Abstract
Riboswitches are structured non-coding RNA elements that control the expression of their embedding messenger RNAs by sensing the intracellular concentration of diverse metabolites. As the name suggests, riboswitches are dynamic in nature so that studying their inherent conformational dynamics and ligand-mediated folding is important for understanding their mechanism of action. Single molecule fluorescence energy transfer (smFRET) microscopy is a powerful and versatile technique for studying the folding pathways and intra- and intermolecular dynamics of biological macromolecules, especially RNA. The ability of smFRET to monitor intramolecular distances and their temporal evolution make it a particularly insightful tool for probing the structure and dynamics of riboswitches. Here, we detail the general steps for using prism-based total internal reflection fluorescence (TIRF) microscopy for smFRET studies of the structure, dynamics and ligand binding mechanisms of riboswitches.
Keywords: Riboswitches, smFRET, conformation, dynamics, folding
1. INTRODUCTION
Riboswitches are present in up to 4% of all bacterial mRNAs, usually in the 5’-untranslated regions (5’-UTRs) (Breaker, 2011; Breaker, 2012; Serganov & Nudler, 2013; Winkler & Breaker, 2005). They are structured domains that regulate gene expression in response to a physiological signal. This physiological signal is most commonly a change in the concentration of a metabolite; but riboswitches that sense temperature, pH and metal ions have also been discovered (Bastet, Dube, Masse & Lafontaine, 2011; Peselis & Serganov, 2014). Many different classes of riboswitches have been identified that bind metabolites—such as nucleobases and their derivatives, amino acids, coenzymes, second messengers and specific metal ions—to control the expression of proteins involved in essential cellular pathways (Barrick & Breaker, 2007; Peselis et al., 2014; Serganov et al., 2013). Riboswitches control gene expression largely through intrinsic transcription termination or inhibition of translation initiation, although some that modulate mRNA splicing, mRNA degradation, and Rho protein mediated termination have also been discovered (Bastet et al., 2011). Riboswitches consist of a highly conserved aptamer domain that is involved in ligand sensing, followed by a variable expression platform (or gene regulatory element) that undergoes a structural change in response to ligand binding by the aptamer. Both domains share a common sequence referred to as the ‘switching’ sequence that communicates the ligand bound state of the aptamer domain to the expression platform (Garst & Batey, 2009; Serganov et al., 2013). Currently, more than 20 classes of riboswitches are known that bind chemically diverse ligands (Peselis et al., 2014; Serganov et al., 2013). In certain cases, multiple classes of riboswitches, with distinct secondary and tertiary structures, have been identified that recognize a common ligand. Examples include the more than five classes of riboswitches recognizing the coenzyme SAM and three classes of preQ1 binding riboswitches (Serganov et al., 2013, McCown, Liang, Weinberg & Breaker, 2014).
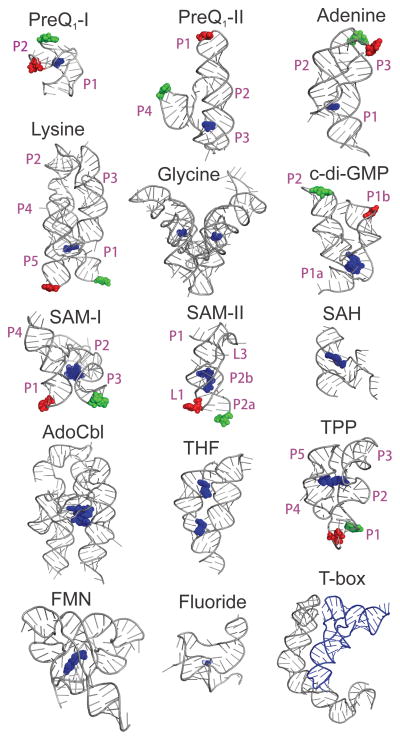
Over the past decade, high-resolution crystal and NMR structures of various classes of ligand-bound riboswitch aptamer domains have been solved that provide insight into the molecular recognition principles used by RNA (Fig. 16.1) (Peselis et al., 2014; Serganov et al., 2013). Structures range from simple pseudoknots to large RNAs with multi-helix junctions. These structures highlight the diversity in architecture and size of riboswitch aptamer domains and show how RNAs can utilize a limited repertoire of functional groups to achieve high specificity and affinity for their typically small cognate ligands. For RNA, like all other biological macromolecules, structural dynamics are crucial for proper biological function (Al-Hashimi & Walter, 2008; Dethoff, Chugh, Mustoe & Al-Hashimi, 2012). Due to a rugged free energy landscape and inherent flexibility, RNA structures can adopt multiple conformations that interconvert on a range of timescales (Al-Hashimi et al., 2008; Marek, Johnson-Buck & Walter, 2011; Mustoe, Brooks & Al-Hashimi, 2014; Solomatin, Greenfeld, Chu & Herschlag, 2010; Zhuang, Kim, Pereira, Babcock, Walter & Chu, 2002). Riboswitch RNAs are intrinsically dynamic in nature, and their structural dynamics play a critical role in the ligand-mediated folding process (Haller, Souliere & Micura, 2011). Riboswitches couple changes in the conformational ensemble caused by ligand binding to effect gene regulation. Although the large number of crystal structures (Fig. 16.1) show atomic details of the ligand recognition mode, these static models do not provide information on the dynamics or the ligand-mediated folding pathways that are critical for riboswitch function. In addition, ligand-free riboswitches sample multiple closely related and, in some instances, transient conformations, which are non-trivial to detect using methods that average across ensembles, and often across time. Due to the challenges in studying ligand-free riboswitch structure and dynamics, our understanding of the ligand-mediated folding process that forms the basis of gene regulation by riboswitches is far from complete (Liberman & Wedekind, 2012).
Figure 16.1. Structures of different riboswitch classes.
Structures of the preQ1-I (PDB: 2L1V (Kang, Peterson & Feigon, 2009)), preQ1-II (PDB: 4JF2 (Liberman, Salim, Krucinska & Wedekind, 2013)), adenine (PDB: 1Y26 (Serganov, Yuan, Pikovskaya, Polonskaia, Malinina, Phan, et al., 2004)), lysine (PDB: 3D0U (Garst, Heroux, Rambo & Batey, 2008)), glycine (PDB: 3P49 (Butler, Xiong, Wang & Strobel, 2011)), c-di-GMP (PDB: 3IWN (Kulshina, Baird & Ferre-D'Amare, 2009)), SAM-I (PDB: 3IQR (Stoddard, Montange, Hennelly, Rambo, Sanbonmatsu & Batey, 2010)), SAM-II (PDB: 2QWY (Gilbert, Rambo, Van Tyne & Batey, 2008)), SAH (PDB: 3NPQ (Edwards, Reyes, Heroux & Batey, 2010)), AdoCbl (PDB: 4GMA (Johnson, Reyes, Polaski & Batey, 2012)), THF (PDB: 4LVV (Trausch & Batey, 2014) ), TPP (PDB: 2GDI (Serganov, Polonskaia, Phan, Breaker & Patel, 2006)), FMN (PDB: 2YIE (Vicens, Mondragon & Batey, 2011)), Fluoride (PDB: 4ENC (Ren, Rajashankar & Patel, 2012)) and the T-box (PDB: 4LCK (Zhang & Ferre-D'Amare, 2013)) riboswitches are shown. Riboswitch RNAs are shown in cartoon representation in grey with their ligands in blue. Green and red spheres indicate nucleotides labeled with Cy3 and Cy5 fluorophores, respectively, in the preQ1-I (Suddala et al., 2013), preQ1-II (Souliere et al., 2013), adenine (Lemay et al., 2006), lysine (Fiegland et al., 2012), c-di-GMP (Wood et al., 2012), SAM-I (Heppell et al., 2011), SAM-II (Haller et al., 2011) and TPP (Haller et al., 2013) riboswitches that were studied using smFRET microscopy. The xpt guanine riboswitch (Brenner, Scanlan, Nahas, Ha & Silverman, 2010, not shown) is very similar to the adenine riboswitch. In smFRET studies of riboswitches where multiple constructs were used, only one of them is shown here for clarity. Structures are not drawn to scale.
In recent years, a number of biophysical studies using nuclear magnetic resonance (NMR) spectroscopy, molecular dynamics (MD) simulations, small angle x-ray scattering (SAXS) and single molecule approaches have provided details of the conformation, dynamics and effect of ligand on the folding kinetics of different riboswitch classes (Brenner et al., 2010;; Chen, Zuo, Wang & Dayie, 2012; Feng, Walter & Brooks, 2011; Fiegland, Garst, Batey & Nesbitt, 2012; Frieda & Block, 2012; Haller, Altman, Souliere, Blanchard & Micura, 2013; Haller, Rieder, Aigner, Blanchard & Micura, 2011; Lemay, Penedo, Tremblay, Lilley & Lafontaine, 2006; Noeske, Buck, Furtig, Nasiri, Schwalbe & Wohnert, 2007; Reining, Nozinovic, Schlepckow, Buhr, Furtig & Schwalbe, 2013; Rieder, Kreutz & Micura, 2010; Souliere, Altman, Schwarz, Haller, Blanchard & Micura, 2013; Suddala, Rinaldi, Feng, Mustoe, Eichhorn, Liberman, et al., 2013; Whitford, Schug, Saunders, Hennelly, Onuchic & Sanbonmatsu, 2009). These studies point to a general mechanism where in the presence of Mg2+, the ligand-free riboswitch exists in multiple interconverting conformations including ‘folded-like’ states that become stabilized upon ligand binding. Divalent metal ions (mainly Mg2+) are known to be crucial for RNA folding by stabilizing tertiary interactions (Johnson-Buck, McDowell & Walter, 2011; Misra & Draper, 1998). In the case of many riboswitches, Mg2+ was shown to be essential for the RNA to sample ‘folded’-like conformations in the absence of ligand. Although Mg2+ is not required for ligand binding, it was shown to generally accelerate ligand dependent folding and slow down the unfolding rate (Brenner et al., 2010; Haller et al., 2011; Lemay et al., 2006; Santner, Rieder, Kreutz & Micura, 2012). However, despite a wealth of structural knowledge on ligand-bound conformations, there is a scarcity of information on ligand-free riboswitch conformations. Crystal structures of a few classes of ligand-free riboswitches are available and they resemble the ligand-bound ones with only local conformational differences around the ligand binding site (Peselis et al., 2014). However, when studied in solution under ambient conditions, ligand-free conformations are generally observed to be more extended than in the (frozen) crystal. Biophysical methods such as SAXS and NMR spectroscopy have been used to interrogate the ligand-free conformations of riboswitches (Baird & Ferre-D'Amare, 2010; Chen et al., 2012; Haller et al., 2011; Reining et al., 2013; Santner et al., 2012). However, due to the difficulties in probing dynamic and lowly populated conformations using such ensemble methods, ligand-free conformations are often recalcitrant to structural inquiry. Consequently, single molecule fluorescence resonance energy transfer (smFRET) has become an increasingly popular tool to avoid ensemble averaging and study ligand-free riboswitch conformations in solution, as well as their dynamics and ligand dependent folding pathways (Liberman et al., 2012; Savinov, Perez & Block, 2014; St-Pierre, McCluskey, Shaw, Penedo & Lafontaine, 2014; Suddala et al., 2013).
1.1. Single molecule FRET
1.1.1 Advantages of single molecule methods
Conventional experiments on biological macromolecules are performed in bulk, where a large number of molecules (typically ~1010–1015) provide an average signal for an observable parameter of interest, such as the catalytic rate constant kcat for enzymes, or the size dimensions or diffusion constant of a molecular species. While such bulk methods are valuable in providing information on the general behavior of a molecule and will continue to be useful, they suffer from an important problem – ensemble and time averaging. Ensemble methods provide only limited information on the distribution of the observable parameter across molecules in the sample but typically only a single average value, thus leading to a loss of valuable information (Deniz, Mukhopadhyay & Lemke, 2008; Tinoco & Gonzalez, 2011; Walter & Bustamante, 2014; Walter, Huang, Manzo & Sobhy, 2008). For example, when ensemble methods are used to study a biomolecule (such as a riboswitch) that exists in equal populations of two distinct states, they often report an average state that is not a real conformation. (The only exceptions are cases where the timescale of the measurement is significantly shorter than the interconversion speed of the states such that two distinct state signals are detected, for example, using certain NMR and ensemble FRET techniques (Bothe, Nikolova, Eichhorn, Chugh, Hansen & Al-Hashimi, 2011; Walter, Burke & Millar, 1999). When more than two states are involved, even this possibility becomes remote. In addition, the presence of any lowly populated, transient states (sometimes – ambiguously – referred to as ‘excited’ states) is extremely challenging to detect using ensemble methods (Dethoff, Petzold, Chugh, Casiano-Negroni & Al-Hashimi, 2012; Tinoco et al., 2011; Walter et al., 2014; Walter et al., 2008). Therefore, single molecule methods are ideally suited for studying dynamic biomolecular systems, such as riboswitch RNAs that generally exist in multiple distinct conformations (Brenner et al., 2010; Fiegland et al., 2012; Haller et al., 2013; Haller et al., 2011; Heppell, Blouin, Dussault, Mulhbacher, Ennifar, Penedo, et al., 2011; Lemay et al., 2006; Suddala et al., 2013; Wood, Ferre-D'Amare & Rueda, 2012; Zhuang, 2005; Zhuang, Bartley, Babcock, Russell, Ha, Herschlag, et al., 2000). In addition, the ability to observe a single molecule for a long period of time using smFRET enables studies of dynamic and static heterogeneity (where molecules do and do not interconvert in their behavior (Marek et al., 2011), respectively, over the available observation time window). Systems can be studied both under equilibrium and non-equilibrium conditions, and the rate constants for conversion between different conformations can be obtained (Paudel & Rueda, 2014; Roy, Hohng & Ha, 2008; Tinoco et al., 2011; Zhao & Rueda, 2009; Zhuang et al., 2002). Furthermore, single molecule methods are usually performed at very low (~pM) concentrations and therefore require little material to work with. This is especially advantageous for studying systems that aggregate or form multimers at higher concentrations. Due to the many advantages, over the past two decades smFRET has been applied to study the structure, folding and dynamics of biomolecules such as DNA, RNA, and proteins, as well as for investigating large macromolecular assemblies (Deniz et al., 2008; Krishnan, Blanco, Kahlscheuer, Abelson, Guthrie & Walter, 2013; Marshall, Aitken, Dorywalska & Puglisi, 2008; Savinov et al., 2014; Schuler & Eaton, 2008; Tinoco et al., 2011; Vafabakhsh & Ha, 2012; Zhuang, 2005).
1.1.2 Fluorescence resonance energy transfer (FRET)
Fluorescence or Förster resonance energy transfer (FRET) refers to the non-radiative energy transfer between donor and acceptor fluorophores that are spatially proximal to each other (Roy et al., 2008; Stryer, 1978). To be suitable as a FRET pair, the emission spectrum of the donor needs to overlap with the excitation spectrum of the acceptor. Energy transfer via FRET occurs only when the fluorophores are present within a certain distance that depends on the identity of the FRET pair and is generally <100 Å. Therefore, FRET can be used as a sensitive spectroscopic ruler to measure intra- or inter-molecular distances in the nm range (~3 – 7 nm) (Fig. 16.2A) (Ditzler, Aleman, Rueda & Walter, 2007; Roy et al., 2008; Schuler, Lipman, Steinbach, Kumke & Eaton, 2005).
Figure 16.2. Prism-based TIRF microscopy setup for smFRET studies of riboswitches, as previously described (Suddala et al., 2013.
(A) FRET efficiency dependence on inter-fluorophore distance. Donor (D) and acceptor (A) fluorophores are shown in green and red, respectively. R0 refers to the Förster radius. The linear range of the FRET measurements is shown in grey. (B) Photograph of the quartz slide with microfluidic channel for smFRET experiments. (C) Schematic of the smFRET experiment for studying dynamics of the Bsu preQ1-I riboswitch (Suddala et al., 2013). The secondary structure of the riboswitch is shown in black and the ligand (preQ1) is shown in yellow. (D) Prism-based TIRF microscopy setup. M1, M2, M3, mirrors; D1, D2, dichroic mirrors; L1, L2, lenses; CCD, ICCD camera.
The excitation energy of the donor is transferred to the acceptor through a dipole-dipole coupling interaction that is distance dependent. The FRET efficiency, E, is given by the equation:
| (1) |
where R is the distance between the donor and the acceptor fluorophores. R0, known as the Förster radius, refers to the distance between the fluorophores where the energy transfer efficiency is 50% (Fig. 16.2A). The value of R0 depends on the spectroscopic properties of the donor and acceptor fluorophores and the local environment. Generally, R0 is constant for a given FRET pair under similar buffer conditions. The R0 values for commonly used FRET pairs range from 40–60 Å. For example, the R0 value for the Cy3 (donor) - Cy5 (acceptor) pair of fluorophores is ~54 Å, with a roughly linear dependency of the FRET efficiency from ~30–70 Å (Roy et al., 2008). Therefore, distance changes below 30 Å and above 75 Å cannot be easily distinguished and will yield FRET values close to 1 and 0, respectively (Fig. 16.2A).
FRET results in a decrease in the intensity of the donor fluorophore with a simultaneous increase in the acceptor fluorophore intensity. Combining FRET with single molecule detection results in a powerful technique known as single molecule FRET (smFRET), which has been used to probe the conformational dynamics of a variety of biomolecules and large complexes (Haller et al., 2013; Krishnan et al., 2013; Marshall et al., 2008; Roy et al., 2008; Savinov et al., 2014; Schuler et al., 2008; Suddala et al., 2013; Vafabakhsh et al., 2012; Zhuang, 2005). In smFRET experiments, one generally monitors the donor and acceptor fluorophore intensities of individual molecules as the apparent FRET efficiency (E), calculated as:
| (2) |
where ID and IA are the background corrected donor and acceptor intensities, respectively. For molecules that are judiciously labeled with a FRET pair, structural dynamics manifest as anti-correlated changes in the intensities of the donor and acceptor fluorophores. Immobilizing molecules permits long observation times of their FRET changes, which in turn make it possible to study their conformational dynamics and folding (Fig. 16.2C). smFRET investigation of riboswitches is sometimes performed using confocal microscopy, but more commonly employing total internal reflection fluorescence (TIRF) microscopy (Fiegland et al., 2012; Haller et al., 2013; Haller et al., 2011; Savinov et al., 2014; Suddala et al., 2013). Here, we limit our discussion to the general steps in smFRET studies of riboswitches using prism-based TIRF microscopy, due to its simplicity and wide applicability. For detailed protocols of the different sections discussed here, the reader is encouraged to also consult some of the previously published reviews (Joo & Ha, 2012; Joo & Ha, 2012; Rinaldi, 2014; Roy et al., 2008; Zhao et al., 2009).
2. METHODS
2.1. Labeling and purification of riboswitches
For studying riboswitch structure and dynamics using smFRET, molecules have to be labeled with both donor and acceptor fluorophores (Roy et al., 2008; Solomatin & Herschlag, 2009; Walter, 2003; Walter & Burke, 2000). In addition, typically a biotin moiety needs to be present on the molecule for immobilization onto the quartz slide surface using biotin-streptavidin chemistry (Fig. 16.2C). A key aspect of obtaining a doubly fluorophore labeled RNA is the judicious selection of nucleotide positions for labeling (Fig. 16.1). Depending on the structure of the aptamer, the fluorophores should be positioned such that they report large-scale distance changes between functionally important regions forming the key tertiary interactions involved in ‘switching’. For example, in the adenine and guanine binding riboswitches, fluorophores were placed adjacent to the hairpin loops P2 and P3 that form a critical tertiary interaction stabilized by ligand binding (Brenner et al., 2010; Lemay et al., 2006) (Fig. 16.1). Similarly, for riboswitches adopting a pseudoknot fold, such as the preQ1-I and SAM-II classes, the fluorophores should be attached to report formation of the thermodynamically less stable helix (generally P2 or P3, closest to the 3’-end) that is stabilized by the ligand (Haller et al., 2011; Souliere et al., 2013; Suddala et al., 2013) (Figs. 16.1, 16.2C). In this regard, the availability of a high-resolution crystal or NMR structure will significantly aid in choosing which nucleotides to label. Structural knowledge is especially valuable for labeling riboswitches with complex architectures. For simpler riboswitches, RNA secondary structure along with nucleotide conservation data may suffice for the selection of suitable labeling positions. In the absence of a high-resolution structure for the riboswitch, chemical structure probing data using methods such as selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) (Wilkinson, Merino & Weeks, 2006) or in-line probing (Regulski & Breaker, 2008) can reveal the identity of nucleotides that form intramolecular interactions (or inter-molecular interactions with the ligand), as well as those exposed to solvent. As a rule of thumb, nucleotides that are evolutionarily less conserved, solvent exposed and not involved in any intramolecular interactions should be chosen for fluorophores labeling. This strategy will ensure that the presence of the bulky fluorophores (molecular weight approximately equivalent to the size of 1–2 nucleotides) is unlikely to affect the structure, folding and/or ligand binding by the riboswitch. Furthermore, the labeling sites should be positioned such that they report distance changes within the linear FRET range of the fluorophores for maximum sensitivity (Fig. 16.2A). The larger the change in the FRET values, the easier it will be to distinguish them, aiding in the subsequent data analysis. Ideally, the labeling sites should be chosen such that a minimum difference of 0.2 (as defined by equation 2) is obtained between any two FRET states.
There are many ways of achieving site-specific labeling of RNA with fluorophores for smFRET experiments (Rinaldi, 2014; Solomatin et al., 2009; Walter, 2003; Walter et al., 2000). However, the most common and easy way of internally labeling riboswitch RNAs is to conjugate N-hydroxysuccinimide (NHS) ester derivatives of fluorophores to free primary amine functional groups of modified nucleotides. Aminoallyl uridine is incorporated into the sequence in the place of uridine during chemical synthesis of oligoribonucleotide (RNA) and labeled using an NHS ester fluorophore. In recent years, due to an increase in the efficiency of RNA chemical synthesis, ordering custom designed singly or doubly fluorophore labeled RNAs with additional modifications, such as 3’ or 5’ biotin, has become affordable. Currently, this is feasible for RNAs that are <70 nt in length, such as the small preQ1-I riboswitches (Suddala et al., 2013). Internal labeling of large riboswitch RNAs however, is more challenging and often involves multiple steps (Rinaldi, 2014; Solomatin et al., 2009). One commonly used method involves chemically synthesizing two or more short RNAs which are labeled independently and covalently linked by T4 DNA/RNA ligase mediated splinted ligation (Lang & Micura, 2008). This method was used to generate a doubly-labeled construct for smFRET study of the SAM-II and the preQ1-II riboswitches (Haller et al., 2011; Souliere et al., 2013). A similar strategy was used for the pbuE adenine, the TPP and the c-di-GMP riboswitches, where two RNA oligonucleotides with internally labeled fluorophores and a 5’-biotin were simply heat annealed and then ligated using T4 RNA/DNA ligase (Haller et al., 2013; Lemay et al., 2006; Wood et al., 2012). Alternatively, for even larger riboswitches, site-specific labeling is difficult but a doubly-labeled construct can be obtained by annealing fluorescently labeled oligonucleotides to in vitro transcribed RNA. For the large lysine riboswitch, a strategy was successfully used where a biotin-labeled DNA oligonucleotide and a Cy3-Cy5 doubly-labeled RNA oligonucleotide were both annealed to a 179-nt in vitro transcribed lysC RNA to generate the smFRET construct (Fiegland et al., 2012). Sequences can be added to the riboswitch for the purpose of annealing oligonucleotides, but structural interference has to be carefully avoided.
For labeling of RNA at the 3’ or 5’ end, a free amine group can be introduced during chemical synthesis, which can then be reacted with a fluorophore NHS ester (Qin & Pyle, 1999; Rinaldi, 2014). Another means of 3’-end labeling of RNA is oxidization with sodium periodate followed by labeling with a hydrazide derivatized fluorophore or biotin. In addition, the 5’-end with a phosphate group can be activated with EDC (1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride). The product can be reacted with imidazole and ethylenediamine to generate a primary amine that can be subsequently labeled with any NHS-ester conjugated fluorophore or biotin. Alternatively, after reaction with EDC, the 5’-end of the RNA can be directly labeled with a hydrazide derivative of the fluorophore (Qin et al., 1999; Rinaldi, 2014). However, it is important to note that the labeling of RNA using EDC or periodate oxidation involves multiple steps and may result in limited yields. Nevertheless, these methods can be useful to end-label in vitro transcribed large riboswitch RNAs. Labeling is often non-trivial and requires multiple steps of purification to remove unlabeled and singly labeled RNAs. Following labeling, excess unreacted dye can be removed by gel filtration and ethanol precipitation. The labeling efficiency can be assessed by measuring the concentrations of RNA and the two fluorophores using a UV-Vis spectrophotometer. If the labeling efficiency is high enough (>80%), the RNA can be directly used for smFRET experiments without further purification. However, purification of the doubly-labeled RNA using denaturing gel electrophoresis or reverse-phase HPLC is recommended when the labeling yield is lower. This enrichment of doubly-labeled riboswitch will enable imaging of hundreds of molecules during subsequent smFRET experiments, resulting in the faster acquisition of statistically significant data.
A variety of chemically diverse fluorophores can be used for smFRET studies of riboswitches (Roy et al., 2008). In general, the cyanine family of dyes is widely used due to their good photostability and quantum yield. The Cy3-Cy5 FRET pair has been most widely used for studying dynamics of RNA by smFRET. Improved derivatives of these dyes such as Cy3B and Dy547, which are known to exhibit higher quantum yield and better photostability, may be used in place of Cy3 (Cooper, Ebner, Briggs, Burrows, Gardner, Richardson, et al., 2004; Roy et al., 2008). Furthermore, a new class of reportedly more stable “self-healing” cyanine fluorophore-triplet quencher conjugates is now available in which the presence of a proximal protective agent suppresses the formation of triplet and radical states, leading to higher photostability of the fluorophore (Zheng, Juette, Jockusch, Wasserman, Zhou, Altman, et al., 2014). These self-healing dyes can also be used for smFRET studies of riboswitches. It is important to note that some of the fluorophores, such as the cyanine family dyes, are known to stack on the ends of DNA and RNA helices, which can lead to mild orientation dependence of the FRET value (Ouellet, Schorr, Iqbal, Wilson & Lilley, 2011). The interactions of dyes with RNA potentially may also alter the rates of structural transitions. Furthermore, some fluorophores, such as those of the Alexa family, are known to be quenched by neighboring bases, which will result in a reduced signal and also photophysical effects such as intensity fluctuations due to spectral shift and triplet state blinking (Stennett, Ciuba & Levitus, 2014). Although the cyanine dyes are less prone to such effects, it is wise to avoid labeling RNAs near a stretch of particularly guanine nucleotides that have redox potentials suitable to exchange electrons with the excited state of a fluorophore (Walter & Burke, 1997). Naturally, it is important to consider all these factors when selecting fluorophore labeling positions.
2.2. Preparation of quartz slides
smFRET experiments on riboswitches are routinely performed on immobilized molecules in a microfluidic channel made using quartz slides (1” x 3” x 1 mm thick, G. Finkenbeiner Inc.) and glass cover slips (24 x 30 mm, VWR Micro Cover Glasses) (Fig. 16.2B) (Michelotti, de Silva, Johnson-Buck, Manzo & Walter, 2010; Roy et al., 2008; Suddala et al., 2013). To this end, the slide surface needs to be thoroughly cleaned using a multi-step protocol to remove any organic impurities (Krishnan et al., 2013; Zhao et al., 2009). Any fluorescent impurities will impede observation of true single molecules and contribute false background signals. Impurities may also affect the local environment of molecules, which may result in heterogeneities in their behavior. The cleaning process first involves boiling the used slides in deionized water to soften the epoxy for removing the cover slips used to make the microfluid channel. Any remaining debris on the slides should be scrapped off using a razor blade. This is followed by scrubbing the slide surface thoroughly with a thick paste of detergent (Alconox, Inc) or alternatively, by sonicating them in a 10 % (w/v) detergent solution for 30 min. After this, the slides are rinsed thoroughly with double-distilled water and sonicated in it for 10 min. Next, slides are sonicated sequentially in a series of four organic and inorganic solvents: a) acetone, b) methanol, c) 5:1:1 (v/v) mixture of double-deionized water:28 % (w/v) ammonium hydroxide:30 % (w/v) hydrogen peroxide, and finally d) 1 M potassium hydroxide. Each sonication step in solvent is performed for at least 30 min, and after every step the slides are rinsed and sonicated in double-deionized water for 10 min. In the end, the slide surface is flamed thoroughly using a propane torch to destroy any remaining organic impurities. A microfluidic channel is made on the clean slides by sandwiching two strips of double-sided sticky tape ~4–6 mm apart between the quartz slide and a clean rectangular glass coverslip (Michelotti et al., 2010; Roy et al., 2008). The edges of the channel are sealed with epoxy (for example, Hardman DOUBLE/BUBBLE Fast-Setting Epoxy) to prevent leakage of the buffer. The microfluidic channel thus made has a low volume of ~30–50 μL that helps decrease background fluorescence from buffer contaminants. A pair of 1 mm holes drilled into the quartz slide will act as inlet and outlet ports for the flow into the channel of buffers containing different molecules. Pipette tips (2–200 μL; epTIPS, Eppendorf) are cut to a length of ~1 cm from the tip and snugly inserted into the two holes of the quartz slide. Into the open ends of the tips, a long ~10 -15 cm plastic tube (0.02” thick, Tygon Tubing) is inserted, followed by sealing the tips and tubing with epoxy. The slides are left at RT for ~30 min for the epoxy to harden. The tubing can be cut to an appropriate length to create an inlet and outlet channel before using the quartz slide for smFRET experiments and flowing buffers containing different solutes (such as ligands, ions, osmolytes, crowding agents) into the channel using a clean syringe (Fig. 16.2B). Buffer exchange enables the convenient study of riboswitch structure and folding under different conditions on a single slide.
2.3. Surface attachment and oxygen scavenging systems
Due to their negative backbone charge, nucleic acids generally do not adsorb significantly even to a bare slide surface. For riboswitches, surface tethering for smFRET is therefore most easily achieved using biotinylated-BSA (bBSA) and streptavidin on plain quartz slides. By contrast, for studies involving proteins, polyethylene glycol (PEG) passivated slides should be used, and both types of slides have been utilized for smFRET studies of riboswitches (Haller et al., 2013; Haller et al., 2011; Suddala et al., 2013). For PEG passivated slides, the surface should be treated with PEG (molecular weight 5000) before making the microfluidic channel to reduce non-specific binding of molecules, especially proteins to the surface. In this method, the clean slide surface is treated with aminosilane, followed by reaction with an NHS-ester modified PEG containing a fraction (5 %) of biotin-PEG. For surface immobilization via bBSA, the microfluidic channel is first coated by flowing 80 μL of 1 mg/mL solution of bBSA and incubating (let sit at room temperature) for 5 min to allow non-specific adsorbtion onto the slide surface. The excess unbound bBSA is washed out with 100 μL of a 1x smFRET buffer of choice (based on experimental requirements), followed by flowing 80 μL of 0.2 mg/mL streptavidin into the channel. After incubation for 5 min, the unbound streptavidin is washed off with 100 μL of 1x buffer. Next, 20–100 pM of doubly-labeled RNA is flowed onto the slide until an optimal density is obtained (~300–400 molecules per field of view) and the unbound molecules are washed away with the 1x buffer.
For imaging the immobilized molecules, a suitable 1x smFRET buffer supplemented with an enzymatic oxygen scavenging system (OSS) should be used that will increase the longevity of the fluorophores before photobleaching, permitting long observation times. A widely used OSS consists of protocatechuic acid (PCA), protocatechuate-3,4-dioxygenase (PCD) enzyme and a triplet state quencher such as Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) to slow down fluorophore photobeaching and reduce dye blinking, respectively (Aitken, Marshall & Puglisi, 2008; Krishnan et al., 2013). Alternatively, a glucose oxidase/catalase based enzymatic system is also widely used (Roy et al., 2008). However, it is important to note that, due to the production of carboxylic acids in both of these systems, the pH of the buffer may decrease over time and needs to be carefully checked (Shi, Lim & Ha, 2010). Therefore, a sufficient strength of buffering species (≥50 mM) should be used when studying riboswitch RNAs, particularly if any putative pH sensitive elements such as non-canonical base pairs are present. Recently, a pyranose oxidase/catalase system was proposed as a better alternative for the smFRET study of such pH sensitive systems (Swoboda, Henig, Cheng, Brugger, Haltrich, Plumere, et al., 2012). In addition, degassing the buffers will aid in decreasing the free molecular oxygen, thereby further prolonging the fluorescence signal.
2.4. smFRET using prism-based TIRF microscopy
smFRET microscopy for studying riboswitch structure and dynamics is generally done using wide-field illumination (Brenner et al., 2010; Haller et al., 2013; Haller et al., 2011; Lemay et al., 2006; Souliere et al., 2013; Suddala et al., 2013; Wood et al., 2012). In this method, fluorescence signal from hundreds of molecules can be recorded simultaneously using a sensitive CCD camera. The molecules are generally immobilized onto the surface of a microfluidic channel for enabling the long observation times (on the order of tens of seconds to minutes) needed for studying the often slow conformational dynamics and folding of riboswitches and/or to record significant numbers of conformational transitions for accurate rate determination (Fig. 16.2C). The immobilized molecules can be excited either by direct illumination as in confocal microscopy or by using an evanescent field in TIRF microscopy (Roy et al., 2008; Walter et al., 2008). The evanescent field is generated using total internal reflection (TIR) of the excitation laser at the quartz-water interface. The TIR of the incident laser can be achieved by using an objective (objective-TIRF microscopy), but more easily by using a prism (prism-based TIRF microscopy, Figs. 16.2C,D). The intensity of the evanescent field decays exponentially with distance from the reflecting surface and therefore only excites immobilized molecules that are within ~100 nm of the surface. The advantage of using TIR microscopy is that it eliminates the background fluorescence from solvent and any free labeled molecules in solution, resulting in a high signal-to-noise (S/N) ratio that is critical for single molecule detection. The prism-based TIRF microscope (Fig. 16.2D) is relatively simple to assemble and therefore has been widely used for studying the conformational dynamics of various riboswitch classes (Haller et al., 2013; Haller et al., 2011; Joo et al., 2012; Roy et al., 2008; Savinov et al., 2014; Suddala et al., 2013).
Fig. 16.2D shows a general schematic for the prism-based TIRF microscopy setup. The quartz slide rests on top of a 1.2 numerical aperture (NA) 60x water immersion objective (e.g., Olympus UplanApo) of an inverted microscope (IX 71, Olympus). The quartz prism is placed on top of the slide containing the microfluidic channel, where it is used to achieve TIR of the excitation laser to generate an evanescent field that excites the surface tethered molecules in the channel. A 532-nm (diode-pumped Nd:YAG, CW) laser is used to excite Cy3 (donor) for FRET measurements and intermittent excitation with a 638-nm red diode laser can be used to check for the presence of Cy5 (acceptor), which helps differentiate donor-only molecules from those showing large-distance zero-FRET states. Mineral oil with a refractive index that matches the quartz slide is placed between the prism and the slide surface to fill any optical gaps. The oil also enables smooth movement of the slide for imaging different regions in the microfluidic channel and prevents scratching the prism surface. The fluorescent emission from the molecules is collected through the objective, and the scattered excitation light is removed using a dichroic mirror. Additionally, the image of the surface-tethered molecules is split into donor and acceptor emissions using a pair of dichroic mirrors (D1, D2, Fig. 16.2D) and two mirrors (M1, M2) to project both colors side-by-side onto two halves of a CCD camera. More details of the prism-based TIRF microscope we use can be found in several references (Michelotti et al., 2010; Roy et al., 2008; Zhao et al., 2009).
2.5. Heat-annealing of riboswitch RNAs
Before immobilization onto the slide, riboswitch RNAs need to be folded into their native conformations. RNA is generally stored at low temperatures (−20 °C or −80 °C) in autoclaved double-distilled water or in near-neutral pH buffers without any divalent cations to prevent its degradation over time. However, under such low-temperature, low-ionic strength conditions and due to repeated freeze-thawing, RNA molecules may misfold or form non-specific aggregates. Heating will break the aggregates and unfold misfolded RNA molecules, allowing them to fold into their native conformations. There is no standard procedure for folding of RNAs, and diverse heat-annealing protocols are used. Commonly, RNA is heated in a buffer with monovalent ions such as Na+ or K+ but without Mg2+ to 60–80 °C for 2–5 min. The RNA is then either rapidly cooled by placing on ice or slowly by allowing to equilibrate to room temperature to fold into its secondary structure. This is followed by the addition of Mg2+ to the required concentration to facilitate the formation of tertiary interactions. For smFRET experiments, heat annealing after diluting the RNA to the required low concentrations of 20–100 pM will prevent the formation of dimers/aggregates, which can be easily identified by multi-step photobleaching in the smFRET experiment. Higher temperatures such as 90 °C should be tried in cases where heating at 75 °C is not effective in breaking the aggregates. As a control, a few different temperature and time regimes should be tried to optimize the folding protocol and to test if the folding protocol has any significant effect on the smFRET results.
3. PRACTICAL EXPERIMENTAL CONSIDERATIONS
Before starting to image single molecules, the slide needs to be coated with bBSA and streptavidin for immobilizing the RNA. For PEG passivated slides, only streptavidin has to be flowed onto the slide as it binds to the biotin-PEG. It is important to visualize the slide surface after flowing the bBSA and/or streptavidin to make sure that it is clean with minimal impurities before immobilizing doubly-labeled riboswitch molecules. Organic fluorescent impurities often look similar to the donor labeled molecules and therefore cannot be distinguished easily. Even minor contamination of any stock solution will render the surface dirty; therefore, all the buffers and solutions for smFRET should be passed through a 0.2 μm filter. For immobilization of riboswitch molecules using a newly labeled stock, start with a low concentration (~20 pM) of the labeled RNA. Optimize the concentration needed to achieve an ideal density of ~300–400 molecules per field of view (256 x 512 pixels, or half of a typical CCD chip). Higher densities will lead to overlap of signals from neighboring spots that makes the identification of single molecules difficult. By contrast, a low density will require more movies to be taken to collect sufficient data. The laser power should be optimized both to maximize the S/N ratio and to prolong the lifetime of the dyes. Higher power will give better signal but will also lead to faster photobleaching. The laser intensity can be controlled using neutral density (ND) filters to reach a compromise between the S/N ratio and the photobleaching time. Different kinds of CCD cameras such as an intensified CCD (ICCD) and the newer electron multiplying CCD (EMCCD) are used for TIRF based smFRET studies of riboswitches. EMCCD cameras, in particular, can be cooled to very low temperatures and may have better sensitivity at faster time resolution (currently ~33 ms for full frame) (Roy et al., 2008; Zhao et al., 2009). The time resolution can be further increased by reading out only part of the chip or by using pixel binning. Choosing the right frame rate for imaging is important and, if possible, molecules should be imaged at multiple time resolutions to capture a range of dwell times. A faster time resolution generally requires using high laser intensities to increase the signal. This will lead to increased photobleaching and therefore slower transitions (with longer dwell times) will be missed. Similarly, faster dynamics will be missed when imaging at a slower camera frame rate. Therefore, for riboswitches with heterogeneous kinetics, the rates of structural dynamics calculated could depend on the choice of the camera frame rate. In addition, when imaging riboswitches by smFRET, it is important to excite at the end with the red laser to check for emission of the acceptor fluorophore. This will help distinguish low-FRET states from donor-only species. Although such low-FRET states may be caused by blinking of the acceptor fluorophore, observation of long-lived states in the presence of a triplet state quencher such as Trolox likely indicates genuine conformations with distances >75 Å. Use of alternative FRET pairs with larger R0 can help in resolving these conformations (Roy et al., 2008). For kinetic measurements, titrations can be performed sequentially on the same slide to minimize variability between experiments. Ligand titrations should sample a few concentrations below the dissociation equilibrium constant (Kd) and a few above it up to saturating conditions (>10 Kd).
4. DATA ANALYSIS
Using wide-field TIRF microscopy, a large number of immobilized molecules are imaged simultaneously and analyzed using custom written Matlab or Visual C++ code, or other programs such as Micro-Manager (plugin for ImageJ, open source) or Metamorph (Molecular Devices, Inc., commercial imaging software) to obtain the raw FRET movies (Blanco & Walter, 2010). Individual smFRET time traces are extracted from the movies using image analysis programs written in IDL (Research Systems, Boulder) or Matlab (MathWorks, Inc). This is done by taking an average image of the first 30 frames for each movie to identify individual spots in both halves of the image (Cy3 and Cy5 spots). After identification of individual spots in one channel, their corresponding spots in the other channel are located and intensities recorded over time. For accurate co-localization of the donor and acceptor spots in both channels, a slide made with surface immobilized red fluorescent beads is used to calibrate the correspondence of the donor and the acceptor images (Churchman, Okten, Rock, Dawson & Spudich, 2005; Roy et al., 2008). Generally, depending on the labeling efficiency and the density of spots on the surface, at least 40–100 smFRET traces can be obtained from each movie. Ergodic behavior is observed when the long time average of one molecule represents the ensemble average of snapshots from many molecules. In most cases, non-ergodic behavior is observed instead, and significant numbers of smFRET traces are needed to characterize the equilibrium properties of a riboswitch, and for direct comparison with ensemble-averaged benchmark measurements (Marek et al., 2011).
4.1. FRET histograms
The smFRET time traces from multiple movies taken under a given condition are combined to generate a data set containing ideally several hundred molecules. These traces provide direct information on the conformational states sampled by individual molecules and the dwell times spent in each of the FRET states before transitioning to other FRET states (Fig. 16.3A). smFRET traces can be analyzed using a number of methods to yield information on the structure and dynamics of the RNA (Blanco et al., 2010). One straightforward way of analyzing the data is to generate a population FRET histogram by binning the first 50 or 100 frames of each time trace (to ensure that the dataset is not biased towards a few longer-lived molecules). Such a histogram directly shows the ensemble FRET distribution of all riboswitch molecules included (Fig. 16.3B). By fitting the histogram with a sum of Gaussian functions, the minimum number of distinct conformations sampled by the riboswitch and their equilibrium distribution can be obtained under a given condition. In addition, the mean value and the width of each Gaussian function can yield more information about the corresponding conformations sampled. Rough distance estimates between the fluorophores in each conformation are estimated from the mean apparent FRET value of the Gaussian curve using equation 1. Depending on the labeling strategy, generally the shortest distance (corresponding to the highest FRET state) should be similar to the distance observed in the ligand-bound crystal structure of the riboswitch. Distances for the other FRET states will provide information on the extent of compactness (degree of foldedness) of those conformations. In general, most riboswitches studied using smFRET have displayed two-state behavior, corresponding to ligand-bound and ligand-free conformations with different properties (Savinov et al., 2014). However, a few riboswitches with more than two states have also been documented (Lemay et al., 2006; Reining et al., 2013).
Figure 16.3. Analysis of smFRET data.
(A) Raw single molecule time trace showing anti-correlated donor (green) and acceptor (red) intensities for the Bsu preQ1-I riboswitch in the presence of 100 nM preQ1 and no Mg2+ (60 ms time resolution, unpublished data). The FRET trace (black) idealized with a two-state HMM fit (cyan) is shown in the lower plot. (B) FRET histogram showing two major populations and their equilibrium distribution. (C) Transition Density Plot (TDP) showing as heat maps the transitions from initial to final FRET states and their frequency. The dwell times of all the molecules in a given FRET state (~0.9 in this example, red broken box) can be extracted from this plot. (D) Cumulative dwell time distribution of the dwell times in the ~0.9 FRET state fit with a single-exponential function. (E) An smFRET trace of the ligand-free Bsu preQ1-I riboswitch with very fast dynamics (33 ms time resolution, unpublished data). (F) Cross-correlation curve of the trace shown in panel E fit with a single-exponential to obtain the combined rate of transitions (k1,2 + k2,1 = 1/τ, for a two-state process).
Folding studies on riboswitches using smFRET routinely probe the effects of ligand and different ions on the FRET distribution and dynamics (Brenner et al., 2010; Fiegland et al., 2012; Haller et al., 2013; Haller et al., 2011; Lemay et al., 2006; Souliere et al., 2013; Suddala et al., 2013; Wood et al., 2012). As a first step, the FRET histogram of the riboswitch is obtained in the absence of both ligand and Mg2+, but in the presence of monovalent cations. Later, the folding of the riboswitch can be probed in the presence of ligand and/or Mg2+ to delineate their effect(s), either individually or together, on the conformational dynamics of the riboswitch. Titrations of Mg2+ or ligand can be performed on the same slide by adjusting the composition of the input solution of the microfluidic flow cell. Such experiments can be used to monitor the effects of increasing ligand or Mg2+ concentrations on the FRET distributions and conformational dynamics of the riboswitch. The occupancy (as seen from the FRET histograms) of one of the conformations, corresponding to the ligand-bound folded state, is expected to increase as a function of ligand concentration. The increasing fraction of the ligand-bound state (typically of high FRET) can be fit with a standard Hill-equation (with n = 1) from which a half-saturation value (K1/2) value can be estimated that relates to the Kd obtained from biochemical methods. Any large discrepancies (>10-fold) between these two values could indicate misfolding of the RNA or fluorophore interference with proper ligand binding and needs to be investigated.
The mean and width of the individual Gaussian peaks can provide additional information on the compactness and dynamic nature of the underlying conformations. The width of individual peaks depends on the instrument noise (shot noise) but also on the conformational behavior of the corresponding structural ensembles (Suddala et al., 2013). A larger width indicates a broad and dynamic conformational ensemble, while a smaller width suggests a stable structure. Following the changes in the mean FRET value and width of the different Gaussian peaks at varying ligand and/or Mg2+ concentrations can also provide additional details on the conformational ensemble and folding of the riboswitch (Suddala et al., 2013).
4.2. Kinetic analysis
The more interesting data from smFRET experiments are the dwell times of individual molecules in different FRET states that can be used to kinetically characterize the ligand-dependent folding pathways of riboswitches (Savinov et al., 2014). For riboswitches exhibiting two distinct states with slow interconversion dynamics, the smFRET traces can be idealized using a defined FRET threshold to obtain the transition kinetics (Blanco et al., 2010). More often, the smFRET traces may be noisy and/or display rapid fluctuations between closely spaced states that need advanced methods to analyze. Statistical methods such as Hidden Markov Modeling (HMM) are used to idealize such smFRET traces in an unbiased manner, to detect transitions between FRET states, and to extract dwell times of individual molecules in the different states (Fig. 16.3A) (Blanco et al., 2010; Qin & Li, 2004). Freely available programs such as QuB, vbFRET and HaMMy, along with custom Matlab scripts are generally used for HMM analysis of smFRET data (Bronson, Fei, Hofman, Gonzalez & Wiggins, 2009; McKinney, Joo & Ha, 2006; Qin et al., 2004). Recently, an extensive data analysis package with a graphical interface named SMART has been described for the objective analysis of complex smFRET data and can be freely downloaded at https://simtk.org/home/smart (Greenfeld, Pavlichin, Mabuchi & Herschlag, 2012).
Using the idealized traces (Fig. 16.3A), a transition density plot (TDP) can be generated that shows the different kinds of transitions and their frequency in all molecules as heat maps (Fig. 16.3C) (Blanco et al., 2010). Dwell times of all molecules in each FRET state before transitioning to a different state can be extracted from the TDP. The dwell times are then plotted as a cumulative distribution plot and fit with an exponential function to extract the rate constants of conformational dynamics between the different states (Fig. 16.3D) (Blanco et al., 2010). For a simple two-state process, the dwell time distributions can be fit with a single-exponential function. However, heterogeneity is often observed in single molecule measurements so that the dwell time distributions are best fit with a sum of exponential functions that suggests the presence of multiple similar structures with different kinetic properties. Careful investigation of the heterogeneities can provide more information about the conformational and dynamic properties of the different FRET states. The heterogeneities in conformational dynamics can also be visualized using a scatter plot of average dwell times in different FRET states for every time trace, and the effect of ligand on this distribution can be studied (Lemay et al., 2006). For a detailed review on the analysis of complex multi-state smFRET data, the reader is referred to a reference (Blanco et al., 2010).
4.3. Cross-correlation analysis
HMM analysis of smFRET traces will fail to detect all transitions in cases where the conformational dynamics of a riboswitch are very fast (Fig. 16.3E) and close to the time resolution of the camera (typically 10–100 ms) (Roy et al., 2008; Suddala et al., 2013). This will result in an underestimation of the rates of structural changes. Imaging the molecules at the fastest achievable frame rate of the camera can help resolve the faster dynamics, but will also lead to a decrease in the S/N ratio. Alternatively, one may switch to a confocal system that uses a point detector to achieve higher time resolution, but only one molecule can be investigated at a time, limiting throughput. In such situations, cross-correlation analysis can be performed on the wide-field smFRET traces to quantify the anti-correlation between the donor and acceptor intensities (Kim, Nienhaus, Ha, Orr, Williamson & Chu, 2002; Ragunathan, Liu & Ha, 2012; Suddala et al., 2013). Cross-correlation (CC) analysis measures the decay in the extent of anti-correlation between the donor and acceptor intensities (auto-correlation of the FRET trace yields the same information) as a function of increasing time-lag between the two traces. In essence, one of the intensity traces is kept fixed while the other trace is moved in the calculation at small time (Δt) increments and the CC value at each step is calculated. The CC value plotted against the time lag is fit with a single-exponential function to obtain the lifetime of the decay (τ, Fig. 16.3F). The inverse of the lifetime yields the sum of rates for transitions between different FRET states (Kim et al., 2002; Ragunathan et al., 2012; Suddala et al., 2013). For individual traces, the rate of transition to a given FRET state can then be obtained by multiplying the fraction of time spent in that state with the sum of rate constants (k1,2 + k2,1 = 1/τ), effectively averaging over any molecular heterogeneity. Multiple smFRET traces with fast dynamics can be analyzed this way to obtain a distribution of rates for riboswitch folding and unfolding. Analyzing how increasing concentrations of ligand affect the rates of conformational dynamics between different FRET states can potentially reveal the mechanism of ligand binding, which is extremely difficult to investigate using ensemble methods (Kim, Lee, Jeon, Choi, Lee, Hohng, et al., 2013).
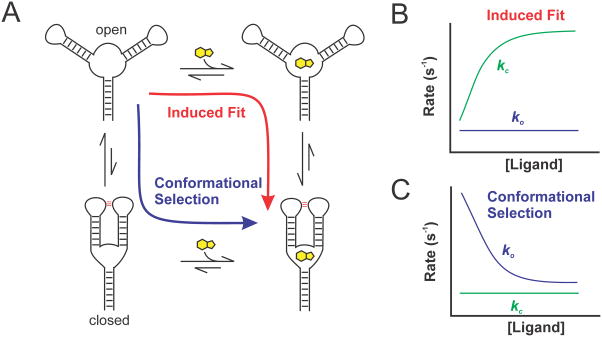
5. INDUCED FIT VERSUS CONFORMATIONAL SELECTION
Kinetic analysis of ligand-dependent conformational dynamics using smFRET can reveal the major folding mechanism of a riboswitch (Hatzakis, 2014; Savinov et al., 2014; Suddala et al., 2013). Traditionally, the ligand-mediated folding pathways for biomolecules such as riboswitches have been classified into two contrasting mechanisms – induced fit and conformational selection (Fig. 16.4A) (Hammes, Chang & Oas, 2009; Kim et al., 2013). In the classical definition of induced fit mechanism, the riboswitch (or receptor) exists predominantly in an “open” ligand-free state and does not sample ligand-bound-like or “closed” conformations in the absence of ligand. Ligand binding to the apo (or open) conformation induces the riboswitch into the folded conformation, which is generally similar to the ligand-bound crystal structure. In conformational selection (also referred to as conformational capture or population shift), the ligand-free ensemble samples a small, but significant population of folded-like (or closed) conformations, albeit transiently. Ligand specifically recognizes such conformations and stabilizes them upon binding. Therefore, induced-fit and conformational selection mechanisms are also sometimes referred to as “binding first” and “folding first” processes (Hammes et al., 2009; Suddala et al., 2013). However, studies on proteins have shown that the two mechanisms are not mutually exclusive, but rather co-exist to different extents depending on the buffer conditions and the relative concentrations of the receptor and ligand (Daniels, Tonthat, McClure, Chang, Liu, Schumacher, et al., 2014; Kim et al., 2013). Therefore, kinetic assays (either ensemble or single molecule) only reveal the major pathway under a chosen set of experimental conditions (Hammes et al., 2009). Calculation of relative flux through each pathway may offer quantitative insight into how ligand concentrations and buffer conditions affects the partitioning of folding through either mechanisms (Daniels et al., 2014). Recently, the process of ligand binding to an open conformation of a receptor that can sample ligand-free, folded-like conformations has been referred to as adaptive induced-fit mechanism, to distinguish it from the classical definition. In addition, binding of ligand to folded-like states followed by local adjustments in the structure has been classified as extended conformational selection mechanism (Csermely, Palotai & Nussinov, 2010).
Figure 16.4. Schematic for the kinetic analysis of ligand-mediated folding mechanism of a riboswitch.
(A) Induced-fit (red) and conformational selection (blue) models of ligand-mediated riboswitch folding. Expected rate constant dependence on ligand concentration for riboswitches folding via (B) induced fit and (C) conformational selection mechanisms. Idealized curves depict the net forward folding, or closing, reaction (kc, green) and the net reverse unfolding, or opening, reaction (ko, blue).
While the absence of any ligand-free closed conformations suggests that folding should proceed via induced-fit like mechanism, the converse is not true. That is, the presence of ligand-free closed conformations does not rule out the possibility that an induced-fit mechanism is in action (Hammes et al., 2009). The differentiating step for both the mechanisms is the conformation to which the ligand preferentially binds. Induced fit and conformational selection can be differentiated by ligand binding to the open and closed conformations, respectively, which generate distinct signatures of ligand-dependent kinetics of conformational dynamics (Fig. 16.4) (Hammes et al., 2009; Kim et al., 2013; Suddala et al., 2013). In the induced-fit mechanism, the rate of closing or folding (kc) increases with ligand concentration and the opening rate (ko) may be slowed or remain unaffected (Fig. 16.4B). In contrast, in conformational selection, the ligand binding has no effect on the closing rate while the opening rate decreases (Fig. 16.4C). This distinction is based on the assumption of a two-state model of riboswitch folding, which is mostly true for many riboswitches (Savinov et al., 2014). For some riboswitches that display more than two conformations, such as the pbuE adenine riboswitch, the kinetic data may be still more heterogeneous and complex and hard to interpret (Lemay et al., 2006; Reining et al., 2013). Taken together, probing riboswitch structure and dynamics using smFRET not only provides valuable information on the conformations sampled but also on the ligand-mediated folding mechanism, which is difficult to probe with other methods (Savinov et al., 2014). Therefore, following the methods described in this article, especially the kinetic analysis of the smFRET data (section 4.2 and Fig. 16.4), will enable a detailed investigation of the ligand mediated folding mechanisms of riboswitches.
6. SUMMARY AND CONCLUSIONS
Understanding gene regulation by riboswitches requires characterizing their structural and dynamic properties. Here, we have provided details of investigating riboswitch conformation and dynamics using smFRET based on TIRF microscopy. The steps described apply to current state-of-the-art studies of dual fluorophore-labeled riboswitch RNAs and the effect of ligand on them using two-color smFRET experiments. In the near future, we anticipate that more complex riboswitches and folding mechanisms will be studied, and increasingly advanced techniques such as three-color smFRET (Hohng, Joo & Ha, 2004; Kim et al., 2013) will be applied to correlate ligand binding and riboswitch folding in real time. In addition, ligand-dependent co-transcriptional riboswitch folding, for which smFRET studies are decidedly non-trivial, may be pursued to gain insight into the increasingly complex nature of cellular folding landscapes of riboswitches (Dangkulwanich, Ishibashi, Bintu & Bustamante, 2014).
Acknowledgments
This work was supported by NIH grant GM062357 and a sub-award on GM063162 (PI Joseph E. Wedekind) to N.G.W.
References
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophysical Journal. 2008;94(5):1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Current Opinion in Structural Biology. 2008;18(3):321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferre-D'Amare AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis (vol 16, pg 598, 2010) RNA. 2010;16(7):1447–1447. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biology. 2007;8(11):R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet L, Dube A, Masse E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Molecular Microbiology. 2011;80(5):1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods in Enzymology. 2010;472:153–178. doi: 10.1016/S0076-6879(10)72011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe JR, Nikolova EN, Eichhorn CD, Chugh J, Hansen AL, Al-Hashimi HM. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nature Methods. 2011;8(11):919–931. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Prospects for riboswitch discovery and analysis. Molecular Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA world. Cold Spring Harbor Perspectives in Biology. 2012;4(2) doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49(8):1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson JE, Fei JY, Hofman JM, Gonzalez RL, Wiggins CH. Learning Rates and States from Biophysical Time Series: A Bayesian Approach to Model Selection and Single-Molecule FRET Data. Biophysical Journal. 2009;97(12):3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chemistry & Biology. 2011;18(3):293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zuo X, Wang YX, Dayie TK. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Research. 2012;40(7):3117–3130. doi: 10.1093/nar/gkr1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(5):1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M, Ebner A, Briggs M, Burrows M, Gardner N, Richardson R, et al. Cy3B (TM): Improving the performance of cyanine dyes. Journal of Fluorescence. 2004;14(2):145–150. doi: 10.1023/b:jofl.0000016286.62641.59. [DOI] [PubMed] [Google Scholar]
- Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends in Biochemical Sciences. 2010;35(10):539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C. Molecular mechanisms of transcription through single-molecule experiments. Chemical Reviews. 2014;114(6):3203–3223. doi: 10.1021/cr400730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KG, Tonthat NK, McClure DR, Chang YC, Liu X, Schumacher MA, et al. Ligand concentration regulates the pathways of coupled protein folding and binding. Journal of the American Chemical Society. 2014;136(3):822–825. doi: 10.1021/ja4086726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: at the interface of biology, physics and chemistry. Journal of the Royal Society, Interface. 2008;5(18):15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482(7385):322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Visualizing transient low-populated structures of RNA. Nature. 2012;491(7426):724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Aleman EA, Rueda D, Walter NG. Focus on function: single molecule RNA enzymology. Biopolymers. 2007;87(5–6):302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- Edwards AL, Reyes FE, Heroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16(11):2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Walter NG, Brooks CL., 3rd Cooperative and directional folding of the preQ1 riboswitch aptamer domain. Journal of the American Chemical Society. 2011;133(12):4196–4199. doi: 10.1021/ja110411m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegland LR, Garst AD, Batey RT, Nesbitt DJ. Single-molecule studies of the lysine riboswitch reveal effector-dependent conformational dynamics of the aptamer domain. Biochemistry. 2012;51(45):9223–9233. doi: 10.1021/bi3007753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338(6105):397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochimica et Biophysica Acta. 2009;1789(9–10):584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. Journal of Biological Chemistry. 2008;283(33):22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nature Structural & Molecular Biology. 2008;15(2):177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Greenfeld M, Pavlichin DS, Mabuchi H, Herschlag D. Single Molecule Analysis Research Tool (SMART): an integrated approach for analyzing single molecule data. PLoS ONE. 2012;7(2):e30024. doi: 10.1371/journal.pone.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A, Altman RB, Souliere MF, Blanchard SC, Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(11):4188–4193. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nature Chemical Biology. 2011;7(6):393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- Haller A, Souliere MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Accounts of Chemical Research. 2011;44(12):1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzakis NS. Single molecule insights on conformational selection and induced fit mechanism. Biophysical Chemistry. 2014;186:46–54. doi: 10.1016/j.bpc.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nature Chemical Biology. 2011;7(6):384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophysical Journal. 2004;87(2):1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Buck AE, McDowell SE, Walter NG. Metal ions: supporting actors in the playbook of small ribozymes. Met Ions Life Sci. 2011;9:175–196. doi: 10.1039/9781849732512-00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Jr, Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492(7427):133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Ha T. Preparing sample chambers for single-molecule FRET. Cold Spring Harb Protoc. 2012;2012(10):1104–1108. doi: 10.1101/pdb.prot071530. [DOI] [PubMed] [Google Scholar]
- Joo C, Ha T. Prism-type total internal reflection microscopy for single-molecule FRET. Cold Spring Harb Protoc. 2012;2012(12) doi: 10.1101/pdb.prot072041. [DOI] [PubMed] [Google Scholar]
- Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Molecular Cell. 2009;33(6):784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Kim E, Lee S, Jeon A, Choi JM, Lee HS, Hohng S, et al. A single-molecule dissection of ligand binding to a protein with intrinsic dynamics. Nature Chemical Biology. 2013;9(5):313–318. doi: 10.1038/nchembio.1213. [DOI] [PubMed] [Google Scholar]
- Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nature Structural & Molecular Biology. 2013;20(12):1450–1457. doi: 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferre-D'Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nature Structural & Molecular Biology. 2009;16(12):1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K, Micura R. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nature Protocols. 2008;3(9):1457–1466. doi: 10.1038/nprot.2008.135. [DOI] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chemistry & Biology. 2006;13(8):857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nature Chemical Biology. 2013;9(6):353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state. Wiley Interdisciplinary Reviews RNA. 2012;3(3):369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek MS, Johnson-Buck A, Walter NG. The shape-shifting quasispecies of RNA: one sequence, many functional folds. Physical Chemistry Chemical Physics. 2011;13(24):11524–11537. doi: 10.1039/c1cp20576e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annual Review of Biochemistry. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- McCown PJ, Liang JJ, Weinberg Z, Breaker RR. Structural, Functional, and Taxonomic Diversity of Three PreQ1 Riboswitch Classes. Chemistry & Biology. 2014;21(7):880–889. doi: 10.1016/j.chembiol.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical Journal. 2006;91(5):1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti N, de Silva C, Johnson-Buck AE, Manzo AJ, Walter NG. A bird's eye view tracking slow nanometer-scale movements of single molecular nano-assemblies. Methods in Enzymology. 2010;475:121–148. doi: 10.1016/S0076-6879(10)75006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra VK, Draper DE. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48(2–3):113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Mustoe AM, Brooks CL, Al-Hashimi HM. Hierarchy of RNA functional dynamics. Annual Review of Biochemistry. 2014;83:441–466. doi: 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of 'induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Research. 2007;35(2):572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet J, Schorr S, Iqbal A, Wilson TJ, Lilley DM. Orientation of cyanine fluorophores terminally attached to DNA via long, flexible tethers. Biophysical Journal. 2011;101(5):1148–1154. doi: 10.1016/j.bpj.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel B, Rueda D. RNA folding dynamics using laser-assisted single-molecule refolding. Methods in Molecular Biology. 2014;1086:289–307. doi: 10.1007/978-1-62703-667-2_17. [DOI] [PubMed] [Google Scholar]
- Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochimica et Biophysica Acta. 2014 doi: 10.1016/j.bbagrm.2014.02.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Li L. Model-based fitting of single-channel dwell-time distributions. Biophysical Journal. 2004;87(3):1657–1671. doi: 10.1529/biophysj.103.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. Site-specific labeling of RNA with fluorophores and other structural probes. Methods. 1999;18(1):60–70. doi: 10.1006/meth.1999.0757. [DOI] [PubMed] [Google Scholar]
- Ragunathan K, Liu C, Ha T. RecA filament sliding on DNA facilitates homology search. Elife. 2012;1:e00067. doi: 10.7554/eLife.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods in Molecular Biology. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- Reining A, Nozinovic S, Schlepckow K, Buhr F, Furtig B, Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499(7458):355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486(7401):85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(24):10804–10809. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi AJ, Suddala KC, Walter NG. Native purification and labeling of RNA for single molecule fluorescence studies. Methods in Molecular Biology. 2014 doi: 10.1007/978-1-4939-1896-6_6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner T, Rieder U, Kreutz C, Micura R. Pseudoknot preorganization of the preQ1 class I riboswitch. Journal of the American Chemical Society. 2012;134(29):11928–11931. doi: 10.1021/ja3049964. [DOI] [PubMed] [Google Scholar]
- Savinov A, Perez CF, Block SM. Single-molecule studies of riboswitch folding. Biochimica et Biophysica Acta. 2014 doi: 10.1016/j.bbagrm.2014.04.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Current Opinion in Structural Biology. 2008;18(1):16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(8):2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152(1–2):17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441(7097):1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chemistry & Biology. 2004;11(12):1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Lim J, Ha T. Acidification of the oxygen scavenging system in single-molecule fluorescence studies: in situ sensing with a ratiometric dual-emission probe. Analytical Chemistry. 2010;82(14):6132–6138. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomatin S, Herschlag D. Methods of site-specific labeling of RNA with fluorescent dyes. Methods in Enzymology. 2009;469:47–68. doi: 10.1016/S0076-6879(09)69003-0. [DOI] [PubMed] [Google Scholar]
- Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463(7281):681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souliere MF, Altman RB, Schwarz V, Haller A, Blanchard SC, Micura R. Tuning a riboswitch response through structural extension of a pseudoknot. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(35):E3256–3264. doi: 10.1073/pnas.1304585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre P, McCluskey K, Shaw E, Penedo JC, Lafontaine DA. Fluorescence tools to investigate riboswitch structural dynamics. Biochimica et Biophysica Acta. 2014 doi: 10.1016/j.bbagrm.2014.05.015. [DOI] [PubMed] [Google Scholar]
- Stennett EMS, Ciuba MA, Levitus M. Photophysical processes in single molecule organic fluorescent probes. Chemical Society Reviews. 2014;43(4):1057–1075. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18(7):787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annual Review of Biochemistry. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- Suddala KC, Rinaldi AJ, Feng J, Mustoe AM, Eichhorn CD, Liberman JA, et al. Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Research. 2013;41(22):10462–10475. doi: 10.1093/nar/gkt798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda M, Henig J, Cheng HM, Brugger D, Haltrich D, Plumere N, et al. Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments. ACS Nano. 2012;6(7):6364–6369. doi: 10.1021/nn301895c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I, Jr, Gonzalez RL., Jr Biological mechanisms, one molecule at a time. Genes & Development. 2011;25(12):1205–1231. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Batey RT. A disconnect between high-affinity binding and efficient regulation by antifolates and purines in the tetrahydrofolate riboswitch. Chemistry & Biology. 2014;21(2):205–216. doi: 10.1016/j.chembiol.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafabakhsh R, Ha T. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science. 2012;337(6098):1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Mondragon E, Batey RT. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Research. 2011;39(19):8586–8598. doi: 10.1093/nar/gkr565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NG. Probing RNA structural dynamics and function by fluorescence resonance energy transfer (FRET) Current Protocols in Nucleic Acid Chemistry. 2003;Chapter 11(Unit 11):10. doi: 10.1002/0471142700.nc1110s11. [DOI] [PubMed] [Google Scholar]
- Walter NG, Burke JM. Fluorescence assays to study structure, dynamics, and function of RNA and RNA-ligand complexes. Methods in Enzymology. 2000;317:409–440. doi: 10.1016/s0076-6879(00)17027-2. [DOI] [PubMed] [Google Scholar]
- Walter NG, Burke JM. Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA. 1997;3(4):392–404. [PMC free article] [PubMed] [Google Scholar]
- Walter NG, Burke JM, Millar DP. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nature Structural Biology. 1999;6(6):544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- Walter NG, Bustamante C. Introduction to single molecule imaging and mechanics: seeing and touching molecules one at a time. Chemical Reviews. 2014;114(6):3069–3071. doi: 10.1021/cr500059w. [DOI] [PubMed] [Google Scholar]
- Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nature Methods. 2008;5(6):475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophysical Journal. 2009;96(2):L7–9. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Protocols. 2006;1(3):1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annual Review of Microbiology. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Wood S, Ferre-D'Amare AR, Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chemical Biology. 2012;7(5):920–927. doi: 10.1021/cb300014u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ferre-D'Amare AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500(7462):363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Rueda D. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods. 2009;49(2):112–117. doi: 10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Juette MF, Jockusch S, Wasserman MR, Zhou Z, Altman RB, et al. Ultra-stable organic fluorophores for single-molecule research. Chemical Society Reviews. 2014;43(4):1044–1056. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X. Single-molecule RNA science. Annual Review of Biophysics and Biomolecular Structure. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288(5473):2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296(5572):1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]