Significance
Antimicrobial peptides exert antimicrobial, antifungal, antiviral, and antiprotozoan activity. They are expressed at high concentrations at the intestinal mucosal surface, where they play a crucial role in intestinal homeostasis. Therefore, approaches aiming to boost expression of antimicrobial peptides represent a future therapeutic strategy to treat infections and dysbiosis-driven diseases in humans at a time of increasing incidence of antibiotic resistance.
Keywords: colonic mucosa, epithelial cells, antimicrobial peptides, epigenetic, acetylation
Abstract
Antimicrobial peptides (AMP) are defense effectors of the innate immunity playing a crucial role in the intestinal homeostasis with commensals and protection against pathogens. Herein we aimed to investigate AMP gene regulation by deciphering specific characteristics allowing their enhanced expression among innate immune genes, particularly those encoding proinflammatory mediators. Our emphasis was on epigenetic regulation of the gene encoding the AMP β-defensin 2 (HBD2), taken as a model of possibly specific induction, upon challenge with a commensal bacterium, compared with the proinflammatory cytokine IL-8. Using an in vitro model of colonic epithelial cells challenged with Escherichia coli K12, we showed that inhibition of histone deacetylases (HDAC) by trichostatin A dramatically enhanced induction of HBD2 expression, without affecting expression of IL-8. This mechanism was supported by an increased phosphorylation of histone H3 on serine S10, preferentially at the HBD2 promoter. This process occurred through activation of the IκB kinase complex, which also led to activation of NF-κB. Moreover, we demonstrated that NF-κB was modified by acetylation upon HDAC inhibition, partly by the histone acetyltransferase p300, and that both NF-κB and p300 supported enhanced induction of HBD2 expression. Furthermore, we identified additional genes belonging to antimicrobial defense and epithelial restitution pathways that showed a similar pattern of epigenetic control. Finally, we confirmed our finding in human colonic primary cells using an ex vivo organoid model. This work opens the way to use epigenetic pharmacology to achieve induction of epithelial antimicrobial defenses, while limiting the deleterious risk of an inflammatory response.
Antimicrobial peptides (AMP) are efficient defense components of the innate immunity. They play a crucial role in the mucosal homeostasis and protection against microbes. In the colon, AMP are produced and secreted by epithelial cells. Most genes encoding these defense peptides are inducible in response to various signals. Regulation of inducible genes encompasses genetic and epigenetic mechanisms taking place at the chromatin level; among them, remodeling of chromatin between relatively “open” and “closed” forms plays a key role. Such remodeling results from the modification of nucleosomal structures. Nucleosomes constitute the fundamental unit of chromatin; they comprise approximately two turns of DNA wound around a histone octamer. A range of modifications of the amino-terminal “tails” of histone proteins are involved in this process, including methylation, phosphorylation, or acetylation (1). These modifications also occur within the “globular” domain of histones, which establish extensive contacts with DNA strands. The consequence of such modifications on gene expression depends on the amino acid residues targeted and their close environment. Perturbing the balance between these modifications leads to changes in gene expression (2). Recent publications provide evidence for the effect of histone modifications on regulation of the innate immune response and expression of associated defense genes (3, 4).
Among histone modifications, acetylation and deacetylation play a crucial role in transcriptional regulation of genes (5). The acetylation status of histone proteins is determined by the opposing actions of histone acetyl-transferases (HAT) and histone deacetylases (HDAC). HAT add acetyl groups to the ε-amino group of lysine residues of nucleosomal histones, whereas HDAC remove these acetyl groups. In most cases, a positive correlation can be established between the level of histone acetylation and transcriptional activity. Acetylation of histones by HAT promotes a relaxed structure of the chromatin by decreasing the positive charges interacting with negatively charged DNA strands, thereby facilitating transcriptional activation. Conversely, HDAC act as transcriptional repressors, because of histone deacetylation, and consequently promote chromatin condensation. In humans, 18 HDAC have been identified and classified based on their homology to yeast HDAC (6). Most of them are zinc-dependent proteins and their enzymatic activity can be inhibited by compounds, such as trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA) (7, 8). On the other hand, HAT have been classified by families, based on their cellular localization and primary structure homology, and include the well-known p300 family (9).
Despite their name, a large number of nonhistone proteins have been identified as substrates for both HAT and HDAC. Many of these proteins are transcription factors involved in the regulation of gene expression, including the transcription factor NF-κB, which regulates a wide range of genes involved in the host innate immune response (10, 11). Reversible acetylation of the p65 subunit regulates diverse functions of NF-κB, including DNA binding and transcriptional activity, as well as its ability to associate with the cytoplasmic inhibitor IκBα (12). Seven acetylated lysines have been identified within p65 (residues K122, K123, K218, K221, K310, K314, and K315). The majority of these residues are acetylated by the HAT p300 (13). For example, acetylation of K310 is required for full transcriptional activity of NF-κB (14). Conversely, several HDAC, including HDAC1, HDAC3, and SIRT1, have been found to specifically deacetylate p65, thereby negatively regulating the transcriptional activity of NF-κB (12).
Most genes involved in the innate immune response are inducible genes whose expression needs to be tightly regulated and rapidly and specifically activated in response to diverse stimuli (15). This is the case at the human colonic mucosal surface. Colonic epithelial cells, being the first line of interaction with microbes, are endowed with innate immune functions encompassing the balanced expression of an array of genes, including those encoding AMP and proinflammatory cytokines. These two groups of genes are generally considered to be synchronously expressed under the necessity to protect the epithelium against pathogenic microbes and keep commensal bacteria at bay, away from the epithelial surface. We hypothesized, however, that these two groups of genes might also obey differential regulatory rules that do not necessarily imply their synchronous expression: for example, in the case of the response to a pathogenic microbe that necessitates the mobilization of both arms of the innate immune system, or in the case of controlling and tolerating the microbiota that will rather mobilize expression of antimicrobial molecules (pathological versus physiological inflammation).
In an attempt to identify the additional regulatory circuit that may disconnect the expression of these two groups of genes when epithelial cells are engaged by a bacterium, we hypothesized that the expression of some of the genes encoding AMP, such as the β-defensin–encoding gene HBD2, may be “protected” by epigenetic mechanisms, in comparison with the bona fide proinflammatory genes. We accordingly conducted an in-depth analysis aiming to compare the epigenetic regulation of a restricted set of genes that are characteristic of the two groups—genes encoding inducible AMP (β-defensins HBD2, HBD3, and cathelicidin LL37) and genes encoding proinflammatory cytokines (IL-1B, IL-8, and chemokine CCL20 or TNF)—in the human colonic epithelial cell line Caco-2, subclone TC7, and in human colonic primary cells. Using the Escherichia coli K12 commensal bacterium or purified flagellin as inducers, we reveal that expression of HBD2 is greatly enhanced in vitro upon HDAC inhibition by TSA, whereas expression of IL-8 is not modified. We investigate the molecular mechanism underlying this observation and identify an increased phosphorylation of the histone H3 protein on the serine S10 residue that occurs preferentially at the HBD2 promoter. We show that this HDAC inhibition-dependent process happens through activation of the IκB kinase (IKK) complex, which ultimately leads to activation of NF-κB. Moreover, we demonstrate that NF-κB is posttranslationally acetylated on the p65 K310 lysine residue, partly by the HAT p300, and that both NF-κB and p300 mediate the enhanced induction of HBD2 expression. Furthermore, we identify other genes from the antimicrobial defense and epithelial restitution pathways that show a similar expression pattern upon HDAC inhibition, using an RNA sequencing approach. Finally, we validate ex vivo the enhanced expression of AMP genes upon HDAC inhibition in human colonic primary cells, using organoids from normal colon tissues. This work highlights the existence of a differential regulatory mechanism occurring in colonic epithelial cells between antimicrobial and proinflammatory genes, which takes place at the chromatin level, through an acetylation-dependent process.
Results
HDAC Inhibition Enhances AMP Expression in Cells Challenged with E. coli.
To analyze the expression of inducible genes of the innate immune response, such as genes encoding AMP (HBD2, HBD3, LL37) and proinflammatory cytokines (IL-1B, IL-8, CCL20), we challenged human colonic epithelial cells Caco-2, subclone TC7 (16), with the commensal strain E. coli K12. Experiments were performed on confluent cell monolayers using a ratio of 10 bacteria per cell. RNA was extracted at different time points after challenge and analyzed by quantitative RT-PCR (qRT-PCR). In nonchallenged cells, we detected a basal expression level for all of the studied genes. Alternatively, challenge of cells with E. coli was followed by strong transcriptional induction of the HBD2, IL-8, IL-1B, and CCL20 genes (Fig. 1A and Fig. S1A). In contrast, expression of HBD3 and LL37 was not induced throughout the time course (Fig. S1A). As a control, cellular viability was not influenced by bacterial challenges, as assessed by lactate dehydrogenase assay (Fig. S1B). Collectively, these results demonstrate that most genes encoding AMP and proinflammatory cytokines are simultaneously expressed and induced in colonic epithelial cells exposed to a commensal bacterium.
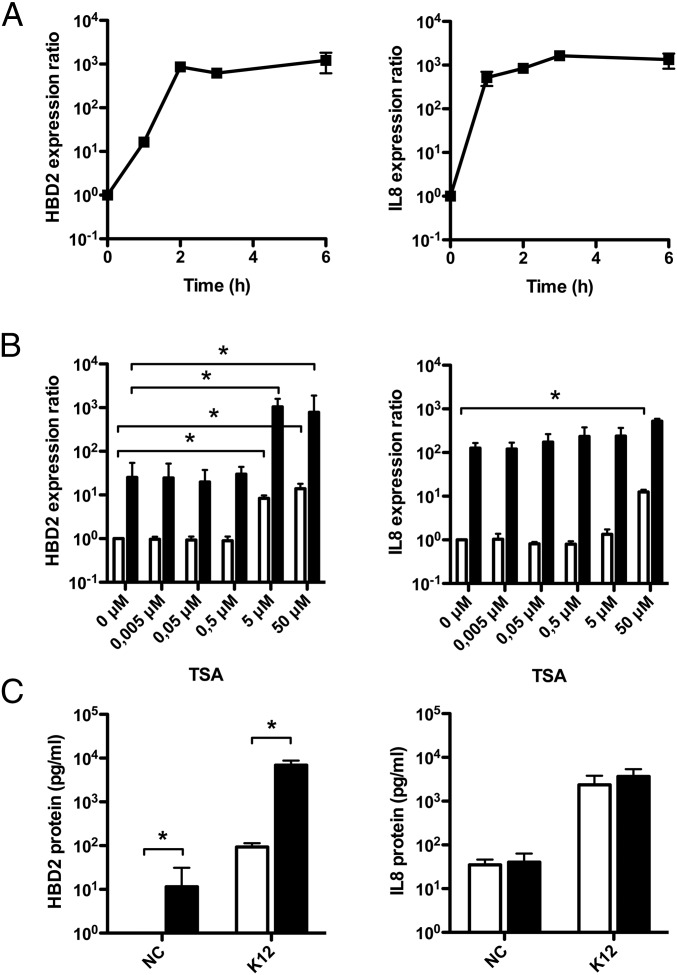
Fig. 1.
HDAC inhibition enhances expression of the beta-defensin HBD2 upon bacterial challenge. (A) Transcriptional expression of genes encoding HBD2 and IL-8 in cells challenged with the E. coli K12 commensal strain. Values are presented on a logarithmic scale as the ratio of gene expression in challenged cells compared with nonchallenged cells. (B) Transcriptional expression of the HBD2 and IL-8 genes in cells pretreated for 16 h with increasing concentrations of the HDAC inhibitor TSA (0–50 μM), and then challenged for 2 h with E. coli. Values are presented on a logarithmic scale as the ratio of gene expression in pretreated and challenged cells (black bars), or pretreated and nonchallenged cells (white bars), compared with nontreated and nonchallenged cells. *P < 0.01 evaluated by Mann–Whitney u test. (C) ELISA dosage of the HBD2 and IL-8 peptides in supernatants of cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not for 6 h with E. coli. Bacterial challenges were stopped by addition of gentamicin, and supernatants were collected 24 h after the beginning of the challenge. Values are presented on a logarithmic scale in picogram of peptide per milliliter. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain. Black bars, 5 μM TSA pretreated cells; white bars, nontreated cells. *P < 0.01 evaluated by Mann–Whitney u test.
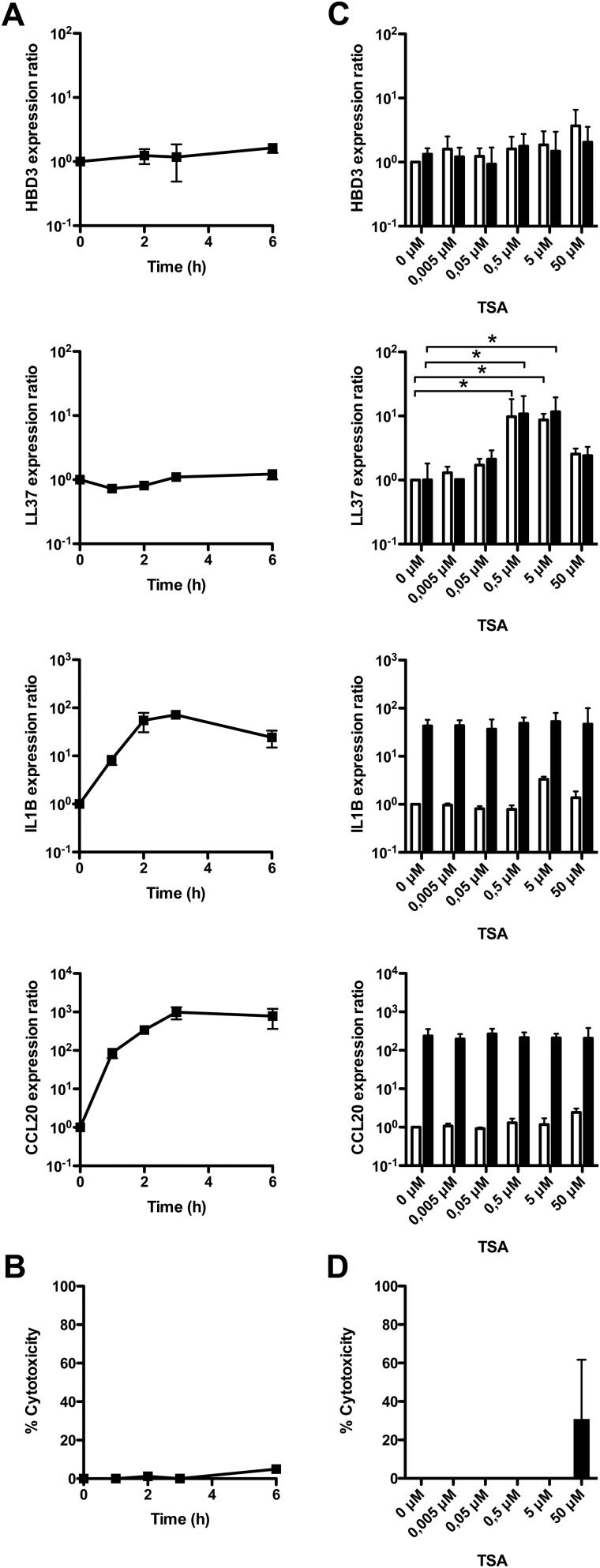
Fig. S1.
TSA-mediated HDAC inhibition enhances expression of AMP upon bacterial challenge. (A) Transcriptional expression of the AMP HBD3 and LL37 genes, and proinflammatory IL-1B and CCL20 genes, in cells challenged with the Escherichia coli K12 commensal strain. Values are presented on a logarithmic scale as the ratio of gene expression in challenged cells compared with nonchallenged cells. (B) Cellular viability of cells challenged with E. coli. Lactate dehydrogenase assays were performed using supernatants of challenged cells. Values represent the percentage of cell death. (C) Transcriptional expression of HBD3, LL37, IL-1B, and CCL20, in cells pretreated for 16 h with increasing concentrations of TSA (0–50 μM), and challenged for 2 h with E. coli. Values are presented on a logarithmic scale as the ratio of gene expression in pretreated and challenged cells (black bars), or pretreated and nonchallenged cells (white bars), compared with nontreated and nonchallenged cells. *P < 0.01 evaluated by Mann–Whitney u test. (D) Cellular viability of cells pretreated for 16 h with increasing concentrations of TSA (0–50 μM), and then challenged for 2 h with E. coli. Lactate dehydrogenase assays were performed using supernatants from pretreated and challenged cells. Values represent the percentage of cell death.
To investigate the role of epigenetic modifications in the mechanisms regulating expression of inducible AMP and proinflammatory genes, we tested the effect of inhibiting several chromatin-modifying enzymes. Dose–response assays were carried out overnight on confluent cell monolayers using inhibitors of DNA methylation (azacytidine), histone demethylation (pargyline), or histone deacetylation (TSA, SAHA). The inhibitor-containing medium was removed and E. coli challenge was performed to induce expression of the studied genes. RNA was extracted at 2 h after challenge and analyzed by qRT-PCR. In these experimental conditions, we found that inhibition of DNA methylases or histone demethylases had no effect on the expression of AMP and proinflammatory genes, regardless of the inhibitor concentration used. In contrast, inhibition of HDAC with TSA or SAHA had a significant impact on the induction of gene expression in a group-specific manner. Induction of the antimicrobial genes HBD2 (Fig. 1B and Fig. S2A) and LL37 (Figs. S1C and S2A) was remarkably enhanced in a concentration dependent manner, whereas proinflammatory genes did not respond to the inhibitors (Fig. 1B and Figs. S1C and S2A). Quantitatively, bacterial-dependent induction of HBD2 expression was close to 10,000-fold after pretreatment with 5 μM TSA, compared with 100-fold without inhibitor (Fig. 1B). In contrast, induction of IL-8 expression remained the same for all TSA concentrations (Fig. 1B). Interestingly, this enhancement of AMP gene expression upon HDAC inhibition was also observed in nonchallenged cells (Fig. 1B and Figs. S1C and S2A), in agreement with previous reports on stimulation of LL37 expression with sodium butyrate, which has been shown to express HDAC inhibitory function (17). As a control, cellular viability was not affected by the inhibitors, except at the highest concentration (Figs. S1D and S2B). Taken together, these results show that HDAC inhibition has a differential impact on the expression of AMP and proinflammatory genes in colonic epithelial cells.
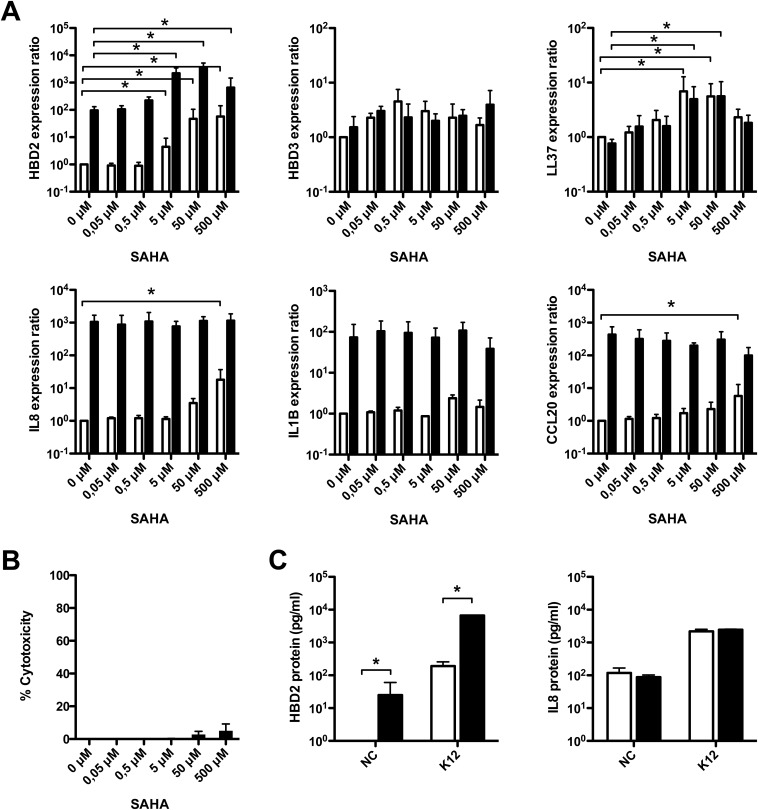
Fig. S2.
SAHA-mediated HDAC inhibition enhances expression of AMP upon bacterial challenge. (A) Transcriptional expression of the AMP HBD2, HBD3, and LL37 genes, and the proinflammatory IL-8, IL-1B, and CCL20 genes, in cells pretreated for 16 h with increasing concentrations of SAHA (0–500 μM SAHA), and then challenged for 2 h with the E. coli K12 strain. Values are presented on a logarithmic scale as the ratio of gene expression in pretreated and challenged cells (black bars), or pretreated and nonchallenged cells (white bars), compared with nontreated and nonchallenged cells. *P < 0.01 evaluated by Mann–Whitney u test. (B) Cellular viability of cells pretreated for 16 h with increasing concentrations of SAHA (0–500 μM SAHA), and then challenged for 2 h with E. coli. Lactate dehydrogenase assays were performed using supernatants from pretreated and challenged cells. Values represent the percentage of cell death. (C) ELISA dosage of the HBD2 and IL-8 peptides in supernatants of cells pretreated or not for 16 h with 5 μM SAHA, and then challenged or not for 6 h with E. coli. Bacterial challenges were stopped by addition of gentamicin, and supernatants were collected 24 h after the beginning of the challenge. Values are presented on a logarithmic scale in picograms of peptide per milliliter. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain. Black bars, 5 μM SAHA pretreated cells; white bars, nontreated cells. *P < 0.01 evaluated by Mann–Whitney u test.
To further characterize the enhanced induction of the HBD2 gene expression upon HDAC inhibition, we investigated secretion of HBD2 peptide. For this purpose, cells were pretreated with TSA or SAHA and challenged for 6 h with E. coli. Then, bacteria were killed by gentamicin treatment, and ELISAs were performed on cell supernatants collected 24 h later to quantify HBD2 peptide secretion. In supernatants of nonchallenged cells, the HBD2 peptide was only detected after TSA or SAHA pretreatment, showing that the 10-fold induction of the HBD2 gene expression observed subsequently to HDAC inhibition was followed by peptide production and secretion (Fig. 1C and Fig. S2C). In supernatants of challenged cells, dosage of the HBD2 peptide revealed a concentration reaching 10,000 pg/mL after pretreatment with TSA or SAHA, compared with 100 pg/mL for nontreated cells (Fig. 1C and Fig. S2C). In contrast, secretion of IL-8 was similar in both conditions (Fig. 1C and Fig. S2C). Collectively, these results demonstrate that HDAC inhibition stimulates transcription of AMP genes—as well as secretion of these defense peptides—without modification of proinflammatory cytokine expression, in colonic epithelial cells.
HDAC Inhibition Activates the IKK Complex and Increases Phosphorylation of Histone H3 on the Serine S10 at the HBD2 Promoter.
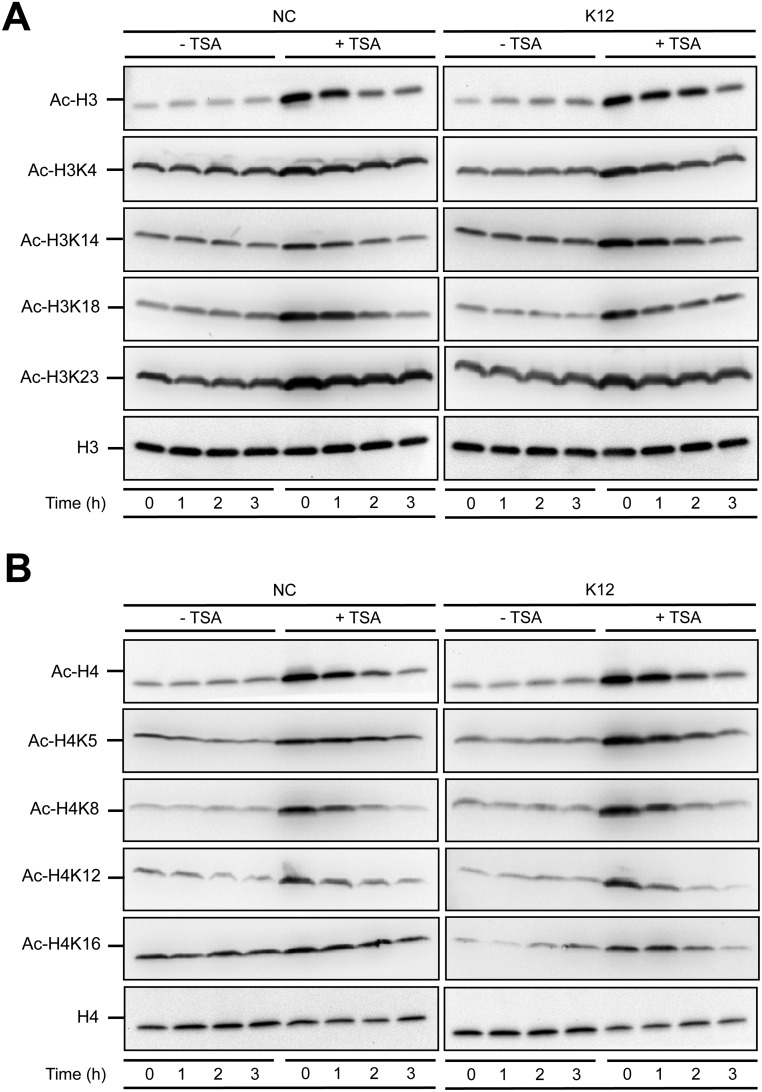
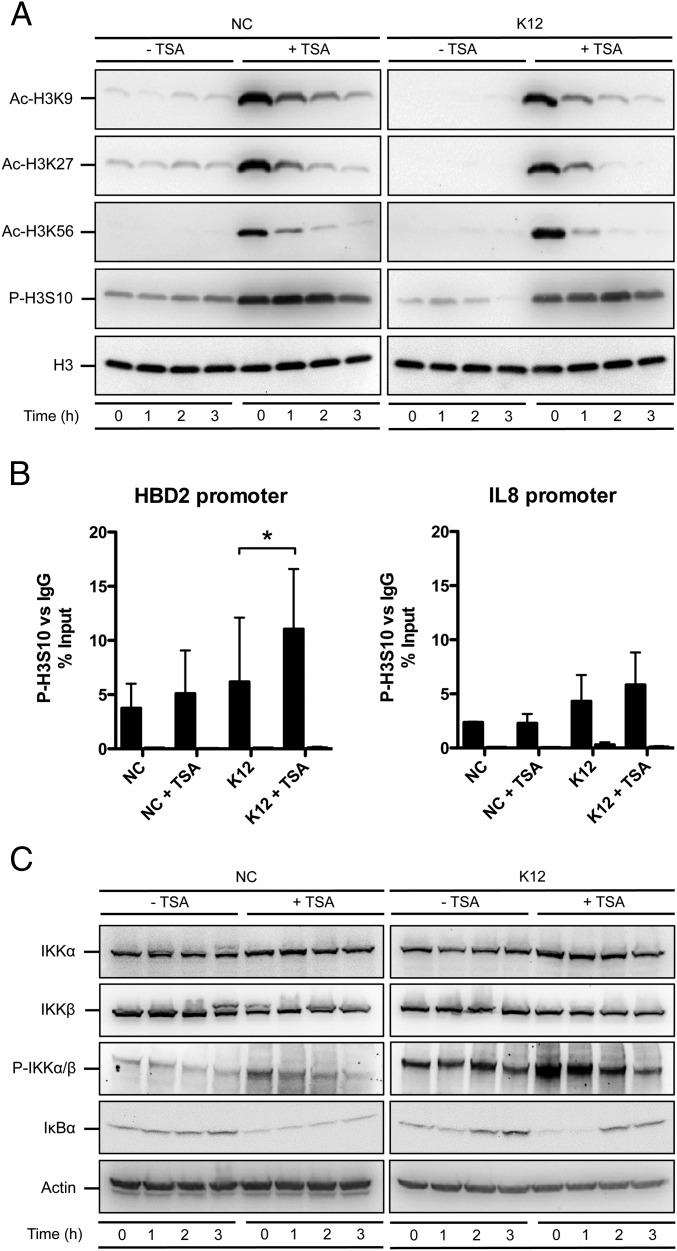
Based on these observations, we sought to determine the molecular mechanisms leading to enhance induction of HBD2 expression upon HDAC inhibition. Several studies have suggested that accessibility of transcription factors to chromatin, especially on the promoters of innate immunity genes, requires acetylation of histones H3 and H4 on several lysine residues, and phosphorylation of histone H3 at serine S10 (H3S10) (3, 18). We therefore investigated the consequence of HDAC inhibition on the occurrence of these histone marks. By immunoblot, we identified two classes of lysine residues that were differentially acetylated in an E. coli-independent manner following TSA pretreatment. The first class included lysines from histones H3 (residues H3K4, H3K14, H3K18, and H3K23) or H4 (residues H4K5, H4K8, H4K12, and H4K16) that were constitutively acetylated and poorly affected by TSA (Fig. S3). The second class encompassed lysines that were only weakly acetylated at basal state, but whose acetylation level was strongly enhanced after TSA pretreatment, such as residues H3K9, H3K27, and H3K56 (Fig. 2A). In addition, using an antibody against phosphorylated H3S10, we observed a strong increase of this phosphorylation mark following TSA pretreatment in nonchallenged and in challenged cells (Fig. 2A). Together, these results indicate that HDAC inhibition by TSA pretreatment has a differential impact on the acetylation status of histone lysine residues, and dramatically enhances the phosphorylation level of the H3S10 residue.
Fig. S3.
HDAC inhibition modifies the acetylation pattern of histone H3 and H4 proteins. (A) Immunoblot analysis of histone H3 protein acetylation marks on lysine residues K9+K14+K18+K23+K27, K4, K14, K18, and K23, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. After lysis of cells at the indicated time points, Western blots were performed using specific antibodies directed against histone posttranslational modification marks. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; “Ac” prefix, acetylation. (B) Immunoblot analysis of histone H4 protein acetylation marks on lysine residues K5+K8+K12+K16, K5, K8, K12, and K16, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. After lysis of cells at the indicated time points, Western blots were performed using specific antibodies directed against histone posttranslational modification marks. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; “Ac” prefix, acetylation.
Fig. 2.
HDAC inhibition activates the IKK complex and induces phosphorylation of the histone H3 protein. (A) Immunoblot analysis of histone H3 acetylation on lysine K9, K27 and K56, and phosphorylation on serine S10, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. After lysis of cells at the indicated time points, Western blots were performed using antibodies directed against histone posttranslational modification marks. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; “Ac” prefix, acetylation; “P” prefix, phosphorylation. (B) ChIP analysis of the phosphorylated H3S10 protein at the HBD2 and IL-8 promoters, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not for 1 h with E. coli. Enrichment in chromatin was detected using anti–P-H3S10 antibody or rabbit IgG as control, and quantified by qRT-PCR using specific primers matching the HBD2 or IL-8 promoters. Values are presented as the percentage of signal relative to the input. NC, nonchallenged cells; NC+TSA, nonchallenged cells pretreated with 5 μM TSA; K12, cells challenged with the E. coli K12 strain; K12+TSA, cells pretreated with 5 μM TSA and challenged with the E. coli K12 strain. Black bars, signal detected using the P-H3S10 antibody; white bars, signal detected using the IgG control. *P < 0.05 evaluated by Mann–Whitney u test. (C) Immunoblot analysis of the IKKα and IKKβ proteins, the IKKα/β complex phosphorylation, and the IκBα protein, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. After lysis of cells at the indicated time points, Western blots were performed using antibodies directed against proteins or posttranslational modification marks. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; “P” prefix, phosphorylation.
We next investigated whether HDAC inhibition increased the level of H3S10 phosphorylation, preferentially at the HBD2 gene promoter. We pretreated cells overnight with 5 μM TSA, followed by 1-h challenge with E. coli, and carried out ChIP assays using either antibody directed against phosphorylated H3S10 (P-H3S10), or an irrelevant IgG of the same isotype as a control. The size of sonicated chromatin fragments was ∼150–900 bp, corresponding to one to five nucleosomes. Data were analyzed by qRT-PCR, using the ribosomal protein RPL30 housekeeping gene as a control. In nonchallenged cells, we observed a weak increase of the P-H3S10 signal at the HBD2 promoter in TSA pretreated cells, compared with nontreated cells (Fig. 2B). Interestingly, in cells challenged with E. coli, we found a twofold enrichment of P-H3S10 at the HBD2 promoter following TSA pretreatment, compared with nontreated and challenged cells (Fig. 2B). In contrast, the P-H3S10 signal remained unchanged in both conditions at the IL-8 promoter (Fig. 2B). Collectively, these data show that the impact of HDAC inhibition on the level of H3S10 phosphorylation is gene-specific.
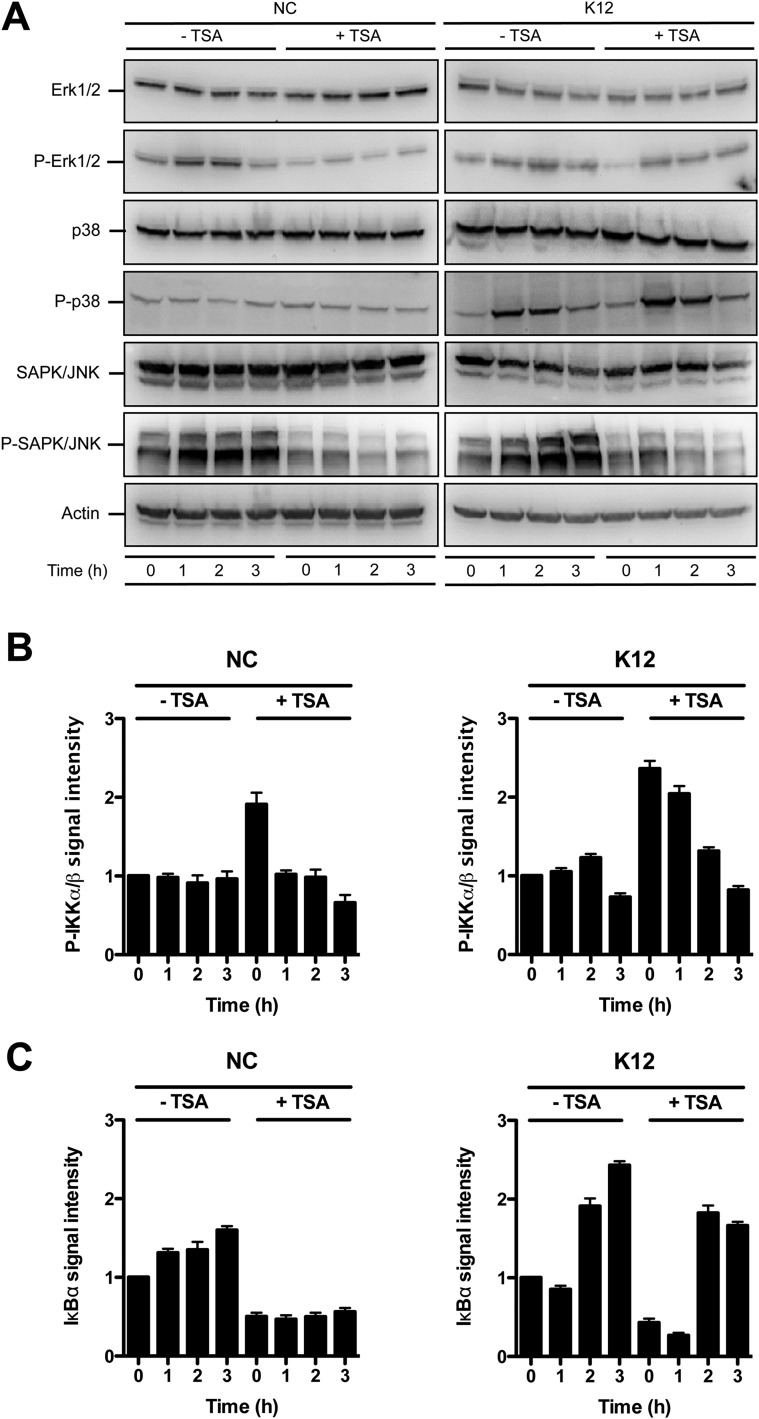
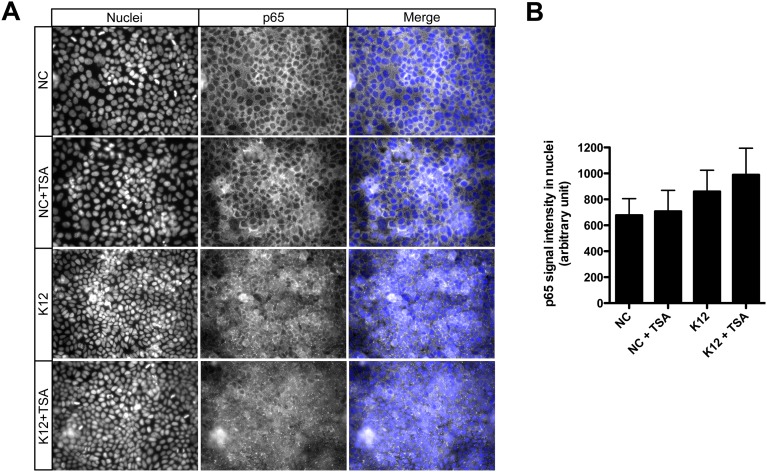
We then investigated the signaling pathway by which TSA-mediated HDAC inhibition induces phosphorylation of H3S10. We analyzed the canonical phosphorylation of histone H3 through activation of the MAPK signaling pathway. However, TSA failed to induce activation of Erk1/2, p38, or SAPK/JNK, as determined by phosphorylation level (Fig. S4A). We therefore considered the alternative IKKα/β complex pathway. Indeed, several studies have demonstrated that IKKα functions as a H3S10 kinase regulating chromatin structure and facilitating gene expression (19, 20). By immunoblot, using an antibody detecting phosphorylated IKKα/β, we observed an increased phosphorylation of the IKK complex in nonchallenged cells following TSA pretreatment (Fig. 2C and Fig. S4B). This TSA-dependent phosphorylation of IKKα/β was further enhanced in cells challenged with E. coli (Fig. 2C and Fig. S4B). Furthermore, the phosphorylated IKK complex phosphorylates the NF-κB inhibitor IκBα, resulting in its degradation. This process releases NF-κB and allows its translocation into the nucleus (21). In agreement, following TSA pretreatment, we observed a strong decrease of the IκBα protein by immunoblot (Fig. 2C and Fig. S4C) and an increase of the NF-κB p65 subunit signal in nuclei by immunofluorescence (Fig. S5). Collectively, these results indicate that TSA-mediated HDAC inhibition promotes induction of H3S10 phosphorylation, preferentially at the HBD2 promoter, and activates the NF-κB pathway through the IKK complex.
Fig. S4.
Phosphorylation status of the Erk, p38, and SAPK/JNK MAPK upon HDAC inhibition. (A) Immunoblot analysis of Erk1/2, phosphorylated Erk1/2, p38, phosphorylated p38, SAPK/JNK, and phosphorylated SAPK/JNK, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. After lysis of cells at the indicated time points, Western blots were performed using specific antibodies directed against proteins or posttranslational modification marks. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; “P” prefix, phosphorylation. (B and C) Quantification of the signal intensity from immunoblot analysis of the IKKα/β complex phosphorylation, and the IκBα protein, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. Quantification was performed using the gel analysis method from the ImageJ software. Signal detected at 0 h in nontreated and nonchallenged cells was used as reference to determine the relative intensities. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain.
Fig. S5.
Translocation of the NF-κB p65 subunit in nuclei upon HDAC inhibition. (A and B) Detection and quantification of the p65-associated signal in nuclei by immunofluorescence experiments, in cells pretreated for 16 h with 5 μM TSA, and then challenged for 30 min with E. coli. Magnification in A: 40×. NC, nonchallenged cells; NC+TSA, nonchallenged cells pretreated with 5 μM TSA; K12, cells challenged with the E. coli K12 strain; K12+TSA, cells treated with TSA and challenged with the E. coli K12 strain.
NF-κB Is Acetylated upon HDAC Inhibition and Mediates the Enhanced Induction of HBD2 Expression.
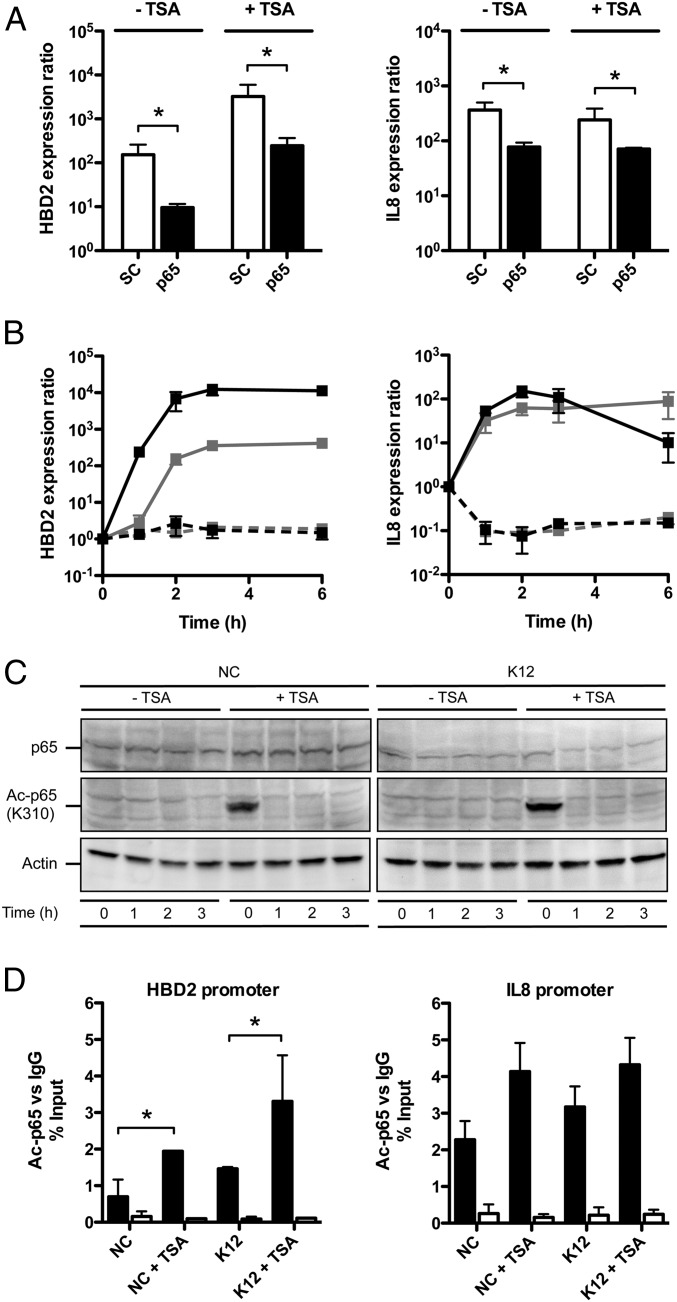
A current model suggests that H3S10 phosphorylation accounts for a histone structure that favors the binding of chromatin-remodeling enzymes, thereby increasing promoter accessibility of NF-κB on a specific set of innate immune genes (18). Both the HBD2 and IL-8 promoters harbor NF-κB binding sites (22, 23). Therefore, we investigated its involvement in the enhanced induction of HBD2 expression upon TSA-mediated HDAC inhibition. Using a pool of siRNA silencing expression of the NF-κB p65 subunit, we found that transcription of HBD2 and IL-8 was dependent on NF-κB upon challenge with E. coli in TSA pretreated and nontreated cells. In both conditions, with an efficacy of p65 silencing reaching 80%, expression of HBD2 and IL-8 was reduced at least 10-fold, compared with cells transfected with the scrambled siRNA negative control (Fig. 3A). To confirm these results by an alternative approach, we tested the effect of the chemical inhibitor BMS-345541 on expression of HBD2 and IL-8. This compound specifically inhibits the NF-κB pathway by targeting IKK complex activation (24). Without BMS-345541, HBD2 expression was enhanced at least 10-fold in TSA pretreated cells, compared with nontreated cells, as soon as 1-h postbacterial challenge (Fig. 3B). In contrast, IL-8 expression showed no difference between TSA pretreated cells and nontreated cells upon bacterial challenge, as previously observed (Figs. 1B and 3B). Strikingly, with BMS-345541, expression of HBD2 and IL-8 was completely abolished, both in TSA pretreated and nontreated cells (Fig. 3B). Collectively, these results demonstrate that NF-κB controls the induction of HBD2 and IL-8 expression and mediates enhancement of HBD2 expression upon TSA-mediated HDAC inhibition.
Fig. 3.
NF-κB is acetylated upon HDAC inhibition and mediates the enhanced induction of the HBD2 gene expression. (A) Transcriptional expression of the HBD2 and IL-8 genes in cells transfected with siRNA silencing the p65 NF-κB subunit, pretreated or not for 16 h with 5 μM TSA, and finally challenged for 2 h with E. coli. Values are presented on a logarithmic scale as the ratio of gene expression in challenged cells, pretreated or not, compared with nonchallenged and nontreated cells. *P < 0.01 evaluated by Mann–Whitney u test. Black bars, p65 siRNA-transfected cells; white bars, scrambled (SC) siRNA-transfected cells. (B) Kinetic of HBD2 and IL-8 expression in cells inhibited for the NF-κB pathway by treatment with BMS-345541, pretreated or not for 16 h with 5 μM TSA, and then challenged with E. coli. Values are presented on a logarithmic scale as the ratio of gene expression in TSA pretreated and challenged cells (black lines) or nontreated and challenged cells (gray lines), compared with nontreated and nonchallenged cells. Dashed lines: with NF-κB inhibitor BMS-345541; solid lines: without NF-κB inhibitor BMS-345541. (C) Immunoblot analysis of the p65 protein, and the p65 K310 acetylated protein, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not with E. coli. After lysis of cells at the indicated time points, Western blots were performed using antibodies directed against the protein with or without its modification mark. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; “Ac” prefix, acetylation. (D) ChIP analysis of the K310 acetylated p65 protein at the HBD2 and IL-8 promoters, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not for 1 h with E. coli. Enrichment in chromatin was detected using an anti–Ac-p65 antibody or rabbit IgG as control, and quantified by qRT-PCR using specific primers matching the HBD2 or IL-8 promoters. Values are presented as the percentage of signal relative to the input. NC, nonchallenged cells; NC+TSA, nonchallenged cells pretreated with 5 μM TSA; K12, cells challenged with the E. coli K12 strain; K12+TSA, cells pretreated with 5 μM TSA and challenged with the E. coli K12 strain. Black bars, signal detected using the Ac-p65 antibody; white bars, signal detected using the IgG control. *P < 0.05 evaluated by Mann–Whitney u test.
Recent studies indicate that posttranslational modifications of NF-κB, especially on the p65 subunit, play a critical role in fine-tuning its activity, adding another level of complexity to the regulation by this transcription factor. Among them, reversible acetylation of p65 on the lysine K310 residue regulates several functions of NF-κB, including its full transcriptional activity at the targeted promoters (12). To determine whether NF-κB is acetylated upon TSA-mediated HDAC inhibition, we performed an immunoblot assay and detected an acetylation of p65 on K310 after TSA pretreatment (Fig. 3C). This acetylation mark was most visible within the first hour following pretreatment of cells, in an E. coli-independent manner. In addition, we investigated the specific recruitment of Ac-p65 at the HBD2 and IL-8 promoters by ChIP. In nonchallenged cells, as well as in cells challenged for 1 h with E. coli, we found a twofold enrichment of Ac-p65 at the HBD2 promoter after TSA pretreatment (Fig. 3D). In contrast, the Ac-p65 signal did not significantly change upon TSA pretreatment at the IL-8 promoter (Fig. 3D). Taken together, these data show that TSA-mediated HDAC inhibition leads to enhanced recruitment of the acetylated NF-κB p65 subunit, preferentially at the HBD2 promoter, compared with the IL-8 promoter.
p300 Acetylates NF-κB and Supports the Enhanced Induction of HBD2 Expression upon HDAC Inhibition.
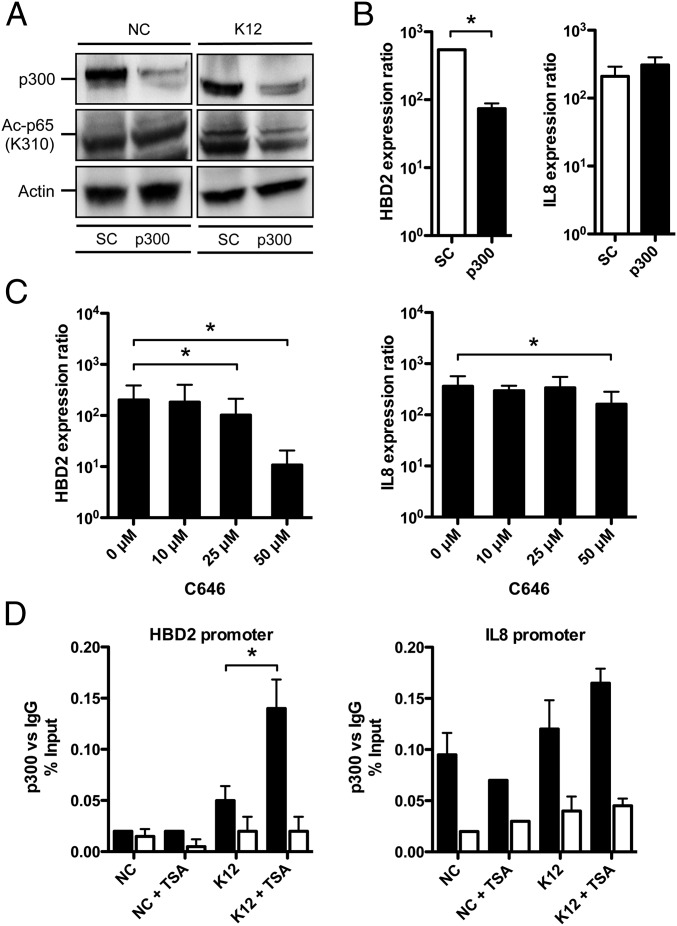
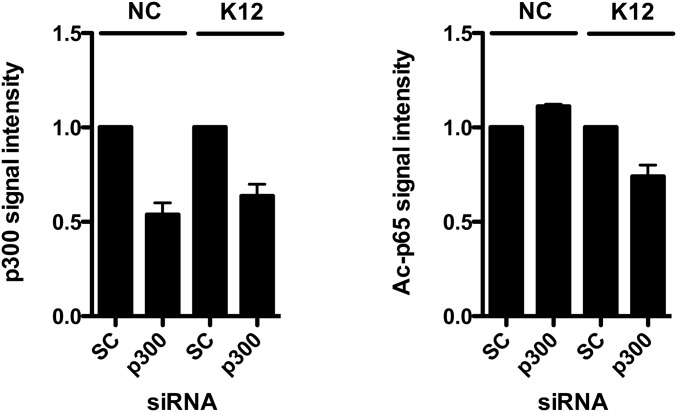
It has recently been demonstrated in human embryonic kidney cells 293T that acetylation of the NF-κB p65 subunit at the lysine K310 residue is performed by the HAT p300 (14, 25). We therefore investigated the involvement of the p300-mediated acetylation of p65 on HBD2 expression. By immunoblot, using a pool of siRNA silencing expression of p300 with 85% efficiency, we found that p65 was less acetylated on the K310 lysine upon E. coli K12 challenge, compared with cells transfected with the scrambled siRNA negative control (Fig. 4A and Fig. S6). As a control, the basal level of p65 acetylation at the K310 residue was found similar in nonchallenged cells, for both silencing conditions (Fig. 4A and Fig. S6). Taken together, these results confirm that p300 takes part in the acetylation process of the NF-κB p65 subunit in colonic epithelial cells.
Fig. 4.
p300 acetylates NF-κB and supports the enhanced induction of the HBD2 gene expression upon HDAC inhibition. (A) Immunoblot analysis of the HAT p300, and p65 protein acetylation on the K310 lysine, in cells transfected with siRNA silencing p300, and challenged for 1 h with E. coli K12. After lysis of cells, Western blots were performed using specific antibodies directed against proteins or posttranslational modification marks. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; SC, scrambled siRNA-transfected cells; p300, p300 siRNA-transfected cells; “Ac” prefix, acetylation. (B) Transcriptional expression of HBD2 and IL-8 in cells transfected with siRNA silencing p300, and challenged for 2 h with E. coli. Values are presented on a logarithmic scale as the ratio of gene expression in challenged cells compared with nonchallenged cells. *P < 0.01 evaluated by Mann–Whitney u test. Black bars, p300 siRNA-transfected cells; white bars, scrambled (SC) siRNA-transfected cells. (C) Transcriptional expression of HBD2 and IL-8 in cells treated with increasing concentrations of the p300 inhibitor C646 (0–50 μM C646), and challenged for 2 h with E. coli. Values are presented on a logarithmic scale, as the ratio of gene expression in challenged cells, compared with nonchallenged cells. *P < 0.01 evaluated by Mann–Whitney u test. (D) ChIP analysis of p300 recruitment at the HBD2 and IL-8 promoters, in cells pretreated or not for 16 h with 5 μM TSA, and then challenged or not for 1 h with E. coli. Enrichment in chromatin was detected using anti-p300 antibody or rabbit IgG as control, and quantified by qRT-PCR using specific primers matching the HBD2 or IL-8 promoters. Values are presented as the percentage of signal relative to the input. NC, nonchallenged cells; NC+TSA, nonchallenged cells pretreated with 5 μM TSA; K12, cells challenged with the E. coli K12 strain; K12+TSA, cells pretreated with 5 μM TSA and challenged with the E. coli K12 strain. Black bars, signal detected using the p300 antibody; white bars, signal detected using the IgG control. *P < 0.05 evaluated by Mann–Whitney u test.
Fig. S6.
Quantification of the signal intensity from immunoblot analysis of the p300 and Ac-p65 proteins. Quantification was performed using the gel analysis method from the ImageJ software. Signal detected in cells transfected with scrambled siRNA was used as reference to determine the relative intensities. NC, nonchallenged cells; K12, cells challenged with the E. coli K12 strain; SC, scrambled siRNA-transfected cells; p300, p300 siRNA-transfected cells; “Ac” prefix, acetylation.
At the transcriptional level, this observation was correlated with a sevenfold decreased induction of HBD2 expression upon p300 silencing, compared with cells transfected with the scrambled siRNA negative control (Fig. 4B). To confirm these results by an alternative approach, we tested the effect of the chemical inhibitor C646 on the expression of HBD2 and IL-8. This compound is a specific inhibitor of p300 (26). Dose–response assays were carried out and showed that induction of HBD2 expression by bacterial challenge was impacted at a concentration of C646 as low as 25 μM, and was reduced more than 10-fold at the highest dose, compared with nontreated cells (Fig. 4C). In contrast, expression of IL-8 was much less affected by the decreased amount or activity of p300 (Fig. 4 B and C). Next, recruitment of p300 at the HBD2 and IL-8 promoters was quantified by ChIP. In nonchallenged cells, the p300-associated signal was measured quite similar in TSA pretreated cells, compared with nontreated cells, at both HBD2 and IL-8 promoters (Fig. 4D). Interestingly, in challenged cells, we found a threefold enhanced recruitment of p300 at the HBD2 promoter after TSA pretreatment, compared with nontreated cells (Fig. 4D). In contrast, the p300-associated signal did not show significant differences between TSA pretreated and nontreated cells at the IL-8 promoter (Fig. 4D). Collectively, these data demonstrate that p300 acetylates p65 and supports the enhanced induction of HBD2 expression upon TSA-mediated HDAC inhibition through its increased recruitment at the HBD2 promoter.
Expression of Additional Genes Belonging to the Antimicrobial Defense and Epithelial Restitution Is Enhanced upon HDAC Inhibition.
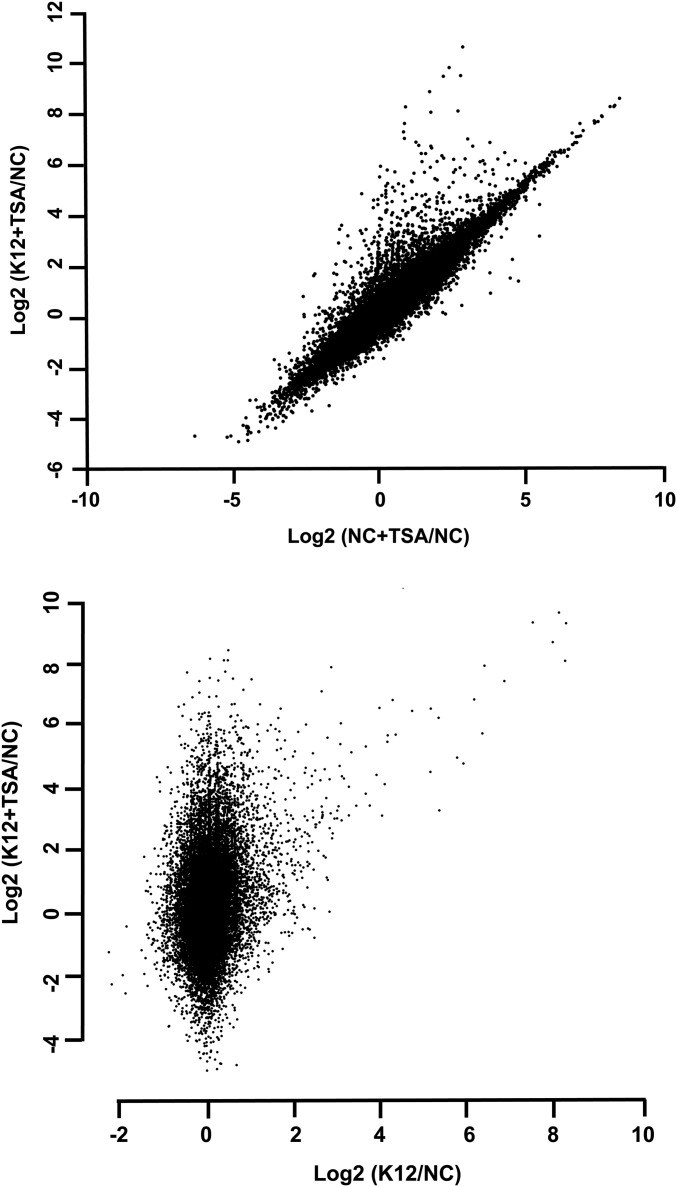
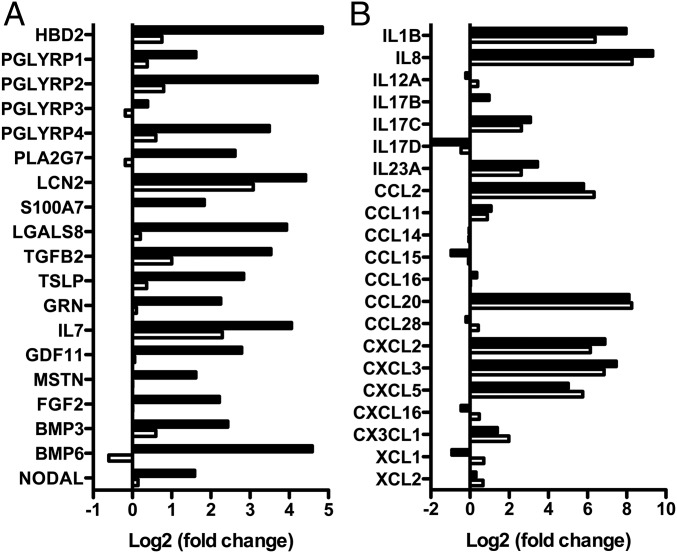
Beside molecular aspects, we tested whether the enhanced induction of HBD2 expression observed upon TSA-mediated HDAC inhibition truly reflected a differential regulatory mechanism existing between antimicrobial and proinflammatory genes. For this purpose, a transcriptomic analysis of cells pretreated or not with 5 μM TSA and challenged for 2 h with E. coli was performed. RNA was extracted and analyzed by RNA sequencing using Illumina technology. The fold-change calculations representative of the whole-gene expression were determined and presented on scatterplots (Fig. S7). For further investigation, we focused on genes from the innate immune pathway, including antimicrobials, cytokines, and chemokines (Fig. 5). From these sets of genes, we identified a total of 58 genes whose expression was significantly modulated in cells upon bacterial challenge. Interestingly, in addition to HBD2, we identified 18 other genes whose expression was enhanced in cells pretreated with TSA compared with nontreated cells (Fig. 5A). Those were genes belonging to the antimicrobial defense, such as: PGLYRP1–4, encoding the four human peptidoglycan recognition proteins, which target the bacterial cell wall; PLA2G7, encoding the phospholipase A2 that hydrolyses bacterial membrane phospholipids; and LCN2, encoding the lipocalin iron sequestration protein. Remarkably, besides these genes encoding antimicrobial defense molecules, we observed the enhanced expression of additional genes belonging to the epithelial restitution machinery upon HDAC inhibition. These genes encompassed TGFB2, TSLP, and IL7 (Fig. 5A). TGFB2 is a member of the TGF-β family of cytokines involved in the promotion of intestinal homeostasis. TSLP is a hemopoietic cytokine promoting T-helper type 2 cell responses that are associated with immunity to moderate intestinal inflammation. IL-7 is produced locally by intestinal epithelial cells and may serve as a regulatory factor for intestinal mucosal lymphocytes. In comparison, the expression of genes belonging to the bona fide proinflammatory pathways was not significantly modified in challenged cells pretreated or not with TSA, as measured for IL-1B and IL-8 and most of the genes encoding proinflammatory cytokines (Fig. 5B). Taken together, these results show that TSA-mediated HDAC inhibition enhances expression of a rather restricted group of innate immunity genes encoding molecules involved in antimicrobial defenses and epithelial restitution, without increasing the proinflammatory gene pathways.
Fig. S7.
HDAC inhibition enhances expression of other genes from the antimicrobial defense and epithelial restitution pathways. Transcriptional analysis of the whole genome of cells pretreated or not for 16 h with 5 μM TSA, and then challenged for 2 h with the E. coli K12 strain. After RNA extraction, RNA sequencing was performed on each sample to determine the number of reads per gene. Values are presented on a logarithmic scale as the ratio of gene expression in TSA pretreated cells compared with nontreated cells (x axis), against gene expression in TSA pretreated and challenged cells compared with nontreated and nonchallenged cells (y axis), and as the ratio of gene expression in challenged cells compared with nonchallenged cells (x axis), against gene expression in TSA pretreated and challenged cells compared with nontreated and nonchallenged cells (y axis).
Fig. 5.
HDAC inhibition enhances expression of other genes from the antimicrobial defense and epithelial restitution pathways. Transcriptional analysis of genes from the antimicrobial defense (A), the epithelial restitution (A), and the proinflammatory (B) pathways, in TC7 cells pretreated for 16 h with 5 μM TSA and challenged for 2 h with E. coli, using an RNA sequencing approach. Values are presented on a logarithmic scale as the ratio of gene expression in challenged cells compared with nonchallenged cells (white bars), or TSA pretreated and challenged cells compared with nontreated and nonchallenged cells (black bars).
Expression of Antimicrobial Peptide Genes Is Enhanced upon HDAC Inhibition in Human Colonic Primary Cells.
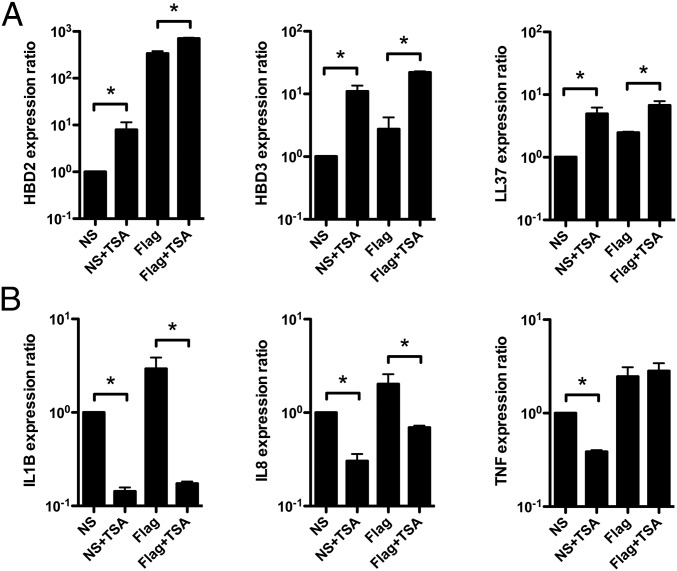
Finally, we investigated the relevance of our finding in human colonic primary cells, using ex vivo cultured organoids of crypts from normal colon tissues. We analyzed expression of a set of genes from the antimicrobial defense and proinflammatory pathways in 6-d-old organoids treated or not with 5 μM TSA, and stimulated or not with 5 μg/mL flagellin to mimic a bacterial challenge, for 24 h (Fig. 6). Crypts were isolated from normal human colons and embedded in Matrigel in the presence of growth factors to induce the formation of 3D structures, termed “organoids.” Organoids recapitulate an intact architecture, harboring an internal lumen, stem cells, which are located in surface protusions that correspond to novel crypts, and the different epithelial lineages, including colonocytes (27). RNA was extracted and analyzed by qRT-PCR using TaqMan assays. In nontreated and nonstimulated organoids, we detected a basal expression level for all of the studied genes. Treatment of organoids with TSA alone was followed by transcriptional induction of the HBD2, HBD3, and LL37 antimicrobial genes, with ratios ranging from 5- to 11-fold (Fig. 6A). In contrast, expression of the IL-1B, IL-8, and TNF proinflammatory genes was decreased (Fig. 6B). Additionally, stimulating organoids with flagellin alone induced expression of all genes from the two classes (Fig. 6). Strikingly, stimulating organoids with flagellin upon inhibition of histone deacetylases with TSA had a differential impact on the gene expression in a group-specific manner. Induction of HBD2, HBD3, and LL37 antimicrobial genes was enhanced, whereas expression of proinflammatory genes was either decreased for IL-1B and IL-8, or not modified in the case of TNF. Interestingly, this decrease of proinflammatory cytokine expression upon HDAC inhibition was previously reported in a work describing the in vivo anti-inflammatory properties of SAHA in LPS-stimulated mice (28). Collectively, these data validate the existence of specific epigenetic regulations allowing the enhancement of antimicrobial peptide gene expression among genes from the innate immune response, particularly in contrast to those encoding proinflammatory mediators, in human colonic primary cells.
Fig. 6.
HDAC inhibition enhances expression of antimicrobial peptide genes in human colonic primary cells. Transcriptional expression of the HBD2, HBD3, and LL37 antimicrobial peptide genes (A), and IL-1B, IL-8, and TNF proinflammatory genes (B), in 6-d-old human colonic organoids treated or not with 5 μM TSA, and stimulated or not with 5 μg/mL flagellin, for 24 h. Values are presented on a logarithmic scale as the ratio of gene expression in organoids treated or not with TSA, stimulated or not with flagellin, compared with nontreated and nonstimulated organoids. NS, nonstimulated organoids; NS+TSA, nonstimulated organoids treated with 5 μM TSA; Flag, organoids stimulated with 5 μg/mL flagellin; Flag+TSA, organoids treated with 5 μM TSA and stimulated with 5 μg/mL flagellin. *P < 0.05 evaluated by Mann–Whitney u test.
Discussion
The human intestinal epithelium maintains a protective barrier function between the host and its microbiota. This barrier protects against invasion and systemic dissemination of colonizing microorganisms. Among the diverse actors that contribute to establishment and maintenance of epithelial homeostasis, AMPs play a crucial regulatory role; they are ubiquitously expressed by epithelial cells throughout the intestinal tract and keep the resident and transient bacterial populations in check (29). These effectors participate in the innate immune response to commensals under steady-state conditions, a situation of tolerance actively maintained by commensals themselves (30). Epithelial cells orchestrate this dialogue mainly through signaling pathways, which result for one part in activation of histone modifying enzymes and remodeling complexes (31). These pathways induce the expression of hundreds of genes with different functions and therefore different regulatory requirements (32). Because these genes are induced by the same pathways, their expression must be regulated by gene-specific rather than signal-specific mechanisms. This is an essential adaptive element of the innate immune response that is based on epigenetic processes (33).
One link between bacterial sensing and its epigenetic consequences has been described for the MAPK cascades, whose activation leads to phosphorylation of H3S10. Both, the Erk and p38 kinases have been shown to induce phosphorylation of H3S10 at the promoter of activated genes (34). Here, we observed an increase of this histone mark in cells challenged with E. coli. Unexpectedly, we also found this increase in nonchallenged cells after TSA-mediated HDAC inhibition; this occurred without significant activation of Erk or p38. Therefore, we speculated that TSA-mediated HDAC inhibition was able to activate an alternative pathway inducing H3S10 phosphorylation. One potential candidate was the IKK pathway that can, on one hand, mediate this process through the IKKα kinase, and on the other hand, activate transcription of NF-κB-responsive genes by phosphorylating and targeting its inhibitor IκBα for proteasomal degradation (19, 20). In agreement, we observed a decrease of IκBα protein levels in nonchallenged cells after TSA pretreatment. The current evidence supports the hypothesis that phosphorylation of H3S10 is a predisposing mark for histone acetylation, a mark for active transcription (35). As such, we observed a strong increase in the acetylation of the H3K9, H3K27, and H3K56 residues after TSA-mediated HDAC inhibition, three hallmarks of active promoters. Therefore, it is tempting to speculate about the role of all of these histone modification marks and their combination. Because we have shown that the increase of H3S10 phosphorylation happens preferentially at the HBD2 promoter upon TSA-mediated HDAC inhibition, it would be of interest to determine whether these histone acetylation marks also occur differently at the HBD2 and IL-8 promoters, which would demonstrate a correlation between their appearance and a gene-specific expression at varying degrees.
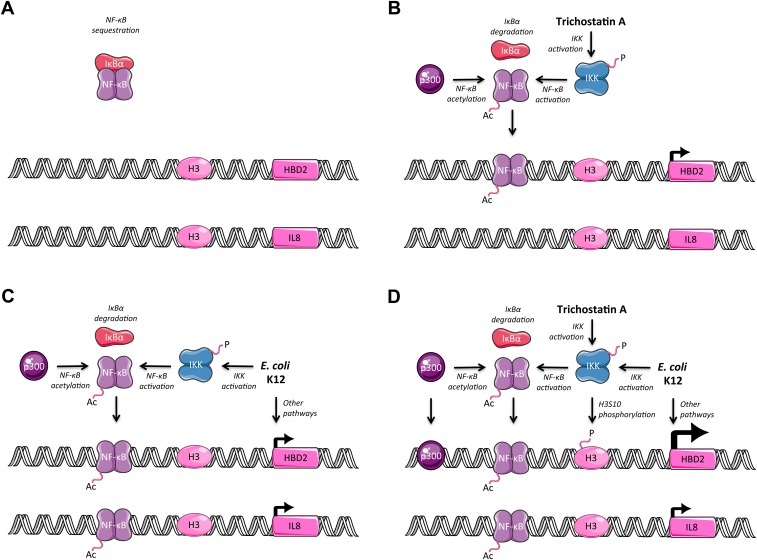
Phosphorylation of H3S10, as well as acetylation of H3 lysines, are highlighted in a current model as discrete modifications promoting chromatin remodeling at the promoter of specific innate immune genes, allowing a precise recruitment of NF-κB (18). Beside this chromatin-conditioned control, NF-κB activity is also subjected to additional regulatory mechanisms; among them, acetylation of NF-κB subunits on specific lysine residues play distinct roles in regulating its DNA-binding ability and transcriptional activity (11). Interestingly, these posttranslational modifications resemble not only those occurring on histone proteins, but also share the same key enzymes, such as HAT and HDAC. Recent studies showed that TSA-mediated HDAC inhibition prolonged the presence of the p65 subunit in the nucleus of cells and enhanced its binding activity to DNA, possibly through its increased acetylation (36, 37). Here, we have observed a strong acetylation of p65 on the K310 lysine upon HDAC inhibition. Therefore, it is tempting to hypothesize that increased acetylation of NF-κB strengthens its nuclear location, and above all its activity; this would be in agreement with the enhanced expression of the HBD2 gene observed at the basal state, as well as its increased induction in cells upon bacterial challenge, after TSA-mediated HDAC inhibition. Moreover, several studies have shown that, once it binds DNA, NF-κB can recruit HAT to targeted promoters where these enzymes change protein acetylation profiles, functioning as coactivators for gene transcription (38, 39); among them, p300 has been shown to interact with the transcriptional activation domain of p65. Because we demonstrated that p300 supports induction of HBD2 expression and takes part in acetylation of p65 at the K310 residue, it would be of interest to investigate whether p300 also acetylates other p65 lysines known to be involved in the NF-κB function. Conversely, looking for other HAT able to acetylate the K310 lysine of p65 would give a better idea about the specific role of this mark in the fine-tuning of the NF-κB activity. This would give more insight into the role of p300, NF-κB, and its acetylation status, in the differential induction of the HBD2 and IL-8 gene expression observed upon HDAC inhibition (Fig. S8).
Fig. S8.
Summarizing figure. (A) In nontreated and nonchallenged cells, the HBD2 and IL-8 promoters are not induced; NF-κB is sequestered by IκBα. (B) In TSA pretreated cells, the IKK complex is phosphorylated, IκBα is degraded, NF-κB is acetylated and detected at the HBD2 promoter at an increased level, and expression of the HBD2 gene is observed. (C) In E. coli K12-challenged cells, the IKK complex is phosphorylated, IκBα is degraded, NF-κB is acetylated and detected at the HBD2 and IL-8 promoters, we speculate that other pathways like MAPK are activated, and expression of the HBD2 and IL-8 genes is detected. (D) In TSA pretreated and E. coli K12-challenged cells, the IKK complex is phosphorylated, IκBα is degraded, NF-κB is acetylated and detected at the IL-8 promoter and at the HBD2 promoter at an increased level, phosphorylation of serine 10 on histone H3 is increased at the HBD2 promoter, along with increased recruitment of p300 at the HBD2 promoter, we speculate that other pathways like MAPK are activated, and expression of the IL-8 gene and enhanced induction of the HBD2 gene are detected.
Several HDAC inhibitors have already been evaluated as therapeutic compounds with activities in cancer therapies (40). Recently, molecules derived from those inhibitors have been shown to have applications beyond cancer therapies, based on their additional properties (41). Apart from applications in oncology, considerable research effort has been aimed at evaluating the potential of these inhibitory molecules as therapeutics for neurodegenerative disorders, cardiac hypertrophy, and asthma. However, except for HIV, hepatitis, and malaria infection, the therapeutic potential of HDAC inhibitors for the treatment of infectious diseases has not been largely investigated. One example is the HDAC inhibitor sodium butyrate, which can stimulate expression of the endogenous AMP cathelicidin in the colon and promote the clearance of the enteropathogen Shigella (42). Along the same line, we have demonstrated the property of the HDAC inhibitor TSA to enhance expression of the AMP β-defensin 2 without increasing the level of IL-8 in vitro and ex vivo, using human colonic primary cells. This observation highlights the possibility to disconnect expression of AMP from proinflammatory cytokines in colonocytes, at the epigenetic level. In addition, we found that HDAC inhibition has a positive impact on expression of other genes from the epithelial defense and restitution pathways upon a bacterial challenge. This finding suggests a potential role of these inhibitors in immune therapies through epigenetic pathways. Understanding the coordinated interplay between epigenetic regulation, gene expression, and environment, will allow translation of this fundamental knowledge into development of innovative pharmacoepigenetic treatments.
Materials and Methods
Antibodies.
We used the rabbit polyclonal antibodies anti–NF-κB p65 subunit (SC-109, Santa Cruz; ab7970, Abcam), anti–NF-κB p65 acetyl K310 (ab52175, Abcam), anti-IκBα (SC-371, Santa Cruz), anti-IKKα (2682, Cell Signaling), anti-IKKβ (2370, Cell Signaling), antiphospho-IKKα (S176)/IKKβ (S177) (2697, Cell Signaling), antihistone H3 (ab1791, Abcam), antiacetyl-histone H3 K9+14+18+23+27 (ab47915, Abcam), antiacetyl-histone H3 K4 (07-539, Millipore), antiacetyl-histone H3 K9 (ab4441, Abcam), antiacetyl-histone H3 K14 (07-353, Millipore), antiacetyl-histone H3 K18 (ab1191, Abcam), antiacetyl-histone H3 K23 (07-355, Millipore), antiacetyl-histone H3 K27 (ab4729, Abcam), antiacetyl-histone H3 K56 (07-677-I, Millipore), antihistone H4 (ab7311, Abcam), antiacetyl-histone H4 K5+8+12+16 (06-866, Millipore), antiacetyl-histone H4 K5 (07-327, Millipore), antiacetyl-histone H4 K8 (07-328, Millipore), antiacetyl-histone H4 K12 (07-595, Millipore), antiphospho-histone H3 S10 (ab5176, Abcam), anti-SAPK/JNK (9252, Cell Signaling), antiphospho-SAPK/JNK (T183/Y185) (4668, Cell Signaling), and antiactin (A2066, Sigma). We used the rabbit monoclonal antibodies anti-p38 (8690, Cell Signaling), antiphospho-p38 (T180/Y182) (4511, Cell Signaling), anti-Erk1/2 (4695, Cell Signaling), antiphospho-Erk1/2 (T202/Y204) (4370, Cell Signaling). We used the mouse monoclonal antibody anti-p300 (ab14984, Abcam), the sheep antibody anti-mouse IgG-POX (NXA931, GE Healthcare), the goat antibody anti-rabbit IgG-POX (GAR/IgG(H+L)/PO, Nordic Immunology), and the Alexa Fluor 488 goat anti-rabbit IgG (H+L) highly absorbed (A11034, Invitrogen).
Cell Culture.
The human colonic epithelial cell line Caco-2, subclone TC7 (16) was cultured with DME (Invitrogen) supplemented with 10% (vol/vol) decomplemented FBS (Invitrogen), 1% nonessential amino acids (NEAA, Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen), at 37 °C and 10% CO2. Cells were split two times per week using Versene solution (Invitrogen).
Bacterial Challenges.
The E. coli K12 bacterial strain was grown in LB medium (Sigma) at 37 °C. For challenge experiments, cells were grown at confluence in six-well plates (1.5 × 106 cells per well) for 48 h at 37 °C and 10% CO2. Bacterial challenges were performed using supplemented DME without antibiotics with overnight bacterial cultures, at a multiplicity of infection of 10 bacteria per cell, for indicated times. For experiments involving pharmacological inhibitors, cells were pretreated overnight for 16 h and washed with supplemented DME without antibiotics.
Inhibitors.
We used the pharmacological inhibitors TSA (T1952, Sigma), SAHA (SC-220139, Santa Cruz Biotechnology), pargyline hydrochloride (P8013, Sigma), 5-azacitidine (A1287, Sigma), BMS-345541 (B9935, Sigma), and C646 (SML0002, Sigma).
Transfections.
The RNAi Max reagent (Invitrogen) was used to transfect cells with a final concentration of 25 nM siGENOME SMARTpool siRNAs (Thermo Scientific) silencing p65 (M-003533-02-0005), p300 (M-003486-04-0005), or with control scrambled siRNAs (D-001210-01-50). Transfections were performed in OptiMEM medium (Invitrogen) supplemented with 1% NEAA and 5% (vol/vol) FBS. Knockdowns were assessed after 48 h using qRT-PCR and immunobloting analysis.
qRT-PCR.
RNA was isolated using the RNeasy Mini kit and the RNase free DNase kit (Qiagen). RT-PCR reactions were performed overnight using the SuperScript II reverse transcriptase (Invitrogen) and the oligo(dT)18 primers (Thermo Scientific), as recommended by the suppliers. Gene-specific primers were designed and purchased from Sigma (HBD2, GCCATGAGGGTCTTGTATCTC/TTAAGGCAGGTAACAGGATCG; HBD3, TTTGGTGCCTGTTCCAGGTCAT/GCCGCCTCTGACTCTGCAATAATA; LL37, AAGGAAGCTGTGCTTCGTGCTA/AATCCTCTGGTGACTGCTGTGT; IL-1B, TACGATCACTGAACTGCACGCT/TCTTTCAACACGCAGGACAGGT; IL-8, AAGAAACCACCGGAAGGAACCA/ATTTCTGTGTTGGCGCAGTGTG; CCL20, AAGAGTTTGCTCCTGGCTGCTT/GCAGTCAAAGTTGCTTGCTGCT; B2M, ATTGCTATGTGTCTGGGTTTCA/AAGACAAGTCTGAATGCTCCAC). The qRT-PCR reactions were carried out in a 20-μL final volume containing 8 μL of cDNA (diluted at 1/100), 2 μL of primers (0.2 μM each), and 10 μL of Power SYBR Green mix (Applied Biosystems). Reactions were run on a QuantStudio 7 (Applied Biosystems) with recommended universal thermal cycling parameters. Each sample reaction was run in duplicate on the same plate. Relative gene expression quantification was performed using the comparative Ct method. Data were normalized to the β-2-microglobulin (B2M) housekeeping gene expression.
ELISA.
We used the ELISA kits for HBD2 (900-K72, PeproTech), and IL-8 (900-K18, PeproTech). Absorbance was measured on a M200PRO fluorimeter (Tecan).
Cytotoxicity Measurement.
Cytotoxic effect of bacterial challenges and pharmacological inhibitor treatments were evaluated by measurement of the lactate dehydrogenase release, using the CytoTox 96 nonradioactive Cytotoxicity Assay (Promega).
Immunoblotting.
Total cell lysates were harvested by removing growth medium and adding Nonidet P-40 lysis buffer [25 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2, 1% Nonidet P-40, 10% (vol/vol) glycerol, 150 mM NaCl] supplemented by a mixture of protease inhibitors [sodium orthovanadate (Sigma), 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma), COMPLETE (Roche)]. Samples were diluted with sample buffer [1 M Tris⋅HCl, 20% (vol/vol) glycerol, 6% (vol/vol) SDS, 0.02% bromophenol blue, 10% (vol/vol) β-mercaptoethanol] and boiled for 5 min. Denatured proteins were loaded on 7.5%, 10%, or 12% acrylamide Mini PROTEAN TGX precast gels (Bio-Rad). Separated proteins were transferred onto a PVDF membrane using the iBlot Gel Transfer System (Invitrogen). Membranes were blocked with 3% (wt/vol) Albumin from Bovine Serum (BSA, Sigma) or 5% (wt/vol) milk (Regilait), at room temperature, before incubation with primary antibodies overnight at 4 °C, in 1% BSA or 5% (wt/vol) milk. Incubation with the secondary horseradish peroxidase-conjugated IgG antibody was performed at room temperature. Blots were developed using the SuperSignal West Dura Extended Duration Substrate solution (Thermo Scientific) and the ChemiDoc XRS System (Bio-Rad). The presented results are representative of two independent experiments. Signal quantification was performed using the gel analysis method from the ImageJ software.
Immunofluorescence Experiments.
Cell monolayers were fixed with 4% (vol/vol) paraformaldehyde (Sigma). Cells were permeabilized for 10 min using permeabilization buffer (1× PBS, 0.1% Triton), followed by an incubation step in saturation buffer [1× PBS, 3% (wt/vol) BSA, 5% (vol/vol) FBS] for 1 h at room temperature. Cells were incubated for 2 h at room temperature with the primary polyclonal antibody. Cells were washed two times with saturation buffer, incubated for 1 h at room temperature with the secondary antibody coupled to Alexa Fluor 488 (Invitrogen), washed two times with saturation buffer, and finally, incubated for few seconds with DAPI (Invitrogen) solution (1× PBS). Coverslips were mounted using Prolong (Invitrogen), and dried overnight before examination with an IX81 inverted microscope (Olympus). Quantification of p65 nuclear signal was performed on five acquisitions per condition with the Fiji image processing software, using the nuclei watershed separation algorithm.
Chromatin Immunoprecipitation.
ChIP weas performed using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling), using magnetic beads, as recommended by the supplier. Chromatin inputs corresponded to 5–10 μg DNA for each individual ChIP assay. The ChIP DNA fractions were quantified by qRT-PCR on a QuantStudio 7 (Applied Biosystems), using the comparative Ct method. Gene-specific primers were designed and purchased from Sigma (HBD2, TTTGGCCAACCCTCCTATTTCCCT/ACCTCTGTAATGAGCATTGCACCC; IL-8, AGGACAAGAGCCAGGAAGAAACCA/AGAGCTGCAGAAATCAGGAAGGCT). All ChIP experiments were performed at 1 h postchallenge because of the time existing between regulatory events occurring at promoters, such as histone posttranslational modifications, recruitment of transcription factors or coregulators, and initiation of the gene transcription.
RNA Sequencing.
RNA libraries were created with Illumina TruSeq stranded PolyA+ mRNA kits and sequenced on two Illumina HiSeq lanes, three samples multiplexed per lane, with paired-end 50-bp reads. Reads were mapped to human genome hg19 (GRCh37) using TopHat 2.0.11, with Gencode 19 human genes as a transcriptome guide and maximum of 50 multihits. Reads mapping to Gencode genes were counted with the featureCounts software of the Subread package. Read counts were normalized using the DESEq. 1.14 estimateSizeFactors function. Genes having fewer than 10 reads in any condition were filtered out, and the remaining transcripts had a pseudocount of 5 added to down-weight poorly expressed transcripts in fold-change calculations. The GEO accession number is GSE80048.
Human Colonic Organoid Culture.
This study was approved by the Institut Pasteur’s ethical and medical committee under the agreement no. 2012–37. Surgically resected human colonic tissues were obtained from the Henri Mondor Hospital. All samples were obtained from patients who provided informed consent before surgery. Normal epithelia were isolated and cultured according to the protocol described by Sato et al., with minor modifications (27). Organoids were cultured with Advanced DMEM/F12 (Invitrogen) supplemented with 10 mM Hepes (Invitrogen), 2 mM GlutaMAX (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen), 1× N2 and B27 supplements (Invitrogen), 1 mM N-acetyl-l-cysteine (Sigma), 10 μM Y-27632 (Sigma), 500 nM A83-01 (Tocris), 10 μM SB202190 (Sigma), 10 mM nicotinamide (Sigma), 10 nM gastrin I (Sigma), 100 ng/mL recombinant human Noggin (R&D Systems), 50 ng/mL recombinant human EGF (R&D Systems), 1 μg/mL recombinant human R-Spondin-1 (Peprotech), 100 ng/mL recombinant human Wnt-3A (R&D Systems), and 10% (vol/vol) FBS (Invitrogen), at 37 °C and 5% CO2. Organoids were cultured in 48-well plates, 100 crypts per well. RNA was isolated from 6-d-old organoids treated or not with 5 μM TSA (Sigma), and stimulated or not with 5 μg/mL flagellin (InvivoGen), for 24 h, using the RNeasy Micro kit and the RNase free DNase kit (Qiagen). Gene expression was analyzed using TaqMan probes from Applied Biosystems: HBD2 (Hs00823638_m1), HBD3 (Hs00218678_m1), LL37 (Hs00189038_m1), IL-1B (Hs01555410_m1), IL-8 (Hs00174103_m1), and TNF (Hs01113624_g1). Data were normalized to the B2M (Hs00984230_m1) housekeeping gene expression.
Statistics.
Statistical analysis was performed on GraphPad Prism 5 (GraphPad software). Results are presented as a mean of at least three independent experiments. Error bars represent the SD. Statistical comparisons were performed using the Mann–Whitney u test. A P value < 0.05 was considered significant.
Acknowledgments
We thank Christian Muchardt and Eric Batsche for helpful discussions; Clément Bonamy for advice; and Mark Anderson for critical reading of the manuscript. The research leading to these results has received funding from the European Union Seventh Framework Program under the Grant Agreement EIMID ITN No 264388. Philippe J. Sansonetti is supported by the European Research Council (DECRYPT project) and by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE80048).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605997113/-/DCSupplemental.
References
- 1.Nightingale KP, O’Neill LP, Turner BM. Histone modifications: Signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16(2):125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;128(4):802. doi: 10.1016/j.cell.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Arbibe L, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8(1):47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 4.Eskandarian HA, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341(6145):1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 5.Lehrmann H, Pritchard LL, Harel-Bellan A. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 6.Marks PA, Dokmanovic M. Histone deacetylase inhibitors: Discovery and development as anticancer agents. Expert Opin Investig Drugs. 2005;14(12):1497–1511. doi: 10.1517/13543784.14.12.1497. [DOI] [PubMed] [Google Scholar]
- 7.Codd R, Braich N, Liu J, Soe CZ, Pakchung AAH. Zn(II)-dependent histone deacetylase inhibitors: Suberoylanilide hydroxamic acid and trichostatin A. Int J Biochem Cell Biol. 2009;41(4):736–739. doi: 10.1016/j.biocel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 8.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T. Acetylation: A regulatory modification to rival phosphorylation? EMBO J. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghizzoni M, Haisma HJ, Maarsingh H, Dekker FJ. Histone acetyltransferases are crucial regulators in NF-κB mediated inflammation. Drug Discov Today. 2011;16(11-12):504–511. doi: 10.1016/j.drudis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Yang X-D, Lamb A, Chen L-F. Posttranslational modifications of NF-kappaB: Another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010;22(9):1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiernan R, et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278(4):2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 14.Chen L-F, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21(23):6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weake VM, Workman JL. Inducible gene expression: Diverse regulatory mechanisms. Nat Rev Genet. 2010;11(6):426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 16.Chantret I, et al. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: Evidence for glucose-dependent negative regulation. J Cell Sci. 1994;107(Pt 1):213–225. doi: 10.1242/jcs.107.1.213. [DOI] [PubMed] [Google Scholar]
- 17.Schauber J, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: Relevance of signalling pathways. Gut. 2003;52(5):735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3(1):69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 19.Anest V, et al. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423(6940):659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto Y, Verma UN, Prajapati S, Kwak Y-T, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423(6940):655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 21.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 22.Wehkamp J, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: A novel effect of a probiotic bacterium. Infect Immun. 2004;72(10):5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72(5):847–855. [PubMed] [Google Scholar]
- 24.Burke JR, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003;278(3):1450–1456. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 25.Yang X-D, Tajkhorshid E, Chen L-F. Functional interplay between acetylation and methylation of the RelA subunit of NF-kappaB. Mol Cell Biol. 2010;30(9):2170–2180. doi: 10.1128/MCB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers EM, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem Biol. 2010;17(5):471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 28.Leoni F, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99(5):2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: A hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19(2):70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Rescigno M, Borrow P. The host-pathogen interaction: New themes from dendritic cell biology. Cell. 2001;106(3):267–270. doi: 10.1016/s0092-8674(01)00454-8. [DOI] [PubMed] [Google Scholar]
- 31.Tato CM, Hunter CA. Host-pathogen interactions: Subversion and utilization of the NF-kappa B pathway during infection. Infect Immun. 2002;70(7):3311–3317. doi: 10.1128/IAI.70.7.3311-3317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294(5543):870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 33.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447(7147):972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 34.Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546(1):51–58. doi: 10.1016/s0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- 35.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103(2):263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 36.Duan J, et al. Nuclear factor-kappaB p65 small interfering RNA or proteasome inhibitor bortezomib sensitizes head and neck squamous cell carcinomas to classic histone deacetylase inhibitors and novel histone deacetylase inhibitor PXD101. Mol Cancer Ther. 2007;6(1):37–50. doi: 10.1158/1535-7163.MCT-05-0285. [DOI] [PubMed] [Google Scholar]
- 37.Katsura T, et al. The effects of trichostatin A on the oncolytic ability of herpes simplex virus for oral squamous cell carcinoma cells. Cancer Gene Ther. 2009;16(3):237–245. doi: 10.1038/cgt.2008.81. [DOI] [PubMed] [Google Scholar]
- 38.Gerritsen ME, et al. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94(7):2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheppard KA, et al. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol Cell Biol. 1999;19(9):6367–6378. doi: 10.1128/mcb.19.9.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 41.Ververis K, Karagiannis TC. Potential non-oncological applications of histone deacetylase inhibitors. Am J Transl Res. 2011;3(5):454–467. [PMC free article] [PubMed] [Google Scholar]
- 42.Raqib R, et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci USA. 2006;103(24):9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]