Significance
Various plant organs respond differentially to environmental signals so that plants can adapt to dynamic environments without movement. Light is a key environmental factor mediating multiple plant developmental processes. For example, it induces cotyledon expansion but inhibits hypocotyl elongation when plants emerge from soil. Although this opposite regulation is crucial for plant survival and has been described for decades, the underlying mechanism is still elusive. In this study, we demonstrated that temporal–spatial expression of a group of Small Auxin Up RNAs (SAURs) is regulated by light through auxin and phytochrome-interacting factors, and these SAURs further mediate the differential growth of cotyledons and hypocotyls. Thus, this study provides a molecular mechanism explaining how light differentially regulates the growth of various plant organs.
Keywords: light signal, seedling development, SAURs, auxin, PIFs
Abstract
During deetiolation of Arabidopsis seedlings, light promotes the expansion of cotyledons but inhibits the elongation of hypocotyls. The mechanism of this differential regulation of cell enlargement is unclear. Our organ-specific transcriptomic analysis identified 32 Small Auxin Up RNA (SAUR) genes whose transcripts were light-induced in cotyledons and/or repressed in hypocotyls. We therefore named these SAURs as lirSAURs. Both overexpression and mutation analyses demonstrated that lirSAURs could promote cotyledon expansion and opening and enhance hypocotyl elongation, possibly by inhibiting phosphatase activity of D-clade type 2C protein phosphatases (PP2C-Ds). Light reduced auxin levels to down-regulate the expression of lirSAURs in hypocotyls. Further, phytochrome-interacting factors (PIFs) were shown to directly bind the genes encoding these SAURs and differentially regulate their expression in cotyledons and hypocotyls. Together, our study demonstrates that light mediates auxin levels and PIF stability to differentially regulate the expression of lirSAURs in cotyledons and hypocotyls, and these lirSAURs further mediate the differential growth of these two organs.
As sessile organisms, plants must optimize their morphologies to adapt to environmental stimuli. Light, one of the most important environmental signals, affects the entire life cycle of plants (1). Dark-grown Arabidopsis seedlings follow the skotomorphogenic developmental program, which is characterized by long hypocotyls, apical hooks, and closed cotyledons; in contrast, light-grown seedlings undergo photomorphogenesis and display short hypocotyls, no apical hooks, and greening and expanded cotyledons (2, 3). It is vital for plants to correctly switch from skotomorphogenesis to photomorphogenesis upon emerging above ground (4). After illumination, the cotyledons quickly open and expand whereas hypocotyls reduce their elongation rate. However, how light oppositely regulates the growth of these two organs is poorly understood.
The opposite effects of light on stem and leaf growth were studied in peas and barley using physiological strategies during the 1940s and 1950s (5–7). At the molecular level, the Arabidopsis tubulin gene TUB1 was demonstrated to be down-regulated by light specifically in hypocotyls (8). Mutants affected in several phytochrome-regulated early light response genes exhibited phenotypic differences only in hypocotyls, which indicated that these genes had specific functions in these organs (9). Microarray analysis of dissected organs from Arabidopsis seedlings grown in the dark or in white light found that the expression of hundreds of light-responsive genes was specific to certain organs (10). Expression of many transcription factors responded differently to light in cotyledons and shoot apices (11). These studies indicated that distinct organs may use different regulatory networks to mediate light responses, but molecular mechanisms linking signals to development remain obscure.
The red range of visible light, which is perceived by the phytochrome family of photoreceptors, is most effective in increasing leaf area and suppressing internode elongation (5, 6). Activated phytochromes directly interact with phytochrome-interacting factors (PIFs) to transduce light signals (12). In Arabidopsis, single or multiple loss-of-function mutants of PIF genes display short hypocotyls in red light and in the dark, indicating that PIFs are repressors of photomorphogenesis (13). Chromatin immunoprecipitation (ChIP)-chip and ChIP-Seq studies have shown that PIFs regulate thousands of downstream genes (14–18). These genes are released from the control of the PIFs upon illumination because light suppresses both the amounts and the activities of the PIFs in various ways (19).
To understand how light differentially regulates the development of cotyledons and hypocotyls, transcriptomic analysis was used to identify organ-specific light-responsive genes. In this study, Small Auxin Up RNAs (SAURs) were found to be enriched in the organ-specific light-responsive genes. SAURs were originally identified as a group of auxin-responsive genes whose expression was induced in elongating hypocotyls within 2–5 min after treatment with the synthetic auxin 2,4-D (20, 21). Based on microarray analysis, about half of SAURs are up-regulated by auxin treatment in Arabidopsis (22). Overexpression of several distinct SAUR genes resulted in increased growth of cotyledons, hypocotyls, or roots, respectively (23–25), whereas a few SAURs have been suggested to negatively regulate cell growth in specific organs (26–28). Recently, it has been found that SAURs could interact with D-clade type 2C protein phosphatase (PP2C) proteins to inhibit their phosphatase activities, therefore activating H+-ATPases to promote cell expansion (29). Although the understanding of the functions and molecular mechanisms of SAURs remains rudimentary, current data suggest that SAURs may play key roles in integrating multiple signals into distinct growth and developmental responses (22).
Here, we identified a group of SAUR genes that are up-regulated in cotyledons and/or down-regulated in hypocotyls during the dark-to-light transition, which we refer to as lirSAURs. These lirSAURs likely promote cell expansion by inhibiting phosphatase activity of PP2C-D proteins. The expression of lirSAURs is regulated by light, possibly through auxin and PIF proteins in an organ-specific manner. Our study suggests that lirSAURs are the key factors determining how light differentially regulates the growth of cotyledons and hypocotyls.
Results
Transcriptomic Analysis of Light-Responsive Genes in Cotyledons and Hypocotyls of Arabidopsis.
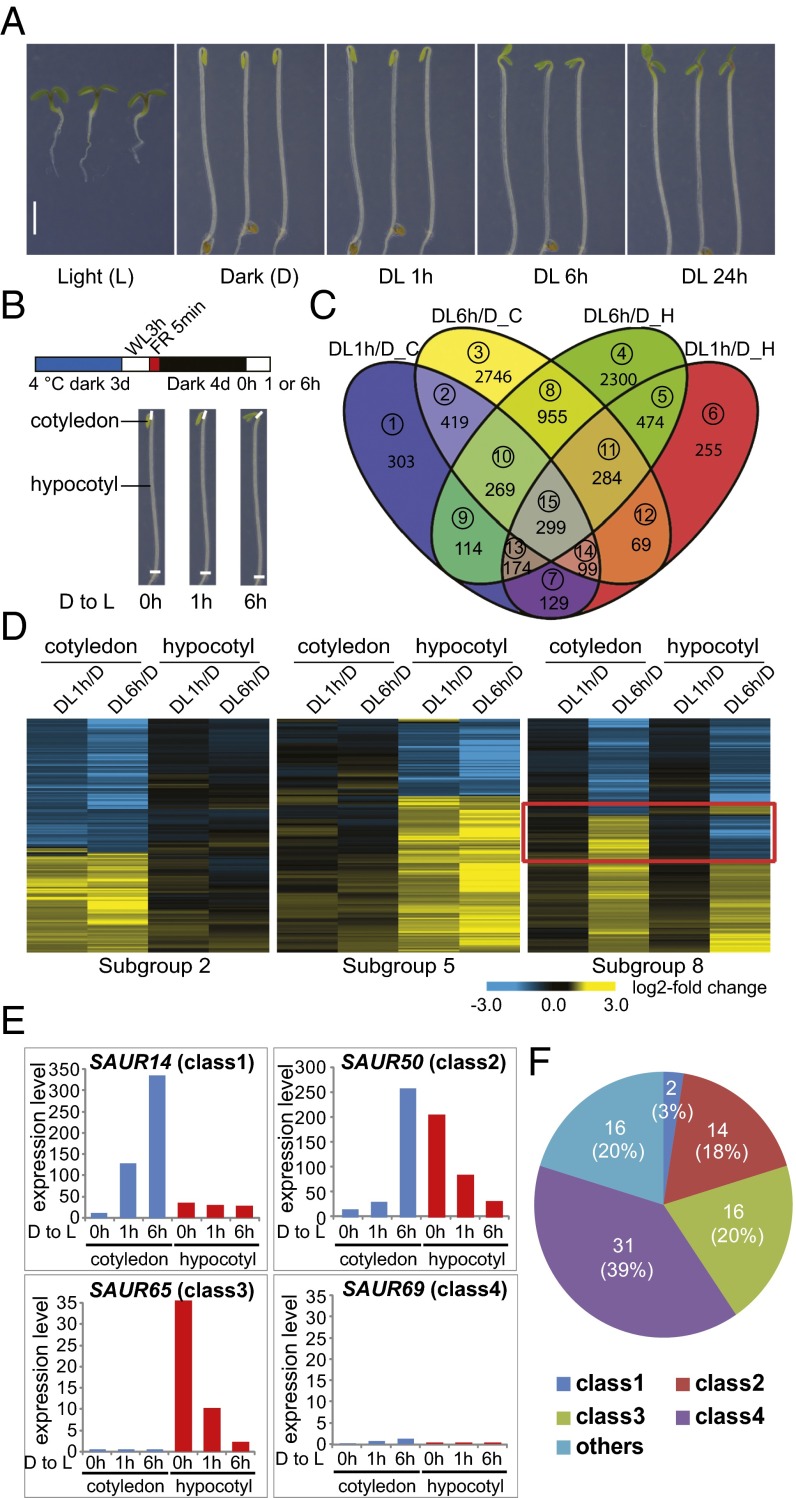
When an Arabidopsis seedling growing in darkness is exposed to light, its cotyledons and hypocotyls display opposite responses to light with regard to cell expansion (Fig. 1A). To study organ-specific light-responsive genes, 4-d-old dark-grown WT (Col, Columbia ecotype type) Arabidopsis seedlings were transferred into white light (WL) for 0, 1, and 6 h (D, DL1h, DL6h), and cotyledons and hypocotyls were dissected for RNA extraction and sequencing at each of these time points (Fig. 1B). Genes that were differentially expressed after 1 h (DL1h/D) and 6 h (DL6h/D) of illumination were divided into 15 subgroups according to the combination of organs and light conditions (Fig. 1C and Dataset S1).
Fig. 1.
Many SAUR family genes are up-regulated in cotyledons and/or down-regulated in hypocotyls during the dark-to-light transition. (A) Morphological comparison of 4-d-old WT Arabidopsis seedlings grown in continuous white light (L), continuous darkness (D), or transferred from darkness to light and then kept in white light for 1 h (DL 1h), 6 h (DL 6h), and 24 h (DL 24h). (Scale bar: 2 mm.) (B) Graphic diagram of RNA-Seq samples. Four-day-old dark-grown seedlings were exposed to white light for 0 h, 1 h, and 6 h, and cotyledons and hypocotyls were dissected at each time point. The Bottom diagram indicates the dissection sites of the seedlings. (C) Venn diagram showing numbers of light-responsive genes at 1 h (DL1h/D) or 6 h (DL6h/D) in cotyledons (C), or hypocotyls (H), respectively. Subgroup numbers are labeled in circles. (D) Heat map of the genes in subgroup 2, subgroup 5, and subgroup 8 of C. The red box marks the genes oppositely regulated by light in cotyledons and hypocotyls. (E) Expression patterns of representatives of four classes of SAUR genes. (F) Pie chart showing the numbers of genes in each class and their percentages in the SAUR gene family.
Organ-Specific Light-Responsive Genes Are Enriched in Auxin Stimulus Genes.
Among the organ-specific light-responsive genes (OLRs), three groups were analyzed in detail: genes that responded to light only in cotyledons (subgroups 1–3; OLR1), genes that responded to light only in hypocotyls (subgroups 4–6; OLR2), and genes that responded oppositely to light in cotyledons and hypocotyls (some genes in subgroups 7, 8, and 15; OLR3) (Fig. 1C). The heat maps of representative subgroups (2, 5, 8) are shown in Fig. 1D.
Gene ontology (GO) analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID) resources was performed to identify the biological processes these three groups of OLRs were involved in (30). Up-regulated and down-regulated genes were analyzed separately. All three groups of OLRs were enriched in genes responding to hormonal stimuli (SI Appendix, Fig. S1 and Dataset S2), and 37% (77/208) of these hormone-responsive genes were involved in the auxin pathway (Dataset S3). In particular, the down-regulated genes of OLR2 were enriched in auxin stimuli-related genes (SI Appendix, Fig. S1B). The GO analysis thus indicated that auxin-related genes may play important roles in organ-specific light responses.
SAUR Family Genes Exhibit Remarkably Organ-Specific Light-Responsive Patterns.
Among the organ-specific light-responsive genes related to auxin, Small Auxin Up RNA (SAUR) genes were remarkably enriched (42%) (Dataset S3). SAUR family genes, consisting of 79 members in Arabidopsis, were originally isolated based on their rapid induction by auxin (20–22, 31).
The expression patterns of all SAUR family genes were analyzed and most belonged to four classes (Fig. 1E and Dataset S4). Genes in class 1 were dramatically up-regulated by light in cotyledons but were not obviously light-responsive in hypocotyls. Genes in class 2 were up-regulated by light in cotyledons but down-regulated in hypocotyls. Genes in class 3 were down-regulated by light in hypocotyls but always showed constitutive low expression in cotyledons. Genes in class 4 showed low constitutive expression in both cotyledons and hypocotyls. Class 2 genes, which were oppositely regulated by light in cotyledons and hypocotyls, were critically interesting, and many of them would have been difficult to identify as light-responsive genes if analyzing whole seedlings.
SAUR14, SAUR50, SAUR65, and SAUR69 were chosen as representatives of the four classes, respectively, and their expression patterns were further confirmed by quantitative real-time PCR (qRT-PCR) (SI Appendix, Fig. S2). Notably, the majority of SAUR family genes (80%) were included in these four classes (Fig. 1F and Dataset S4). We named the 32 SAURs in classes 1–3 as lirSAURs (light-induced in cotyledons and/or repressed in hypocotyls SAURs), whose expression patterns correlated well with the growth changes in cotyledons and hypocotyls during the dark-to-light transition. Given previous reports that SAURs function in regulating cell expansion (22–29), we hypothesized that differential regulation of lirSAUR expression in cotyledons and hypocotyls may play important roles in organ-specific cellular responses to light. In the class “others,” several SAURs were down-regulated by light in cotyledons (Fig. 1F and Dataset S4), and these SAURs may have other unknown functions in seedling development.
lirSAURs Promote Cotyledon Opening and Expansion and Increase Hypocotyl Elongation.
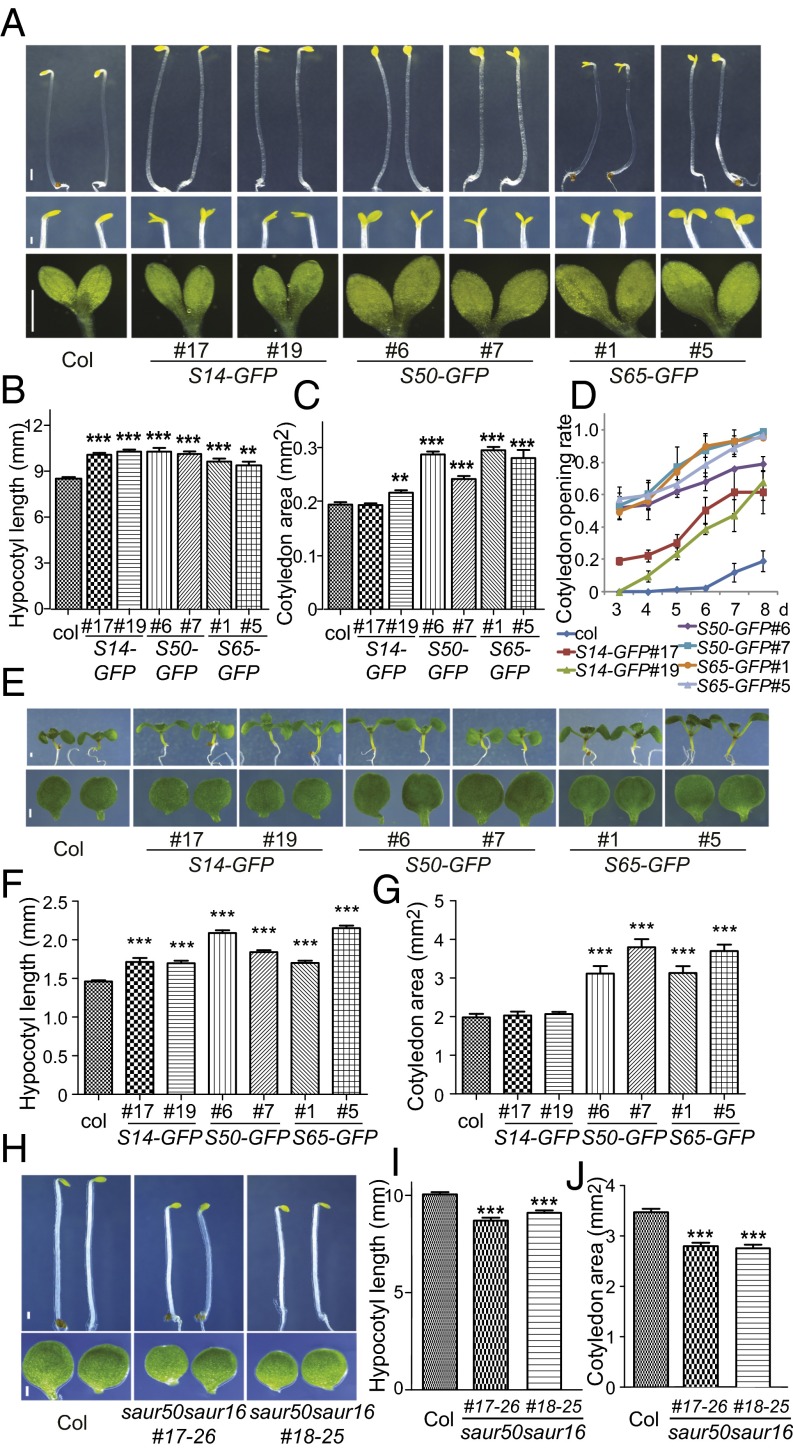
Previous studies have shown that ectopic expression of several SAUR family genes resulted in elongated hypocotyls in the light and that plants overexpressing SAUR19 exhibited opened cotyledons in the dark (23–25). However, several other SAURs have been suggested to inhibit cell expansion in certain organs (26–28). To test the effects of lirSAURs on plant growth, we generated transgenic lines overexpressing SAUR14, SAUR50, and SAUR65, which were selected to represent classes 1, 2, and 3, respectively. qRT-PCR analyses revealed that lirSAURs were overexpressed in these transgenic materials (SI Appendix, Fig. S3).
When grown in the dark for 3 d, 35S:SAUR14-GFP, 35S:SAUR50-GFP, and 35S:SAUR65-GFP seedlings displayed longer hypocotyls than WT plants (Fig. 2 A and B). Moreover, the cotyledon areas of the SAUR50 and SAUR65 overexpression lines were significantly larger than the cotyledon areas of WT (Fig. 2 A and C). Upon extended growth in the dark, a greater percentage of seedlings from these transgenic lines showed opened cotyledons compared with the seedlings of WT at each time point (Fig. 2 A and D). These phenotypes indicate that these three classes of lirSAUR family genes can promote cotyledon opening and expansion and can increase hypocotyl elongation in the dark.
Fig. 2.
Arabidopsis lirSAURs promote cotyledon expansion and opening, and enhance hypocotyl elongation. (A–D) Cotyledon areas, cotyledon opening angles, and hypocotyl lengths of dark-grown Col, 35S:SAUR14-GFP(S14-GFP), 35S:SAUR50-GFP(S50-GFP), and 35S:SAUR65-GFP(S65-GFP) seedlings. (A) Representative 3-d-old seedlings (Top), images of cotyledon opening of 6-d-old seedlings (Middle), and images of pressed cotyledons of 4-d-old seedlings (Bottom). (Scale bars: Top, 0.8 mm; Middle, 0.3 mm; Bottom, 0.5 mm.) (B) Hypocotyl lengths of 3-d-old dark-grown seedlings. (C) Cotyledon areas of 4-d-old seedlings. (D) Cotyledon opening rates of the seedlings growing 3 d to 8 d in the dark. Cotyledons open more than 30 degrees were counted as “opened cotyledon.” (E–G) Hypocotyl lengths and cotyledon areas of 7-d-old Col, S14-GFP, S50-GFP, and S65-GFP seedlings grown under light. (E) Representative 7-d-old seedlings (Top) and cotyledons (Bottom). (Scale bar: 0.5 mm.) (F and G) Bar graphs of hypocotyl lengths and cotyledon areas. (H–J) Hypocotyl lengths of Col and saur50saur16 mutant seedlings grown in darkness and cotyledon areas of these plants grown under light. (H) Representatives of 3-d-old dark-grown seedlings (Top) and cotyledons of 7-d-old light-grown seedlings (Bottom). (Scale bar: 0.5 mm.) (I) Bar graph of the hypocotyl lengths of 3-d-old dark-grown seedlings. (J) Bar graph of the cotyledon areas of 7-d-old light-grown seedlings. For bar graphs, the data are shown as mean ± SEM (n > 20). Statistical significance was calculated using Student’s t test compared with Col. ***P < 0.001, **P < 0.01.
When grown in the light, 35S:SAUR14-GFP, 35S:SAUR50-GFP, and 35S:SAUR65-GFP seedlings all exhibited longer hypocotyls than the seedlings of WT (Fig. 2 E and F). The cotyledons of 35S:SAUR50-GFP and 35S:SAUR65-GFP were significantly larger than the cotyledons of WT (Fig. 2 E and G). Because both the cotyledons and hypocotyls of 35S:SAUR50-GFP and 35S:SAUR65-GFP were significantly larger than WT, they were selected for further cell size examination. Clearly, the size of epidermal cells in cotyledons and hypocotyls in these overexpression lines was larger than in WT (SI Appendix, Fig. S4), which indicated that lirSAURs accelerated growth of cotyledons and hypocotyls by promoting cell expansion. Taken together, although GFP-tagged SAUR proteins may be more stable than endogenous SAURs, these results suggest that lirSAURs promote growth of cotyledons and hypocotyls in both the dark and the light.
To further investigate the function of lirSAURs in seedling development, mutants of representative SAURs were generated. SAUR50 was selected to be the representative because its expression changed dramatically in both cotyledons and hypocotyls after light treatment (Fig. 1E and Dataset S4). Because the SAUR family genes are highly redundant (22), we included another class 2 gene, SAUR16, that showed a similar expression pattern as SAUR50 in the mutation analysis (Dataset S4). Because there were no available mutants, we generated saur50saur16 double mutant lines using the clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9 system (32), and two different alleles were displayed for each (SI Appendix, Fig. S5A).
SAUR50 and SAUR16 were expressed at high levels in the hypocotyls of dark-grown seedlings and cotyledons of light-treated seedlings (Fig. 1E and Dataset S4). In accordance with these expression patterns, the hypocotyls of dark-grown saur50saur16 mutants were shorter than WT (Fig. 2 H and I) whereas the cotyledons of light-grown saur50saur16 mutants were significantly smaller than WT (Fig. 2 H and J). Moreover, during the dark-to-light transition, the opening of cotyledons was slower in saur50saur16 mutants than WT (SI Appendix, Fig. S5B). These results further confirm that lirSAURs can promote cotyledon opening and expansion as well as hypocotyl elongation and that their absence can delay the photomorphogenic response in cotyledons. Moreover, consistent with previous studies, we found that the three classes of lirSAUR proteins could interact with PP2C-Ds and inhibit their phosphatase activities, which supports the general mechanism whereby SAURs promote cell expansion (29) (SI Appendix, Fig. S6).
Light Down-Regulates the Expression of lirSAURs in Hypocotyls Partially by Reducing Auxin Levels.
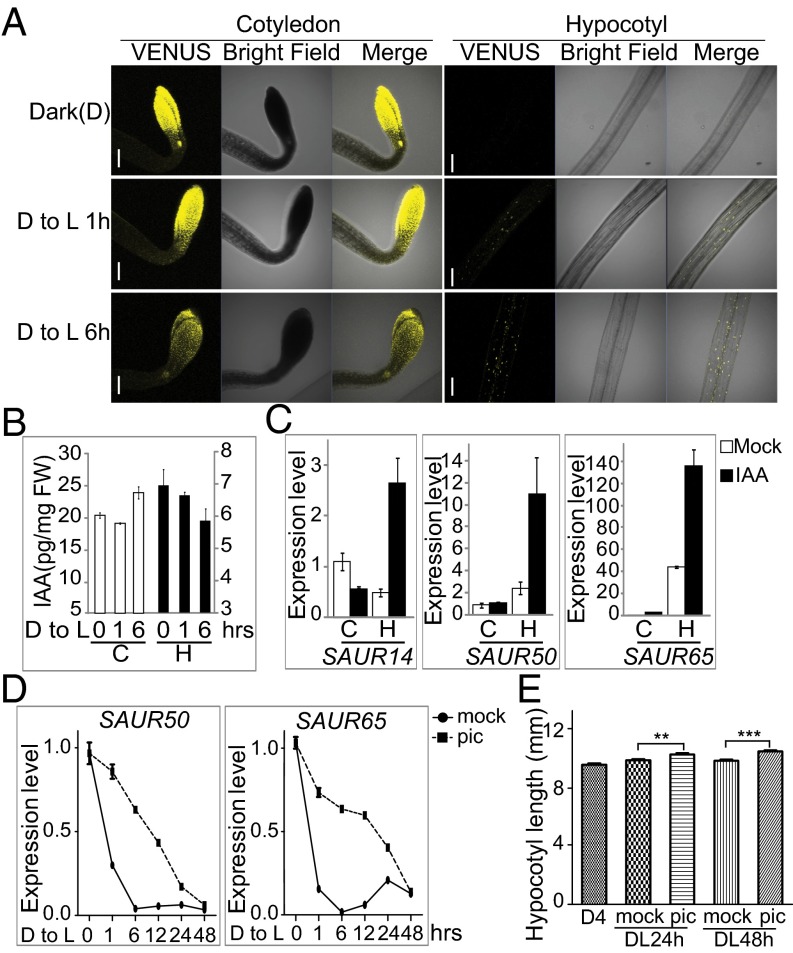
The above analyses showed that differential expression of lirSAURs after light treatment in cotyledons and hypocotyls may determine the differential growth of these two organs. Next, we examined how the expression of lirSAUR genes was regulated by light. Considering that the expression of many SAURs was rapidly induced by auxin (22) and that auxin levels increased in hypocotyls during shade avoidance (33), we hypothesized that light might regulate the expression of SAUR family genes by modulating the auxin level. To monitor changes in auxin levels during the dark-to-light transition, we used DII VENUS transgenic plants in which the fluorescence intensity of VENUS is inversely related to the auxin level (34). The change of fluorescence intensity indicated that auxin levels didn’t obviously change after 1 h of light treatment and slightly increased after 6 h in cotyledons whereas the auxin levels continuously decreased in hypocotyls (Fig. 3A). The changes in auxin levels in cotyledons and hypocotyls were further confirmed by measuring indole-3-acetic acid (IAA) contents (Fig. 3B).
Fig. 3.
Light reduces auxin levels and down-regulates the transcription of lirSAUR family genes in hypocotyls. (A) Fluorescence intensities of VENUS in the cotyledons and hypocotyls of DII-VENUS marker lines kept in the dark (D), after 1 h in the light (D to L 1h), or after 6 h in the light (D to L 6h). (Scale bars: 200 μm.) (B) Free IAA contents in cotyledons and hypocotyls during the dark-to-light transition at D to L 0 h, 1 h, and 6 h. (C) Transcript levels of SAUR14, SAUR50, and SAUR65 in cotyledons and hypocotyls of 4-d-old dark-grown WT seedlings with or without IAA (5 μM) treatment. (D) Transcript levels of SAUR50 and SAUR65 in 4-d-old dark-grown WT seedlings treated with 10 μM picloram (pic) or DMSO as a control (mock) during the dark-to-light transition. (E) Hypocotyl lengths of 4-d-old dark-grown WT seedlings after transfer into white light for 24 h (DL24h) or 48 h (DL48h) with or without 10 μM picloram treatment. The data are shown as mean ± SEM (n > 40). Statistical significance was calculated using Student’s t test. ***P < 0.001, **P < 0.01.
To check how auxin levels influenced the expression of lirSAUR genes in various organs, 4-d-old dark-grown seedlings were treated with 5 μM IAA, and the expression levels of representative lirSAUR genes were measured. Interestingly, IAA treatment induced SAUR expression only in hypocotyls, but not in cotyledons (Fig. 3C). However, IAA treatment under the same condition induced the expression of GUS in DR5:GUS transgenic lines in both cotyledons and hypocotyls, which indicated that treatment with 5 μM IAA for 1 h was enough to induce the expression of auxin-inducible genes and that our treatment worked well (SI Appendix, Fig. S7).
Because the expression of lirSAURs obviously increased in cotyledons after light treatment for even 1 h (Fig. 1E and Dataset S4) whereas the free IAA level showed no obvious change at the same time point (Fig. 3 A and B), we speculated that auxin may not play important roles in acute light induction of lirSAURs in cotyledons. In addition, the expression of SAUR14, SAUR50, and SAUR65 could not be induced by IAA treatment in cotyledons (Fig. 3C), which further indicates that the increased SAUR expression in cotyledons was caused by signals other than auxin.
In hypocotyls, on the other hand, light treatment reduced both the auxin levels and the expression of lirSAURs (Figs. 1E and 3 A and B and Dataset S4), and auxin positively regulated SAUR expression (Fig. 3C). We speculated that light may down-regulate SAUR expression and hypocotyl elongation by reducing auxin levels in the hypocotyls. If true, supplementing with exogenous auxin should weaken the inhibition of hypocotyl elongation by light. To test this hypothesis, we used the auxin analog picloram because it is more effective than IAA at inducing hypocotyl elongation under light (35). SAUR50 and SAUR65 were selected for the test, which are representative lirSAURs whose expression declines in hypocotyls after light treatment. During the dark-to-light transition, the expression of SAUR50 and SAUR65 decreased quickly whereas the picloram treatment decreased the rate of decline in their expression levels over a 2-d period (Fig. 3D) and hypocotyls of the seedlings treated with picloram were significantly longer than the controls (Fig. 3E). This result demonstrates that a reduction in auxin levels is critical for rapid down-regulation of lirSAURs by light, as well as effective inhibition of hypocotyl elongation by light. Interestingly, lirSAUR levels dramatically increased in the hypocotyls after auxin treatment in the dark (Fig. 3C), but their levels declined continuously upon light illumination, even with the auxin treatment (Fig. 3D). This result indicates that light regulates the expression of lirSAURs only partially through auxin levels and other mechanisms that silence specific SAURs in hypocotyl cells must exist.
PIFs Differentially Regulate lirSAUR Transcript Levels in Cotyledons Versus Hypocotyls.
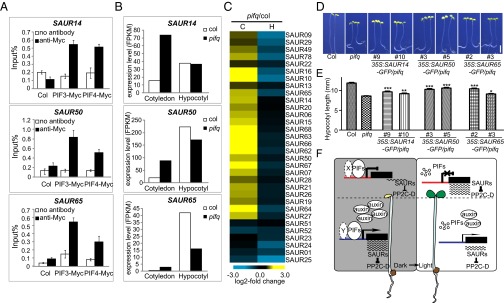
Because auxin levels could not fully explain how light regulates the expression of lirSAUR genes, we looked for other connections between light signal and lirSAUR genes. Upon searching previous ChIP-seq data (16–18), we found that PIF3 and PIF4 may bind the promoters of several SAUR genes. To check whether these PIFs could bind lirSAURs, 35S:PIF3-Myc and 35S:PIF4-Myc were used to perform ChIP-qPCR assays. We found that the promoters of SAUR14, SAUR50, and SAUR65 were all enriched in PIF3-myc and PIF4-myc ChIP samples compared with WT controls, indicating that PIF3 and PIF4 bound these sequences in vivo (Fig. 4A).
Fig. 4.
The regulation of lirSAUR gene expression by PIFs and a working model of how light differentially regulates the development of cotyledons and hypocotyls. (A) ChIP-qPCR analysis of 4-d-old dark-grown seedlings showing specific enrichment of PIF3 and PIF4 on SAUR14, SAUR50, and SAUR65 promoters. Col and no antibody were used as controls. (B) Expression of SAUR14, SAUR50, and SAUR65 in the cotyledons and hypocotyls of 4-d-old dark-grown pifq and WT seedlings. (C) Heat map of lirSAURs in cotyledons (C) and hypocotyls (H) of 4-d-old dark-grown pifq seedlings compared with WT (pifq/col) by RNA-seq analysis. (D) Representative 4-d-old dark-grown seedlings of WT, pifq, and SAUR14, SAUR50, and SAUR65 overexpression transgenic lines in pifq background. (Scale bar: 2 mm.) (E) Hypocotyl lengths of the seedlings shown in D. The data are shown as mean ± SEM (n > 20). Statistical significance was calculated using Student’s t test compared with pifq. ***P < 0.001, **P < 0.01, *P < 0.05. (F) A working model of how light differentially regulates the growth of cotyledons and hypocotyls during deetiolation. X and Y represent possible different PIF cofactor(s) in cotyledons and hypocotyls, respectively. Promoters are labeled in two colors (red in the cotyledons and blue in the hypocotyls) indicating the possible differences in epigenetic modifications of the same lirSAUR gene in these two organs.
To test how PIFs regulate SAUR expression, transcript levels of all of the lirSAUR genes were compared in etiolated WT and pif1pif3pif4pif5 (pifq) mutants using mRNA deep sequencing. Among them, SAUR14, SAUR50, and SAUR65, representing the three classes of lirSAURs, were up-regulated in cotyledons and/or down-regulated in hypocotyls in pifq mutants compared with WT plants (Fig. 4B). Further, the heat map of lirSAUR gene expression showed that almost all lirSAURs were up-regulated in cotyledons and/or down-regulated in hypocotyls in pifq (Fig. 4C and Dataset S5). This result indicates that PIFs repress the expression of lirSAUR genes in cotyledons but promote their expression in hypocotyls in the dark, which is consistent with the phenotype of the pifq mutant showing open and expanded cotyledons and short hypocotyls. To elucidate the genetic relationships between lirSAURs and PIFs, SAUR14, SAUR50, and SAUR65 were overexpressed in the pifq background (SI Appendix, Fig. S8). The short hypocotyl lengths of pifq were partially but significantly rescued by the ectopic expression of lirSAURs (Fig. 4 D and E), which indicates that lirSAURs are key factors acting downstream of PIFs to regulate cell elongation. Taken together, these results show that PIFs can bind directly to the promoters of lirSAUR genes and differentially regulate their expression in cotyledons and hypocotyls in the dark. Thus, PIF degradation after light treatment leads to the differential expression of lirSAURs in these two organs.
Discussion
Light brings about different effects in different organs throughout plant development, including events such as deetiolation and shade avoidance. The opposite effects of light on the growth of leaves and stems have been described for more than 70 y (5–7). Although specific light-responsive characteristics in cotyledons and hypocotyls have been used as common phenotypic indicators to study light signal transduction, our understanding of organ specification itself remains superficial. Deetiolation is an excellent model process to study differential light effects on various organs. Although certain organ-specific light-regulated genes have already been identified (8–10), the core components and molecular mechanisms connecting signals to development were lacking. In our study, by analyzing light-responsive genes in cotyledons and hypocotyls separately, we identified a group of lirSAURs that were oppositely regulated by light in these two organs and that might play key roles in the differential light regulation of various organs (Figs. 1 and 2).
Our results indicate that lirSAURs are a group of core factors involved in organ-specific light responses, so we propose a working model of how light differentially regulates the development of cotyledons and hypocotyls during deetiolation (Fig. 4F). In cotyledons, PIFs bind the promoters of key lirSAUR genes to repress their expression. Upon illumination, PIFs are rapidly degraded and release this repression, so the transcripts of these lirSAUR genes accumulate and promote cotyledon expansion. In hypocotyls, PIFs and high levels of free auxin activate lirSAUR gene expression in the dark, and the abundance of lirSAUR proteins accelerates the quick elongation of hypocotyls. Upon illumination, PIFs are degraded, and free auxin levels decrease; therefore, lirSAUR transcript levels decline and hypocotyl elongation slows. Because previous studies have shown that PIFs positively regulated auxin biosynthesis in response to high temperature or shade (36–38), the degradation of PIFs should cause some of the decrease in auxin levels after light illumination in hypocotyls. The differential regulation of target genes by PIFs may be achieved by their cofactors, and this speculation is supported by previous ChIP-seq and RNA-seq data, which showed that PIFs may both positively and negatively regulate their target genes (16–18). Based on our transcriptomic analysis, thousands of genes are differentially expressed in cotyledons and hypocotyls (subgroup 7 in Dataset S6), and some may work as PIF cofactors. Additionally, it is possible that the epigenetic status of the same SAUR gene varies in different organs. The detailed mechanisms of how PIF cofactors and the epigenetic status of SAUR genes contribute to differential gene expression in various organs need further investigation.
Although SAURs have been identified as early auxin responsive genes for nearly 30 y, their functions have only just begun to be unlocked, due to likely genetic redundancy and tight linkage in the genome (22). Our study and previous studies indicate that SAURs function as a hub to integrate plant hormones and environmental signals. Multiple classes of SAURs with different temporal–spatial expression patterns may provide the plants with the capability to decode the combination of many environmental signals, including light and other signals, and then mediate the differential growth of various organs precisely. Further analysis of the functions and regulation of SAURs will greatly improve our understanding of plant growth and development.
Materials and Methods
Plant materials and growth conditions, generation of transgenic plants, and phenotype analysis are described in SI Appendix, Materials and Methods. The details and procedures of RNA-seq analysis, qRT-PCR analysis, confocal microscopy, measurement of free IAA levels, IAA treatment, picloram treatment, ChIP assays, yeast two-hybrid assays, and in vitro phosphatase assays are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jian-Kang Zhu for CRISPR/Cas9 plasmids. We appreciate the suggestions from Dr. Yuling Jiao. This work was supported by grants (to H.C.) from the National Natural Science Foundation of China (31271294) and the State Key Laboratory of Protein and Plant Gene Research; by grants (to X.W.D.) from the National Natural Science Foundation of China (31330048), the National Program on Key Basic Research Project of China (973 Program: 2012CB910900), the Peking–Tsinghua Center for Life Sciences, and the State Key Laboratory of Protein and Plant Gene Research; and by grants (to N.W.) from the NIH (GM-47850).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. 79576).
See Commentary on page 5774.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604782113/-/DCSupplemental.
References
- 1.Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. Curr Top Dev Biol. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- 2.Nemhauser J, Chory J. Photomorphogenesis. Arabidopsis Book. 2002;1:e0054. doi: 10.1199/tab.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Josse EM, Halliday KJ. Skotomorphogenesis: The dark side of light signalling. Curr Biol. 2008;18(24):R1144–R1146. doi: 10.1016/j.cub.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 4.Zhong S, et al. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc Natl Acad Sci USA. 2014;111(11):3913–3920. doi: 10.1073/pnas.1402491111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Went F. Effects of light on stem and leaf growth. Am J Bot. 1941;28(2):83–95. [Google Scholar]
- 6.Parker M, Hendricks S, Borthwick H, Went F. Spectral sensitivities for leaf and stem growth of etiolated pea seedlings and their similarity to action spectra for photoperiodism. Am J Bot. 1949;36(2):194–204. [Google Scholar]
- 7.Borthwick H, Hendricks S, Parker M. Action spectrum for inhibition of stem growth in dark-grown seedlings of albino and nonalbino barley (Hordeum vulgare) Bot Gaz. 1951;113(2):95–105. [Google Scholar]
- 8.Leu WM, Cao XL, Wilson TJ, Snustad DP, Chua NH. Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in arabidopsis. Plant Cell. 1995;7(12):2187–2196. doi: 10.1105/tpc.7.12.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khanna R, et al. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell. 2006;18(9):2157–2171. doi: 10.1105/tpc.106.042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, et al. Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 2005;138(1):80–91. doi: 10.1104/pp.104.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-Juez E, et al. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell. 2008;20(4):947–968. doi: 10.1105/tpc.107.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leivar P, Quail PH. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16(1):19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106(18):7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21(2):403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28(24):3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14(8):802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9(1):e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant. 2014;7(11):1598–1618. doi: 10.1093/mp/ssu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong J, Choi G. Phytochrome-interacting factors have both shared and distinct biological roles. Mol Cells. 2013;35(5):371–380. doi: 10.1007/s10059-013-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClure BA, Guilfoyle T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol. 1987;9(6):611–623. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- 21.McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ. Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell. 1989;1(2):229–239. doi: 10.1105/tpc.1.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren H, Gray WM. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant. 2015;8(8):1153–1164. doi: 10.1016/j.molp.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spartz AK, et al. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70(6):978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chae K, et al. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71(4):684–697. doi: 10.1111/j.1365-313X.2012.05024.x. [DOI] [PubMed] [Google Scholar]
- 25.Kong Y, et al. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013;54(4):609–621. doi: 10.1093/pcp/pct028. [DOI] [PubMed] [Google Scholar]
- 26.Markakis MN, et al. Characterization of a small auxin-up RNA (SAUR)-like gene involved in Arabidopsis thaliana development. PLoS One. 2013;8(11):e82596. doi: 10.1371/journal.pone.0082596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou K, Wu W, Gan SS. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013;161(2):1002–1009. doi: 10.1104/pp.112.212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J-E, Kim Y-S, Yoon H-K, Park C-M. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007;172(1):150–157. [Google Scholar]
- 29.Spartz AK, et al. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell. 2014;26(5):2129–2142. doi: 10.1105/tpc.114.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Gil P, et al. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 1994;104(2):777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Z, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133(1):164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482(7383):103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 35.Savaldi-Goldstein S, et al. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci USA. 2008;105(39):15190–15195. doi: 10.1073/pnas.0806324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franklin KA, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA. 2011;108(50):20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8(3):e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26(8):785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.