Summary
The central proteins for protection against tuberculosis are attributed to interferon-γ, tumor necrosis factor-α, interleukin (IL)-6 and IL-1β, while IL-10 primarily suppresses anti-mycobacterial responses. Several studies found alteration of expression profile of genes involved in anti-mycobacterial responses in macrophages and natural killer (NK) cells from active and latent tuberculosis and from tuberculosis and healthy controls. This alteration of cellular composition might be regulated by microRNAs (miRNAs). Albeit only 1% of the genomic transcripts in mammalian cells encode miRNA, they are predicted to control the activity of more than 60% of all protein-coding genes and they have a huge influence in pathogenesis theory, diagnosis and treatment approach to some diseases. Several miRNAs have been found to regulate T cell differentiation and function and have critical role in regulating the innate function of macrophages, dendritic cells and NK cells. Here, we have reviewed the role of miRNAs implicated in tuberculosis infection, especially related to their new roles in the molecular pathology of tuberculosis immunology and as new targets for future tuberculosis diagnostics.
Keywords: Mycobacterium tuberculosis, Tuberculosis infection, microRNA, Tuberculosis immunology
1. Introduction
Tuberculosis remains a major global health problem, causes morbidity among millions of people each year and ranks as the second leading cause of death from an infectious disease worldwide [1]. The latest estimates suggest approximately 9 million new cases and 1.4 million tuberculosis deaths in 2011 [1]. Both innate and adaptive immune responses are required for host control of tuberculosis infection [2,3]. In tuberculosis pathogenesis, the host cellular immune response determines whether an infection is arrested as latent tuberculosis infection (LTBI) or progresses to the next stages, active tuberculosis infection. Efficient cell-mediated immunity frequently keeps tuberculosis infection arrested permanently as LTBI, but if an infected person cannot control the initial infection in the lung or if the immune system becomes weakened, Mycobacterium tuberculosis (M. tuberculosis) can cause active pulmonary or extrapulmonary tuberculosis [4]. Approximately 90% of infected individuals will remain asymptomatic with LTBI and only 10% of the individuals infected with M. tuberculosis will develop active disease, suggesting that host genetics factors play an important role to regulate progression of tuberculosis infection [5].
MicroRNAs (miRNAs) have been identified as important regulators of gene expression at posttranscriptional level and influences many biological systems including mammalian immune systems [6]. Hundreds of miRNAs encoded in the human genome and thousands of target mRNAs have been shown to be involved in cell development, differentiation, proliferation, apoptosis, DNA methylation, DNA repair and provide anti-inflammatory or pro-inflammatory stimuli [7,8]. Rapid advancement in new miRNAs discoveries has continued the possibility that miRNAs will be associated with the regulation of almost every aspect of cell physiology. Subsequent reports have identified that miRNAs are associated with non-communicable diseases [9–15] and communicable diseases [16,17]. In addition, miRNAs have potential uses as biomarkers for both non-communicable and communicable diseases [18–23].
Previous studies revealed altered gene expression profiles in macrophages and natural killer (NK) cells from active and latent tuberculosis, tuberculosis-infected and healthy controls [24–28]. This alteration of cellular composition and related gene expression in tuberculosis patients is likely regulated by miRNAs. Several miRNAs have been found to regulate T cell differentiation and function [29–33]. In addition, miRNAs have been found to be important in regulating the innate function of macrophages, dendritic cells (DCs) and NK cells [31,34,35]. Therefore here we will discuss several miRNAs involved in molecular pathology of tuberculosis infection and discuss their potential as tuberculosis biomarkers.
2. Discussion
2.1. The world of microRNAs: biogenesis, mechanism of action and biological functions
The human genome encodes only approximately 20,000 protein coding genes, representing <2% of the total genome sequence [36,37]. However, with advance technology, it was determined that at least 90% of the genome is actively transcribed [38]. The human transcriptome was found to be more complex than a collection of protein-coding genes and their splice variants; showing extensive antisense, overlapping and non-coding RNA (ncRNA) expression [39,40]. NcRNAs are grouped into two major classes based on transcript size; small ncRNAs and long ncRNAs [41]. The small ncRNAs class, which includes miRNAs, ranges in length from 18 to 200 nucleotides (nt) [41]. miRNAs are a class of approximately 1000 bioinformatically predicted 22-nt length ncRNAs found in eukaryotes [41].
All miRNAs are matured and processed through a multistep process that involves several enzymes first in the cell nucleus and finally in the cytoplasm. The primary miRNA transcripts are transcribed as a huge double-stranded primary transcript called primiRNAs by RNA polymerase II which can be several hundreds nt long [31]. Pri-miRNAs fold into hairpin structures which are polyadenylated and capped. The RNase III-type enzymes Drosha convert this precursor into a double-stranded miRNA precursor of 60- to 100-nt hairpins known as pre-miRNAs [42]. Pre-miRNAs contain a local stem-loop structure that encodes miRNA sequences which are exported from nucleus to cytoplasm by exportin 5 [43].
In cytoplasm, pre-miRNAs are further processed by the RNase III Dicer to yield imperfect 22-nt double-stranded miRNA. This unstable duplex consists of the guide strand (miRNA) and the passenger strand (miRNA*). The guide strand miRNA is selected to become a mature miRNA, whereas the miRNA* is degraded [44]. The mature miRNA is incorporated into the RNA-induced silencing complex (RISC), which recognizes specific targets and induces posttranscriptional gene silencing which regulates protein expression [45]. An alternative biogenesis pathway was recently discovered in which miRNA enters RISC by skipping further processing by Dicer. The strand enters RISC by direct loading of the pre-miRNA after Drosha processing [46].
Specific miRNAs function is defined by the genes it targets and its effect on their expression. Harapan et al. [10] summarizes several mechanisms, direct and indirect, in which miRNAs repress gene translation in cells. Direct on translational repression occurs via initiation block or post-initiation block. In initiation block, the miRISC inhibits translation initiation by interfering with eIF4F-cap recognition and 40S small ribosomal subunit recruitment or by antagonizing 60S subunit joining and preventing 80S ribosomal complex formation. Post-initiation block includes premature ribosomal drop-off, where the 40S/60S ribosomes are dissociated from mRNA, stalled or slowed elongation, the 40S/60S ribosomes are prohibited from joining during the elongation process or facilitating proteolysis of nascent polypeptides. The indirect on translational repression occurs via mRNA deadenylation and degradation. In addition, it has been reported that the majority of miRNA binding sites are in the 3′ untranslated region (3′UTR) of target mRNA molecules and this binding leads to the inhibition of mRNA translation and subsequent degradation [47].
Malfunction of miRNA regulation is contributed to the fundamental role biological processes and associate with a variety of human diseases. Importantly, miRNA dysregulation may contribute to the broader understanding of the human pathologies. Abnormalities can occur by the following ways: (1) loss or downregulation of miRNA expression due to mutation, epigenetic inactivation, transcriptional downregulation or abnormality processing [48], (2) overexpression of miRNA due to gene amplification or transcriptional upregulation may result in the suppressed production of its target proteins [49], (3) a mutation in 3′UTR of an mRNA may affect a miRNA binding site and the miRNA may no longer be able to bind [50], and (4) a mutation in 3′UTR of a gene may generate a new miRNA binding site. [51] A huge number of studies reported that miRNAs dysregulation associated to a wide spectrum of diseases such as chronic kidney disease [9], liver cirrhosis [14], systemic sclerosis [15], cardiac fibrosis [13], diabetes [11], pregnancy-related diseases [10,52], and most notably cancer [12]. Recent studies have shown the regulation of miRNA in human diseases only understood and explained by genetic (deletions, mutations and translocation), epigenetic mechanisms (methylation) or abnormalities in the miRNA processing machinery. The contributions of this understanding have been implicated in several infectious diseases, providing the prospective use of miRNA as clinical biomarkers [53].
In bacterial infections, miRNAs have different mechanisms depending on the characteristic of the bacteria. For example, Helicobacter pylori (H. pylori) infection alters the expression of oncogenes, tumor suppressor genes and miRNA in the host. Interestingly, a study showed that treat of the infection with anti-H. pylori regiment restored decreased expression of several miRNAs [16]. In Salmonella infection, treatment of immune cells with bacterial lipopolysaccharide (LPS) led to induction of several miRNAs. These results indicate that miRNAs play a role in activation of inflammatory factors when mammalian cells are target by bacterial pathogens [54].
2.2. Immunology aspect of tuberculosis
Toll-like receptors (TLRs) are the primary receptors in macrophage used to recognize M. tuberculosis and their signals can activate nuclear factor κ beta (NF-κB) to induce proinflammatory cytokines release [55,56]. Interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) are the main pro-inflammatory cytokines central for protection against to tuberculosis infection [26,57,58]. TNF-α plays a crucial in controlling M. tuberculosis growth in macrophages through several mechanisms and has a central role in the establishment and maintenance of LTBI [59–62]. IFN-γ is critical for innate and adaptive immunity to infection, especially intracellular bacterial infections [63]. Indeed, the crucial role of IFN-γ is well established in the protection against M. tuberculosis infection in both mouse models and human [64,65]. Moreover, studies suggested for a protective role of interleukin-6 (IL-6) and IL-1β in host resistance to M. tuberculosis infection [66,67], while IL-10 primarily suppresses anti-mycobacterial responses [68]. Therefore, M. tuberculosis lately inhibits production of proinflammatory cytokines as a survival mechanism [69].
In addition, the ability of M. tuberculosis to reside and replicate within phagocytes allow M. tuberculosis to survive in the cell. This mechanism avoids immune response of the cell [70]. M. tuberculosis successfully invades and parasitizes macrophages by inhibiting phagolysosome fusion and neutralizing the acidic environment of the phagolysosomal compartment [71]. With a neutralized acidic environment, T cells do not respond to antigens [72].
2.3. The role of microRNA in tuberculosis
2.3.1. microRNA regulation of immune response to tuberculosis
Several studies revealed altered gene expression profiles in macrophages and NK cells from active and latent tuberculosis and from tuberculosis and healthy controls [24–28]. This alteration of cellular composition and related gene expression in tuberculosis patients is likely regulated by miRNAs. Several miRNAs have been found to regulate T cell differentiation and function [29–33]. In addition, miRNAs have been found to be important in regulating the innate function of macrophages, DCs and NK cells [31,34,35]. Here we reviewed the roles of several miRNAs in the molecular pathology of tuberculosis.
2.3.1.1. miRNA-29
In vitro and clinical studies documented that miR-29 is overexpressed after virulent Mycobacterium species infection in several human cell types [11,73,74]. miR-29 suppresses immune responses to M. tuberculosis by downregulating IFN-γ [75]. Beside targeting 3′UTR IFN-γ mRNA, miR-29a promotes the association of IFN-γ mRNA with Argonaute 2 (Ago2) protein to form an RNA-induced silencing complex and subsequently suppressed the IFN-γ expression posttranscriptionally (Figure 1). Moreover, several reports have indicated that miR-29 also targets the antiapoptotic proteins B-cell lymphoma 2 (Bcl-2) and myeloid cell leukemia-1 (Mcl-1), the kinase p85α and the GTP-binding protein Cdc42 [76,77], thus suggesting a central role for miR-29 in regulating the apoptotic pathway in immune cells. Therefore, overexpressed miR-29 in tuberculosis infection partly explain one mechanism bywhich M. tuberculosis avoids macrophage digestion through inhibition of IFN-γ and increasing apoptosis of cells involved anti-tuberculosis responses.
Figure 1.
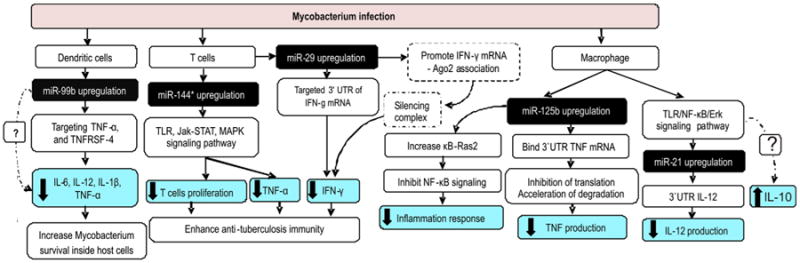
The roles of miRNAs in pathology tuberculosis infection. Several studies revealed evidence that Mycobacterium species infection in several cell types upregulated miR-99b, miR-144*, miR-29 and miR-125b. These miRNAs target several genes that are important in tuberculosis immunity. Upregulated of these miRNAs help Mycobacterial to survival inside host cells and enhance anti-tuberculosis immunity by inhibiting production of proinflammatory cytokines and inducing IL-10 through several mechanisms. Abbreviation: 3′UTR: 3′-untranslated region, Ago2: Argonaute 2, IFN-γ: Interferon-γ, IL-12: Interleukin 12, IL-1β: Interleukin 1β, IL-6: Interleukin 6, Jak-STAT: Janus kinase/signal transducers and activators of transcription, MAPK: mitogen-activated protein kinase, NF-κB: nuclear factor κ beta, TLR: Toll-like receptor, TNFRSF-4: tumor necrosis factor receptor superfamily, member 4, TNF-α: tumor necrosis factor-α.
Interestingly, a contrasting phenomenon occurred in a non-virulent Mycobacterium species infection model. Mycobacterium bovis Bacillus Calmette-Guerin (BCG) downregulated miR-29 expression and induced IFN-γ expression in NK cells and T cells [75]. This result indicates miR-29 inhibition may have facilitated IFN-γ production by these T cells and expression of miR-29 is influenced by Mycobacterium species-specific virulence.
2.3.1.2. miRNA-147
Previous study shows that miR-147 is induced upon TLRs/NF-κB signaling pathway in macrophages and attenuates expression of proinflammatory cytokines, such as TNF-α and IL-6 [78]. These results indicate that miR-147 has potent anti-inflammatory properties. Studies found that TNF-α and IL-6 from serum or peripheral blood mononuclear cells (PBMCs) were higher in active tuberculosis compared healthy control [79–81]. A key study found that miR-147 was overexpressed in sputum obtained from active tuberculosis compared with controls [74]. Interestingly, this study found the levels of TNF-α and IL-6 in sputum did not differ significantly between active tuberculosis group and controls, indicating cytokines dysregulation is mainly occurring in the bloodstream as opposed to the lungs. Altogether, these facts indicate that miR-147, together with miR-29 enhance anti-tuberculosis immunity and act as a negative regulator of immune response against tuberculosis.
2.3.1.3. miRNA-21
Previous study suggests miR-21 inhibited proinflammatory cytokines expression and promoted an anti-inflammatory cytokine, IL-10, production [82]. Recent study found that after M. bovis BCG infection, both in vitro and in vivo; miR-21 is upregulated in un-sensitized DCs and macrophages through TLR/Erk/NF-κB pathway [83]. miR-21 is also upregulated following challenge of macrophages with M. tuberculosis early secreted antigenic target 6 kDa protein (ESAT-6) antigen [84]. miR-21 inhibited IL-12 expression by directly targeting 3′UTR IL-12 mRNA, and thus suppressed host Th1 responses (Figure 1). In addition, this study also found that miR-21 downregulated the gene of protective cytokines in M. tuberculosis infection (TNF-α and IL-6) but these cytokines levels did not change significantly after ESAT-6 exposure. Interestingly, miR-21 also promoted DC apoptosis by targeting Bcl-2, which corroborates previous findings suggesting that M. tuberculosis can induce apoptosis of infected cells [85]. However, the exact mechanism by which miR-21 regulated Bcl-2 expression remains unclear. Furthermore, inhibitors of miR-21 induced IL-12 production and triggered more potent anti-mycobacterial responses. Therefore, miR-21 may be an effective strategy Mycobacteria use to escape the host immune response and establish chronic infection.
2.3.1.4. miRNA-99b
In murine DCs from MyD88-deficient mice, exposure to M. tuberculosis but not to LPS induced overexpression of miR-99b [86]. Further analysis found miR-99b blocking (by antagomirs and knockdown approach) resulted in significantly reduced M. tuberculosis growth and significantly upregulated proinflammatory cytokines such as TNF-α, IL-6, IL-12, and IL-1β. Inhibition of M. tuberculosis growth might be due to an increase in the production of those proinflammatory cytokines. In addition, this study found that miR-99b directly targeted tumor necrosis factor receptor superfamily, member 4 (TNFRSF-4) and TNF-α mRNA, to regulate expression of various cytokines and transcription factors involved in T cell differentiation pathways and M. tuberculosis clearance (Figure 1). Moreover, treatment of anti-miR-99b-transfected DCs with anti-TNF-α antibody resulted in increased bacterial death. These data confirmed that miR-99b has important role in M. tuberculosis growth in DCs by inhibiting TNF-α production, which allows the bacteria to evade host protective immune responses and to survive within host phagocytes.
2.3.1.5. miRNA-125b
Rajaram et al. [87] found that human macrophages incubated with M. tuberculosis and it's component, lipomannan, induce high miR-125b expression with correspondingly low TNF production. miR-125b directly targets 3′UTR of TNF mRNA transcript, contributing to the inhibition of translation and possibly its accelerated degradation and downregulates TNF production. This study also found that only virulent Mycobacterium species limit activation of mitogen-activated protein kinase (MAPK) p38 and Akt, two components that contribute significantly to TNF production in mycobacterial-infected macrophages compared with avirulent Mycobacterium species [62,88]. miR-125b also enhances the stability of κB-Ras2, an inhibitor of NF-κB signaling in human macrophages, thereby decreasing the inflammatory response [89]. Taken together, these results revealed that M. tuberculosis blocks TNF biosynthesis by upregulating miR-125b (Figure 1). In contrast, avirulent Mycobacterium species infection and it's lipomannan exposure downregulated miR-125b expression which correlated with high TNF production. Further analysis found that down-regulation of miR-125b increased TNF mRNA stability and promote proinflammatory response.
2.3.1.6. miRNA-155
miR-155 has been identified as a multifunctional miRNA involved in a number of biologic processes, including infection, inflammation, and immunity [90]. Previous study showed knockdown miR-155 mouse model, more IL-4 and less IFN-γ were produced, suggesting that miR-155 play an important role in regulating T cell–dependent responses [91]. One study found debatable results regarding dysregulation of miR-155 in human macrophages that are infected by different Mycobacterium species [87]. In virulent Mycobacterium species infection model, study found that M. tuberculosis infection and it's lipomannan exposure to human macrophages downregulated miR-155 corresponding low TNF production. This downregulation mediated upon TLR-MAPK/Akt pathway. Further analysis revealed that M. tuberculosis leads to reduced translation, by inhibiting initiation, and stability of TNF mRNA. However, these result contradict previous study that found miR-155 targeted the 3′UTR of the inositol phosphatase SH2-containing inositol 5-phosphatase (SHIP1) mRNA, a negative regulator of TNF production, leading to its degradation, therefore increased TNF production [92]. The other study also found the expression of miR-155 was shown to increase TNF production through increasing mRNA stability and half-life [93].
Results conflicting with Rajaram et al. [87] from another study [88] suggests M. tuberculosis infection (ESAT-6 antigen exposure) in murine macrophage also upregulated miR-155. Furthermore, miR-155 attenuates SHIP1 and the transcriptional repressor BTB and CNC homology 1 (Bach1). SHIP1 regulate negatively the activation of the serine/threonine kinase AKT while Bach1 is a transcriptional repressor of heme oxygenase-1 (HO-1). AKT is required for the survival of M. tuberculosis in macrophages [94,95], and HO-1 is an activator of the M. tuberculosis dormancy. Moreover, upregulation of miR-155 inhibit the expression of two modulators of the innate immune response - IL-6 and cyclooxygenase-2 (Cox-2). Altogether, these evidences revealed that virulent Mycobacterium species infection decreased proinflammatory cytokines. In line with this findings, previous study found evidence that hypervirulent M. tuberculosis strain elicit reduced levels of proinflammatory cytokines, including TNF-α and IL-6 [96]. These results corroborate that miR-155 was upregulated in virulent Mycobacterium species and likely offers a survival advantage to Mycobacterium within its host (Figure 2). In summary, virulent Mycobacterium species infection in human macrophages downregulate miR-155 expression, but infection in murine macrophages upregulate miR-155 expression. This controversy needs more comprehensive work to understand the similarities and differences in the response of human and murine macrophages with regard to miRNA regulation.
Figure 2.
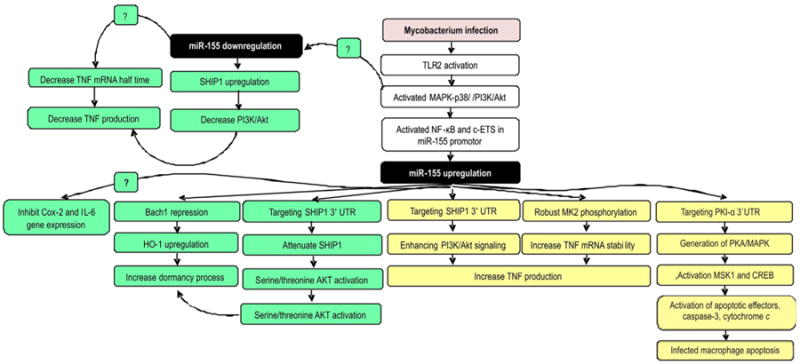
The role of miR-155 on virulent (green) and avirulent (yellow) Mycobacterium species infection in macrophage. Avirulent Mycobacterium species infection upregulates of miR-155 by triggering the TLR2/PI3K/PKCδ/MAPK signaling pathways then recruit of NF-κB and c-ETS to miR-155 promoter. miR-155 induces apoptosis of infected macrophage by targeting PKI-α 3′UTR thus increases PKA signaling and finally activates of apoptotic effectors, caspase-3, cytochrome c translocation. miR-155 also increases TNF production by increasing TNF mRNA stability. To meet this role, miR-155 directly targeting SHIP1 3′UTR, a negative regulator of the PI3K/Akt pathway, a pathway involves in TNF biosynthesis and enhances MK2 phosphorylation which is a key molecule for TNF mRNA stability. Conflict results come from virulent Mycobacterium infections. Infection in murine macrophage induces miR-155 expression then attenuates the SHIP1 and Bach1 expressions therefore promote mycobacterium dormancy and survival in macrophages. Moreover, upregulation of miR-155 inhibits the expression IL-6 and cyclooxygenase-2. However, virulent Mycobacterium infection in human macrophage downregulates miR-155. Briefly, this condition decrease TNF production by decreasing TNF mRNA half time and decreases PI3K/Akt pathway. Abbreviation. 3′UTR: 3′ -untranslated region, Cox-2: cyclooxygenase-2, CREB: cyclic AMP response element binding protein, MAPK: mitogen-activated protein kinase, MK2: MAPK activated protein kinase 2, MSK1: mitogen and stress-activated protein kinase-1, NF-κB: nuclear factor κ beta, PI3K: phosphatidylinositol 3-kinase, PKCδ: protein kinase Cδ, PKI-α: protein kinase inhibitor alpha, SHIP1: SH2-containing inositol 5-phosphatase, TLR: Toll-like receptor. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In contrast with virulent Mycobacterium, infection of macrophage with avirulent Mycobacterium species enhance miR-155 and low miR-125 expression with high TNF production [87]. They found that miR-155 increased TNF mRNA stability by directly targeting inositol phosphatase SHIP1 3′UTR, a negative regulator of the PI3K/Akt pathway – a pathway involved in TNF biosynthesis [92,93], thereby enhancing PI3K/Akt-mediated signaling and TNF mRNA stability [87]. Further analysis found that avirulent Mycobacterium species stimulated robust and prolonged MAPK activated protein kinase 2 (MK2) phosphorylation, a key molecule for TNF mRNA stability [97]. Activated MK2 translocates to the cytoplasm and maintains TNF mRNA stability through tristetraprolin (TTP) phosphorylation [98].
Another comprehensive study revealed evidences regarding the protective role of miR-155 in avirulent Mycobacterium species [99]. This study demonstrated that M. bovis BCG upregulated miR-155 by activating TLR2, phosphatidylinositol 3-kinase (PI3K), protein kinase Cδ (PKCδ), and MAPK pathways, which resulted in the participation of NF-κB and c-ETS in transcriptional activation of miR-155 promoter. Further study demonstrated that miR-155 regulates protein kinase A (PKA) signaling by directly targeting 3′UTR of protein kinase inhibitor alpha (PKI-α), an endogenous inhibitor of PKA activity. PKA is one of the effector kinases that regulate a wide array of cellular responses [84]. Consequently, enhanced activation of PKA signaling directs the generation of PKA C-α, mediates activation of MAPKs and phosphorylation of mitogen and stress-activated protein kinase-1 (MSK1) and cyclic AMP response element binding protein (CREB). This complex mechanism modulated expression of apoptosis effectors, activation of caspase-3 and translocation of cytochrome c, thus leading to apoptosis of infected macrophages (Figure 2). It is mean that avirulent Mycobacterium species could stimulate the macrophage to undergo apoptosis. However, virulent mycobacteria are likely able to inhibit apoptosis by altering the Bcl-2 pathway [100]. Previous studies showed that apoptosis is an innate defense function of macrophages against M. tuberculosis infection. However, virulent Mycobacterium species caused macrophage death by a process that proceeds to necrosis, which produces a permeable cell membrane that enables bacteria to escape and spread [72,101].
2.3.1.7. miRNA-144*
miR-144* is overexpressed in active tuberculosis patients [102]. miR-144* target genes in Janus kinase/signal transducers and activators of transcription (Jak-STAT) signaling pathway, MAPK signaling pathway, TLR signaling pathway and cytokine–cytokine receptor interactions. Further transfection of T cell with miR-144* precursor found that miR-144* inhibit TNF-α and IFN-γ production. Since TNF-α and IFN-γ have important role in protective immunity, it is possible that miR-144* influence the development and outcome of tuberculosis. miR-144* possibly in regulates anti-tuberculosis immunity through modification of cytokine production and cell proliferation of T cells.
2.3.1.8. miRNA-223 and miRNA-424
Wang et al. [28] found that miR-223 and miR-424 were highly expressed in PBMCs from patient with active tuberculosis. A miR-223 lost function mouse study showed an increased proration of granulocytes, which are morphologically hypermature and hypersensitive to activating stimuli, and have more fungicidal activity [103]. miR-424 promotes monocyte differentiation and subsequently downregulates expression of the transcription factor NFI-A78 [104]. Wang et al. [28] found basic leucine zipper transcription factor 2/BTB and CNC homology 1 (BACH2) and B-cell CLL/lymphoma 7A (BCL7A) are targeted by multiple miRNAs that overexpressed in active tuberculosis. Wang et al. [28] hypothesize the reduced expression level of these two genes regulated by multiple miRNAs such as miR-223 and miR-424 may lead to a disorder in the proportions of Tcells and B cells in active tuberculosis patients, which may disturb the delicate balance of immune control in M. tuberculosis infection. Wang et al. [28] also found that miR-424 targets BACH2, BCL2, BCL7A, and forkhead box O1 (FOXO1) that are involved in cellular differentiation and development, to regulate the differentiation of B cells to plasma cell [105].
2.3.2. microRNA as biomarker for tuberculosis
Early diagnosis is essential for effective tuberculosis control and therapy. Current diagnostic approaches relies on the detection of the pathogen in clinical specimens. However, due to the heterogeneous clinical presentations of M. tuberculosis infection (active tuberculosis disease/asymptomatic LTBI; pulmonary and/or extrapulmonary tuberculosis), the development of affordable diagnostic tests based on host biomarkers is urgent in order to improve the quality of the TB diagnostic process inpaucibacillary or not microbiologically confirmed cases (e.g. children, HIV-positive individuals, extrapulmonary cases) [106].
In particular, the mechanisms of LTBI and its transition to active tuberculosis remain elusive. Many studies give evidences that this transition occur if it cell-mediated immunity fails [107,108]. Previous study revealed altered gene expression profiles in macrophages and NK cells from active tuberculosis and LTBI [25]. Considering that miRNAs havea central role in controlling gene expression, they could represent valuable markers to distinguish active tuberculosis and LTBI. Despite several studies focused on cellular miRNAs in in vitro infection experiments, only recently extracellular (usually referred as “circulating”) miRNAs have raised attention as tuberculosis biomarkers. Sputum, serum, plasma, or other body fluid specimens are readily available and amenable to noninvasive analyses for miRNAs. Indeed, circulating miRNAs have been extensively investigated as novel and noninvasive diagnostic and prognostic biomarkers for several different diseases, including cancer and infections [19–23,109,110]. These routinely taken specimens and robust methods of detection make miRNAs ideal biomarkers for disease diagnosis. Here we provide an overview on the potential role of both cellular and circulating miRNAs as biomarkers in tuberculosis.
2.3.2.1. miRNA profiling in blood samples
Among the studies evaluating miRNA profiles from blood as clinical specimens in tuberculosis, two main categories can be defined: serum-based studies and cells-based studies.
a Serum-based study
Serum miRNAs are present in a stable form that is protected from endogenous RNase activity [19]. The first serum-based study found that 92 miRNAs were differentially dysregulated in active pulmonary tuberculosis patients compared with healthy controls, of which 59 were upregulated and 33 were downregulated [73]. Further validation analysis found that miR-3125 was down-regulated while miR-93* and miR-29a were upregulated in active tuberculosis patients. In addition, receiver operating characteristic (ROC) analysis was performed to examine if these miRNAs could be used as a diagnostic biomarker for active pulmonary tuberculosis. Analysis found that area under the ROC curve (AUC) of miR-29a was 0.831 (Table 1), which reflected that miR-29a has great potential as biomarker to detect active pulmonary tuberculosis infection.
Table 1.
Receiver operating characteristic analysis of several microRNAs as tuberculosis biomarker to discriminate tuberculosis patient and healthy person.
| MicroRNAs | Specimen | AUC | Author |
|---|---|---|---|
| miR-292 | Blood (serum) | 0.831 | Fu et al. [73] |
| miR-361 | Blood (serum) | 0.848 | Qi et al. [111] |
| miR-889 | Blood (serum) | 0.765 | Qi et al. [111] |
| miR-576-3p | Blood (serum) | 0.711 | Qi et al. [111] |
| Combination (miR-361, miR-889 and miR-576-3p) | Blood (serum) | 0.863 | Qi et al. [111] |
| miR-155 | Blood (PBMC) | 0.897 | Wu et al. [116] |
| miR-155* | Blood (PBMC) | 0.794 | Wu et al. [116] |
AUC, area under curve; PBMC, peripheral blood mononuclear cell.
Another study found that 97 miRNAs were differentially expressed in active pulmonary tuberculosis patient sera compared with healthy control individuals (90 upregulated and 7 down-regulated) [111]. Validation study showed that the expression levels of miR-361-5p, miR-889, miR-576-3p, miR-210, miR-26a, miR-432, and miR-134 were upregulated in tuberculosis infected individuals. ROC analysis among these miRNAs, three miRNAs (miR-361-5p, miR-889 and miR-576-3p) were shown to distinguish active tuberculosis patients from uninfected individuals with moderate sensitivity and specificity. In addition, combination of these three miRNAs showed an enhanced ability to discriminate between active tuberculosis and healthy individuals with AUC value of 0.863 (Table 1). However, the differences between miR-144* and miR-29a expression reported by this study did not significantly differ from the expression previously reported [73].
Only a recent study compared the serum levels of miRNAs from active tuberculosis patients and patients infected by Bordetella pertussis, varicella-zoster virus and enterovirus [111]. This study documented that three miRNAs (miR-361-5p, miR-889, and miR-576-3p) were overexpressed in tuberculosis patients compared to other infection groups. The combination of these miRNAs is suitable biomarkers to differentiate tuberculosis from other microbial infections. Further analysis found that these miRNAs target several genes involved in immune system development. For instance, miR-361-5p target SP-1 transcription factor (SP1) that was a key signaling pathway for IL-10 expression in the lung. [112]
b Cells-based study
The first study profiling miRNA expression in PBMCs from active pulmonary tuberculosis patients and healthy controls found that 28 miRNAs were upregulated and 2 miRNAs downregulated in active tuberculosis patients [102]. miR-144* was one of the miRNA upregulated in active tuberculosis patients. miR-144 was the highest upregulated miRNA observed in comparison between active tuberculosis and uninfected individuals in whole-blood cellular miRNA analysis [113].
Wang et al. [28] conducted a study to compare miRNA expression from PBMCs of active tuberculosis patients and healthy controls and found that four miRNAs (miR-144, miR-365, miR-133a and miR-424) were upregulated and three miRNAs (miR-500, miR-661 and miR-892b) were downregulated in active tuberculosis patients. Further experimentation confirmed that gene expression levels of miR-365 and miR-424 remain significantly different between tuberculosis infected and healthy control individuals. Using the software package BRB Array Tool, they found four miRNAs (miR-130a*, miR-493*, miR-520d-3p, miR-661) upregulated and miR-296-5p downregulated in LTBI compared with healthy controls. This study assessed also miRNA expression in PBMCs from active pulmonary patients and LTBI [28]. They found that 6 miRNAs (miR-21*, miR-223, miR-302a, miR-424, miR-451, miR-486-5p) were upregulated and miR-130b* was downregulated in active tuberculosis. However, further testing and validation are needed to determine whether miRNAs are useful markers to discriminate active tuberculosis and LTBI.
Maertzdorf et al. [113] compared miRNAs expression in serum between active tuberculosis and sarcoidosis patients. This study found 145 miRNAs expressed in both diseases. Further analysis documented that four miRNAs (miR-182, miR-355, miR-15b*, and miR-340) were differentially expressed between these two diseases. From this study we can see, both tuberculosis and sarcoidosis revealed highly similar miRNA profiles. Interestingly, the level of miRNA expression significantly differed between these two diseases and healthy control groups that is consistent with the view that miRNAs are primarily responsible for fine tuning of responses rather than on/off switch genes regulating expression [114].
T cells tend to be at rest in tuberculosis patients, but can be activated by stimulation of specific antigens. Previously, a study documented that Mycobacterium antigen can activate sensitized lymphocytes, leading to elevated inflammatory cytokine production for instance IFN-γ in tuberculosis patients after M. tuberculosis purified protein derivative (PPD) exposure [115]. A study comparing the expression profile PPD-induced miRNAs in PBMCs from tuberculosis patients and healthy controls using polymerase chain reaction (PCR) found that only miR-155 and miR-155* were upregulated in PBMCs from tuberculosis patients [116]. In addition, this study found that the AUC value was 0.897 for miR-155 and 0.794 for miR-155* (Table 1). Based on the highest likelihood ratio, the elicited sensitivity and specificity were 47.62% and 94.74% for miR-155, and 42.86% and 94.74% for miR-155*. This implies that miR-155 and miR-155* may specifically be upregulated in the early immune response during resensitization by tuberculosis-specific antigens. A recent in vitro study found that miR-155 and miR-146a were significantly elevated in human macrophage after M. tuberculosis infection compared to uninfected cells [117]. miR-155 and miR-146a are induced following stimulation of TLR ligands and release of TNF-α and both TLR and TNF-α signaling are important in mounting the immune response to tuberculosis. Previous study also found miR-146a is upregulated following challenge of macrophages with M. tuberculosis ESAT-6 antigen [84]. In contrast, regarding miR-146a, a recent clinical study found that the expression of miR-146a appeared to be decreased in PBMCs of active tuberculosis (pulmonary and extrapulmonary) patients compared with healthy controls [81]. miR-146a has been previously described as a negative regulator of the immune response [118,119] and its systemic downregulation may be associated with the exacerbated inflammatory response observed in tuberculosis patients [120]. In addition this study also found a statistically significant increase in the levels of miR-424 in PBMCs from pulmonary tuberculosis patients compared with healthy individuals [81].
2.3.2.2. miRNA profiling in sputum specimens
Several studies have shown that miRNAs are stably present in sputum [121,122]. Moreover, different studies revealed unique dysregulation of miRNAs in sputum in non-infectious lung diseases and has role in the assessment of lung disease [123,124]. Yi et al. [74] conducted a study to compare miRNA expression in sputum between active pulmonary tuberculosis patients and healthy individuals. This study found that a total of 95 miRNAs (43 miRNAs were overexpressed and 52 miRNAs were underexpressed) were differentially expressed between tuberculosis infected and healthy individuals, which was further supported by cluster analyses indicating a clear distinction between tuberculosis group and healthy individuals. Further validation analysis found that miR-19b-2* was underex-pressed while miR-3179 and miR-147 were overexpressed in the tuberculosis group compared with controls. Fu and colleagues [73] together with serum miRNA profiles, analyzed sputum specimens. In this study miR-93* and miR-29a were present in higher abundance in tuberculosis sputum in comparison to in healthy individuals, but only miR-29a overexpression was validated via PCR [73].
2.3.2.3. miRNA profiling in pleural fluids
A recent study found that was no alteration in measured miRNAs level between pleural fluids mononuclear cells (PFMC) of tuberculosis patients and healthy individuals [81]. In addition, expression of miRNAs were significantly downregulated in PFMCs in comparison to PBMCs. The summary of several miRNAs candidate as possible biomarker to discriminate tuberculosis patients and healthy person is provided in Table 2.
Table 2.
Dysregulation of miRNAs to discriminate tuberculosis patient and healthy person.
| Specimens | Dysregulation pattern in tuberculosis compare to healthy person | Author |
|---|---|---|
| Blood (serum) | Up: miR-144* | Liu et al. [102] |
| Blood (serum) | Up: miR-361-5p, miR-889 and miR-576-3p | Qi et al. [111] |
| Blood (serum) | Up: miR-93* and miR-29a | Fu et al. [73] |
| Blood (serum) | Down: miR-3125 | Fu et al. [73] |
| Blood (PBMC) | Up: miR-424 and miR-365 | Wang et al. [28] |
| Sputum | Down: miR-19b-2* | Yi et al. [74] |
| Sputum | Up: miR-3179 and miR-147 | Yi et al. [74] |
| Sputum | Up: miR-29a | Fu et al. [73] |
| Blood (PBMC) and PFMC | Down: miR-146a | Spinelli et al. [81] |
2.3.3. The role of microRNA to evaluate tuberculosis treatment
Previous study found that H. pylori altered several cellular miRNAs and anti-H. pylori treatment restored miRNAs expression to normal level [16]. The first study to understand the miRNAs expression profile throughout the course of anti-tuberculosis treatment recently was published [81]. In the beginning of treatment, miR-424 was upregulated and miR-164a was downregulated in mononuclear cells of tuberculosis patients. This study found that miR-424 and miR-164a was within normal values after two months of tuberculosis treatment. In addition, blood cytokines analysis indicated that active tuberculosis patients had elevated amounts of circulating IL-1β and IL-6 in the beginning of treatment which decreased upon tuberculosis treatment, as recorded in previous studies [79,80]. Positive association between miR-424 and IL-6 may be explained because this miRNA promotes monocyte-macrophage differentiation [125]. Additionally, the negative relationship between miR-146a and IL-6 in this interaction is consistent with the known role of miR-146a downregulating NF-κB [35]. Data on the effect of anti-tuberculosis therapy on circulating miRNAs or miR-NAs in sputum samples are not available yet.
3. Conclusion
There is significant scientific evidence implicating the central role of miRNAs to modulate a new molecular mechanism of pathogenesis in tuberculosis infection. In addition, blood, sputum and pleural fluid-based studies revealed evidence for the potential roles of specific miRNAs to discriminate tuberculosis infected and healthy individuals, active and latent tuberculosis infections, tuberculosis and other infections and pulmonary diseases, and as biomarkers to evaluate tuberculosis treatment outcomes. Therefore, miRNAs stand to increase our understanding in pathogenesis of tuberculosis, and might be used as new diagnostic strategies for tuberculosis treatment and control in the next decade.
Acknowledgments
Funding: NAH acknowledges support from NIH Biomedical Informatics training grant 2T15LM009451-06.
Footnotes
Competing interests: All authors have none to declare.
Ethical approval: Not required.
Contributor Information
Harapan Harapan, Email: personal@harapanharapan.com.
Fitra Fitra, Email: afalfitra@gmail.com.
Ichsan Ichsan, Email: Ichsanmd_aceh@yahoo.com.
Mulyadi Mulyadi, Email: mul.0862@gmail.com.
Paolo Miotto, Email: miatto.paolo@hsr.it.
Nabeeh A. Hasan, Email: hasann@njhealth.org.
Marta Calado, Email: mrtcalado@gmail.com.
Daniela M. Cirillo, Email: cirillo.daniela@hsr.it.
References
- 1.WHO. Global tuberculosis report. Vol. 2012. World Health Organization; 2012. 2012. [Google Scholar]
- 2.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natarajan K, Kundu M, Sharma P, Basu J. Innate immune responses to M. tuberculosis infection. Tuberculosis. 2011;91:427–31. doi: 10.1016/j.tube.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16:463–96. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalilullah SA, Harapan H, Winardi W, Ichsan I, Mulyadi M. Host genome polymorphisms: implications in tuberculosis susceptibility, severity and development. Unpublished manuscript 2013 [Google Scholar]
- 6.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–62. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belver L, de Yébenes VG, Ramiro AR. MicroRNAs prevent the generation of autoreactive antibodies. Immunity. 2010;33:713–22. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small EM, Olson EN. Pervasive roles of miRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dussaule JC, Guerrot D, Huby AC, Chadjichristos C, Shweke N, Boffa JJ, Chatziantoniou C. The role of cell plasticity in progression and reversal of renal fibrosis. Int J Exp Pathol. 2011;92:151–7. doi: 10.1111/j.1365-2613.2011.00760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harapan H, Andalas M, Mudhakir D, Pedroza NC, Laddha SV, Anand JR. Micro RNA: new aspect in pathobiology of preeclampsia? Egypt J Med Hum Genet. 2012;13:127–31. [Google Scholar]
- 11.Rome S. Are extracellular microRNAs involved in type 2 diabetes and related pathologies. Clin Biochem. 2013 doi: 10.1016/j.clinbiochem.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Krenning G, Zeisberg EM, Kalluri R. The origin of fibroblasts and mechanisms of cardiac fibrosis. J Cell Physiol. 2010;225:631–7. doi: 10.1002/jcp.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinzani M, Rosselli M, Zuckermann M. Liver cirrhosis. Best Pract Res Clin Gastroenterol. 2011;25:281–90. doi: 10.1016/j.bpg.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Wei J, Bhattacharyya S, Tourtellotte WG, Varga J. Fibrosis in systemic sclerosis: emerging concepts and implications for targeted therapy. Autoimmun Rev. 2011;10:267–75. doi: 10.1016/j.autrev.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signature in Helicobacter pylori infected gastric mucosa. Int J Cancer. 2011;128:361–70. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 17.Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68. doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 20.Cui M, Yue L, Fu Y, Yu W, Hou X, Zhang X. Association of microRNA-181c expression with the progression and prognosis of human gastric carcinoma. Hepatogastroenterology. 2013;60(127) doi: 10.5754/hge121333. [DOI] [PubMed] [Google Scholar]
- 21.Dettmer MS, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. Comprehensive microRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid. 2013 Feb 21; doi: 10.1089/thy.2012.0632. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberger N, Walker RC, Kim CH, Winter S, Hunter KW. Inherited variation in miR-290 expression suppresses breast cancer progression by targeting the metastasis susceptibility gene Arid4b. Cancer Res. 2013;73(8):2671–81. doi: 10.1158/0008-5472.CAN-12-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo M, Shen D, Zhou X, Chen X, Wang W. MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery. 2013;12:751–9. doi: 10.1016/j.surg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O'Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–U998. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, Walzl G, Kaufmann SH. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 26.Marín ND, París SC, Rojas M, García LF. Functional profile of CD4+ and CD8+ T cells in latently infected individuals and patients with active TB. Tuberculosis. 2013;93(2):155–66. doi: 10.1016/j.tube.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 2011;6:e20258. doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Yang S, Sun G, Tang X, Lu S, Neyrolles O, Gao Q. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS One. 2011;6(10):e25832. doi: 10.1371/journal.pone.0025832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 30.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–9. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 32.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stittrich AB, Haftmann C, Sgouroudis E, Kühl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, Heinz GA, Fang Z, Li N, Bissels U, Hatam F, Jahn A, Hammoud B, Matz M, Schulze FM, Baumgrass R, Bosio A, Mollenkopf HJ, Grün J, Thiel A, Chen W, Höfer T, Loddenkemper C, Löhning M, Chang HD, Rajewsky N, Radbruch A, Mashreghi MF. The micro-RNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–62. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 34.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DG, Lanier LL. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol. 2010;185:3835–46. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19(R2):R162–8. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein LD. Human genome: end of the beginning. Nature. 2004;431(7011):915–6. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 38.Costa FF. Non-coding RNAs: meet thy masters. Bioessays. 2010;32(7):599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 39.Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13(8):894–7. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 40.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 41.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 43.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 46.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam MM, O'Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol. 2012;41(9):2482–5. doi: 10.1002/eji.201141740. [DOI] [PubMed] [Google Scholar]
- 48.Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia micro-RNAs as new players with clinical significance. Semin Oncol. 2006;33:167–73. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Lawrie CH. MicroRNAs and haematology: small molecules, big function. Br J Haematol. 2007;137:503–12. doi: 10.1111/j.1365-2141.2007.06611.x. [DOI] [PubMed] [Google Scholar]
- 50.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific target-ing of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–34. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prieto DMM, Markert UR. MicroRNAs in pregnancy. J Reprod Immunol. 2011;88:106–11. doi: 10.1016/j.jri.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Ling YK. MicroRNAs in the regulation of immune response against infections. J Zhejiang Univ Sci B. 2013;14:1–7. doi: 10.1631/jzus.B1200292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–89. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011;2011:405310. doi: 10.1155/2011/405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moynagh PN. The NF-kappaB pathway. J Cell Sci. 2005;118:4589–92. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 57.Bruns H, Meinken C, Schauenberg P, Harter G, Kern P, Modlin RL, Antoni C, Stenger S. Anti-TNF immunotherapy reduces CD8+ T cell-mediated antimicrobial activity against Mycobacterium tuberculosis in humans. J Clin Invest. 2009;119:1167–77. doi: 10.1172/JCI38482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–52. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Algood HM, Lin PL, Flynn JL. Tumor necrosis factor and chemokine interactions in the formation and maintenance of granulomas in tuberculosis. Clin Infect Dis. 2005;41(Suppl. 3):S189e93. doi: 10.1086/429994. [DOI] [PubMed] [Google Scholar]
- 60.Jacobs M, Togbe D, Fremond C, Samarina A, Allie N, Botha T, Carlos D, Parida SK, Grivennikov S, Nedospasov S, Monteiro A, Le Bert M, Quesniaux V, Ryffel B. Tumor necrosis factor is critical to control tuberculosis infection. Microb Infect. 2007;9:623e8. doi: 10.1016/j.micinf.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Stenger S. Immunological control of tuberculosis: role of tumour necrosis factor and more. Ann Rheum Dis. 2005;64(Suppl. 4):iv24–8. doi: 10.1136/ard.2005.042531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yadav M, Roach SK, Schorey JS. Increased mitogen-activated protein kinase activity and TNF-α production associated with Mycobacterium smegmatis- but not Mycobacterium avium-infected macrophages requires prolonged stimulation of the calmodulin/calmodulin kinase and cyclic AMP/protein kinase A pathways. J Immunol. 2004;172:5588–97. doi: 10.4049/jimmunol.172.9.5588. [DOI] [PubMed] [Google Scholar]
- 63.Schoenborn JR, Wilson CB. Regulation of interferon-γ during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 64.North RJ, Jung YJ. Immunity to tuberculosis. Annu Rev Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 65.Redford PS, Boonstra A, Read S, Pitt J, Graham C, Stavropoulos E, Bancroft GJ, O'Garra A. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur J Immunol. 2010;40:2200–10. doi: 10.1002/eji.201040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper AM, Mayer-Barber KD, Sher A. Role of innate cytokines in mycobacterial infection. Mucosal Immunol. 2011;4:252–60. doi: 10.1038/mi.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, Kugler D, Hieny S, Caspar P, Núñez G, Schlueter D, Flavell RA, Sutterwala FS, Sher A. Caspase-1 independent IL-1beta production is critical for host resistance to Mycobacterium tuberculosis and does not require TLR signaling in vivo. J Immunol. 2010;184:3326–30. doi: 10.4049/jimmunol.0904189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redford PS, Murray PJ, O'Garra A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011;4:261–70. doi: 10.1038/mi.2011.7. [DOI] [PubMed] [Google Scholar]
- 69.Fremond CM, Togbe D, Doz E, Rose S, Vasseur V, Maillet I, Jacobs M, Ryffel B, Quesniaux VF. IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J Immunol. 2007;179:1178–89. doi: 10.4049/jimmunol.179.2.1178. [DOI] [PubMed] [Google Scholar]
- 70.Kaufmann SH, McMichael AJ. Annulling a dangerous liaison: vaccination strategies against AIDS and tuberculosis. Nat Med. 2005;11:S33–44. doi: 10.1038/nm1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 72.Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 2011;4:279–87. doi: 10.1038/mi.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. 2011;49:4246–51. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One. 2012;7(8):e43184. doi: 10.1371/journal.pone.0043184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–9. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 76.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat Struct Mol Biol. 2009;16:23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 77.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–45. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 78.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009;106:15819–24. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bongiovanni B, Díaz A, D'Attilio L, Santucci N, Dídoli G, Lioi S, Nannini LJ, Gardeñez W, Bogue C, Besedovsky H, del Rey A, Bottasso O, Bay ML. Changes in the immune and endocrine responses of patients with pulmonary tuberculosis undergoing specific treatment. Ann N Y Acad Sci. 2012;1262:10–5. doi: 10.1111/j.1749-6632.2012.06643.x. [DOI] [PubMed] [Google Scholar]
- 80.Santucci N, D'Attilio L, Kovalevski L, Bozza V, Besedovsky H, del Rey A, Bay ML, Bottasso O. A multifaceted analysis of immune-endocrine-metabolic alterations in patients with pulmonary tuberculosis. PLoS One. 2011;6(10):e26363. doi: 10.1371/journal.pone.0026363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spinelli SV, Diaz A, D'Attilio L, Marchesini MM, Bogue C, Bay ML, Bottasso OA. Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol Immunol. 2013;53:265–9. doi: 10.1016/j.molimm.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 82.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11(2):141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 83.Wu Z, Lu H, Sheng J, Li L, Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012;586:2459–67. doi: 10.1016/j.febslet.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 84.Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, Ghosh Z, Sharma P, Kundu M, Basu J. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol. 2012;14(10):1620–31. doi: 10.1111/j.1462-5822.2012.01827.x. [DOI] [PubMed] [Google Scholar]
- 85.Riendeau CJ, Kornfeld H. THP-1 cell apoptosis in response to mycobacterial infection. Infect Immun. 2003;71:254–9. doi: 10.1128/IAI.71.1.254-259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, Suar M, Kaer LV, Bishai WR, Das G. Mycobacterium tuberculosis controls MicroRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. 2013;288:5056–61. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rajaram MVS, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A. 2011;108:17408–13. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, Clark AR. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol. 2006;26:2408–18. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-kappaB activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J Immunol. 2010;184:5029–37. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. 2009;9:514–20. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 92.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Upregulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2010;286:1436–44. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van den Eeden SJ, Geluk A, Poot A, van der Marel G, Beijersbergen RL, Overkleeft H, Ottenhoff TH, Neefjes J. Intracellular bacterial growth is controlled by a kinase network around PKB/Akt1. Nature. 2007;450:725–30. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- 95.Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140(5):731–43. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 96.Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., 3rd A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–7. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 97.Neininger A, Kontoyiannis D, Kotlyarov A, Winzen R, Eckert R, Volk HD, Holtmann H, Kollias G, Gaestel M. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J Biol Chem. 2002;277:3065–8. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- 98.Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol. 2006;26:2399–407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil CA, Balaji KN. Micro-RNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol Cell Biol. 2012;32(12):2239–53. doi: 10.1128/MCB.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Jiang R, Takayama H, Tanaka Y. Survival of virulent Mycobacterium tuberculosis involves preventing apoptosis induced by Bcl-2 upregulation and release resulting from necrosis in J774 macrophages. Microbiol Immunol. 2005;49:845–52. doi: 10.1111/j.1348-0421.2005.tb03673.x. [DOI] [PubMed] [Google Scholar]
- 101.Gan H, Lee J, Ren F, Chen M, Kornfeld H, Remold HG. Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat Immunol. 2008;9(10):1189–97. doi: 10.1038/ni.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011;48:1084–90. doi: 10.1016/j.molimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 103.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 104.Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104(50):19849–54. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Igarashi K, Ochiai K, Muto A. Architecture and dynamics of the transcription factor network that regulates B-to-plasma cell differentiation. J Biochem. 2007;141:783–9. doi: 10.1093/jb/mvm106. [DOI] [PubMed] [Google Scholar]
- 106.Wallis RS, Pai M, Menzies D, Doherty TM, Walzl G, Perkins MD, Zumla A. Biomarkers and diagnostics for tuberculosis: progress, needs, and translation into practice. Lancet. 2010;375(9729):1920–37. doi: 10.1016/S0140-6736(10)60359-5. [DOI] [PubMed] [Google Scholar]
- 107.Elbek O, Uyar M, Aydin N, Börekçi S, Bayram N, Bayram H, Dikensoy O. Increased risk of tuberculosis in patients treated with antitumor necrosis factor alpha. Clin Rheumatol. 2009;28:421–6. doi: 10.1007/s10067-008-1067-x. [DOI] [PubMed] [Google Scholar]
- 108.Sichletidis L, Settas L, Spyratos D, Chloros D, Patakas D. Tuberculosis in patients receiving anti-TNF agents despite chemoprophylaxis. Int J Tuberc Lung Dis. 2006;10:1127–32. [PubMed] [Google Scholar]
- 109.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, Zhuang R, Zhang Y, Hu H, Liu C, Xu L, Wang J, Shen H, Zhang J, Zen K, Zhang CY. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–8. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 110.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–61. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X, Yang D, Yu H, Cui L, Shan Y, Zhou M, Wang H, Lu Z. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis. 2012;12:384. doi: 10.1186/1471-2334-12-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brunagel G, Ruecker VA, Muller SC, Ellinger J. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maertzdorf J, Weiner J, Mollenkopf HJ, Network TB, Bauer T, Prasse A, Müller-Quernheim J, Kaufmann SH, Bellinger O, Diel R, Ehlers S, Heykes-Uden H, Hölscher C, Kaufmann SH, Lange C, Meyer C, Niemann S, Nöthlings U, Reiling N, Schaberg T, Stenger S. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A. 2012;109:7853–8. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 115.Stern JN, Keskin DB, Romero V, Zuniga J, Encinales L, Li C, Awad C, Yunis EJ. Molecular signatures distinguishing active from latent tuberculosis in peripheral blood mononuclear cells, after in vitro antigenic stimulation with purified protein derivative of tuberculin (PPD) or Candida: a preliminary report. Immunol Res. 2009;45:1–12. doi: 10.1007/s12026-008-8024-2. [DOI] [PubMed] [Google Scholar]
- 116.Wu J, Lu C, Diao N, Zhang S, Wang S, Wang F, Gao Y, Chen J, Shao L, Lu J, Zhang X, Weng X, Wang H, Zhang W, Huang Y. Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Hum Immunol. 2012;73(1):31–7. doi: 10.1016/j.humimm.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 117.Walter ND, Bemis L, Edwards M, Ovrutsky A, Ramamoorthy P, Bai X, Geraci M, Chan ED. Differential expression of microRNA in Mycobacterium tuberculosis-infected human macrophages. Am J Respir Crit Care Med. 2012;185:A1011. [Google Scholar]
- 118.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 119.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–51. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 120.Bottasso O, Bay ML, Besedovsky H, del Rey A. The immuno-endocrine component in the pathogenesis of tuberculosis. Scand J Immunol. 2007;66:166–75. doi: 10.1111/j.1365-3083.2007.01962.x. [DOI] [PubMed] [Google Scholar]
- 121.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–6. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–8. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oglesby IK, McElvaney NG, Greene CM. MicroRNAs in inflammatory lung disease – master regulators or target practice? Respir Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pottelberge GR, Mestdagh P, Bracke KR, Thas O, Durme YM, Joos GF, Vandesompele J, Brusselle GG. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- 125.Rahimian A, Soleimani M, Kaviani S, Aghaee-Bakhtiari SH, Atashi A, Arefian E, Nikougoftar M. Bypassing the maturation arrest in myeloid cell line U937 by over-expression of microRNA-424. Hematology. 2011;16:298–302. doi: 10.1179/102453311X13085644680140. [DOI] [PubMed] [Google Scholar]


