Significance
Prion proteins provide the best-understood mode for protein-based molecular memory. Since their discovery in mammals, prions have been identified in diverse organisms including fungi, Aplysia, and Drosophila, but not in the plant kingdom. Applying methods we used to uncover yeast prions, we identified nearly 500 Arabidopsis proteins that harbor potential prion-like domains (PrDs). At least one of these domains, Luminidependens PrD, had some of the classical characteristics of prion proteins when tested experimentally in yeast, making it, to our knowledge, the first protein from the plant kingdom with bona fide prion attributes. Importantly, Luminidependens is involved in the process of flowering, a crucial development course that integrates several internal and external cues, including memories of winter, for its regulation.
Keywords: prions, Luminidependens, plant prion domains
Abstract
Prion proteins provide a unique mode of biochemical memory through self-perpetuating changes in protein conformation and function. They have been studied in fungi and mammals, but not yet identified in plants. Using a computational model, we identified candidate prion domains (PrDs) in nearly 500 plant proteins. Plant flowering is of particular interest with respect to biological memory, because its regulation involves remembering and integrating previously experienced environmental conditions. We investigated the prion-forming capacity of three prion candidates involved in flowering using a yeast model, where prion attributes are well defined and readily tested. In yeast, prions heritably change protein functions by templating monomers into higher-order assemblies. For most yeast prions, the capacity to convert into a prion resides in a distinct prion domain. Thus, new prion-forming domains can be identified by functional complementation of a known prion domain. The prion-like domains (PrDs) of all three of the tested proteins formed higher-order oligomers. Uniquely, the Luminidependens PrD (LDPrD) fully replaced the prion-domain functions of a well-characterized yeast prion, Sup35. Our results suggest that prion-like conformational switches are evolutionarily conserved and might function in a wide variety of normal biological processes.
Prions are proteins capable of adopting profoundly different structures that possess strong functional distinction in which at least one conformation is self-perpetuating (1). Although prions were first identified as the cause of a transmissible neurodegenerative disease in mammals, of greater interest has been their function as a protein-based system of memory (1–3). In yeast, a dozen prion proteins are known (4). Most of these are regulators of transcription, translation, or RNA processing (5). Their self-propagating conformations serve as elements for the inheritance of diverse traits that are passed through the cytoplasm of mother to daughter cells (6, 7). The heritable conformations of prions impart profound phenotypic consequences, which can be beneficial, benign, or detrimental, depending on the genetic background of the strain and the environmental conditions (6).
Each prion has a unique function depending upon the biological context, with self-templating of the protein-conformational state being the unifying characteristic (8). In mammals, prion-like proteins are involved in signal transduction mechanisms in innate immunity and inflammation (9, 10). In Aplysia and Drosophila, an evolutionarily conserved prion conformation of the cytoplasmic polyadenylation element binding protein serves as a “molecular memory” for the long-lasting maintenance of neuronal synapses (11–13). The diversity of these traits and the conservation of prion formation capability from yeast to human suggests not only that prions are biologically important and frequently beneficial, but also that a world of other prions remains to be discovered.
Plants also form memories, recording previous exposure to drought, heat, prolonged cold, and pathogens (14). The basis for these memories is epigenetic, and they are remarkably stable. For instance, the memory of wintering (known as vernalization) is formed after prolonged exposure to cold and promotes flowering in the spring (15, 16). This memory can persist in a plant grown from the callus of a plant exposed to cold, even if the new plant never experiences the cold (17). The molecular mechanisms underlying these remarkable memory phenomena are not fully understood and could involve prion-like mechanisms. However, no prions have been identified in plants. Using a computational algorithm developed to find yeast prions (5, 18), we identified ∼500 Arabidopsis proteins that have distinct prion-like domains (PrDs). These proteins showed a functional enrichment for involvement in flowering.
We focused further investigation on proteins belonging to the autonomous flowering pathway: Luminidependens (LD), Flowering Locus PA (FPA), and Flowering Locus CA (FCA) (19, 20). To investigate the prion-forming capacity of these proteins in living cells, we took advantage of a comprehensive suite of biochemical and genetic tools previously established in yeast. Most of the well-characterized yeast prions adopt a variety of related, but distinct, self-templating conformations, known as “prion strains.” In genetically identical yeast strains, these prion strains create a spectrum of heritable prion elements, with each exhibiting a different phenotypic strength (21). Because prion proteins in their prion conformations efficiently convert nonprion conformers into the prion state, prion traits are dominant in genetic crosses (denoted by capital letters). They are also generally inherited cytoplasmically, segregating to most or all meiotic progeny after sporulation (22) (denoted by brackets). Thus, a prion phenotype is denoted as [PRION+], whereas the corresponding nonprion state is denoted as [prion−]. Because it is based on a protein conformation-based genetic element, prion inheritance is sensitive to changes in activity of the proteostasis machinery (23). Collectively, all of these properties distinguish prion-based traits from changes caused by genetic mutations. By testing for the presence of these properties, we show that the prion domain of the plant protein LD can function as a prion capable of forming and perpetuating molecular memory.
Results
Identification and Functional Classification of Arabidopsis Prion Domains.
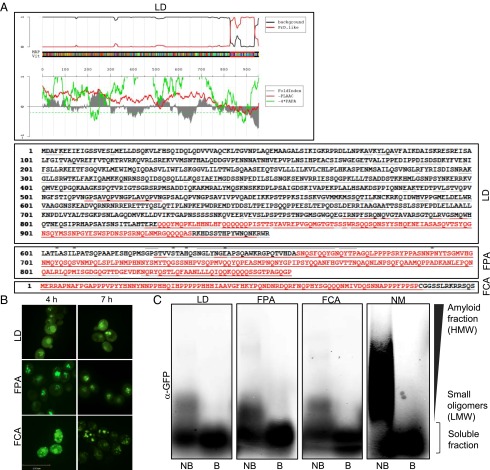
We previously developed a computational algorithm, PLAAC, to identify prion domains. The algorithm uses a hidden Markov model (HMM) to compare the similarity of a given sequence to the known sequence characteristics of previously identified yeast prions (18). We scored the entire Arabidopsis proteome using this algorithm and identified 474 Arabidopsis proteins that contained PrDs (Fig. 1A and Dataset S1).
Fig. 1.
Characterization of the candidate Arabidopsis PrDs in S. cerevisiae. (A) HMM prediction of the cArabPrDs. The Arabidopsis proteome was searched for candidate prion domains by using the HMM. Predicted prion domains of LD, FPA, and FCA are highlighted in red. (B) Fluorescent microscopy images of yeast cells expressing the GFP-tagged Arabidopsis cPrDs. Samples were collected at 4 and 7 h. (C) Western blot analysis of a semidenaturing agarose gel of the cArabPrD-GFPs using α-GFP. Amyloid fractions are marked as high-molecular-weight fraction (HMW); small oligomers are marked as low-molecular-weight fraction (LMW). The Sup35 PrD (NM) was used as control. B, boiled fraction; NB, not boiled.
Gene Ontology (GO) annotation analysis of the candidate proteins (Dataset S2) showed enrichment in proteins categorized as regulators of transcription (P = 2.96 × 10−31), RNA binding (P = 2.28 × 10−21), and RNA metabolic processes (P = 3.73 × 10−12). Other enriched categories included reproductive development processes (P = 1.16 × 10−7) and flower development (P = 1.41 × 10−7). Strikingly, four of eight proteins in the autonomous flowering pathway contained PrDs: LD, FPA, FCA, and Flowering Locus Y (FY). LD is a transcriptional factor; FPA and FCA are RNA-binding proteins; and FY affects mRNA processing. Thus, these proteins (i) have predicted prion domains; (ii) are involved in transcription or regulation of RNA stability; and (iii) regulate flowering time, an important developmental course that involves “memory” of previous environmental exposures. Given these intriguing connections, we chose to further characterize the PrDs of these four proteins from the autonomous pathway.
Expression of Arabidopsis Prion Domains in Yeast.
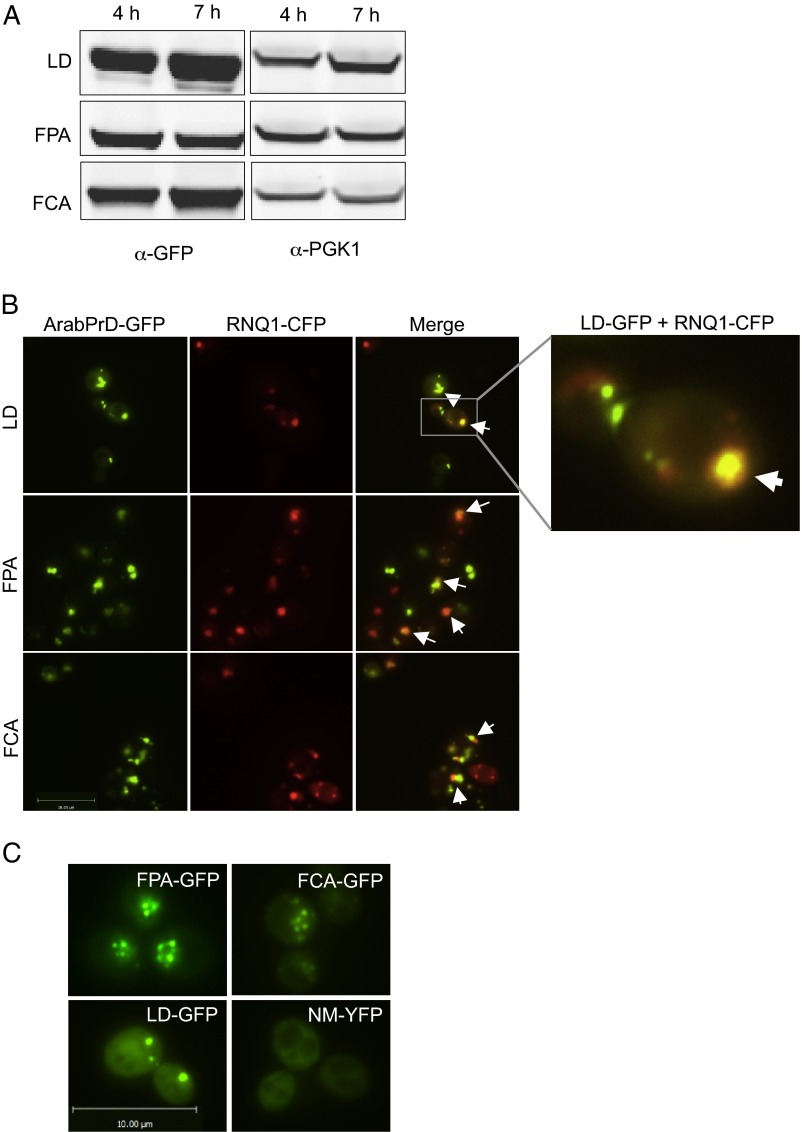
We fused the PrDs of LD, FPA, FCA, and FY to GFP and expressed the fusion proteins from a galactose-inducible promoter. FY expression was not detectable, but LD, FPA, and FCA were expressed at similar, readily detectable levels (Fig. S1A). All three chimeric proteins formed distinct foci, a common characteristic of prion proteins (Fig. 1B). For each protein, multiple initial foci coalesced into one or two larger foci over time (Fig. 1B). As is common for yeast prions, candidate Arabidopsis PrD (cArabPrDs) foci colocalized with Rnq1 aggregates, indicating their localization to the cytoplasmic site known as the “insoluble protein deposit” (24) (SI Results; Fig. S1B). However, the [RNQ+] prion was not required for their localization (Fig. S1C).
Fig. S1.
Characterization of Arabidopsis prion domains in S. cerevisiae. (A) Immunoblot analysis of cArabPrD-GFPs. The constructs were expressed for various time points and analyzed by α-GFP. α-PGK1 was used as the loading control. (B) Some cArabPrD-GFP foci localize in the insoluble protein deposit (IPOD). Fluorescence microscopy of yeast cells with the cArabPrD-GFP (green) and Rnq1-CFP (red) after 7 h of coexpression is shown. The colocalized aggregates are marked with arrows. (C) Foci formation is not dependent on [RNQ+]. Fluorescence microscopy images of cArabPrD-GFPs in a [rnq-] strain background are shown. NM-YFP is shown as a control.
Many proteins form foci upon overexpression. The SDS resistance of protein complexes on semidenaturing detergent-agarose gels (SDD-AGE) can distinguish nonspecific aggregates and cellular organelles from prion-like foci (25). Indeed, the cArabPrD-GFPs formed SDS-resistant oligomers, which were eliminated when the samples were boiled. However, unlike known prions that typically form high-molecular-weight amyloid fibrils, the oligomers formed by cArabPrD-GFPs are low molecular weight (Fig. 1C).
LDPrD Provides a Faithful, Protein-Based Phenotypic Switch.
As a defining prion characteristic, alternate prion conformations must self-perpetuate, templating other proteins of the same type to the prion state. In yeast cells, this process produces a heritable phenotypic switch that is stably perpetuated across hundreds of generations. To assess whether the cArabPrDs have this characteristic, we took advantage of the modular nature of prion domains (26) and the well-characterized [PSI+] phenotype (6, 27). When the N-terminal PrD of Sup35 is replaced by the PrD of another prion, the resulting chimeras can form a heritable element that behaves like [PSI+]. Any such chimeras (PrD-Sup35C) can be readily assessed for their prion-forming ability in strains carrying an ade1-14 allele, which harbors a premature stop codon. In the nonprion state, the chimeric Sup35 protein is fully functional, causing ribosomes to stop translation at the stop codon. Such cells are unable to grow in the absence of adenine and form red colonies on complete medium (due to the accumulation of a synthetic by-product). When the chimeric protein switches to the prion state, the C-terminal termination activity is sequestered, ribosomes are read through the stop codon, and functional Ade1 is produced. Cells can then grow in the absence of adenine and produce white colonies (5, 10, 28–30).
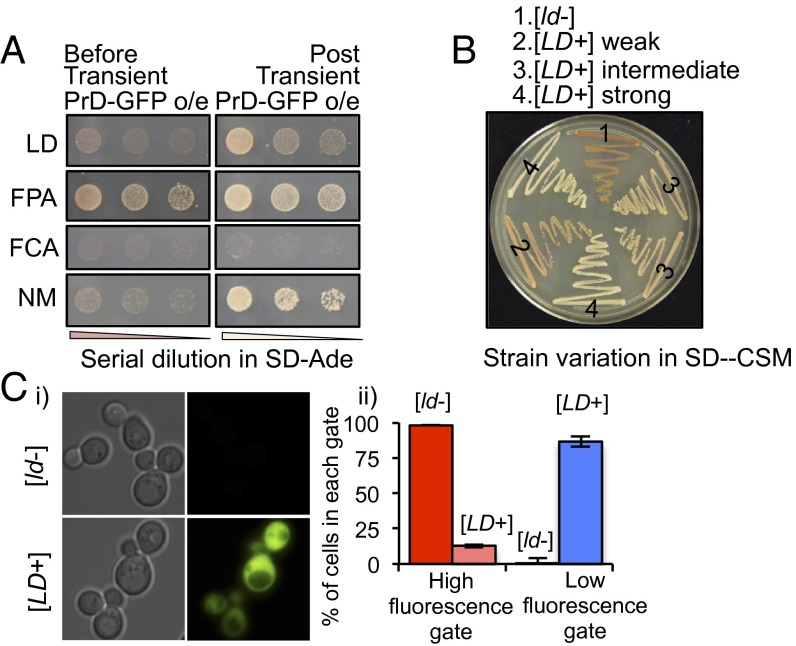
To test this prion characteristic, cArabPrD-Sup35 chimeras were individually expressed in cells lacking endogenous Sup35. The strains harboring cLDPrD- or cFCAPrD-Sup35C did not grow in medium without adenine (Fig. 2 A, Left) and produced red colonies in complete media, indicating that these chimeric Sup35 proteins were functional. However, cells harboring cFPAPrD-Sup35C grew in adenine-deficient medium and formed white colonies, indicating that most of this chimeric protein was already in a nonfunctional state. We were therefore unable to test its prion-forming ability any further.
Fig. 2.
Phenotypic switch to [PRION+] state. (A) Sup35C assay of the different cArabPrDs. Shown is the ability of the different cArabPrD-Sup35C to grow on medium lacking adenine post (Right) or before (control; Left) transient overexpression of the corresponding cArabPrD-GFP proteins. The Sup35 PrD (NM) was used as control. (B) Strain variation [assessed in complete medium (SD-CSM)] of the [LD+]-state as indicated. (C) GFP read-through assay. [LD+] and [ld−] strains were transformed with a fusion protein containing a GFP marker preceded by a stop codon. (C, i) The two strains were analyzed with fluorescence microscopy. Read-through indicated by GFP expression was observed only in the [LD+] cells. (C, ii) Multiple replicates of the [LD+] and [ld−] strains were analyzed by flow cytometry. High (blue) and low (red) GFP gates were set such that >95% of [ld−] cells were included in the low GFP gate. Percentages of cells in these gates are shown.
To test the capacity of cLDPrD- and cFCAPrD-Sup35C to undergo a prion switch, we took advantage of the fact that transient overexpression of prion proteins is sufficient to induce the conformational switch (31). This switch in conformation then self-perpetuates, even when expression levels are returned to normal. Transient overexpression of cLDPrD-GFP switched the strain harboring cLDPrD-Sup35C into a prion state, as evidenced by the ability to grow on adenine-deficient medium (Fig. 2A, Right). Moreover, we observed a spectrum of prion strains with this protein, ranging from strong (white) to weak (dark pink) in complete medium (Fig. 2B). However, cFCAPrD did not provide a prion switch because it was unable to grow on adenine-deficient medium, even after overexpression (Fig. 2 A, Right). Because LDPrD-Sup35C could stably exist in either a distinguishable [prion−] or a variety of [PRION+] states, we henceforth focused our efforts on characterizing the [PRION+] states provided by LDPrD ([LD+]).
In addition to the ade1-14 read-through, we confirmed that [LD+] cells had the ability to read through other premature stop codons. We introduced a GFP construct with a premature stop-codon in both [LD+] and [ld−] strains. GFP expression was obtained only in the [LD+] population. This result was assessed by fluorescence microscopy and FACS analysis (Fig. 2C).
We confirmed that the phenotype obtained in the [LD+] strain was solely dependent upon the LDPrD (SI Results; Fig. S2 A and B). Further, [LD+] was maintained with high fidelity through sequential transformations, successive streaks on plates, and hundreds of generations of growth in liquid medium, although we observed spontaneous but rare loss of [LD+] (<5%; Fig. S2C). These data firmly established that the LDPrD could maintain a heritable self-perpetuating state.
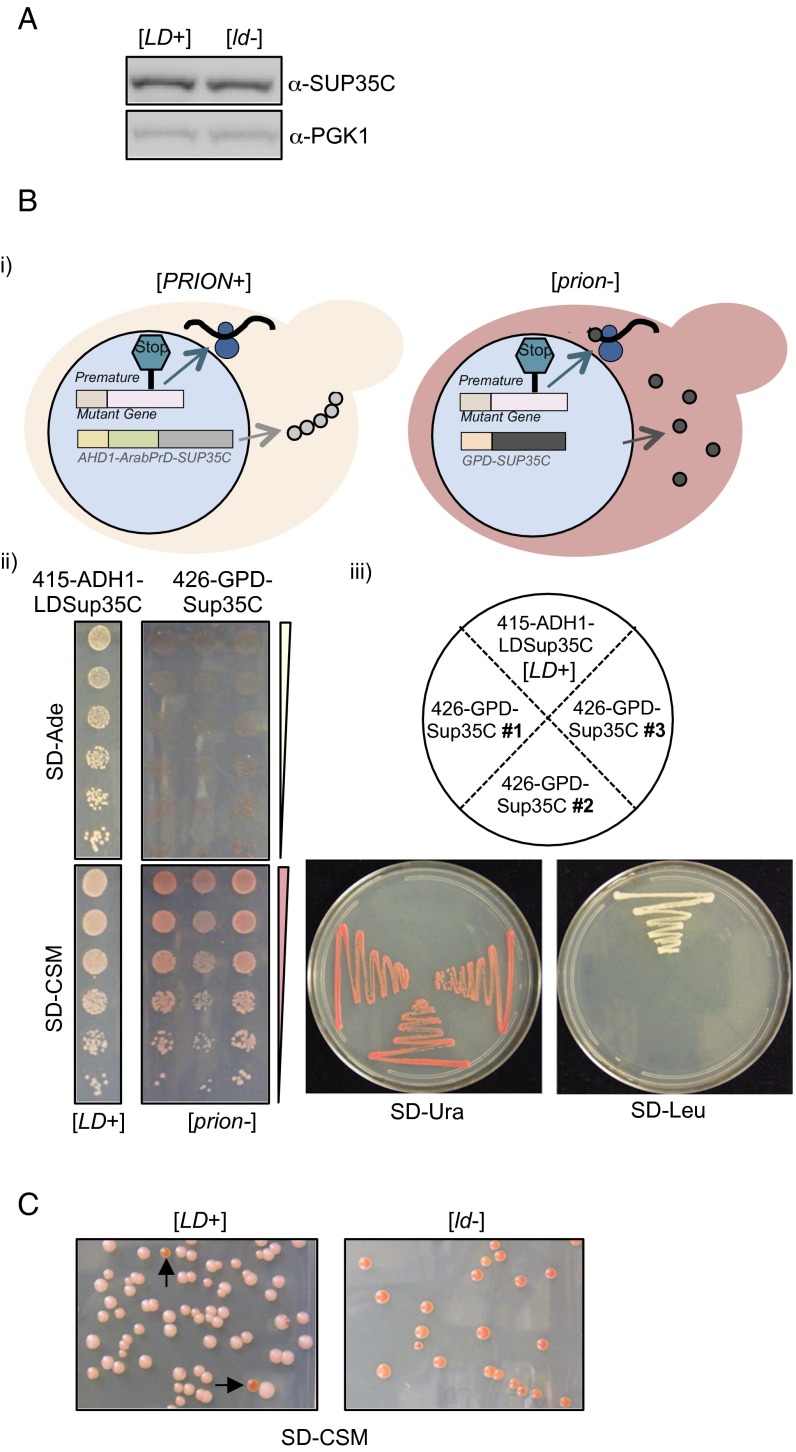
Fig. S2.
Phenotypic switch into the [PRION+]-state. (A) Levels of the LDPrD-Sup35C protein are the same in [LD+] and [ld−] cells. The soluble protein levels of LDPrD-Sup35C were similar in both strains as assessed by α-Sup35C antibody. α-PGK1 antibody was used as loading control. (B) The [LD+] phenotype is dependent on the LDPrD. (B, i) Cartoon representation. A reverse plasmid shuffle was performed by which the untagged GPD-Sup35C was transformed into the [LD+] strains and the LDPrD-Sup35C plasmid was removed. This led to faithful translation termination due to the availability of soluble Sup35C protein. (B, ii) The [LD+] strain reverts back into a [psi−]-state, and therefore there is no growth in medium lacking adenine and cells appear red in complete medium compared with the [LD+] strain. (B, iii) The strains were checked for accuracy by growing on respective selective medium, SD-Ura for the presence of GPD-Sup35C and SD-Leu for LDPrD-Sup35C. (C) The [LD+] phenotype is lost randomly at a very low frequency. This result was apparent from the appearance of red color colonies as marked by the arrows among [LD+] cells.
[LD+] Strains Have Strong Prion-Like Characteristics.
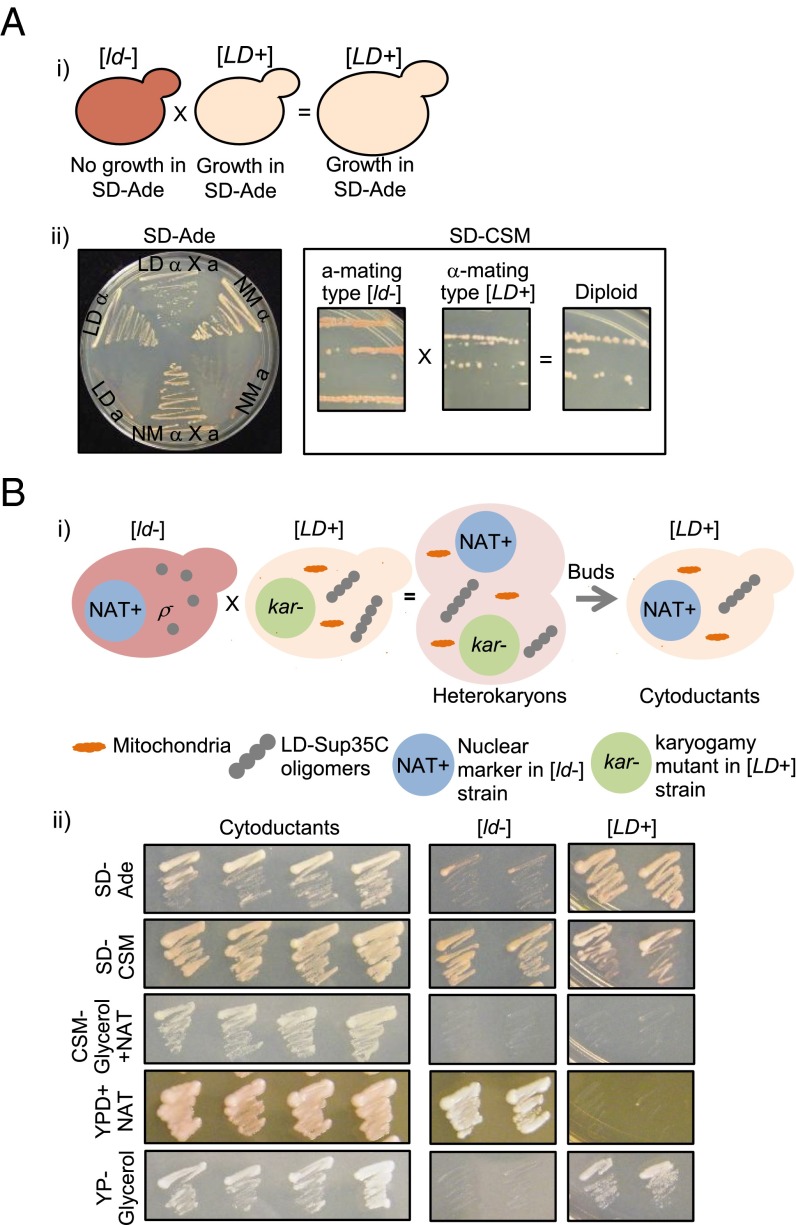
We next examined the dominance of the [LD+] phenotype by analyzing matings and the cytoplasmic heritability of [LD+]. Indeed, both the diploids, formed by mating [LD+] with [ld-] cells, and the [ld-] haploids that were cytoduced with [LD+] cells grew on adenine-deficient medium and appeared white in complete medium (Fig. 3 A and B). Because the diploids exhibited sporulation defects, our ability to analyze any inheritance patterns in their meiotic progeny was restricted. However, our results established that the [LD+] phenotype is indeed protein-based, fulfilling a key criterion of prion biology.
Fig. 3.
The [LD+] state is dominant and can be inherited through the cytoplasm. (A) Dominance of the [LD+] state. (A, i) Cartoon depiction of the crosses. The α-mating type of [LD+] was crossed with the a-mating type of [ld−] and the resulting diploids were assessed for Ade-read-through in adenine-deficient medium and color phenotype in complete medium (A, ii). (B) [LD+] phenotype can be cytoduced. (B, i) Cartoon depiction of generation of cytoductants. Various modifications were made to both the α-mating type of [LD+] and the a-mating type of [ld−], as indicated in the figure and described in Materials and Methods. (B, ii) The buds from the heterokaryons were grown in different selective media to ensure the strain accuracy: [LD+] were able to grow in glycerol, whereas [ld-] could not due to defective mitochondria; only [ld−] grew on YPD+NAT because [ld−] has an antibiotic marker in the nucleus; only cytoductants grew in medium with both glycerol and NAT. The phenotypes of the cytoductants were assessed in medium lacking adenine and complete medium.
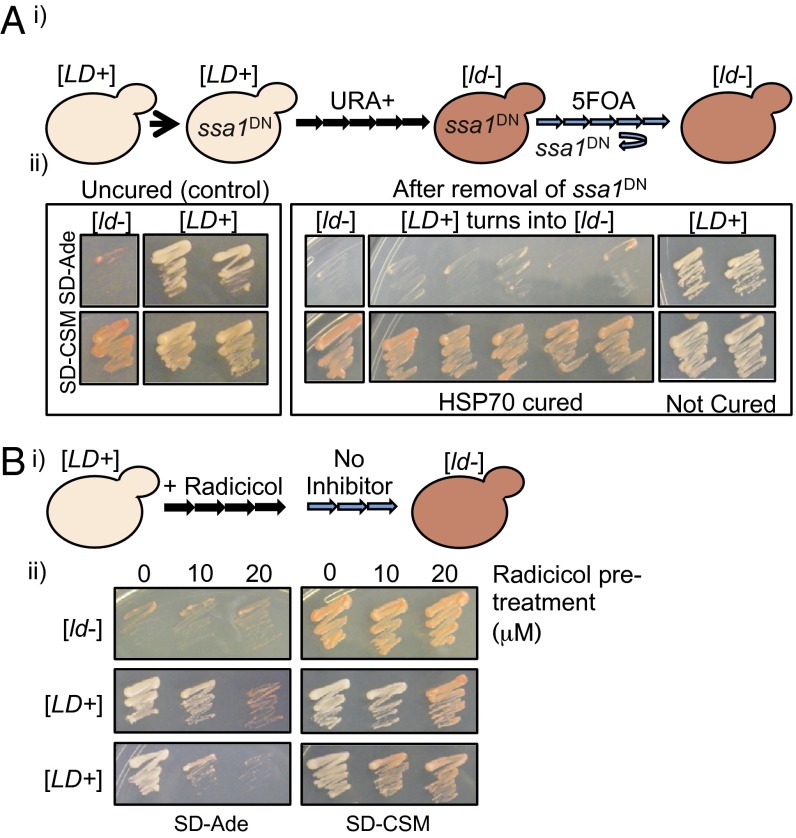
Propagation of prions in fungi relies on the generation of smaller “seeds” that are generated by the disaggregase, Hsp104 (32–35). Traditional manipulations of Hsp104 that cure most prions were ineffective against [LD+]. Transient inhibition of Hsp104 activity using chemical inhibition with 5 mM GdnHCl (Fig. S3A), genetic perturbation using a dominant-negative variant of Hsp104 that interferes with the wild-type Hsp104 activity (Fig. S3B), and overexpression of Hsp104 (Fig. S3C) did not affect the [LD+] phenotype. In contrast, all of these methods eliminated the [PSI+] phenotype generated by Sup35 (Fig. S3 A–C). Although most known yeast prions are dependent on Hsp104, recently emerging evidence indicates that other molecular chaperones like Hsp70 and, potentially, Hsp90 may play Hsp104-independent roles in prion biology (9, 36, 37). Therefore, we exploited a dominant-negative Hsp70 variant (Ssa1-K69M or Hsp70DN) that eliminates the yeast prion [GAR+] (37). We transiently expressed Hsp70DN in [LD+] strains and assayed the [LD+] phenotype after restoring Hsp70 activity (by removing the plasmid harboring Hsp70DN). Most [LD+] strains (65%) reverted to [ld−] strains (Fig. 4A). This phenomenon was especially common among intermediate strains.
Fig. S3.
Perturbations in Hsp104 levels do not affect [LD+] phenotype. (A, i) Cartoon depiction of transient inhibition of Hsp104 activity chemically (5 mM GdnHCl) or genetically (Hsp104 dominant negative, Hsp104DN, or Hsp104 overexpression). (A, ii, B, and C) Sup35C assay. Transient inhibition of Hsp104 activity chemically or genetically did not affect the [LD+] phenotype because the cells appeared white in complete medium. In all cases, the [PSI+] phenotype reverted to a [psi−] phenotype.
Fig. 4.
The [LD+] state can be perturbed by transient inhibition of molecular chaperones. (A) Transient inhibition of Hsp70 by expression of Hsp70DN (ssa1DN). (A, i) Cartoon depiction of transient inhibition of Hsp70. (A, ii) Sup35C assay. Transient inhibition of Hsp70 affected some [LD+] strains because they no longer grew in medium lacking adenine and appeared red in complete medium. (B) Transient inhibition of Hsp90 by radicicol. (B, i) Cartoon depiction of transient inhibition of Hsp90. (B, ii) Sup35C assay. Transient inhibition of Hsp90 using 10 and 20 μM radicicol affected most [LD+] strains because they no longer grew in medium lacking adenine and appeared red in complete medium.
Hsp90 has been implicated in a prion-like phenotype underlying innate immunity: Aggregates of the prion-like MAVS protein were eliminated by chemical inhibition of Hsp90 (9). We transiently inhibited Hsp90 activity by passaging [LD+] and [ld−] strains in media containing either a concentration of 10 or 20 μM radicicol. In contrast to dominant-negative Hsp104 overexpression, the majority (80%) of the [LD+] strains altered their phenotype to the [ld−]-state, although some of them showed this effect only at a higher dose of radicicol (Fig. 4B). These results raise the possibility that the inheritance of [LD+] may be blocked by transient inhibition of Hsp70 or Hsp90.
SI Results
Expression of cArabPrDs in Yeast.
We achieved maximal expression of the cArabPrD-GFPs after 7 h of induction, as demonstrated by both fluorescence microscopy and immunoblot analysis (Fig. 1B and Fig. S1A). We used this time point for further experimentation.
GO Annotation Analysis.
Like known yeast prions, proteins categorized as regulating transcriptional (P = 2.97 × 10−31) or RNA metabolic processes (P = 3.72 × 10−12) were highly enriched (Dataset S2). This result suggests that the power of prion-like proteins affecting the phenotype in the absence of genotype alterations may be global, extending this phenomenon even to the plant kingdom.
Foci Formation Is Not Dependent on [RNQ+].
The amyloid assemblies of some yeast prions, like [PSI+], require inducibility factors to acquire these conformations. The protein Rnq1 acts as an inducibility factor in its prion form, [RNQ+]. For example, overexpression of the Sup35 PrD, NM, does not induce foci in a [rnq−] background (Fig. S1C). However, cArabPrD-GFPs formed foci even in the absence of [RNQ+], distinguishing them from yeast prions like [PSI+]. Therefore, although the cArabPrD-GFPs colocalize with the Rnq1-CFP, they are not dependent on Rnq1 for formation of oligomeric species.
LDPrD Is Responsible for the Protein-Based Phenotypic Switch.
We eliminated the remote possibility that the [LD+] phenotype resulted from degradation of the chimeric protein by immunoblot analysis. The LDPrD-Sup35C levels were very similar in both [LD+] and [ld−] cells (Fig. S2A). We also confirmed that the phenotype we obtained was indeed solely due to LDPrD by swapping the LDPrD-Sup35C with an untagged-Sup35C in the [LD+] strain. Without the LDPrD domain, the cells instantly reverted back to a [prion−] state (Fig. S2B).
Discussion
Here, to our knowledge, we report the first evidence that at least one plant protein has the capacity to serve as a prion. The LD protein, which functions in the autonomous flowering pathway, contains a domain that is similar in sequence to the prion domains of yeast proteins and can fully substitute for prion function in yeast. The LD prion domain enters into a higher-order, self-perpetuating structure that can change the function of an associated protein domain in a stable and heritable way. That is, it is capable of forming protein-based molecular memories.
Plants are known to possess the capacity to form memories in response to diverse environmental factors, including changes in temperature and day length (38). One such memory is vernalization, which develops upon exposure to prolonged cold and is the mechanism by which a plant recognizes and remembers the passing winter (15, 16). Epigenetic changes in response to this exposure prime the plant for flowering in the spring (39). The phenomenon is, in a sense, similar to long-term potentiation of a neuronal synapse, which has recently been shown to involve the prion-like assembly of RNA-binding proteins (12, 30, 40).
To identify potential Arabidopsis prions, which might be involved in some forms of plant memory, we scanned the proteome using a computer algorithm trained on known yeast prions. Using yeast cells as “living test tubes” for protein folding in a eukaryotic cytoplasm, we showed that three domains from Arabidopsis proteins (LD, FPA, and FCA) had some prion-like properties. All of these proteins are in the autonomous flowering pathway (39), which participates in flowering by responding to yet-unknown internal cues. All three aggregated and formed SDS-resistant assemblies. However, in our test system, only the candidate prion domain of LD could stably and heritably adopt either a soluble conformation or a higher-order prion conformation. Indeed, as is the case for most yeast prion proteins, the LD PrD formed a variety of self-templating conformations with distinguishable phenotypic consequences.
Unlike classical yeast prions, [LD+] was not curable by Hsp104. Instead, our results suggest that Hsp70 and Hsp90 may play a role in maintaining this prion. Also, instead of high-molecular-weight amyloids, LDPrD-GFP formed low-molecular-weight oligomers. We have previously shown that at least one fungal prion, [GAR+], does not form amyloid or require Hsp104; instead, this prion is Hsp70-dependent (36, 37). Therefore, it is possible that [LD+] belongs to this noncanonical class of prions.
In Arabidopsis, prolonged exposure to cold causes heritable chromatin modifications, including histone methylation and silencing of FLC, a key repressor of flowering (41–45). But how, at a molecular level, does a plant differentiate between a single cold night and winter? Stable, self-perpetuating changes in the conformational states of certain proteins, such as those studied here, might be involved. LD, FCA, and FPA are predicted to bind nucleic acids. FCA and FPA both contain RNA-recognition motifs (46, 47) and are known to play important roles in RNA 3′ processing and transcription termination (48). LD contains a homeodomain that can bind DNA and regulate transcription (20). Because all three of these proteins are able to form SDS-resistant assemblies, altered activities in their assembled forms could influence the chromatin modifications that are known to be involved in flowering decisions.
Whether the LD protein undergoes a biologically significant prion-like conformational change that might play a role in the flowering decision in plants remains to be seen. However, roles for self-perpetuating prion conformations in chromatin remodeling are highly likely. Indeed, at minimum, one yeast chromatin-remodeling factor, the Swi1 subunit of the SWI/SNF chromatin-remodeling complex, can form a prion that epigenetically alters its chromatin remodeling functions and thereby imparts heritable changes in the biological state of the organism (49). The role that prion-like conformational changes might play in plant biology and, in particular in the many forms of plant memory, is currently unknown but highly intriguing.
On another note, it is interesting to reflect on the fact that in the mid-20th century, some Soviet scientists used the plant memory phenomenon of vernalization as a strong argument against the “gene-based” concept of inheritance (50). Given this fact, it is ironic that the concept of vernalization may, indeed, have a heritable non-DNA element involved in its perpetuation. Of course, the capacity for proteins to stably switch states in a somatically heritable way is itself dictated by DNA sequences that encode proteins capable of such unusual, autonomous, self-perpetuating biological switches. Our work suggests that much broader roles for prions exist and that this is a subject ripe for further exploration.
Materials and Methods
Computational Detection of Arabidopsis Prion Candidates.
Each Arabidopsis protein was searched for the presence of PrDs by using a HMM that parsed each protein into prion-like and non-prion-like (background) regions. Any protein containing a PrD of at least 60 consecutive prion-like amino acid residues received a nonzero COREscore (Dataset S1). This COREscore is the sum of the individual log-likelihood ratios for each amino acid residue contained within this domain. The COREscores of the candidate 474 Arabidopsis PrDs are presented in Dataset S1.
GO Annotation Analysis.
We performed GO annotation analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource 6.7 platform. We used Arabidopsis proteome as the background for functional category enrichment (Dataset S2).
Yeast Strains, Vectors, and Antibodies.
Yeast strains used in this study were YMJ509 and YMJ584 (5). They were grown on standard synthetic medium lacking standard amino acids/bases and containing either d-glucose (SD) or d-galactose as carbon source. The PrDs from LD, FPA, FCA, and FY were synthesized from GeneArt (Life Technologies). These domains were then used to generate yeast entry clones by using the p221 donor plasmid (Addgene).
The above entry clones were used to generate various destination plasmids by using 424GAL1ccdBGFP and 415ADH1ccdBSup35C (51): 424GALArbdPrD-GFP (cArabPrD-GFP) and 415ADH1ArabPrD-SUP35c (cArabPrD-SUP35C). Expression of cArabPrD-GFP was controlled by using 2% (wt/vol) galactose for the indicated time points. The insoluble protein deposit (IPOD) structure was marked by overexpressing the fusion protein Rnq-CFP (24).
GFP expression was assessed by either fluorescent microscopy or by standard immunoblot procedures using anti-GFP antibody at 1:3,000 dilution (anti-mouse; Roche). Sup35C chimeras were detected by using a specific antibody against only the C-terminal domain of Sup35 at 1:2,000 dilution (anti-mouse; Cocalico Biologicals). To confirm equal loading, α-PGK1 antibody was used at 1:1,000 (anti-rabbit; Acris Antibodies).
SDD-AGE.
SDD-AGE was performed as detailed in ref. 5. A final concentration of 1% SDS was used in the loading buffer.
Cell Cytometer Analysis.
Yeast cultures were grown in synthetic dropout medium to log phase and then transferred to a 96-well plate for analysis in an EasyCyte flow cytometer (Guava Technologies). GFP fluorescence was monitored in the green channel. High and low GFP gates were set such that > 95% of [ld−] cells were included in the low-GFP gate. Percentage of cells in these gates were compared for all strains.
Read-Through Assays.
ade1-14 read-through was assessed as detailed in ref. 5. Briefly, the ArabPrD-Sup35C chimeras were individually expressed in the YRS100 strain (a derivative of YMJ584), where the endogenous Sup35 protein has been deleted, but the activity of this essential protein is covered by 416GPDSup35C. Then a plasmid shuffle was performed by plating the cells in 5-fluoroorotic acid (5-FOA). Therefore, the cArabPrD-Sup35C (LD) was the only source of Sup35C activity in these cells.
To assess GFP read-through, [LD+] and [ld−] strains were transformed with a fusion protein containing a GFP marker preceded by a stop codon (52).
Generating α-Mating Type Strains.
Yeast have two mating types: MATa and MATα. Only crosses from opposite mating type yield diploids. To mate [LD+] strain with [ld−] strain, we switched the mating type of [LD+] strain from a-type to α-type. To do so, we transiently overexpressed a Homothallic switching endonuclease (HO) gene (on a Ura3 marker) for 2–4 h. The HO plasmid was counterselected by plating single colonies on 5-FOA. α-type colonies were then identified by crossing with mating type tester strains (D588a or M84α).
Generating Strains for Cytoduction.
The [LD+] strain was made kar1 (karyogamy) mutant, so that it was unable to undergo karyogamy during mating. Next, we introduced an antibiotic-resistant (NAT+, resistant to nourseothricin) nuclear marker in the [ld−] strain by disrupting the nonessential HO locus. This strain was then converted into respiration-deficient petite strain (ρ−), so that it was unable to grow in medium containing nonfermentable carbon sources, like glycerol (Fig. 3B). Heterokaryons were established by crossing the resulting “donor” [LD+] and the “recipient” [ld−] strains. Cytoductants were selected by growing the buds from the heterokaryons in medium containing nourseothricin and where glycerol was the only carbon source.
Supplementary Material
Acknowledgments
We thank G. Karras, B. Bevis, L. K. Clayton, P. Narayan, and D. Landgraf, along with other members of the S.L. laboratory for materials, helpful discussions, and comments on the manuscript. This work was also supported by grants from the Howard Hughes Medical Institute, the Harold and Leila Mathers Charitable Foundation, and the Eleanor Schwartz Charitable Foundation (to S.L.). S.L. is a Howard Hughes Medical Institute investigator. S.C. was supported by a Broodbank Fellowship and as a Former Fellow of Hughes Hall, University of Cambridge. G.A.N. was supported by an NSF graduate research fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604478113/-/DCSupplemental.
References
- 1.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6(6):435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 3.Bailey CH, Kandel ER, Si K. The persistence of long-term memory: A molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44(1):49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191(4):1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407(6803):477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 7.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431(7005):184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 8.Newby GA, Lindquist S. Blessings in disguise: Biological benefits of prion-like mechanisms. Trends Cell Biol. 2013;23(6):251–259. doi: 10.1016/j.tcb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156(6):1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Si K, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115(7):893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 12.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140(3):421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Majumdar A, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148(3):515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita T, Seki M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014;55(11):1859–1863. doi: 10.1093/pcp/pcu125. [DOI] [PubMed] [Google Scholar]
- 15.Gassner G. 1918. Beiträge zur Physiologischen Charakteristik Sommer-und Winter annueller Gewächse Insbesondere der Getreidepflanzen. Z Bot 10:417. German.
- 16.Chouard P. Vernalization and its relations to dormancy. Annu Rev Plant Physiol. 1960;11:191–238. [Google Scholar]
- 17.Whelan ED, Schaalje GB. Vernalization of embryogenic callus from immature embryos of winter wheat. Crop Sci. 1992;32:78–80. [Google Scholar]
- 18.Lancaster AK, Nutter-Upham A, Lindquist S, King OD. PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30(17):2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229(1):57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 20.Lee I, et al. Isolation of LUMINIDEPENDENS: A gene involved in the control of flowering time in Arabidopsis. Plant Cell. 1994;6(1):75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435(7043):765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165(1):23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20(3):125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyedmers J, et al. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010;107(19):8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp. 2008;20–22(17):838. doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287(5453):661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 27.Liebman SW, Derkatch IL. The yeast [PSI+] prion: Making sense of nonsense. J Biol Chem. 1999;274(3):1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 28.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5(1):163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 29.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106(2):183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24(3):268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 32.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268(5212):880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 33.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15(12):3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 34.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304(5678):1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 35.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23(3):425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown JCS, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23(19):2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarosz DF, et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell. 2014;158(5):1083–1093. doi: 10.1016/j.cell.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz RJ, Amasino RM. Vernalization: A model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim Biophys Acta Gene Struct Expr. 2007;1769:269–275. doi: 10.1016/j.bbaexp.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Michaels SD, Amasino RM. Memories of winter: Vernalization and the competence to flower. Plant Cell Environ. 2000;23:1145–1153. [Google Scholar]
- 40.Heinrich SU, Lindquist S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB) Proc Natl Acad Sci USA. 2011;108(7):2999–3004. doi: 10.1073/pnas.1019368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell. 2004;16(Suppl):S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107(4):525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 43.Greb T, et al. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol. 2007;17(1):73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz RJ, Sung S, Amasino RM. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(2):411–416. doi: 10.1073/pnas.0710423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung S, et al. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006;38(6):706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 46.Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13(6):1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macknight R, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89(5):737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 48.Sonmez C, et al. RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc Natl Acad Sci USA. 2011;108(20):8508–8513. doi: 10.1073/pnas.1105334108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Z, Park K-W, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40(4):460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chernoff YO. Mutation processes at the protein level: Is Lamark back? Mutat Res Rev Mutat Res. 2001;488:36–64. doi: 10.1016/s1383-5742(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 51.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24(10):913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6(11):e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.