50 mM NaCl, 10 mM Tris, pH 7.5) and incubated with 50 μL of 2× Urea sample buffer for 10 min at 50 °C. The samples were then loaded on a polyacrylamide gel. Following transfer to nitrocellulose, immunoblotting with an anti-ubiquitin or anti-GFP/anti-HA antibody was performed.
In Vivo Hrd1p Ubiquitination.
It was assayed by immunoprecipitating Hrd1 from pdr5Δ null strains after treatment of log-phase cultures (A600 > 0.25) with the proteasome inhibitor MG132 (benzyloxycarbonyl-Leu-Leu-aldehyde; Sigma) for 2.25 h before lysis. Four OD of the treated cells were pelleted and lysed in SUME with protease inhibitors. IP was performed as mentioned above by using anti Hrd1 antibody. The samples were run on 8% SDS/PAGE, and after transfer to nitrocellulose, immunoblotting with an anti-ubiquitin or anti-myc antibody was performed.
In Vitro Ubiquitination.
Ubiquitination of Hmg2-GFP or Hrd1-5myc was essentially done as described (17).
In Vivo Cross-Linking.
The in vivo cross-linking assay was essentially done as described (12). Briefly 10 OD cells of each strain were spheroplasted by using zymolyase and were treated with vehicle (DMSO) or DSP [dithiobis(succinimidyl propionate)] for 40 min. The spheroplasts were pelleted and solubilized. Hmg2-GFP was immunoprecipitated by using GFP antibodies.
Native Co-IP.
The native co-IP assay was adapted (12). Microsomes were harvested from various strains in MF buffer (20 mM Tris, pH 7, 100 mM NaCl, 300 mM sorbitol). Once centrifuged, microsomes were resuspended in 1 mL of Tween IP buffer (500 mM NaCl, 50 mM Tris, pH 7.5, 100 mM EDTA, 1.5% Tween-20) and incubated on ice for 15–30 min. Lysates were then centrifuged for 30 min at 14,000 × g, and the supernatant was incubated with 15 μL of anti-Hrd1 antibody overnight at 4 °C followed by 2 h with Protein-A Sepharose beads. The beads were then washed with the Tween-20 IP buffer and incubated with 70 μL of 2× urea sample buffer for 10 min at 50 °C. The samples were then analyzed on 8% SDS/PAGE.
Antibodies and Immunoblotting.
Subsequent to electrophoresis and transfer, immunoblotting was carried out by using the following antibodies. The blocking solution for all antibodies was 5% milk in TBST [10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% (vol/vol) Tween 20], except for antiubiquitin, which requires blocking with 20% heat-inactivated calf-serum in TBSHT [TBST with 0.45% (vol/vol) Tween 20]. Additionally, to readily detect ubiquitin, the nitrocellulose was microwaved (3 min). Anti-hemagglutinin was obtained as an ascites fluid from Covance monoclonal anti-Myc antibody was obtained from Genscript. Monoclonal anti-GFP was obtained from Clontech. Monoclonal antiubiquitin was obtained from Fred Hutchinson Cancer Center, Seattle. For immunoprecipitating Hmg2p-GFP, 15 μL of polyclonal anti-GFP antiserum (obtained from C. Zuker; University of California at San Diego) was used for each sample. For immunoprecipitating Hrd1p, 5 μL of rabbit polyclonal anti-myc antiserum was added to each sample. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit was obtained from Bio-Rad. HRP-conjugated goat anti-mouse was obtained from Jackson ImmunoResearch. Light-chain–specific HRP-conjugated mouse anti-rabbit was obtained from Jackson ImmunoResearch and was used in the detection following immunoprecipitation experiments.
Supplementary Material
Acknowledgments
We thank Davis Ng (National University of Singapore), Randy Schekman (University of California, Berkeley), Thomas Sommer (Max-Delbruck Center for Molecular Medicine), and Dieter Wolf (University of Stuttgart) for providing plasmids, strains, and antibodies; and the R.Y.H. laboratory members for discussions, an enthusiastic environment, and many ice cream bets. N.V. thanks Gaurav Agrawal for incisive late night discussions and tenacious support, and R.Y.H. thanks running partner Carol Galeno for many early morning miles all over San Diego. These studies were supported by NIH Grants 5R37DK051996-18 (to R.Y.H.) and 5F32GM111024-02 (to S.E.N.), and Cell and Molecular Genetics Training Program 5T32GM007240-36 (to S.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
†Abbreviations: CPY, carboxypeptidase Y; Hmg2, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, isozyme 2; Hrd1/Hrd3, HMG-CoA reductase degradation 1/HMG-CoA reductase degradation 3; PDR5, pleiotropic drug resistance 5; PGK, phosphoglycerate kinase; Sec-61, secretory pathway 61; Ubc7, ubiquitin-conjugating enzyme 7; Usa1, U1-Snp1 associating.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603079113/-/DCSupplemental.
References
- 1.Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: Lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109(4):1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife. 2013;2:e00953. doi: 10.7554/eLife.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3(1):24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 4.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120(1):73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443(7113):827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 6.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165(1):41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plemper RKR, Egner R, Kuchler K, Wolf DHD. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem. 1998;273(49):32848–32856. doi: 10.1074/jbc.273.49.32848. [DOI] [PubMed] [Google Scholar]
- 8.Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9(1):209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126(2):361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 10.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16(1):77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 11.Mehnert M, et al. The interplay of Hrd3 and the molecular chaperone system ensures efficient degradation of malfolded secretory proteins. Mol Biol Cell. 2015;26(2):185–194. doi: 10.1091/mbc.E14-07-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner RG, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151(1):69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34(2):212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plemper RKR, et al. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112(Pt 22):4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143(4):579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll SM, Hampton RY. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem. 2010;285(8):5146–5156. doi: 10.1074/jbc.M109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem. 2009;284(22):14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158(6):1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126(2):349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 20.Horn SC, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36(5):782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8(8):849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 22.Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1997;94(24):12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15(9):2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 24.Flury I, et al. INSIG: A broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J. 2005;24(22):3917–3926. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malkus P, Jiang F, Schekman R. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J Cell Biol. 2002;159(6):915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun S, et al. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc Natl Acad Sci USA. 2014;111(5):E582–E591. doi: 10.1073/pnas.1318114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.