Significance
Ubiquitin-specific protease 6 (USP6) is a deubiquitylase that is overexpressed by chromosome translocation in two human neoplasms, aneurysmal bone cyst and nodular fasciitis. The relevant substrates of this ubiquitin-specific protease are not clear. Here, we identify the Wnt receptor Frizzled (Fzd) as a key target of the USP6 oncogene. Increased expression of USP6 increases the membrane abundance of Fzd, and hence increases cellular sensitivity to Wnts. USP6 opposes the activity of the ubiquitin ligase and tumor suppressor ring finger protein 43 (RNF43). This study identifies a new mechanism for pathological Wnt pathway activation in human disease and suggests a new approach to regulate Wnt activity therapeutically.
Keywords: Wnt signaling, ubiquitin-specific protease, USP6, Frizzled, ubiquitin
Abstract
The Wnt signaling pathways play pivotal roles in carcinogenesis. Modulation of the cell-surface abundance of Wnt receptors is emerging as an important mechanism for regulating sensitivity to Wnt ligands. Endocytosis and degradation of the Wnt receptors Frizzled (Fzd) and lipoprotein-related protein 6 (LRP6) are regulated by the E3 ubiquitin ligases zinc and ring finger 3 (ZNRF3) and ring finger protein 43 (RNF43), which are disrupted in cancer. In a genome-wide small interfering RNA screen, we identified the deubiquitylase ubiquitin-specific protease 6 (USP6) as a potent activator of Wnt signaling. USP6 enhances Wnt signaling by deubiquitylating Fzds, thereby increasing their cell-surface abundance. Chromosomal translocations in nodular fasciitis result in USP6 overexpression, leading to transcriptional activation of the Wnt/β-catenin pathway. Inhibition of Wnt signaling using Dickkopf-1 (DKK1) or a Porcupine (PORCN) inhibitor significantly decreased the growth of USP6-driven xenograft tumors, indicating that Wnt signaling is a key target of USP6 during tumorigenesis. Our study defines an additional route to ectopic Wnt pathway activation in human disease, and identifies a potential approach to modulate Wnt signaling for therapeutic benefit.
Wnts are a family of secreted proteins that regulate key developmental processes during embryogenesis as well as the homeostasis of adult tissues, including bone, skin, and intestine (1). Wnt signaling is a tightly regulated process, and its deregulation is implicated in various diseases, including cancer and inflammatory and vascular diseases (2). Wnt ligands interact with multiple receptors and coreceptors, and the specific combination of ligand and receptor determines which downstream pathways are regulated. Wnts can signal both through β-catenin–dependent and β-catenin–independent pathways (3). In the β-catenin–dependent pathway, Wnt binding to its receptors, Frizzleds (Fzds) and low-density lipoprotein-related protein 5/6 (LRP5/6), leads to the inhibition of ubiquitylation and proteasomal degradation of cytosolic β-catenin (4). β-catenin is thus stabilized and enters the nucleus, where it binds to the TCF/LEF family of transcription factors to activate transcription of a multitude of target genes (5).
In addition to controlling β-catenin protein stability, ubiquitylation regulates the subcellular localization and/or stability of multiple proteins within the Wnt signaling network, most notably membrane-proximal signaling events. For example, the cytoplasmic effector protein Dishevelled (DVL) is ubiquitylated by the KLHL12-CUL3, ITCH, and PDZRN3 ligases, resulting in its degradation or the endocytosis of Dvl/Fzd complex (6, 7). The turnover of Wnt receptors Fzd and LRP6 is regulated by the transmembrane E3 ubiquitin ligases ring finger protein 43 (RNF43) and zinc and ring finger 3 (ZNRF3) that promote their ubiquitylation, leading to endocytosis and lysosomal degradation. RNF43 and ZNRF3 are also Wnt/β-catenin target genes, and thereby function as negative feedback regulators of Wnt signaling (8, 9). The activity of RNF43 and ZNRF3 is regulated, in turn, by the extracellular signaling proteins, R-spondins (RSPO1–4), which form a ternary complex with RNF43/ZNRF3 and the coreceptors LGR4–6 (8, 10). Either loss-of-function RNF43 mutations (11, 12) or overexpression of R-spondin, arising from its translocation, leads to increased surface levels of Wnt receptors, β-catenin activation, and tumorigenesis (13, 14). Thus, diverse and tightly regulated ubiquitin ligases play a pivotal role in controlling Wnt activity.
Like ubiquitylation, regulated deubiquitylation plays a crucial role in governing information flow within the cell. Ubiquitin-specific proteases (USPs), one of the most abundant groups of deubiquitylation enzymes (DUBs), have fundamental roles in the ubiquitin system by specifically deconjugating ubiquitin from targeted proteins. Several DUBs have been shown to regulate components of the Wnt signaling pathway directly. CYLD functions as a negative regulator of Wnt signaling by deubiquitylating the cytoplasmic effector DVL (15), and USP4 modulates Wnt signaling through interaction with Nemo-like kinase (NLK) and transcription factor 7 like 2 (TCF7L2) (16). USP34 regulates AXIN1/2 stability and opposes its tankyrase (TNKS)-dependent ubiquitylation, hence functioning as a positive regulator of Wnt signaling (17). TRABID also positively regulates Wnt signaling by deubiquitylating adenomatous polyposis coli (APC) and regulating the stability of the β-catenin/TCF transcription complex (18). USP47 and USP4 can directly regulate β-catenin deubiquitylation (19, 20). USP8 deubiquitylates endosome-tethered Fzd, resulting in the recycling of Fzd to the plasma membrane (21). Although genetic alterations in the ubiquitylation machinery have been identified in cancers (i.e., RNF43/ZNRF3, RSPO3), to date, no mutations in DUBs that activate Wnt receptors have been documented in human tumors.
Using genome-wide small interfering RNA (siRNA) screens coupled with focused gain-of-function validation, we identified USP6 as a potent activator of the Wnt pathway. USP6, which is overexpressed upon translocation in aneurysmal bone cyst (ABC) and nodular fasciitis, potentiates Wnt signaling by increasing the abundance of Fzds and LRP6 at the plasma membrane. We show that USP6 functions by deubiquitylating Fzds and, as such, antagonizes the activity of the E3 ubiquitin ligases RNF43 and ZNRF3. Wnt signaling is a functionally important target of USP6, because pharmacological inactivation of Wnt signaling using a Porcupine (PORCN) inhibitor significantly attenuates the growth of xenografts driven by high USP6 expression. Finally, activation of Wnt signaling is observed in human tumors harboring USP6 translocation, supporting its clinical relevance in regulating Wnt activity. Our study uncovers a mechanism that controls Wnt receptor abundance on the cell membrane, and thus provides new targets for modulating Wnt signaling.
Results
Functional Genomic Screen of β-Catenin–Dependent WNT Signaling.
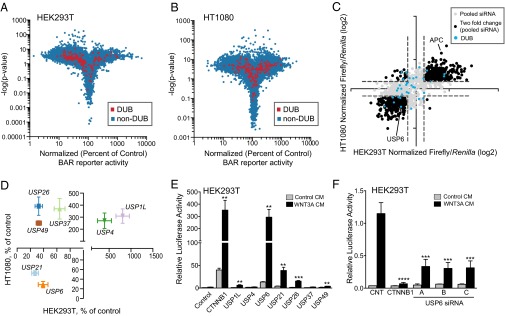
To identify novel regulators of Wnt signaling, an extensively validated and near-saturation genome-wide siRNA screen in HEK293T human embryonic kidney cells and HT1080 human sarcoma cells was performed. A similar strategy was previously used in DLD1 cells, a colorectal cancer cell line that expresses a mutant form of APC that disrupts the β-catenin destruction complex (22). In the current screen, exogenous Wnt ligand was used to activate receptor-mediated signaling. HEK293T and HT1080 cell lines with an integrated Wnt/β-catenin–activated firefly luciferase reporter and cytomegalovirus-driven Renilla luciferase reporter were screened in the presence of WNT3A-conditioned medium in 1,536-well plates with three nonoverlapping gene-specific siRNAs in each pool. Of 28,124 siRNA pools targeting 20,042 messenger RNAs, 1,877 increased or decreased Wnt/β-catenin reporter expression threefold or greater in both cell lines, with a P value less than 0.01 (Fig. 1 A and B and Dataset S1). A secondary validation screen of 1,172 hits from the primary screen was performed by independently evaluating three to nine nonoverlapping single siRNAs. Hit-calling criteria for the secondary screen included an increase or decrease in the Wnt/β-catenin reporter activity of at least twofold with a Student’s t test P value <0.01. Additionally, at least two independent siRNAs and the repeat test of the pool had to meet a statistically significant twofold change. We identified 186 gene products that have an impact on Wnt signaling in both HEK293T and HT1080 cells (Fig. 1C and Dataset S1). Compiled and cross-listed genome-wide primary screens from DLD1, HEK293T, and HT1080 and secondary screen data from HEK293T and HT1080 are provided in Dataset S1. The DLD1 primary and secondary screen data are reprinted with permission from AAAS (from ref. 22).
Fig. 1.
Genome-wide siRNA screen of WNT/β-catenin signaling. (A and B) Volcano plot of the primary siRNA screen. HEK293T (A) and HT1080 (B) cells are shown; red dots represent deubiquitylases. Full cross-referenced primary screen data are provided in Dataset S1. The HT1080 screen was published previously (43). (C) Scatter plot of the secondary siRNA screen. Data points represent genes targeted by a pool and at least two single siRNAs that increased or decreased the reporter by twofold or greater. Full secondary screen data are provided in Dataset S1. (D) Scatter plot of the secondary siRNA screen. Selected deubiquitylases and variance across replicates are illustrated. Normalized luciferase activity (firefly/Renilla) of DUBs whose loss of function regulated the Wnt/β-catenin reporter activity greater than twofold is depicted. Error bars represent mean ± SD. (E) Focused gain-of-function screen. HEK293T cells were cotransfected with the Wnt/β-catenin reporter, CMV-Renilla luciferase, and indicated expression constructs. The cells were treated with either control- or WNT3A-conditioned media overnight. Relative luciferase reporter activity is shown as mean ± SD (n = 3). (F) Loss of function of USP6 negatively regulates Wnt/β-catenin signaling. HEK293T cells stably expressing the Wnt/β-catenin reporter were transfected with the indicated siRNAs. Sixty hours after transfection, the cells were treated with control- or WNT3A-conditioned medium for 16 h. siRNAs targeting β-catenin (CTNNB1) serve as controls. Data represent mean ± SD (n = 4).
Of the 86 DUBs in the human genome, the genome-wide pooled siRNA screens identified 28 DUBs as putative regulators of Wnt signaling in both HT1080 and HEK293T cells. Secondary screens revealed seven DUBs that have an impact on the Wnt pathway activity (Fig. 1D). Silencing USP21 and USP6 significantly down-regulated Wnt/β-catenin reporter activity in both HEK293T and HT1080 cell lines, whereas loss of function of USP4 and USP1L increased pathway activity (Fig. 1D). These findings partially overlap with previously reported DUBs regulating Wnt signaling, although several, including CYLD and USP8, were not identified in our screen.
To confirm and extend these findings further, we performed the converse experiment, testing whether overexpression of selected DUBs affects Wnt/β-catenin reporter activity. HEK293T cells were transfected with the indicated expression constructs, along with the Wnt/β-catenin firefly luciferase reporter and Renilla luciferase reporter. USP6 overexpression strongly potentiated WNT3A-induced reporter activity, comparable to β-catenin overexpression (Fig. 1E). In agreement with the siRNA screen data, USP21 and USP26 also potentiated Wnt signaling, albeit significantly less than USP6. In addition, because high-throughput siRNA screening is highly susceptible to artifact and false-positive discovery, HEK293T cells were transfected with three nonoverlapping USP6-specific siRNAs that were distinct from the siRNAs used in the primary and secondary siRNA screens; all siRNAs tested suppressed Wnt signaling (Fig. 1F).
USP6 Activates Wnt/β-Catenin Signaling in Diverse Cell Types in a USP-Dependent Manner.
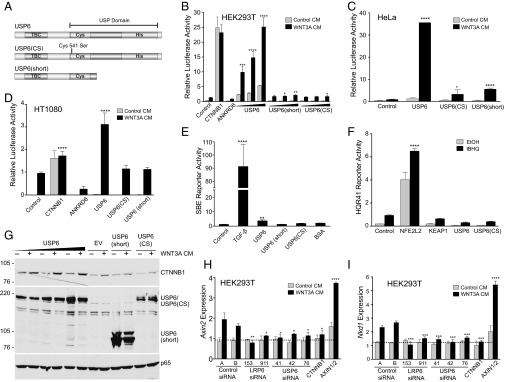
The full-length USP6 isoform contains the USP domain at its C terminus and a TBC (Tre2/Bub2/Cdc16) domain at its N terminus (Fig. 2A). A naturally occurring C-terminally truncated splice variant of USP6, USP6(short), lacks an essential portion of the USP domain and is therefore catalytically inactive (23). To determine whether USP function is required for Wnt activation by USP6, we examined Wnt/β-catenin reporter activation by USP6(short), as well as USP6 with a point mutation in a key catalytic residue, Cys541→Ser, USP6(CS). As shown in Fig. 2B, expression of USP6 in HEK293T cells enhanced Wnt signaling in a dose-dependent manner. In contrast, USP6(short) and USP6(CS) did not have a strong impact on the the Wnt/β-catenin reporter activity. Similar results were observed in HeLa and HT1080 cells (Fig. 2 C and D). Furthermore, using various dilutions of WNT3A-conditioned medium, USP6 was found to activate Wnt responsiveness at all doses of treatment (Fig. S1A). In sum, these results demonstrate that the ubiquitin protease activity of USP6 is essential for activation of β-catenin–dependent Wnt signaling, and that USP6 overexpression activates Wnt signaling in multiple cell types of diverse tissue origin.
Fig. 2.
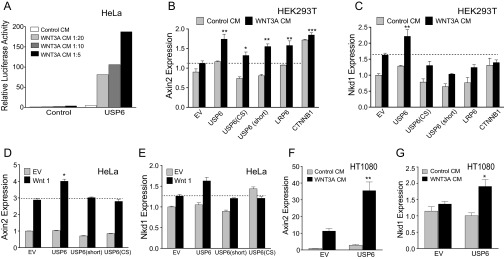
USP6 activates WNT signaling. (A) Model depicting USP6 alleles. Cysteine (Cys) and histidine (His) subdomains of the USP domain are shown. Ser, serine; TBC, TBC domain. USP6(CS) harbors a point mutation in the cysteine subdomain. (B) Dose-dependent activation of Wnt signaling by USP6. HEK293T cells with an integrated Wnt/β-catenin reporter were transfected with increasing amounts of plasmids encoding USP6 or its variants. Cells were incubated with WNT3A- or control-conditioned media for 16 h before analysis. Data represent mean ± SD (n = 3). (C) USP6 activates Wnt signaling in HeLa cells. HeLa cells were transfected with plasmids expressing USP6 or its variants and Wnt/β-catenin reporter. The cells were incubated with WNT3A- or control-conditioned medium for 24 h before analysis. Data represent mean ± SD (n = 7). (D) USP6 activates Wnt signaling in HT1080 cells. HT1080 cells were transfected with Wnt/β-catenin–firefly luciferase reporter, Renilla luciferase reporter, and the indicated plasmids. Cells were incubated with WNT3A- or control-conditioned media for 12 h before analysis. Data represent mean ± SD (n = 3). (E and F) USP6 does not activate TGF-β or NRF2 signaling. HEK293T cells expressing SBE (TGF-β) reporter or antioxidant response element (hQR41) reporter were transfected with USP6 or its mutants. Tert-butylhydroquinone (tBHQ; 50 μM) and recombinant human TGF-β1 (10 pM) are agonists for the SBE and hQR41 reporters, respectively. Renilla-normalized relative luciferase activity was plotted. Error bars represent mean ± SD (n = 3). (G) USP6 expression stabilizes cytosolic β-catenin. HeLa cells were transfected with plasmids expressing USP6-HA or its mutant isoforms and then incubated overnight with WNT3A- or control-conditioned medium. Cell lysates were analyzed for levels of endogenous β-catenin by immunoblotting. NF-κB p65 subunit was used as a loading control. (H and I) Loss of function of USP6 down-regulates expression of WNT-responsive genes. HEK293T cells were transfected with three independent siRNAs targeting USP6 transcript. Forty-eight hours after siRNA transfection, the cells were treated with control- or WNT3A-conditioned medium. Total RNA was isolated, and expression of Wnt target genes AXIN2 (H) and NKD1 (I) was analyzed by quantitative RT-PCR. Data were normalized to transcript abundance for GAPDH, and represent mean ± SD for biological triplicates.
Fig. S1.
Regulation of Wnt target gene expression by USP6. (A) USP6 activates Wnt signaling in HeLa cells: HeLa cells were transfected with plasmids encoding USP6 and a Wnt/β-catenin reporter. The cells were incubated with indicated dilutions of WNT3A-conditioned media, and luciferase activity was measured after 24 h. (B and C) Activation of AXIN2 and NKD1 expression in HEK293 cells transfected with USP6: HEK293 cells were transfected with plasmids encoding full-length USP6 or its variants and incubated with WNT3A CM or control media for 24 h. AXIN2 and NKD1 expression was measured by quantitative RT-PCR (qRT-PCR). Data are presented as mean ± SD (n = 3). (D and E) Activation of AXIN2 and NKD1 expression in HeLa cells transfected with USP6: HeLa cells were transfected with plasmids encoding full-length USP6 or its variants and WNT1. After 24 h, AXIN2 and NKD1 expression was measured by qRT-PCR. Data are presented as mean ± SD (n = 3). (F and G) Activation of AXIN2 and NKD1 expression in HT1080 cells transfected with USP6: HT1080 cells were transfected with a USP6 expression plasmid and incubated with WNT3A CM for 24 h. AXIN2 and NKD1 expression was measured by qRT-PCR. Data are presented as mean ± SD (n = 3). EV, empty vector.
To test if USP6 is a general activator of transcription, we examined whether USP6 overexpression had an impact on the expression of a TGF-β–responsive reporter [SMAD-binding element (SBE)] or an antioxidant responsive reporter (hQR41). USP6 overexpression weakly activated the SBE reporter (threefold), compared with 90-fold activation by TGF-β1 ligand (Fig. 2E). Moreover, USP6 did not increase transcription from the NFE2L2 transcription factor-driven hQR41 reporter (Fig. 2F), indicating that USP6 shows specificity for the Wnt pathway and is not a general transcriptional activator.
We next evaluated whether USP6 activates endogenous Wnt signal transduction. Binding of Wnt ligands to Fzd and LRP5/6 receptor complexes transiently inhibits the β-catenin destruction complex, leading to increased steady-state levels of β-catenin in the cytoplasm and nucleus. As shown in Fig. 2G, basal as well as WNT3A-induced total β-catenin protein levels were elevated following USP6 overexpression. This increase in endogenous β-catenin by USP6 was dependent on its catalytic activity because no increase was observed with the catalytically inactive USP6 variants (Fig. 2G). The effect of USP6 is not cell type-specific, because USP6 increased the expression of the endogenous Wnt-responsive genes NKD1 and AXIN2 in three cell lines of diverse origins: HEK293, HeLa, and HT1080 (Fig. S1 B–G). Conversely, silencing of USP6 using three nonoverlapping siRNAs down-regulated WNT-induced expression of AXIN2 and NKD1 (Fig. 2 H and I). Although these experiments required ectopic expression of USP6, taken together with the USP6 siRNA data in multiple cell lines, they support the identification of USP6 as a novel potentiator of endogenous Wnt/β-catenin signaling.
USP6 Functions Upstream of the Destruction Complex and Requires Wnt Ligand–Receptor Interactions.
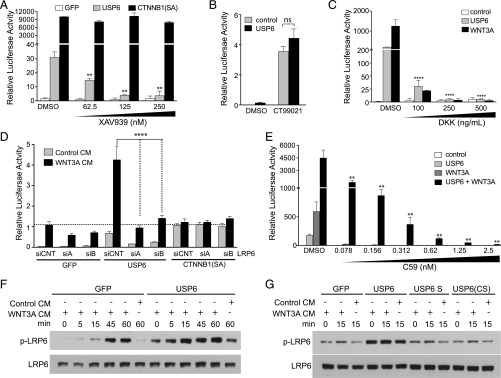
To elucidate the mechanism of Wnt/β-catenin activation by USP6 further, we conducted a series of epistasis experiments. We first sought to determine whether USP6 functions upstream of the β-catenin destruction complex. The β-catenin destruction complex catalyzes β-catenin phosphorylation by AXIN-bound glycogen synthase kinase 3 (GSK3) and casein kinase 1α (CSNK1A), leading to its ubiquitylation and degradation. AXIN protein levels are negatively regulated by the poly(ADP)-ribosylation activity of the TNKS1/2 enzyme. Inhibiting TNKS1/2 with a small-molecule inhibitor, XAV939, results in increased AXIN protein levels and, consequently, decreased β-catenin activity. USP6-induced Wnt signaling was dramatically inhibited by XAV939 (Fig. 3A) indicating that USP6 activates Wnt signaling upstream of the β-catenin destruction complex. In contrast, XAV939 had no effect on Wnt signaling driven by a stabilized, constitutively active β-catenin mutant (Fig. 3A), confirming the epistatic specificity of the drug. Inhibition of GSK3 activity increases β-catenin abundance by preventing β-catenin phosphorylation and its subsequent degradation. With near-complete repression of GSK3 activity by the GSK3 inhibitor CT99021, USP6 failed to enhance Wnt signaling further (Fig. 3B). Together, these data support a model wherein USP6 potentiates Wnt signaling upstream of the β-catenin destruction complex.
Fig. 3.
USP6 regulates WNT signaling at the level of the Wnt ligand and receptor complex. (A) Inhibition of USP6 induced Wnt signaling by XAV939. HEK293T cells with integrated Wnt/β-catenin reporter were transfected with USP6 expression plasmid. Following treatment with increasing concentrations of XAV939 for 16 h, luciferase activity was measured. β-catenin(SA) serves as a control; it is constitutively active and resistant to degradation by the destruction complex. Data represent mean ± SD. (B) USP6 does not potentiate Wnt/β-catenin signaling induced by GSK3 inhibitor. HEK293T cells stably expressing the Wnt/β-catenin reporter were transfected with control or USP6 expression plasmids. The cells were treated with 1 μM GSK inhibitor CT99021 for 16 h. Luciferase reporter activity as measured is expressed as mean ± SD (n = 3). ns, not significant. (C) Inhibition of USP6-induced Wnt signaling by DKK1. HEK293 cells with an integrated Wnt/β-catenin reporter were transfected with USP6 and/or WNT3A and incubated with varying amounts of DKK1. Data represent mean ± SD (n = 3). (D) LRP6 silencing decreases USP6-induced Wnt signaling. HEK293T cells were transfected with the indicated siRNAs. Forty-eight hours after siRNA transfection with two independent LRP6 siRNAs (siA and siB) or control siRNA (siCNT), plasmids expressing GFP, USP6, or β-catenin(SA) were transfected and cells were treated with control- or WNT3A-conditioned medium. Luciferase reporter activity is represented as mean ± SD (n = 4). (E) C59 inhibits USP6-induced Wnt signaling. HEK293 cells with an integrated Wnt/β-catenin reporter were transfected with USP6 and WNT3A expression plasmids and incubated with the indicated concentrations of C59. Data represent mean ± SD (n = 3). (F and G) USP6 expression increases LRP6 phosphorylation. HEK293T cells were transfected with control or plasmids expressing USP6 or its variants before treatment with control- or WNT3A-conditioned media for the indicated duration. Total cellular LRP6 levels and phosphorylated LRP6 (p-LRP6) levels were determined by Western blot analysis.
We next tested whether the interaction of Wnts with their receptors is required for activation of the pathway by USP6. Dickkopf-1 (DKK1) is a Wnt antagonist that prevents the interaction of Wnts with their coreceptors, LRP5/6. USP6-induced Wnt signaling was blocked by recombinant DKK1 treatment (Fig. 3C), suggesting that Wnt-LRP5/6 interaction is required for USP6 function. Further supporting a requirement for a Wnt ligand engaged receptor complex, knockdown of LRP6 with two independent siRNAs inhibited USP6-induced Wnt/β-catenin reporter activity in HEK293 cells (Fig. 3D). Importantly, subnanomolar amounts of the PORCN inhibitor C59, which effectively blocks Wnt secretion (24), inhibited basal USP6-stimulated β-catenin signaling, as well as the synergistic activation obtained upon cotransfection of WNT3A and USP6 expression plasmids (Fig. 3E).
Binding of Wnts to their cell surface receptors leads to phosphorylation of LRP6, which triggers a cascade of events leading to stabilization of β-catenin. We observed that compared with the control, LRP6 was more rapidly phosphorylated in USP6-expressing cells upon treatment with WNT3A CM (Fig. 3F). Consistent with the observed effect of USP6 in enhancing Wnt signaling, basal as well as Wnt-induced LRP6 phosphorylation was elevated in USP6-expressing cells. This activity was dependent on the protease activity of USP6 because the USP6(short) and USP6(CS) variants were not active (Fig. 3G). These data suggest that USP6 regulates Wnt signaling at the membrane, and that USP6 functions only in the presence of Wnt ligand secretion and Wnt receptor engagement.
USP6 Regulates Cell Surface Abundance of LRP6 and Fzds.
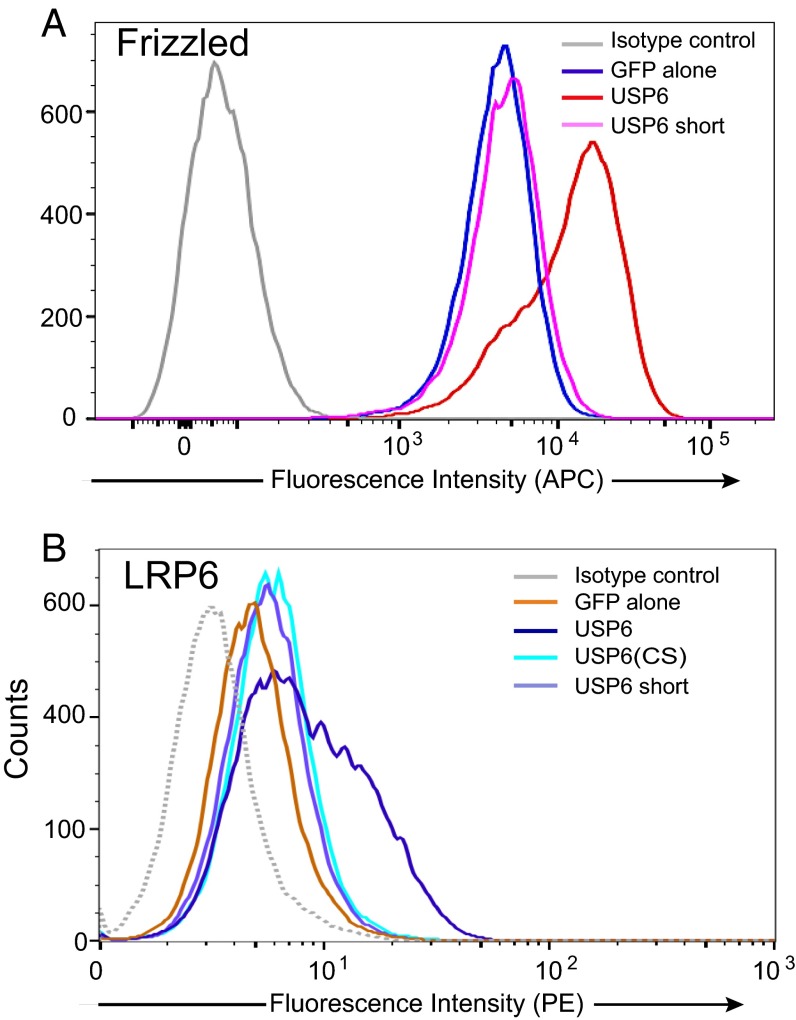
Because ubiquitylation is a key mechanism regulating Fzds and LRP5/6 membrane abundance, we tested if USP6, as a deubiquitylase, increased the abundance of the Wnt receptors on the cell surface. A USP6-GFP fusion protein was expressed in HEK293 cells, and plasma membrane levels of endogenous Fzds were measured by flow cytometry using the anti-Fzd monoclonal antibody OMP18R5 (25). Wild-type, but not inactive, USP6 markedly increased the cell surface abundance of Fzds, because 69.5% cells expressing USP6-GFP had high Fzd levels compared with 4.5% of GFP control cells (Fig. 4A). Wild-type USP6 similarly increased the cell surface abundance of endogenous LRP6, with 37.8% of cells showing high LRP6 levels compared with 3% of the controls (Fig. 4B). Expression of the catalytically inactive USP6(CS) and USP6(short) variants had no significant effect on surface LRP6 abundance.
Fig. 4.
USP6 increases abundance of Wnt receptors on the cell surface. (A) USP6 overexpression regulates Fzd abundance. Flow cytometric analysis of endogenous Fzd levels in HEK293 cells expressing pEGFP (GFP alone) or USP6-GFP and its variants. Fzd levels in GFP-positive cells are shown. Data are representative of three independent experiments. (B) USP6 overexpression regulates LRP6 abundance. A flow cytometric analysis of LRP6 levels in HEK293T cells transfected with USP6 and USP6 variants is shown. Data represent three biological replicate experiments.
USP6 Does Not Potentiate Wnt Signaling in RNF43 Mutant Cells.
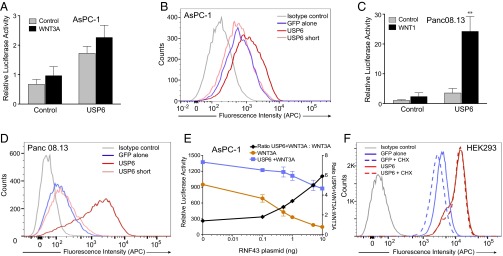
E3 ubiquitin ligases RNF43 and ZNRF3 negatively regulate Wnt signaling by promoting ubiquitylation and degradation of LRP6 and Fzds (8). AsPC-1 cells derived from pancreatic ductal adenocarcinoma (PDAC) have an inactivating mutation in RNF43, which presumably abrogates Fzd ubiquitylation, leading to Wnt-driven proliferation in vivo (14). We compared AsPC-1 with Panc 08.13 cells, a PDAC cell line with wild-type RNF43. Consistent with the loss of the RNF43 ubiquitylation activity, expressing the ubiquitin protease USP6 in the ubiquitin ligase-deficient AsPC-1 cells caused only a twofold increase in Wnt signaling (Fig. 5A); this residual effect may be due to reversal of some ZNRF3 activity. Unlike the synergy observed in RNF43 wild-type cells, in AsPC-1 cells, there was no significant enhancement of signaling in the presence of WNT3A (Fig. 5A). Consistent with the twofold activation of Wnt signaling, USP6 expression drove only a modest increase in Fzd membrane abundance (Fig. 5B). In contrast, in Panc 08.13 cells, both Wnt signaling (Fig. 5C) and Fzd cell surface abundance (Fig. 5D) were markedly enhanced by USP6 to a degree comparable to the degree measured in HEK293, HT1080, and HeLa cells.
Fig. 5.
USP6 does not potentiate Wnt signaling in RNF43 mutant cells. (A) USP6 does not synergize with WNT3A in RNF43 mutant pancreatic cancer cells. AsPC-1 cells were cotransfected with Wnt/β-catenin reporter and USP6 and WNT3A expression plasmids 24 h before analysis. Relative luciferase activity is plotted; error bars represent SD from the mean (n = 3). (B) Fzd levels in AsPC-1 cells expressing USP6. AsPC-1 cells were transfected with GFP or USP6-GFP and its variants. Cell surface levels of Fzds in GFP-positive cells as analyzed by flow cytometry are shown. Data are representative of three independent experiments. (C) USP6 synergizes in pancreatic cancer cells with WT RNF43. Panc 08.13 cells were transfected as in A. Relative luciferase activity is represented as mean ± SD (n = 3). (D) Fzd levels in Panc 08.13 cells expressing USP6. Panc 08.13 cells were transfected with GFP or USP6-GFP and its variants. Cell surface levels of Fzds in GFP-positive cells as analyzed by flow cytometry are depicted. Data are representative of four independent experiments. (E) Restoration of USP6 synergy in RNF43 mutant cells. AsPC-1 cells were transfected with Wnt/β-catenin reporter and WNT3A and indicated concentrations of RNF43 expression plasmids. Luciferase activity was measured after 24 h. Data represent mean ± SD (n = 3). (F) USP6 prevents down-regulation of Fzd receptors. HEK293 cells were transfected with GFP or USP6-GFP. After 48 h, new protein synthesis was blocked by addition of 40 μg/mL cycloheximide. Ninety minutes later, cell surface levels of Fzds in GFP-positive cells were assessed by flow cytometry. Data are representative of four independent experiments.
These results suggest that USP6 enhances Wnt signaling by counteracting the effects of the ubiquitin ligases RNF43 and ZNRF3. We therefore tested if titrated restoration of RNF43 activity in AsPC-1 (RNF43 mutant) cells rescued the synergistic activation of Wnt signaling by USP6. As indicated by the ratio of Wnt-stimulated β-catenin reporter activity in the absence or presence of USP6, restoring the expression of RNF43 modestly decreased overall signaling but markedly increased the synergistic activation by USP6 (Fig. 5E). These data are consistent with a model in which deubiquitylation of Fzds by USP6 increases their cell surface abundance by reversing their RNF43/ZNRF3-mediated ubiquitylation.
Because RNF43 increases Fzd endocytosis, we tested if USP6 decreased the clearance of Fzd from the cell surface. HEK293 cells have abundant cell surface Fzd, and when protein synthesis was blocked with cycloheximide, the Fzd abundance decreased, presumably due to endocytosis (Fig. 5F, purple lines). However, when USP6 was overexpressed, cell surface Fzd markedly increased, and its rate of clearance from the membrane decreased (Fig. 5F, brown lines). These data are consistent with USP6 working to oppose Fzd endocytosis.
USP6 Regulates Fzd Deubiquitylation.
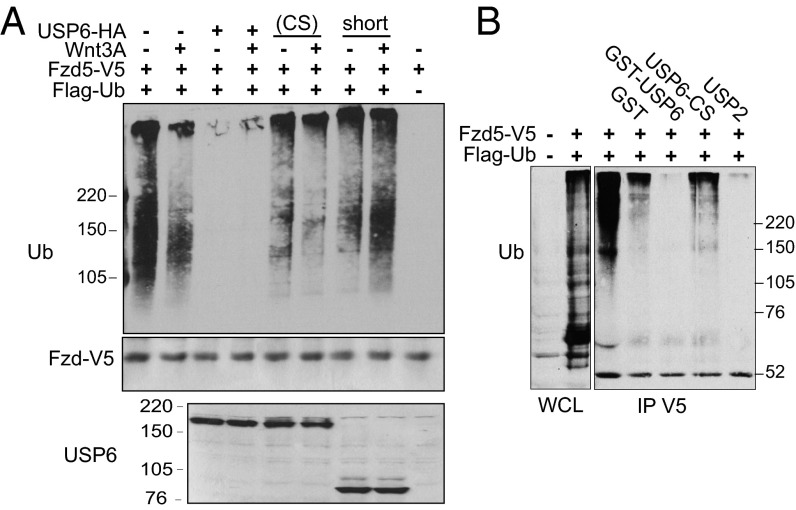
We next tested if USP6 directly deubiquitylates Fzds. HEK293T cells were cotransfected with Fzd-V5, FLAG-ubiquitin (Ub), and HA-USP6 expression plasmids, and treated with control- or WNT3A-conditioned media. Fzd-V5 was immunoprecipitated using anti-V5 antibody, and levels of ubiquitylation were monitored by FLAG immunoblotting. In immunoprecipitates from control cells, a smear of high-molecular-weight species corresponding to ubiquitylated Fzd was detected (Fig. 6A). Overexpression of USP6, but not catalytically inactive USP6(CS) or USP6(short), strongly reduced Fzd ubiquitylation.
Fig. 6.
USP6 promotes Fzd deubiquitylation. (A) USP6 promotes Fzd5 deubiquitylation in vivo. HEK293T cells were transfected with plasmids encoding Fzd5-V5, Flag-Ub, and the indicated USP6 constructs, and then treated with control or WNT3A CM. (Top) Fzd5 was immunoprecipitated using V5, and its ubiquitylation was detected with anti-Flag antibody. Whole-cell extracts were blotted with anti-V5 (Middle) or anti-USP6 (Bottom) antibodies. (B) In vitro deubiquitylation of Fzds by USP6. HeLa cells were transfected with plasmids encoding Fzd5-V5 and Flag-Ub. Fzd5 was purified using anti-V5 and then incubated with GST-tagged recombinant USP6 or USP6(CS). Fzd5 ubiquitylation was detected by immunoblotting with anti-FLAG. GST and USP2 were used as negative and positive controls, respectively. Data are representative of two independent experiments. IP, immunoprecipitation; WCL, whole cell lysate.
To determine if USP6 can directly deubiquitylate Fzd, in vitro deubiquitylation assays were performed. Fzd5-V5 was immunoprecipitated from HeLa cells cotransfected with Fzd-V5 and FLAG-Ub expression plasmids. The immunoprecipitated Fzd-V5 was incubated with recombinant GST-tagged USP6 (wild type or a catalytically inactive point mutant). As shown in Fig. 6B, ubiquitylation of Fzd-V5 was dramatically diminished by USP6, but not its catalytically inactive variant USP6(CS). USP2, a highly active nonspecific deubiquitylase, was used as a positive control. The effect of USP6 on ubiquitylation of endogenous Fzd5 could not be monitored due to lack of antibodies that efficiently immunoprecipitate Fzd5. Nevertheless, these results suggest that Fzd is a direct substrate of USP6, consistent with the model in which USP6 activates Wnt/β-catenin signaling by deubiquitylating Fzd receptors.
Human Tumors Harboring USP6 Translocation Have a Wnt/β-Catenin Transcriptional Signature.
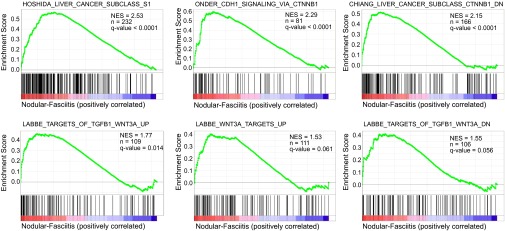
USP6 is implicated in human nonmalignant neoplastic disorders, with chromosomal translocation-driven overexpression of USP6 occurring in two independent tumor types, ABC and nodular fasciitis, in ∼70% and 90% of cases, respectively (26, 27). To determine if a Wnt/β-catenin gene signature is activated in these tumors, gene expression profiling by microarray analysis was performed on nodular fasciitis tumors with confirmed USP6 translocation/overexpression. The cell of origin in nodular fasciitis has yet to be defined, but is of mesenchymal origin. We compared the nodular fasciitis transcriptome with an averaged expression profile generated from 27 predominantly mesenchymal tumors lacking USP6 translocation. This strategy was used to exclude genes that are general mesenchymal markers or common indicators of the transformed state, and instead identify those genes selectively induced by USP6 in nodular fasciitis. Gene set enrichment analysis demonstrated strong positive correlation with multiple independent Wnt/β-catenin signatures (28–32), further supporting the model that overexpression of USP6 drives Wnt/β-catenin signaling in human tumors (Fig. 7 and Dataset S2).
Fig. 7.
Wnt/β-catenin–responsive gene signature in nodular fasciitis. Gene set enrichment analysis (GSEA) plots evaluating Wnt/β-catenin–dependent transcriptional responses in USP6-translocated nodular fasciitis (details of analysis are provided in Materials and Methods). FDR, false discovery rate; n, number of genes in gene set; NES, normalized enrichment score.
Inhibition of Wnt Signaling Prevents Growth of Tumors Overexpressing USP6.
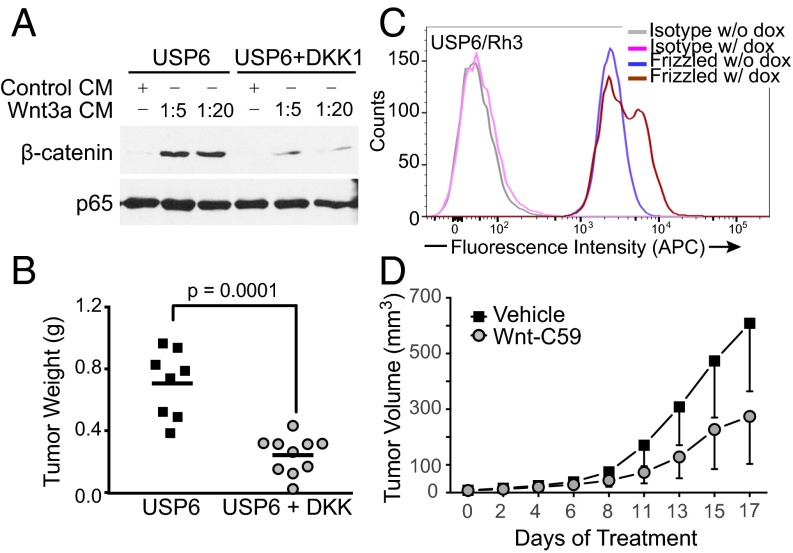
Because USP6 regulates multiple cellular signaling pathways (23, 33, 34), we assessed the functional importance of Wnt/β-catenin pathway activation in USP6-driven tumor formation. Because no immortalized ABC or nodular fasciitis cell lines exist, it was necessary to express USP6 alleles ectopically in heterologous cells. Previous studies have shown that mesenchymal NIH 3T3 fibroblasts stably overexpressing USP6 (USP6/NIH 3T3) serve as a useful cellular model for ABC and nodular fasciitis. When xenografted into immunodeficient mice, USP6/NIH 3T3 cells, but not control NIH 3T3 cells, formed highly vascularized, hemorrhagic tumors, recapitulating features of the human neoplasms, particularly ABC (34). To test whether USP6-mediated tumor formation was dependent on Wnt signaling, we introduced into USP6/NIH 3T3 cells DKK1, a secreted protein that blocks Fzd receptor activation. As shown in Fig. 8A, USP6 induction of β-catenin was significantly attenuated by DKK1, compared with cells expressing USP6 alone. The s.c. xenografting of these cells into NOG-SCID mice revealed that USP6-dependent tumor growth was significantly inhibited by DKK1 expression (Fig. 8B).
Fig. 8.
Inhibition of Wnt signaling prevents growth of tumors with USP6 activation. (A) Coexpression of DKK-1 reduces β-catenin activation in USP6-expressing NIH 3T3 cells. NIH 3T3 cells transiently expressing USP6 alone or with DKK-1 were treated with various dilutions of WNT3A CM. Cell lysates were immunoblotted for endogenous β-catenin and p65 as a control. (B) Expression of DKK-1 inhibits tumorigenesis of USP6-expressing NIH 3T3 cells. NIH 3T3 cells stably expressing doxycycline-inducible USP6 or USP6 plus DKK1 were injected s.c. into NOD-SCID mice. Tumors were harvested after 14 d, and tumor weights were recorded. (C) USP6 expression in ARMS cells increases surface abundance of Fzds. Flow cytometric analysis of cell surface Fzds in Rh3 cells upon induction of USP6 expression by doxycycline (dox). (D) PORCN inhibitor prevents growth of ARMS with inducible USP6 expression. NSG mice were injected s.c. with Rh3 cells expressing doxycycline-inducible USP6. Following establishment of tumors, the mice were gavaged with vehicle or 20 mg/kg C59, and tumor weights were recorded on study termination. Data are represented as mean ± SD (n = 14). P = 0.0003 as determined by two-way ANOVA.
The functional role of Wnt activation by USP6 was assessed further in a second tumorigenesis model. A high percentage of primary alveolar rhabdomyosarcoma (ARMS) patient samples express high levels of USP6. Interestingly, however, in all immortalized ARMS cell lines examined, USP6 expression was down-regulated. Thus, to determine the effects of USP6 in the context of this cancer, it was expressed under a doxycycline-inducible promoter in the ARMS cell line, Rh3 (USP6/Rh3) at levels comparable to the levels in primary ARMS samples. Induction of USP6 expression significantly increased the abundance of endogenous Fzds on the cell surface (Fig. 8C), indicating activation of Wnt signaling. The s.c. injection of USP6/Rh3 cells into NOD-SCID mice led to the rapid growth of tumors. Treatment of tumor-bearing mice with the PORCN inhibitor C59 inhibited tumor growth by 45% when dosed at 20 mg/kg once daily by oral gavage (Fig. 8D). Taken together, these data indicate that USP6 overexpression works, in large part, through activation of the Wnt/β-catenin pathway to drive tumorigenesis. Pharmacological agents targeting the Wnt/β-catenin pathway may therefore have therapeutic efficacy in USP6-overexpressing cancers.
Discussion
Aberrant activation of Wnt signaling promotes the initiation and progression of diverse tumor types. Accumulating evidence reveals that dynamic ubiquitylation/deubiquitylation of Fzd and LRP6 is a pivotal regulatory mechanism regulating Wnt/β-catenin signaling in a subset of human cancers. Our study further highlights the importance of maintaining appropriate surface levels of Wnt receptors for cellular homeostasis. We describe a major role for the poorly understood human oncogene, USP6, as a novel potentiator of Wnt signaling in multiple cell types. USP6 functions by deubiquitylating Fzds and perhaps LRP6 receptors, thereby increasing the abundance of both Fzds and LRP6 at the cell surface and rendering cells exquisitely sensitive to Wnt ligands. USP6 activity appears to reverse the ubiquitylation of Fzds and LRP6 by RNF43/ZNRF3. Our data show that USP6 stabilizes the membrane pool of Fzd. Ongoing and future experiments will test the hypothesis that increase Fzd at the membrane occurs through altered ubiquitin-dependent endocytosis of Fzd. In human tumors harboring USP6 translocations, the consequence is activation of Wnt/β-catenin signaling. Confirming the functional importance of USP6-mediated Wnt pathway activation, inhibition of Wnt signaling either by expression of DKK1 or treatment with the PORCN inhibitor C59 suppresses USP6-mediated tumor formation in murine xenograft models. Prior studies have shown that loss-of-function mutations in RNF43/ZNRF3 and translocation-mediated overexpression of its inhibitor, R-spondin, also drive pathological activation of Wnt signaling in human tumors. Together, these findings reinforce the critical role of this regulatory node of the Wnt pathway in human tumorigenesis.
We have previously reported that USP6 promotes deubiquitylation of cargo proteins to prevent their lysosomal targeting and increase their surface levels through enhanced recycling (35). USP8/UBPY is also shown to deubiquitylate Fzd, rescuing it from lysosomal degradation and promoting its recycling to the plasma membrane (21). USP8 was not identified in our siRNA screen for Wnt pathway activators, perhaps due to subphenotypic silencing by the siRNAs. Thus, USP6 and USP8 may possess both redundant and unique functions in regulating trafficking and Wnt signaling, with cell type and subcellular localization of both the USPs and cargoes contributing to their specificity. In this context, it is important to note that the USP6 gene only exists in hominoids, and thus cannot readily be studied in model organisms (36).
The current analysis demonstrates the importance of USP6’s deubiquitylating activity in the regulation of Wnt/β-catenin signaling. However, USP6 also contains a TBC domain, which binds to the ADP ribosylation 6 (ARF6) GTPase and promotes its activation in vivo independent of its USP domain (37). Recent studies have shown that ARF6 can activate Wnt signaling by stimulating MAPK1/ERK activation and phosphatidylinositide production to promote LRP6 phosphorylation (38, 39). In addition, ARF6 is reported to regulate endocytosis of Fzd receptors in the context of noncanonical Wnt signaling (40). Hence, both the deubiquitylase activity and the TBC/ARF6 interaction may contribute to the USP6-mediated activation of Wnt. This role of TBC/Arf6 interaction in regulating Frizzled endocytosis is consistent with our observation that in certain cell types, USP6(CS), an inactive point mutant of USP6 with an intact TBC domain, retains residual activity on β-catenin reporters (Fig. 2). Future studies will examine the role of USP6’s ability to regulate Arf6 in Wnt receptor trafficking and β-catenin activation.
USP6 translocations are the key etiological agent in two human tumors, ABC and nodular fasciitis (26, 27). In both, the USP6 rearrangement leads to overexpression of wild-type USP6 protein in a mesenchymal cell of origin. Although these tumors share some histological features, they exhibit distinct clinical behaviors (41): ABC is a locally aggressive tumor that most commonly arises in bone, whereas nodular fasciitis is a more benign s.c. lesion. Our recent studies in cellular and mouse models for these tumors revealed that NF-κB (23, 34) and signal transducer and activator of transcription 3 (STAT3) are two critical effectors of USP6 during tumorigenesis. Here, we identify Wnt signaling as an additional key pathogenic mediator of USP6, supported by the finding that inhibition of Wnt function markedly slows the growth of USP6-high tumors. Furthermore, microarray analysis of nodular fasciitis samples compared with other mesenchymal tumors supports the model in which USP6 activates a Wnt/β-catenin response in vivo. Consistent with the regulation of multiple pathways by USP6, nodular fasciitis tumors showed activation of gene signatures associated with Wnt/β-catenin and TGF-β signaling (Fig. 7 and Dataset S2), as well as NF-κB and STAT3 activation. The dysregulated Wnt/β-catenin pathway is also associated with a subset of cranial fasciitis, further supporting a key role for Wnt signaling in fibroproliferative lesions (42). Together, these results implicate the Wnt/β-catenin pathway as a potential therapeutic target for the treatment of human neoplasms with translocation-driven overexpression of USP6. Given the role of USP6 in regulating NF-κB, STAT3, and Wnt signaling pathways, inhibition of these pathways or inhibition of USP6 by small molecules may benefit patients with proliferative disorders, such as ABC or nodular fasciitis, that are driven by overexpression of USP6.
Materials and Methods
Tissue Culture.
HEK293T, HT1080, L-, HeLa, and Panc 08.13 cells were obtained from the American Type Culture Collection (ATCC) and grown in DMEM supplemented with 10% (vol/vol) FBS and 2 mM GlutaMAX (Life Technologies) in a 37 °C humidified incubator with 5% CO2. AsPC-1 cells from the ATCC were grown in RPMI supplemented with 10% FBS in a 37 °C humidified incubator with 5% CO2. Selection and passage of HEK293T and HT1080 stable cell lines harboring the Wnt/β-catenin reporter were performed with 1 μg/mL puromycin.
Primary and Secondary siRNA Screens.
Near-saturation genome-wide primary siRNA screens and secondary siRNA validation screens were performed as previously described (22, 43, 44). For both screens, cells were transfected with siRNAs for 96 h; 16 h before cell lysis, cells were treated with WNT3A-conditioned media. The HT1080 primary screen data presented in Fig. 1 have been previously reported (43).
Luciferase Reporter Assays.
For reporter assays in AsPC-1 and Panc 08.13 cells, cells were plated in 24-well plates and transiently transfected with a β-catenin reporter plasmid, Super 8xTOPFlash reporter, and 50 ng of control or WNT3A expression plasmid. Twenty-four hours after transfection, the cells were washed with PBS and lysed in 0.6% Nonidet P-40 in PBS containing protease inhibitors; β-catenin reporter activity was measured using firefly luciferase substrate (Promega). Cell viability was determined using Lactate Dehydrogenase assay (LDH), which was then used for normalizing the β-catenin reporter activity. For HEK293T and HT1080 cells, cells were transiently transfected in 48-well plates with 10 ng of Wnt/β-catenin reporter construct and 5 ng of control plasmid with Renilla luciferase driven by a constitutive cytomegalovirus promoter, using Lipofectamine 2000 following the manufacturer’s protocol (Life Technologies). Twenty-four to 36 h posttransfection, activation of the reporter was measured as the ratio of firefly to Renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). The hQR41 reporter was a kind gift from Jeffery Johnson (University of Wisconsin, Madison, WI). For siRNA experiments, cells with integrated Wnt/β-catenin firefly luciferase reporter and Renilla control reporter were transiently transfected with siRNA using RNAiMax or Dharmafect. Wnt/β-catenin reporter activity was measured 48–60 h posttransfection. GCmed1 and GCmed2 control stealth siRNAs were obtained from Life Technologies. siRNA sequences are listed in Table S1.
Table S1.
List of PCR primer and siRNA sequences
| Target | Sequence |
| siRNAs | |
| USP6-A siRNA | GGG AGA CAU AUA AGC ACA GUA GCA A |
| USP6-B siRNA | AUA AGU AUG UCC UUC CGC UCC UGU G |
| USP6-C siRNA | GAG AUU CCU GGA GUG GAA GUC AAU G |
| LRP6-A siRNA | ACG CAG CAU UGA GCG UGC CAA CAA A |
| LRP6-B siRNA | GAU CCC AUG GUU GGG UAC AUG UAU U |
| Control siRNA | CGU ACG CGG AAU ACU UCG ATT |
| Control siRNA | UCG AAG UAU UCC GCG UAC GTT |
| ZNRF3 siRNA | UCAAGAGGCCGGUGGUGUA |
| RNF43 siRNA | GCAGAACAGAAAGCUAUUA |
| CTNNB1 siRNA | CUA UCU GUC UGC UCU AGU ATT |
| AXIN1 siRNA | GGU GUU GGC AUU AAA GGU GTT |
| AXIN2 siRNA | GGG AGA AAU GCG UGG AUA CTT |
| PCR primers | |
| AXIN2 F1 | 5′-CTCCCCACCTTGAATGAAGA-3′ |
| AXIN2 R1 | 5′-TGGCTGGTGCAAAGACATAG-3′ |
| AXIN2 F2 | 5′- GCGATCCTGTTAATCCTTATCAC-3′ |
| AXIN2 R2 | 5′- AATTCCATCTACACTGCTGTC-3′ |
| RNF43 F | 5′-CTGACAGCAGTGGATCTGGAG-3′ |
| RNF43 R | 5′-CGTGCAGTTGACCACAGAGTC-3′ |
| HPRT F | 5′-CCTCACTGCTTTCCGGAGCGG-3′ |
| HPRT R | 5′-ATCGCTAATCACGACGCTGGGA-3′ |
| NKD1 F | 5′-TCGCCGGGATAGAAAACTACA-3′ |
| NKD1 R | 5′-CAGTTCTGACTTCTGGGCCAC-3′ |
| ZNRF3 F | 5′-TGAGAGTGTGACATTGTTGGAA-3′ |
| ZNRF3 R | 5′-GTAAAATCTGTGTGCAATTATCATGT-3′ |
| NKD1 F | 5′-GGAGAGAGTGAGCGAACCCT-3′ |
| NKD1 R | 5′-CTTGCCGTTGTTGTCAAAGTC-3′ |
| GAPDH F | 5′ATGGGGAAGGTGAAGGT-3′ |
| GAPDH R | 5′-AAGCTTCCCGTTCTCAG-3′ |
F, forward; R, reverse.
Immunoblot.
For Western blot analysis of LRP6, cells were transfected 48 h before lysis. Lysis buffer contained 0.1% Nonidet P-40, 0.1% SDS, 10% glycerol, 25 mM Tris⋅HCl, 0.25% sodium deoxycholate, 150 mM NaCl, 2 mM EDTA (pH 8.0), and protease inhibitor mixture (Roche). Equal amounts of proteins were resolved on a 10% SDS polyacrylamide gel and transferred to PVDF membranes. Western blots were performed according to standard methods. The pLRP6 (catalog no. 2568S), LRP6 (catalog no. 3395S), anti-rabbit IgG-HRP (catalog no. P0448), and anti-mouse IgG-HRP (catalog no. P0447) antibodies were obtained from Cell Signaling Technology, and the GSK3β and β-catenin antibodies were obtained from BD Transduction Labs (catalog no. 610201). Anti-p65 was obtained from Santa Cruz Biotechnology (sc-372). USP6 antibody was previously described (23). The blots were developed using SuperSignal West Dura substrate (Thermo Scientific) or enhanced chemiluminescence (Life Sciences). The images were captured digitally using the LAS-3000 Life Science Imager (Fujifilm).
RNA Isolation and Quantitative RT-PCR.
Total RNA was isolated from the cell lines using an RNeasy kit (Qiagen). RNA was reverse-transcribed with iScript reverse transcriptase (BioRAD) or RevertAid (Thermo Fisher Scientific). Real-time quantitative PCR was performed with SsoFast EvaGreen assay Supermix from BioRad. HPRT and GAPDH were used as housekeeping genes. The primers used are listed in Table S1.
Flow Cytometry.
For flow cytometric analysis of Fzds and LRP6, cells were seeded in six-well plates and transfected with pEGFP plasmid or plasmids expressing USP6 or its catalytically inactive variants. Forty-eight hours after transfection, cells were harvested and washed with PBS containing 2–5% FBS. Nonpermeabilized cells were then stained with anti-Fzd antibody (OMP18R5) (25) or 5 μg/mL LRP6 antibody (MAB1505; R&D Systems) for 45 min. Cells were washed with PBS, followed by incubation with goat anti-human IgG-APC (catalog no. 109-135-098; The Jackson Laboratory) or anti-mouse phycoerythrin (PE) (catalog no. 115-116-146; The Jackson Laboratory) secondary antibodies for Fzds and LRP6, respectively. The following antibodies were used as isotype controls: MAB003 (R&D Systems) for LRP6 and human IgG2 (AbD264; AbD Serotec) for Fzds. Samples were acquired on a BD Fortessa (BD Biosciences), and the data were analyzed using FlowJo software, version 10.0.7.
Ubiquitylation Assays.
For in vitro deubiquitylase assays, HeLa cells were cotransfected with Fzd5-V5 and FLAG-Ub. After overnight incubation with anti-V5 antibody (catalog no. A7345; Sigma), immunoprecipitates were washed three times in the lysis buffer [50 mM Tris (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 10% glycerol, 0.7 μg/mL pepstatin, 1 μg/mL leupeptin, 2 μg/mL aprotinin, 10 mM NEM, and 10 μM MG132] and with DUB assay buffer [20 mM Tris (pH 7.5), 100 mM NaCl, 0.05% Tween-20, 0.5 mg/mL BSA, and 5 mM β-mercaptoethanol]. The sample was divided into equal portions. One aliquot was immediately boiled in sample buffer, and the other aliquots were incubated with GST or GST-USP6 or its catalytically inactive mutant (catalog no. 64-0045-050; Ubiquigent) for 1 h at 37 °C. Samples were washed once in lysis buffer, resolved on a polyacrylamide gel, and blotted with anti-FLAG antibody (catalog no. F1804; Sigma).
To monitor Fzd5 ubiquitylation in vivo, 293T cells were cotransfected with Fzd5-V5 and FLAG-Ub, with or without USP6-HA plasmids. After lysis, Fzd was immunoprecipitated with anti-V5 beads overnight at 4 °C. Immunoprecipitated Fzd-V5 was resolved by SDS/PAGE, and the immunoblots were probed for Ub with anti-Flag antibody.
Animal Care.
BALB/c nude mice and NOD-SCID-gamma (NSG) mice were purchased from InVivos or The Jackson Laboratory. Animals were housed in standard cages and were allowed access ad libitum to food and water. The Duke-NUS Institutional Animal Care and Use Committee (IACUC) and Children’s Hospital of Philadelphia IACUC approved all of the animal studies.
Tumor Implantation and Treatment of Mice.
NIH 3T3 (ATCC) and Rh3 (a gift from Peter Houghton, University of Texas Health Science Center, San Antonio) cells were stably transfected with doxycycline-inducible USP6 expression plasmids as previously described (23). For the NIH 3T3 lines, 2 × 106 cells were s.c. injected into NOD-SCID mice; for the Rh3, 2 × 106 cells were suspended in 50% Matrigel and injected s.c. into flanks of NSG mice. Following development of palpable tumors, mice were treated with Wnt-C59 formulated in 50% PEG400 (vol/vol) in water administered by oral gavage at a dosing volume of 10 μL/g of body weight as previously described (24). Mice were maintained on 1 mg/mL doxycycline-containing water during the study duration.
Microarray and Gene Set Enrichment Analysis.
Microarray analysis was performed on nine nodular fasciitis tumors with USP6 translocation and 27 additional tumors, including three each of ARMS, dermatofibroma or benign fibrous histiocytoma, dermatofibrosarcoma protuberans, synovial sarcoma, embryonal rhabdomyosarcoma, melanoma, neurofibroma, and malignant peripheral nerve sheath tumor; one gastrointestinal stromal tumor; and two schwannomas. Total RNA was extracted using a miRNeasy FFPE kit (Qiagen), and microarray analysis was performed using a Human WG-DASL assay with Human HT12 v4.0 BeadChips (Illumina), containing 29,377 probes. Data filtering, normalization, and analysis were performed using Illumina GenomeStudio and Partek Genomics Suite software. A total of 20,818 probes remained after filtering. Differentially expressed genes in nodular fasciitis compared with other tumor types were analyzed using ANOVA. P values were corrected for multiple comparison. According to source of variance analyses, the microarray batch (array slide of 12 samples) was also included as a variable in the ANOVA. Gene set enrichment analysis was performed using the Hallmark gene set from the Broad Institute.
Data Analysis.
Data were analyzed using Prism, version 5.0 (GraphPad). Significance for all tests was set at P ≤ 0.05 unless otherwise stated (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 in all instances).
Other Supporting Information Files
Supplementary Material
Acknowledgments
This research is supported, in part, by the National Research Foundation Singapore and administered by the Singapore Ministry of Health’s National Medical Research Council under the Singapore Translational Research Investigator (STAR) Award Program (to D.M.V.). The University of North Carolina (UNC) Flow Cytometry Core Facility is supported, in part, by a National Cancer Institute Center Core Support Grant (P30CA016086) to the UNC Lineberger Comprehensive Cancer Center. M.B.M. is supported, in part, by the NIH (New Innovator Award 1-DP2-OD007149-01). M.P.W. received support from the NIH (Grant T32-CA009156-35). M.M.C. is supported, in part, by NIH Grants RO1 CA168452 and R21 CA178601. R.T.M. is an investigator of the Howard Hughes Medical Institute. A.M.O. is supported, in part, by Department of Laboratory Medicine and Pathology Mayo Clinic Award 2014.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605691113/-/DCSupplemental.
References
- 1.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 3.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13(12):767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb Perspect Biol. 2012;4(12):1–23. doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4(11):1–22. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sewduth RN, et al. The ubiquitin ligase PDZRN3 is required for vascular morphogenesis through Wnt/planar cell polarity signalling. Nat Commun. 2014;5:4832. doi: 10.1038/ncomms5832. [DOI] [PubMed] [Google Scholar]
- 7.Wei W, Li M, Wang J, Nie F, Li L. The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol Cell Biol. 2012;32(19):3903–3912. doi: 10.1128/MCB.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao H-X, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 9.Koo B-K, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 10.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108(28):11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ong CK, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44(6):690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2013;110(31):12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madan B, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. August 10, 2015 doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tauriello DVF, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37(5):607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Schlesiger C, Masucci MG, Lindsten K. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J Cell Mol Med. 2009;13(8B):1886–1895. doi: 10.1111/j.1582-4934.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lui TTH, et al. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/β-catenin signaling. Mol Cell Biol. 2011;31(10):2053–2065. doi: 10.1128/MCB.01094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran H, Hamada F, Schwarz-Romond T, Bienz M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22(4):528–542. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun S-I, et al. Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol Oncol. 2015;9(9):1834–1851. doi: 10.1016/j.molonc.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, et al. Deubiquitinase USP47/UBP64E Regulates β-Catenin Ubiquitination and Degradation and Plays a Positive Role in Wnt Signaling. Mol Cell Biol. 2015;35(19):3301–3311. doi: 10.1128/MCB.00373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukai A, et al. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29(13):2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Major MB, et al. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal. 2008;1(45):ra12. doi: 10.1126/scisignal.2000037. [DOI] [PubMed] [Google Scholar]
- 23.Ye Y, et al. TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF-kappaB. Oncogene. 2010;29(25):3619–3629. doi: 10.1038/onc.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proffitt KD, et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73(2):502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- 25.Gurney A, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci USA. 2012;109(29):11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson-Johnson MR, et al. Nodular fasciitis: A novel model of transient neoplasia induced by MYH9-USP6 gene fusion. Lab Invest. 2011;91(10):1427–1433. doi: 10.1038/labinvest.2011.118. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira AM, et al. USP6 (Tre2) fusion oncogenes in aneurysmal bone cyst. Cancer Res. 2004;64(6):1920–1923. doi: 10.1158/0008-5472.can-03-2827. [DOI] [PubMed] [Google Scholar]
- 28.Kenny PA, Enver T, Ashworth A. Receptor and secreted targets of Wnt-1/beta-catenin signalling in mouse mammary epithelial cells. BMC Cancer. 2005;5:3. doi: 10.1186/1471-2407-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labbé E, et al. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67(1):75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 30.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang DY, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68(16):6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onder TT, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 33.Lau AW, et al. TRE17/ubiquitin-specific protease 6 (USP6) oncogene translocated in aneurysmal bone cyst blocks osteoblastic maturation via an autocrine mechanism involving bone morphogenetic protein dysregulation. J Biol Chem. 2010;285(47):37111–37120. doi: 10.1074/jbc.M110.175133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pringle LM, et al. Atypical mechanism of NF-κB activation by TRE17/ubiquitin-specific protease 6 (USP6) oncogene and its requirement in tumorigenesis. Oncogene. 2012;31(30):3525–3535. doi: 10.1038/onc.2011.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funakoshi Y, Chou MM, Kanaho Y, Donaldson JG. TRE17/USP6 regulates ubiquitylation and trafficking of cargo proteins that enter cells by clathrin-independent endocytosis. J Cell Sci. 2014;127(Pt 21):4750–4761. doi: 10.1242/jcs.156786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulding CA, Ruvolo M, Haber DA. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc Natl Acad Sci USA. 2003;100(5):2507–2511. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinu L, et al. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol Cell Biol. 2004;24(22):9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellon-Cardenas O, Clancy J, Uwimpuhwe H, D’Souza-Schorey C. ARF6-regulated endocytosis of growth factor receptors links cadherin-based adhesion to canonical Wnt signaling in epithelia. Mol Cell Biol. 2013;33(15):2963–2975. doi: 10.1128/MCB.01698-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim W, et al. ADP-ribosylation factors 1 and 6 regulate Wnt/β-catenin signaling via control of LRP6 phosphorylation. Oncogene. 2013;32(28):3390–3396. doi: 10.1038/onc.2012.373. [DOI] [PubMed] [Google Scholar]
- 40.Onishi K, et al. Antagonistic functions of Dishevelleds regulate Frizzled3 endocytosis via filopodia tips in Wnt-mediated growth cone guidance. J Neurosci. 2013;33(49):19071–19085. doi: 10.1523/JNEUROSCI.2800-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira AM, Chou MM. USP6-induced neoplasms: The biologic spectrum of aneurysmal bone cyst and nodular fasciitis. Hum Pathol. 2014;45(1):1–11. doi: 10.1016/j.humpath.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Rakheja D, et al. A subset of cranial fasciitis is associated with dysregulation of the Wnt/beta-catenin pathway. Mod Pathol. 2008;21(11):1330–1336. doi: 10.1038/modpathol.2008.112. [DOI] [PubMed] [Google Scholar]
- 43.Conrad W, et al. FAM129B is a novel regulator of Wnt/β-catenin signal transduction in melanoma cells. F1000Res. 2013;2:134. doi: 10.12688/f1000research.2-134.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung N, et al. A 1,536-well ultra-high-throughput siRNA screen to identify regulators of the Wnt/β-catenin pathway. Assay Drug Dev Technol. 2010;8(3):286–294. doi: 10.1089/adt.2009.0262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.