Abstract
Despite the importance of siRNA delivery systems, understanding of their intracellular fate remains elusive. We recently developed a multi-component siRNA nanocomplex to deliver siRNA to hepatic stellate cells (HSCs). The objective of this study is to study post internalization trafficking of this siRNA nanocomplex and its multiple components like siRNA, protamine, and streptavidin, in HSCs. After internalization, the nanocomplex entrapped in early endosomes undergoes three possible routes including endosomal escape, exocytosis, and entrapment in lysosomes. Significant amount of siRNA dissociates form the nanocomplex to exert silencing activity. After escaping from endosomes, protamine dissociates from the nanocomplex and stays inside the cytoplasm. Golgi complex plays an important role in exocytosis of the nanocomplex. We also demonstrate that exocytosis is one of the major reasons accounting for the transient silencing activity of nonviral siRNA delivery. Incorporation of exocytosis inhibitors in nonviral siRNA delivery systems may extend the silencing activity of siRNA.
Keywords: siRNA delivery, nanocomplex, hepatic stellate cells, intracellular trafficking, exocytosis
Graphical Abstract
Although endocytosis of nanocarriers has been extensively studied, intracellular trafficking and exocytosis of nanocarriers have always been elusive and ignored. Previously, we developed a multi-component streptavidin-based siRNA nanocomplex to deliver the PCBP2 siRNA into rat hepatic stellate cells (HSCs). The study aims to utilize confocal microscopy and flowcytometry to precisely study the intracellular trafficking of each of the major components, including the siRNA, protamine, and streptavidin of the nanocomplex. Our studies particularly indicate the importance of exocytosis in nonviral siRNA delivery systems. We demonstrate that exocytosis is one of the major reasons for the transient silencing activity of nonviral siRNA delivery. Therefore, further study and understanding of the exocytosis of nanocarriers may provide new insights for improving nanoscale drug delivery systems.
Background
Over the past decade, RNA interference (RNAi) has emerged as a powerful therapeutic modality to knockdown genes associated with various diseases that are un-targetable by small molecule drugs.1–3 Despite tremendous progress and interest in this field, the therapeutic promise of RNAi in humans has been limited due to the absence of effective delivery systems.4, 5 To exert its therapeutic action, siRNA needs to overcome a number of biological barriers before it can enter cell cytoplasm.6–8 Non-viral delivery systems, such as bio-conjugates, liposomes, cationic peptides/polymers, and several other types of nanoparticles have been extensively investigated because of their safety.9–11
Most recently, we developed a streptavidin-based nanocomplex for the delivery of the PCBP2 siRNA and named it as SSCP (siRNA, Streptavidin, Cholesterol and Protamine) nanocomplex.12, 13 However, the precise mechanism underlying the SSCP mediated siRNA delivery is not fully understood. We previously observed that cellular uptake of the SSCP in hepatic stellate cells (HSCs) depends on the presence of positively charged protamine and cholesterol, which facilitates LDLR (Low Density Lipoprotein)-mediated endocytosis. In addition, a cleavable disulfide bond between siRNA and biotin is essential for its silencing activity. It has been proven that disassembly of multi-component siRNA nanocomplex inside cells is critical to facilitate the assembly of siRNA into RNAi machinery.14 We therefore raised several questions, such as what are the respective fates of the different components of the SSCP nanocomplex inside the cells. Once internalized, how efficiently the siRNA is released from the nanocomplex? Whether the nanocomplex is able to escape from endosomes? Answering these important queries could provide vital clues for further improvement of the streptavidin-based siRNA nanocomplex for liver fibrosis.
The importance of delivery in the therapeutic application of siRNA has long been realized, but analysis of the intracellular fate of siRNA delivery systems still remains elusive. Recently, the role of exocytosis on siRNA delivery has been explored.15, 16 Internalized siRNA nanoparticles could be exocytosed by non-secretory exosomes, and there are a number of possible recycling routes for siRNA nanoparticles.16 Several strategies such as elemental analysis have been utilized to study the intriguing phenomenon of endocytosis and exocytosis of nanoparticles.17, 18 By contrast, fluorescence microscopy and fluorescence tracking are the most accepted methods that can provide both qualitative and quantitative analysis 16.
In this study, we utilized confocal microscopy and flowcytometry to precisely study the intracellular trafficking of the major components, including the siRNA, protamine, and streptavidin of the siRNA nanocomplex in HSC-T6 cells. Each of the components were labeled with a different fluorescent dye. Exocytosis of the siRNA nanocomplex was also studied.
Methods
Materials
Streptavidin and BCA protein assay kit were obtained from Pierce (Rockford, IL). Protamine sulfate (salmon X grade) was purchased from Sigma-Aldrich (St. Louis, MO). Alexa Fluor® 350 succinimidyl esters (NHS esters) and Streptavidin-Alexa Fluor 350 conjugate was purchased from Life Technologies (Grand Island, NY). Cell culture medium, Lipofectamine-2000, PCBP2 siRNA (sense sequence 5′-GUCAGUGUGGCUCUCUUAU-3′), negative control siRNA, and Alexa Fluor 647-siRNA were purchased from Invitrogen (Carlsbad, CA). Sephadex G-15 was obtained from GE healthcare life sciences (Pittsburgh, PA). Biotin siRNA was purchased from Gene Pharma (Shanghai, China). Alexa Fluor 647 was conjugated to the 5′ end of the siRNA antisense strand, while biotin was conjugated to the 3′ end of the siRNA sense strand. RAB5 antibody (rabbit IgG), goat anti-rabbit IgG (H+L) secondary antibody (Alexa Fluor® 488 conjugate) and BODIPY® FL C5-Ceramide complexed to BSA were purchased from Thermo Fisher Scientific (Waltham, MA).
Fluorescent Labeling of Protamine
Protamine was labeled with Alexa Fluor 350 dye using succinimidyl ester (NHS) chemistry according to manufacturer’s instruction. Briefly, protamine was diluted in 0.1 M sodium bicarbonate (pH ~8) and reacted with the dye in a mole ratio of 1:3 (protein:dye). The labeled protamine was purified by Sephadex G-15 column using PBS as the elution buffer. The concentration of protamine was determined using a BCA assay because protamine lacks the aromatic amino acids like tyrosine, phenylalanine and tryptophan that can absorb UV light at 280 nm19. Labeling efficacy was calculated using the formula:
Where, ε = 19,000 cm−1 M−1 (approximate molar extinction coefficient of Alexa Fluor 350 at 346 nm).
Preparation of Streptavidin-Based siRNA Nanocomplex
The multi-component SSCP nanocomplex was prepared as we reported.12 Briefly, biotin siRNA containing a disulfide linker in the 3’ end of the sense strand and Alexa Fluor 647 in the 5’ end of the antisense strand was mixed with streptavidin and biotin-cholesterol in a 2:1:2 molar ratio. The complex was incubated at room temperature for 10 min and then condensed with protamine to form the SSCP nanocomplex. Particle size and zeta potential of the nanocomplex were measured using a Malven Zetasizer Nano ZS. For intracellular trafficking study, Alexa Fluor 350-labeled streptavidin and protamine were used along with the Alexa Fluor 647-labeled siRNA.
Cell Culture and SSCP Uptake Study
The rat hepatic stellate cell line (HSC-T6) was kindly provided by Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York University). HSC-T6 cells were cultured in DMEM supplemented with 10% FBS, 100 units/mL penicillin streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The cells were seeded in Nunc Lab-Tek II Chambered Cover glasses (Fisher Scientific; Pittsburgh, PA) at a density of 1×104 cells/well as described previously.1, 13, 20 The SSCP nanocomplex was added to each well along with OptiMEM at a final concentration of 100 nM siRNA. The cells were then incubated for different time periods to evaluate cellular uptake. On the completion, the cells were fixed with 10% formalin buffer and examined under a confocal microscope (Leica TCS SP5). To investigate endosome entrapment of the SSCP nanocomplex, the cells were washed with DPBS and incubated with 150 nM lysotracker red in Opti-MEM for 15 min. Images were taken at 40X with 3.0 zoom and 6.0 zoom.
Assessment of Cellular Uptake Using Flow Cytometry
HSC-T6 cells were seeded in 24-well plates at a density of 25,000 cells per well 12 h before transfection. The cells were transfected with the SSCP nanocomplex or Lipofectamine-2000 containing Alexa Fluor 647-labeled siRNA for different time periods as described above. The cells were then washed with DPBS, trypsinized, and subjected to fluorescence analysis using a BD FACS II Flow Cytometer (Bectone Dickinson Instruments, Franklin Lakes, NJ).
Exocytosis and Cellular Recycling of the SSCP Nanocomplex
Alexa Fluor 647-labeled siRNA was utilized to prepare the SSCP complex. In this study, the final siRNA concentration was maintained at 50 nM because the major purpose of this study was to monitor the siRNA recycling and exocytosis and excessive labeled siRNA might give false positives.15 Five thousand HSC-T6 cells per well were seeded in 96 well plates and transfected with the SSCP nanocomplex diluted in OptiMEM. In one group, the medium was collected at 3, 6, 12, and 24 h post-transfection to evaluate exocytosis. In another group, the cells were transfected with the SSCP nanocomplex for 6 h, followed by replacement with fresh medium. The fresh medium was then collected at 1, 3, 6, 18, 21, and 24 h to determine the fluorescence of exocytosed siRNA. Fluorescence of the collect media was measured using a multimode fluorescence detector.
To further investigate the nature of exocytosed siRNA in the medium, we developed a method to determine whether the exocytosed siRNA is still entrapped inside the nanocomplex or simply in a free form. Briefly, the harvested media containing exocytosed siRNA were treated with heparin to release siRNA from the nanocomplex. By comparing the fluorescence intensity before and after the heparin treatment, we can determine the fraction of free siRNA in the exocytosed samples. Briefly, we incubated the nanocomplex prepared in OptiMEM with various heparin concentrations (10, 20 and 40 μM) to understand the extent of fluorescence quenching by complexation and OptiMEM. After optimizing the concentration and incubation time of heparin, we again performed exocytosis study by treating the HSC-T6 cells as described above and collected medium at various time points after 6 h incubation with the SSCP. Followed by replacement of fresh medium and subsequent collection of medium at different time points, we incubated collected medium with or without heparin for 30 min to analyze the nature of siRNA exocytosed. Exocytosis was also evaluated in the presence of two exocytosis inhibitors, Exo1 and Brefeldin A.21
Early Cellular Trafficking of the SSCP Nanocomplex
HSC-T6 cells were transfected with the SSCP nanocomplex containing Alexa Fluor 647-siRNA for 1h. One group of the cells were immediately washed and fixed with 10% formalin, followed by staining with the early endosome marker (Rab5 antibody). For the second group of the cells, the transfection medium was replaced with fresh culture medium and incubate for another 1 h before fixation with 10% formalin and staining with Rab5 antibody. Alexa Fluor® 488 conjugate secondary antibody was added to visualize early endosomes under confocal microscope.
Since Trans-Golgi Network (TGN) plays a vital role in the recycling of nanoparticles, we also stained the Golgi complexes using BODIPY-FL C5 ceramide as per manufacturer’s protocol. Pearson’s correlation coefficient for co-localization of siRNA with early endosomes or Golgi was calculated by using image J software and JACOP plugin.
Results
Preparation of the SSCP Nanocomplex Containing Alexa Fluor Labeled Components
To monitor cellular uptake, intracellular trafficking, endosomal entrapment and release of free siRNA into the cytosol, we prepared the SSCP nanocomplex containing the PCBP2 siRNA labeled with Alexa Fluor 647. The Alexa Fluor dyes were selected because of their pH independence and excellent photo-stability.14 As we reported before, the siRNA, streptavidin, and cholesterol were mixed in a molar ratio of 2:1:2, followed by condensation with protamine (N/P 10:1) to form the SSCP siRNA nanocomplex.12 The nanocomplex has a uniform mean size of 230 nm with a PDI smaller than 0.15. Zeta potential of the nanocomplex is approximately 23 mV. Fluorescent-labeled streptavidin and protamine were used for the SSCP nanocomplex preparation to evaluate cellular uptake and intracellular trafficking.
Cellular Uptake of the SSCP Nanocomplex
Successful delivery of siRNA using the streptavidin-biotin technology has motivated us to further investigate the intracellular fate of the different components in the nanocomplex, which will provide important insight to further improve the efficacy of the system. In our previous study, we hypothesized that the SSCP nanocomplexes, once internalized, can escape from endosomes due to the presence of multiple arginines in protamine.22 After endosomal escape, the SSC (siRNA, Streptavidin and Cholesterol) complex releases siRNA in the cytoplasm upon cleavage of the disulfide linker between siRNA and biotin. The free siRNA is then incorporated in the RISC pathway and exert its silencing activity.
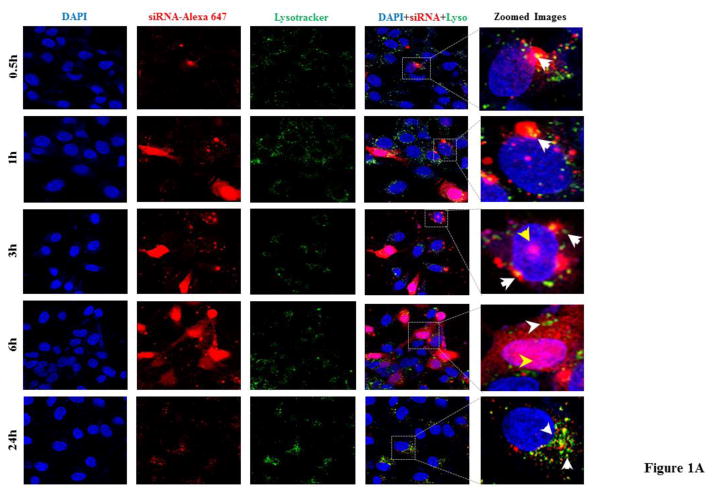
Confocal microscopy and flowcytometry were utilized to investigate uptake of the SSCP nanocomplex. First, HSC-T6 cells were transfected with the SSCP nanocomplex containing Alexa Fluor 647-labeled siRNA for 0.5, 1, 3, 6 and 24 h. As shown in Figure 1A, the SSCP nanocomplex rapidly enters the cells and reaches its highest uptake at 6 h post-transfection. Nearly all the cells were transfected with the SSCP nanocomplex at 6 h. LysoTracker® was used to evaluate lysosome entrapment of the siRNA. LysoTracker® is a highly specific, one-step staining dye for acidic organelles and widely employed to track lysosomes.23 As the merged images illustrated (Figure 1A), only a very small amount of the siRNAs were entrapped in late-endosomes/lysosomes between 0.5 and 6 h post-transfection. Moreover, majority of the siRNAs were localized in the cytoplasm and few translocated into the nucleus at 3 and 6 h post-transfection. However, there was a significant decrease in cellular uptake at 24 h post-transfection where siRNAs were mainly located in the cytoplasm as clustered aggregates at the perinuclear space localized with lysosomes. Pearson correlation coefficients for the co-localization of siRNA and lysosomes are 0.265, 0.432 and 0.683 at 3 h, 6 h and 24 h respectively, indicating time-dependent accumulation of siRNA in lysosomes.
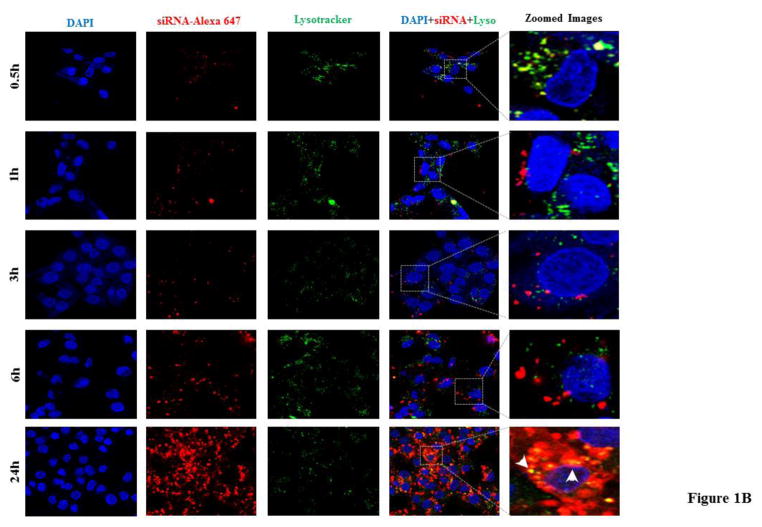
Figure 1. Cellular uptake of the PCBP2 siRNA in HSC-T6 cells.
The Alexa Fluor 647 labeled siRNA is encapsulated in the SSCP nanocomplex (A) or Lipofectamine-2000 (B). The siRNA is incubated with HSC-T6 cells for 0.5, 1, 3, 6, and 24h, followed by fixation and confocal imaging.
Using the same method, we studied cellular uptake of the siRNA using Lipofectamine-2000 (Figure 1B). The resulting confocal images indicate a slow but continuous increase in cellular uptake up to 24 h post-transfection, and only a small fraction of them were entrapped in lysosomes. Compared to Lipofectamine-2000, the SSCP nanocomplex exhibits a faster and more uniform uptake of siRNA in HSC-T6 cells.
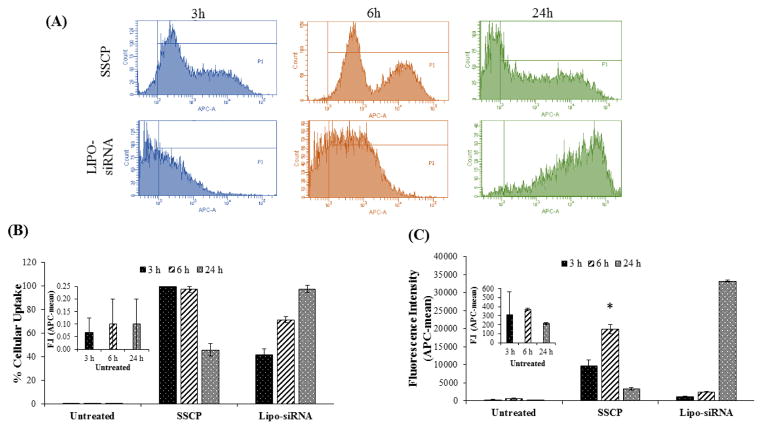
We next compared transfection efficacy of the SSCP nanocomplex and Lipofectamine-2000 in HSC-T6 cells using flowcytometry. As shown in Figure 2A&B, nearly all cells were transfected with the SSCP nanocomplex at 3 and 6 h post-transfection, indicating a fast and efficient cellular uptake of the SSCP. On the contrary, the transfection efficacy of Lipofectamine-2000 was lower at early time-points but higher at 24 h post-transfection as compared to SSCP (Figure 2B). The fluorescence intensity results at 3 and 6 h post-transfection (Figure 2C) reveal that the SSCP nanocomplex is able to deliver siRNAs into the cells at a much faster rate than Lipofectamine-2000. However, the fluorescence intensity in SSCP treated cells decreases at 24 h post-transfection. These results are consistent with our confocal images (Figures 1A&B) where a decrease in fluorescence intensity was observed at 24 h post-transfection. The differences in the uptake kinetics of Lipofectamine-2000 and SSCP could be due to their different chemical nature.
Figure 2. Flow cytometry analysis of the cellular uptake of siRNA encapsulated in the SSCP nanocomplex and Lipofectamine-2000.
Alexa Fluor 647 labeled PCBP2 siRNA was incubated with HSC-T6 cells for 3, 6, and 12h, followed by flow cytometry analysis. (A) Histogram of flow cytometry. Percent of cellular uptake (B) and relative fluorescence intensity (C) are presented as the mean ± SD (n=3).
Subcellular Distribution of Different Components of the SSCP Nanocomplex
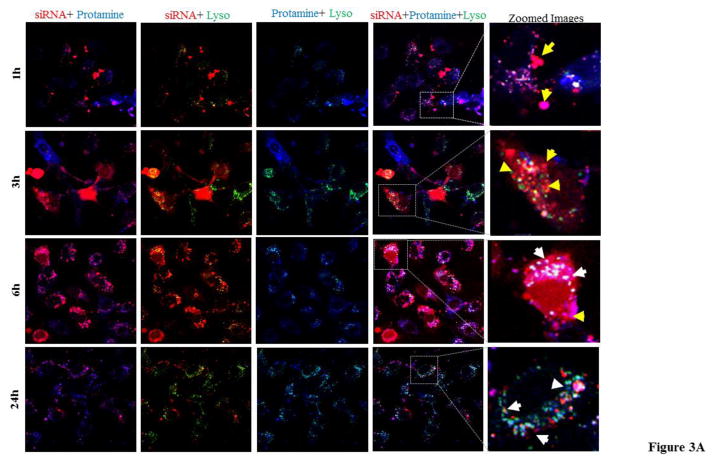
Confocal microscopy was used to further investigate subcellular distribution of different components (streptavidin, siRNA, and protamine) of the SSCP. HSC-T6 cells were transfected with the SSCP nanocomplex containing Alexa Fluor 647-labeled siRNA (far-red) and Alexa Fluor 350-labeled protamine (blue) for 1, 3, 6 and 24 h. As Figure 3A illustrates, a small percentage of the siRNAs in the cytoplasm are co-localized with protamine (indicated with yellow arrows) at early time points, while majority of the siRNA are co-localized with protamine in lysosomes at 24 h post-transfection (indicated with white arrows). We observed that fraction of the siRNA can be delivered into the nucleus without protamine co-localization (Figure 3A). This result indicates that protamine only acts as cell penetrating peptide in internalization. It dissociates from the SSCP inside the cells and slowly translocate to perinuclear space and other parts in the cytoplasm, while a small amount of the siRNAs transit into the nucleus.
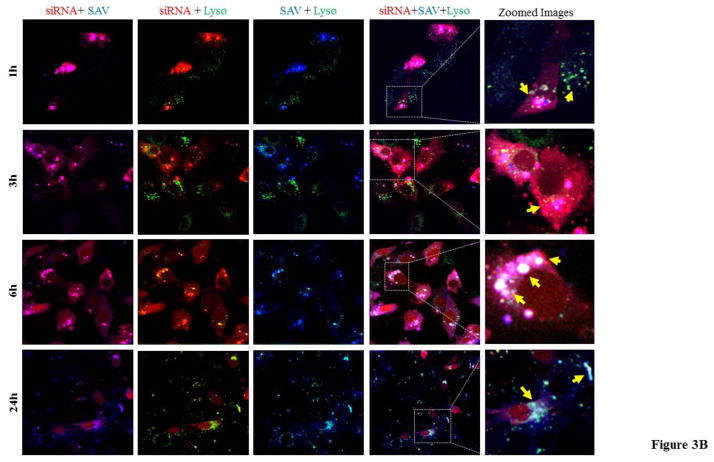
Figure 3. Co-localization of siRNA with protamine (A) and streptavidin (B) after cellular internalization.
The siRNA nanocomplex was incubated with HSC-T6 cells for 1, 3, 6 and 24h, followed by fixation and confocal imaging. White color shows the co-localization of siRNA (red), lysosome (green) and protamine or streptavidin (blue). [Lyso: lysosome].
We next evaluated subcellular trafficking of streptavidin in HSC-T6 cells. Alexa Fluor 350-labeled streptavidin (blue) was used in the SSCP nanocomplex along with Alexa Fluor 647-labeled siRNA (red). As shown in Figure 3B, streptavidin is localized with siRNA in the cell cytoplasm at all time points. However, streptavidin shows variable co-localization with lysosomes. Streptavidin distributes in the cytoplasm at early time points (1 and 3 h) with negligible co-localization with lysosomes, but co-localization increases (indicated with yellow arrows) at 6 and 24 h post-transfection. The merged images of streptavidin and siRNA (Figure 3B) illustrate that majority of siRNAs are in the cytoplasm and few of the siRNAs are co-localized with streptavidin inside lysosomes, suggesting incomplete release of SSCP from endosomes at extended time points or reuptake of nanocomplex by lysosomes (indicated by yellow arrows).
Exocytosis and Cellular Recycling of the SSCP Nanocomplex
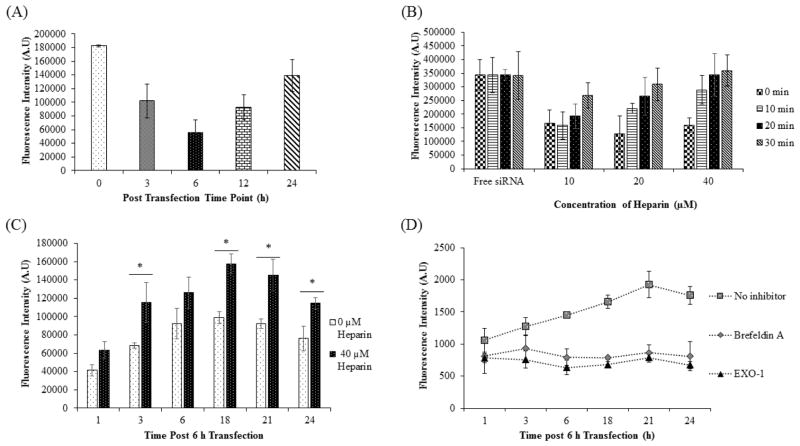
Confocal microscopy and flow cytometry results (Figures 1–4) suggest that SSCP can rapidly and efficiently deliver siRNA into the HSC-T6 cells at early time points, but cellular uptake of the siRNA is reduced at 24 h post-transfection. This is accordance with other reports showing transit silencing activity of synthetic siRNA. Degradation and dilution of internalized siRNAs during cell division are generally believed the major reasons for the transit activity. Recently, the role of exocytosis on siRNA delivery has been explored.15, 16 We hypothesized that the transit activity of synthetic siRNAs could be partially due to exocytosis. Exocytosis of SSCP containing Alexa Fluor 647-labeled siRNA was therefore evaluated in HSC-T6 cells. As depicted in Figure 4A, fluorescence intensity of the medium constantly decreases till 6 h post-transfection, indicating a rapid uptake of the nanocomplex into the cells where the rate of endocytosis exceeds exocytosis. However, fluorescence intensity of the medium starts increasing after 6 h, suggesting that exocytosis of the SSCP nanocomplex dominates the endocytosis/exocytosis kinetics.
Figure 4. Exocytosis of the siRNA nanocomplex.
(A) Fluorescence intensity of the transfection medium at various time points post-transfection. Alexa Fluor 647-labeled siRNA was formulated in the SSCP nanocomplex and incubated with HSC-T6 cells for various time intervals. The extracellular medium was harvested to measure fluorescence of the siRNA. (B) Heparin incubation dissociates siRNA from the nanocomplex and reverse the fluorescence quenching effect. (C) HSC-T6 cells were incubated with Alexa Fluor 647-labeled siRNA nanocomplex for 6h, and the medium was replaced. The medium was then collected at 1, 3, 6, 18, 21 and 24h post-incubation for fluorescence analysis. Fluorescence intensities of the medium samples were determined with and without 40 μM heparin treatment. (n=3). (D) HSC-T6 cells were incubated with Alexa Fluor 647-labeled siRNA nanocomplex for 6h, and the medium was replaced with normal OptiMEM or OptiMEM containing Exo1 (40 μM) or OptiMEM with Brefeldin A (100 ng/ml). The medium was then collected at 1, 3, 6, 18, 21 and 24h post-incubation for fluorescence analysis. (* P< 0.05)
To further investigate whether exocytosed siRNAs are still complexed with the carrier, we treated the medium with heparin to dissociate encapsulated siRNAs. We first studied the fluorescence quench effects of nanocomplex and cell culture medium (OptiMEM) on the fluorescence intensity of Alexa Fluor 647-labled siRNA. As Figure 4B shows, formation of SSCP nanocomplex reduces fluorescence intensity of the siRNA. Similar quench effect was also observed in the addition of OptiMEM medium (data not shown). Incubation of SSCP with 40 μM heparin dissociate siRNAs from the nanocomplex and reverses the quench effect of the nanocomplex and OptiMEM. Figure 4B also shows that incubation of SSCP with 40 μM heparin for 30 min can completely dissociate siRNAs form the nanocomplex. After transfection with the SSCP nanocomplex for 6 h, HSC-T6 cells were washed and incubated with fresh medium. The media were then collected at different time intervals, and the fluorescence intensity was detected after incubation with 40 μM heparin for 30 min (Figure 4C). Without heparin treatment, fluorescence intensity of the medium increases with time and reaches the highest value at 18 h post-incubation. The heparin treated samples exhibit the same trend of the fluorescence intensity. However, heparin treatment significantly increases the fluorescence intensity of all exocytosed samples, indicating that significant amount of the exocytosed siRNA are still complexed with the SSCP nanocomplex. This result suggests a significant amount of intracellular siRNAs are exocytosed, leading to the low concentration of the siRNA in the cells at 24 h post-transfection. We next investigated whether exocytosis of SSCP is mediated the Golgi recycling pathway. Two Golgi recycling inhibitors, Exo-1 and Brefeldin A,21 were used in this study. As Figure 4D illustrates, exocytosis of the siRNA nanocomplex is dramatically inhibited by the treatment of Exo1 or Brefeldin A, suggesting the important role of Golgi recycling pathway in the the exocytosis of the siRNA nanocomplex.
SSCP Mediated Gene Silencing
Figure 5 illustrates gene silencing activity of the SSCP in HSC-T6 cells. Silencing activity of PCBP2 siRNA is compared with a scrambled siRNA (negative control, NC) using the same nanocomplex. SSCP rapidly induces approximately 75% silencing of the PCBP2 gene at 6 hr post-transfection. The silencing activity reaches the highest level (80%) at 12 hr post-transfection and then decreases with time (Figure 5). This trend is in accordance with cellular uptake results (Figure 2) and our previous findings, which exhibit a similar time-dependent silencing activity using Lipofectamime.1, 24 There could be four possible reasons of this decreased silencing activity: i) saturation of cellular uptake and silencing machinery; ii) siRNAs entrapped in late endosomes undergo degradation in lysosomes; iii) cellular recycling of nanocomplex; and iv) cell division as the classic example of transient silencing activity 25.
Figure 5. Time course of the silencing activity.
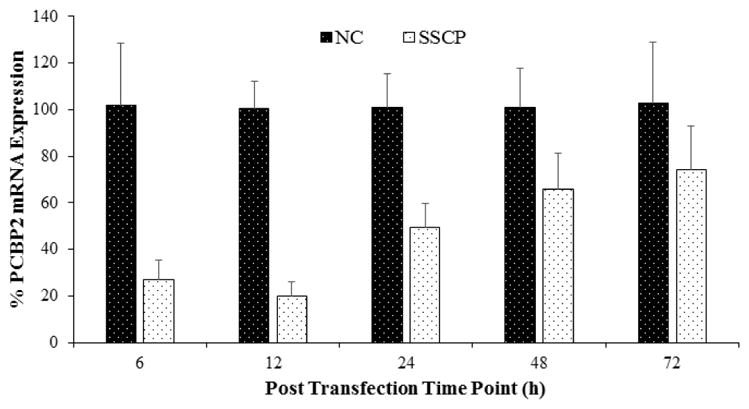
SSCP formulated with PCBP2 siRNA or negative control siRNA was incubated with HSC-T6 cells for various time points (6, 12, 24, 48 and 72h). Total RNA was isolated and the silencing effect was evaluated by qRT-PCR (n=3)
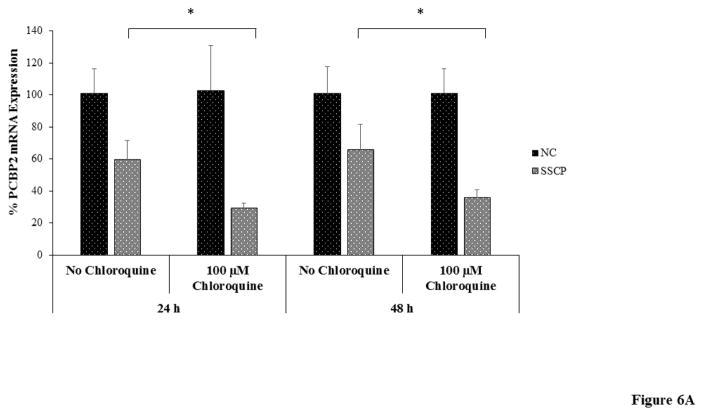
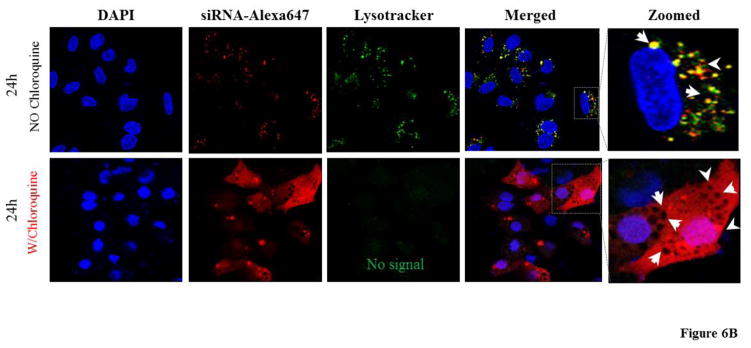
Although only a small amount of siRNA/streptavidin complexes are entrapped in lysosomes at extended time intervals, release of the entrapped siRNA can improve the silencing activity because even a small amount of siRNA in the cytoplasm can exhibit activity.26 We therefore studied the effect of endosome disrupting agents like chloroquine on the silencing activity of SSCP. We transfected the cells with the SSCP nanocomplex in the presence of chloroquine at 100 μM. As shown in Figure 6A, chloroquine treatment increases silencing activity of the nanocomplex, suggesting that lysosomal entrapment is one of the reasons accounting for low silencing activity at 24 and 48 h post-transfection. Chloroquine acts by swelling endosomes and lysosomes and neutralizing their pH. In Figure 6B, confocal images reveal that after chloroquine treatment siRNA intensity increases in the cytoplasm with numerous swollen lysosomes (indicated by arrow).
Figure 6. The effect of chloroquine on the silencing activity (A) and cellular uptake (B) of the siRNA SSCP.
HSC-T6 cells were transfected with siRNA nanocomplex with or without chloroquine for 24h and 48h respectively (n=3). (* P< 0.05)
Early Cellular Trafficking of the SSCP Nanocomplex
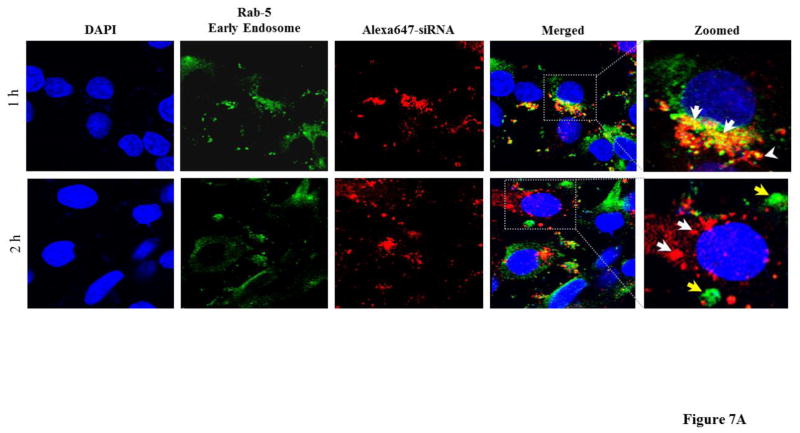
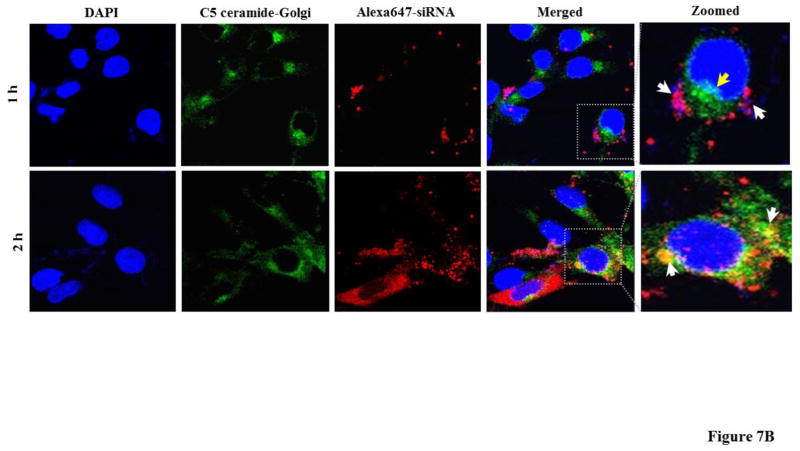
We also examined how the SSCP nanocomplex traffics inside the cells after one-hour transfection. Both early endosomes and Golgi complex were stained to examine how the siRNAs traffic in these organelles. As Figure 7A illustrates, one hour after the transfection, a significant amount of the siRNAs are entrapped inside early endosomes (indicated with white arrows). However, after two hours, nearly all the siRNAs escapes from early endosomes. Pearson’s correlation coefficient for co-localization of siRNA with early endosome was 0.187 and 0.045 at 1 h and 2 h respectively. Figure 7B illustrates co-localization of the siRNAs with Golgi complex inside the cells. One hour after the transfection, negligible siRNAs are localized in the Golgi complex. By contrast, more siRNAs are co-localized with the Golgi complex 2 h post-transfection (indicated with white arrows), suggesting that a fraction of the siRNAs translocate into the Golgi complex for recycling as time elapses. Pearson’s coefficient for co-localization of siRNA with Golgi complex was 0.044 and 0.185 at 1 h and 2 h respectively.
Figure 7. Early cellular trafficking of the SSCP nanocomplex.
(A) Early endosome (Rab5 antibody, Green) and Alexa Fluor 647 siRNA tracking (Red). (B) Golgi complex (BODIPY-FL-C5 Ceramide, Green) and Alexa Fluor 647 siRNA (Red) tracking. HSC-T6 cells were incubated with SSCP for 1h and further incubated with fresh OptiMEM for 0 or 1h before fixation and staining.
Discussion
Despite the tremendous interest in using siRNA as therapeutics, efficient delivery still remains the major obstacle to fully exploit the therapeutic potential of siRNA.5, 8 To overcome the various biological barriers in vivo, an efficient siRNA delivery system should contain multiple components to address these challenges simultaneously.10, 27 The multiple components in the system should not interfere with the silencing activity of siRNA. Our previous work has demonstrated that the SSCP nanocomplex can safely and efficiently deliver siRNA via receptor-mediated uptake.12 However, it is not clear how these multiple components dissociate and traffic inside the cells. Understanding behaviors of the multiple components and siRNA inside the cells is fundamental to further improve its efficacy for future in vivo studies. Recently there has been an increasing interest in studying the extravasation or exocytosis of nanocarriers.15, 16, 28, 29 As a result, we studied intracellular trafficking of each of the components of the siRNA nanocomplex using confocal microscopy and flowcytometry.
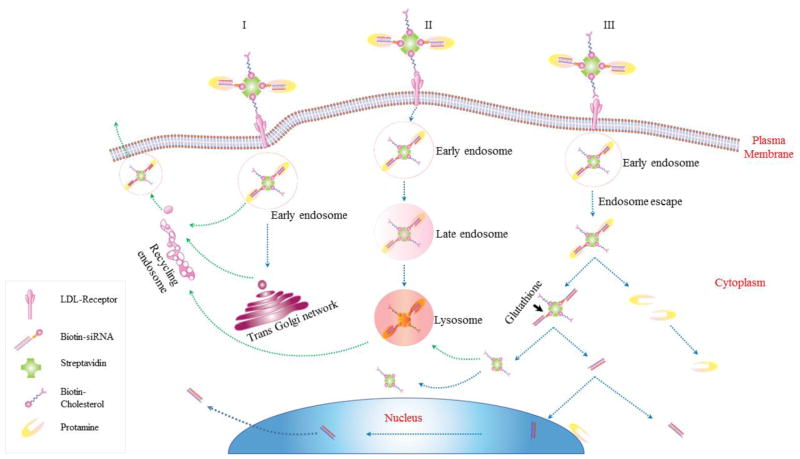
Based on the findings of this study, we hypothesize an intracellular trafficking mechanism of the multi-component SSCP nanocomplex in HSC-T6 cells. As Figure 8 illustrates, SSCP is first entrapped in early endosomes and commutes intracellularly through three possible routes: 1) the early endosomes are recycled through Trans Golgi network or recycling endosomes; 2) the endosomes undergo endosome maturation to lysosomes; 3) SSCP escapes from the early endosomes, and protamine dissociates from the SSCP nanocomplex to form the SSC complex, which releases free siRNA by the action of glutathione reductase in the cytoplasm. This is in accordance with a recent study reporting similar recycling pathways for nanoparticles.30 Significant amount of siRNA rapidly dissociates from the SSCP nanocomplex and distributes in the cell cytoplasm to exert silencing activity. As time elapses, protamine is either co-localized with SSC in lysosomes or stays free in the cytoplasm (Figure 3A). A fraction of the siRNA translocate into the nucleus, which is in accordance with a previous report.31 A very small fraction of the siRNA remains complexed with streptavidin and entrapped in lysosomes. A significant amount of siRNA and nanocomplex are exocytosed, which may explain the transient silencing activity of synthetic siRNA delivered by nonviral vectors.
Figure 8. Schematic diagram of the intracellular trafficking of SSCP nanocomplex.
Once internalized, SSCP commutes intracellularly through three possible routes: 1) SSCP entrapped early endosomes are recycled through Trans Golgi network or recycling endosomes, 2. SSCP entrapped endosomes undergo endosome maturation to lysosomes; 3. SSCP escapes from early endosomes, and protamine dissociates from the SSCP nanocomplex to form the SSC complex, which releases free siRNA by the action of glutathione reductase in the cytoplasm.
Due to its high arginine content (~67%) and inherent characteristics to condense nucleic acids, protamine has been widely used to enhance the complexation of siRNA in delivery systems.12, 32–35 It is known that peptides containing high arginine content promote not only cellular internalization but also nuclear entry of its cargo through the nuclear pore complexes (NPC).36, 37 As Figure 3A shows, protamine dissociates from the SSCP at early time points and slowly accumulates around the nucleus. However, we did not observe nuclear entry of protamine, which is different from the reports postulating nuclear uptake of protamine/DNA or polyarginine/DNA complex.37, 38 In accordance with our results, several other groups also demonstrated that siRNA alone can translocate inside and out of the nucleus.31, 39
Streptavidin is used as a “backbone” to form complex with biotin-conjugated siRNA and ligands in the SSCP nanocomplex.12 Streptavidin distributes in the cytoplasm at 3 h but slowly enters lysosomes at 6 h (yellow arrows) and converts into granular structures at 24 h post-transfection (Figure 3B). Interestingly, these granular structures co-localize with siRNA, suggesting incomplete endosomal escape of the SSCP complexes or reuptake of the SSC complex by lysosomes in the cytoplasm in a similar way as auto-phagosomes.40, 41 Retention of the SSCP in endosomes or reuptake by lysosomes may lead to lysosomal degradation42 or recycling via exocytosis.43 Reports from others also showed that after endosomal escape nanocomplexes or siRNA can again be taken up by either lysosomes or recycling vesicles.40, 41
Although endocytosis of nanocarriers has been extensively studied, exocytosis of nanocarriers has always been elusive and ignored.16, 44 We therefore studied the exocytosis of siRNA and demonstrated the dynamic transport of siRNA across cell membrane with elapse of time. Alexa Fluor 647 is well accepted as a highly stable probe for tracking siRNAs and remains attached to siRNA under physiological conditions.45 In the exocytosis study (Figure 4C), the trend of increasing fluorescence in extracellular medium is in agreement with the basic endocytic machinery and recycling circuit proposed by Huotari.30 Recycling of nanoparticles starts as soon as uptake occurs in early endosomes, but the rate of endocytosis exceeds the rate of exocytosis.30 Our results (Figure 4A&C) demonstrates that exocytosis dominates the endocytosis/exocytosis kinetics at early time points. Heparin treatment shows that a significant amount of the exocytosed siRNA is still entrapped in the nanocomplex, suggesting a similar recycling pathway as described by Langer and his colleagues.15 Efforts have been made to overcome nanocarrier recycling by downregulating NPC1 (Niemann-Pick type C1)15 or lipid recycling regulators.46 Our results suggest that the Golgi recycling pathway may be inhibited to reduce exocytosis, leading extended silencing activity of nonviral siRNA.
Transient gene silencing has been a major shortcoming of synthetic siRNA delivered by nonviral systems. Multiple factors, such as endogenous nucleases mediated degradation,47, 48 dilution due to cell division,49 and inefficient release from delivery systems 50 are considered to be the classic reasons for the transient silencing effect. Here we have demonstrated that exocytosis and recycling circuit (Early endosome, Golgi body and Lysosomes) mediated recycling is another possible pathway that accounts for the transient silencing activity of siRNA nanoparticles. Although exocytosis is one of the reasons accounting for the decreased cellular uptake and silencing activity in vitro in a monolayer cell culture system, it may be beneficial for intercellular transportation.
In conclusion, we elucidate the intracellular trafficking and fate of the streptavidin based siRNA delivery system. By using fluorescent labeled siRNA, streptavidin and protamine, we monitored their distinct behaviors inside the cells. Cytoplasmic and nuclear translocation of the different components of SSCP have been elucidated. Our studies particularly indicate the importance of exocytosis in nonviral siRNA delivery systems. The use of endosomolytic agents or exocytosis inhibitors may further enhance and extend the silencing activity of the siRNA nanocomplex.
Acknowledgments
This work was supported by 1R01AA021510 from the National Institute of Health. We acknowledge use of the confocal microscope in the University Missouri-Kansas City School of Dentistry Confocal Microscopy Core. This facility is supported by the UMKC Office of Research Services, UMKC Center of Excellence in Dental and Musculoskeletal Tissues, and NIH grant S10RR027668.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tai W, Qin B, Cheng K. Inhibition of breast cancer cell growth and invasiveness by dual silencing of HER-2 and VEGF. Mol Pharm. 2010;7(2):543–56. doi: 10.1021/mp9002514. [DOI] [PubMed] [Google Scholar]
- 2.Qin B, Cheng K. Silencing of the IKKepsilon gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast Cancer Res. 2010;12(5):R74. doi: 10.1186/bcr2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahato R, Qin B, Cheng K. Blocking IKKalpha expression inhibits prostate cancer invasiveness. Pharm Res. 2011;28(6):1357–69. doi: 10.1007/s11095-010-0351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, de Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1(1):14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng K, Mahato RI. Biological and therapeutic applications of small RNAs. Pharm Res. 2011;28(12):2961–5. doi: 10.1007/s11095-011-0609-0. [DOI] [PubMed] [Google Scholar]
- 6.Cheng K, Qin B. RNA Interference for Cancer Therapy. In: Lu Y, Mahato RI, editors. Pharmaceutical Perspectives of Cancer Therapeutics. Springer; US: 2009. pp. 399–440. [Google Scholar]
- 7.Bin Qin RSS, Cheng Kun. Delivery of nucleic acids for ocular gene modulation. Advances in Ocular Drug Delivery. 2012:87–114. [Google Scholar]
- 8.Cheng K, Mahato RI. siRNA delivery and targeting. Mol Pharm. 2009;6(3):649–50. doi: 10.1021/mp900094j. [DOI] [PubMed] [Google Scholar]
- 9.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin B, Chen Z, Jin W, Cheng K. Development of cholesteryl peptide micelles for siRNA delivery. J Control Release. 2013;172(1):159–68. doi: 10.1016/j.jconrel.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DC, Cho YA, Li H, Yung BC, Lee RJ. Proteinase K-containing Lipid Nanoparticles for Therapeutic Delivery of siRNA LOR-1284. Anticancer Res. 2014;34(7):3531–5. [PubMed] [Google Scholar]
- 12.Shukla RS, Tai W, Mahato R, Jin W, Cheng K. Development of streptavidin-based nanocomplex for siRNA delivery. Mol Pharm. 2013;10(12):4534–45. doi: 10.1021/mp400355q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla RS, Qin B, Wan YJ, Cheng K. PCBP2 siRNA reverses the alcohol-induced pro-fibrogenic effects in hepatic stellate cells. Pharm Res. 2011;28(12):3058–68. doi: 10.1007/s11095-011-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alabi CA, Love KT, Sahay G, Stutzman T, Young WT, Langer R, Anderson DG. FRET-labeled siRNA probes for tracking assembly and disassembly of siRNA nanocomplexes. ACS Nano. 2012;6(7):6133–41. doi: 10.1021/nn3013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31(7):653–8. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakhtianchi R, Minchin RF, Lee KB, Alkilany AM, Serpooshan V, Mahmoudi M. Exocytosis of nanoparticles from cells: Role in cellular retention and toxicity. Advances in Colloid and Interface Science. 2013;201–202(0):18–29. doi: 10.1016/j.cis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Chithrani BD, Chan WCW. Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes. Nano Letters. 2007;7(6):1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 18.Stayton I, Winiarz J, Shannon K, Ma Y. Study of uptake and loss of silica nanoparticles in living human lung epithelial cells at single cell level. Analytical and Bioanalytical Chemistry. 2009;394(6):1595–1608. doi: 10.1007/s00216-009-2839-0. [DOI] [PubMed] [Google Scholar]
- 19.Yang VC, Fu YY, Teng CL, Ma SC, Shanberge JN. A method for the quantitation of protamine in plasma. Thromb Res. 1994;74(4):427–34. doi: 10.1016/0049-3848(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 20.Qin B, Tai W, Shukla RS, Cheng K. Identification of a LNCaP-specific binding peptide using phage display. Pharm Res. 2011;28(10):2422–34. doi: 10.1007/s11095-011-0469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Yu S, Lasell TK, Jadhav AP, Macia E, Chardin P, Melancon P, Roth M, Mitchison T, Kirchhausen T. Exo1: a new chemical inhibitor of the exocytic pathway. Proc Natl Acad Sci U S A. 2003;100(11):6469–74. doi: 10.1073/pnas.0631766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Sayed A, Futaki S, Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. Aaps j. 2009;11(1):13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez AP, Cosaka ML, Romero EL, Morilla MJ. Uptake and intracellular traffic of siRNA dendriplexes in glioblastoma cells and macrophages. Int J Nanomedicine. 2011;6:2715–28. doi: 10.2147/IJN.S25235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng K, Yang N, Mahato RI. TGF-beta1 gene silencing for treating liver fibrosis. Mol Pharm. 2009;6(3):772–9. doi: 10.1021/mp9000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34(1):322–33. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu JJ, Langer R, Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol Pharm. 2009;6(3):763–71. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Qin B, Cheng K. Delivery of nucleic acids. In: Mitra AK, Lee CH, Cheng K, editors. Advanced Drug Delivery. John Wiley & Sons; 2013. pp. 257–274. [Google Scholar]
- 28.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31(7):638–46. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 29.Park K. Extravascular transport of nanoparticles in solid tumors. J Control Release. 2012;161(3):967. doi: 10.1016/j.jconrel.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Huotari J, Helenius A. Endosome maturation. The EMBO Journal. 2011;30(17):3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirsch M, Helm M. Live cell imaging of duplex siRNA intracellular trafficking. Nucleic Acids Research. 2015;43(9):4650–4660. doi: 10.1093/nar/gkv307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge X, Zhang Q, Cai Y, Duan S, Chen S, Lv N, Jin T, Chen Y, Yuan W. PEG-PCL-DEX polymersome-protamine vector as an efficient gene delivery system via PEG-guided self-assembly. Nanomedicine (Lond) 2014;9(8):1193–207. doi: 10.2217/nnm.13.83. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Xu Z, Guo S, Zhang L, Sharma A, Robertson GP, Huang L. Intravenous delivery of siRNA targeting CD47 effectively inhibits melanoma tumor growth and lung metastasis. Mol Ther. 2013;21(10):1919–29. doi: 10.1038/mt.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YM, Park MR, Song SC. An injectable cell penetrable nano-polyplex hydrogel for localized siRNA delivery. Biomaterials. 2013;34(18):4493–500. doi: 10.1016/j.biomaterials.2013.02.050. [DOI] [PubMed] [Google Scholar]
- 35.Chang LC, Lee HF, Yang Z, Yang VC. Low molecular weight protamine (LMWP) as nontoxic heparin/low molecular weight heparin antidote (I): preparation and characterization. AAPS pharmSci. 2001;3(3):E17. doi: 10.1208/ps030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgado D, del Pozo-Rodriguez A, Solinis MA, Rodriguez-Gascon A. Understanding the mechanism of protamine in solid lipid nanoparticle-based lipofection: the importance of the entry pathway. Eur J Pharm Biopharm. 2011;79(3):495–502. doi: 10.1016/j.ejpb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya Y, Ishii T, Okahata Y, Sato T. Characterization of Protamine as a Transfection Accelerator for Gene Delivery. Journal of Bioactive and Compatible Polymers. 2006;21(6):519–537. [Google Scholar]
- 38.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276(8):5836–40. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 39.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Research. 2006;34(5):1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Zhou L, Zhou Y, Liu J, Xing X, Zhong J, Xu G, Kang Z, Liu J. Mitophagy induced by nanoparticle-peptide conjugates enabling an alternative intracellular trafficking route. Biomaterials. 2015;65:56–65. doi: 10.1016/j.biomaterials.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, Manoharan M, Kirchhausen T, Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol. 2015;33(8):870–6. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erazo-Oliveras A, Muthukrishnan N, Baker R, Wang T-Y, Pellois J-P. Improving the Endosomal Escape of Cell-Penetrating Peptides and Their Cargos: Strategies and Challenges. Pharmaceuticals (Basel, Switzerland) 2012;5(11) doi: 10.3390/ph5111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takano M, Kawami M, Aoki A, Yumoto R. Receptor-mediated endocytosis of macromolecules and strategy to enhance their transport in alveolar epithelial cells. Expert Opinion on Drug Delivery. 2014;12(5):813–825. doi: 10.1517/17425247.2015.992778. [DOI] [PubMed] [Google Scholar]
- 44.Oh N, Park JH. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomedicine. 2014;9(Suppl 1):51–63. doi: 10.2147/IJN.S26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alabi CA, Sahay G, Langer R, Anderson DG. Development of siRNA-probes for studying intracellular trafficking of siRNA nanoparticles. Integr Biol (Camb) 2013;5(1):224–30. doi: 10.1039/c2ib20155k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu XH, Jiang N, Yao PB, Zheng XL, Cayabyab FS, Tang CK. NPC1, intracellular cholesterol trafficking and atherosclerosis. Clin Chim Acta. 2014;429:69–75. doi: 10.1016/j.cca.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 47.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam JKW, Liang W, Lan Y, Chaudhuri P, Chow MYT, Witt K, Kudsiova L, Mason AJ. Effective endogenous gene silencing mediated by pH responsive peptides proceeds via multiple pathways. Journal of controlled release : official journal of the Controlled Release Society. 2012;158(2):293–303. doi: 10.1016/j.jconrel.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Research. 2006;34(1):322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi J, Xu Y, Xu X, Zhu X, Pridgen E, Wu J, Votruba AR, Swami A, Zetter BR, Farokhzad OC. Hybrid lipid-polymer nanoparticles for sustained siRNA delivery and gene silencing. Nanomedicine. 2014;10(5):897–900. doi: 10.1016/j.nano.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]