Summary
Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) expression correlates with malignancy. But its role(s) in pathogenesis remain enigmatic. Here, we interrogated the IGF2BP3-RNA interaction network in pancreatic ductal adenocarcinoma (PDAC) cells. Using a combination of genome-wide approaches we identify 164 direct mRNA targets of IGF2BP3. These transcripts encode proteins enriched for functions such as cell migration, proliferation and adhesion. Loss of IGF2BP3 reduced PDAC cell invasiveness and remodeled focal adhesion junctions. Individual-nucleotide resolution crosslinking immunoprecipitation (iCLIP) revealed significant overlap of IGF2BP3 and miRNA binding sites. IGF2BP3 promotes association of the RNA induced silencing complex (RISC) with specific transcripts. Our results show that IGF2BP3 influences a malignancy-associated RNA regulon by modulating miRNA-mRNA interactions.
Keywords: IGF2BP3, post-transcriptional gene regulation, RNA binding protein, RISC complex, microRNA, cancer
Introduction
RNA binding proteins (RBPs) and micro(mi)RNAs are the central mediators of posttranscriptional gene regulation (Gerstberger et al., 2014). Like transcription factors, RBPs and miRNAs coordinate expression of proteins with related functions (Blackinton and Keene, 2014, Keene, 2007). These regulatory factors often converge on the 3’ untranslated regions (3’UTRs) of mRNAs (Jens and Rajewsky, 2015). Juxtaposition of their binding sites contributes to combinatorial mechanisms of post-transcriptional gene regulation (Glorian et al., 2011, Zhang et al., 2008, Kundu et al., 2012, Young et al., 2012, Kim et al., 2009, Jafarifar et al., 2011, Ho et al., 2013).
IGF2BP1, -2 and -3 are a family of structurally and functionally related RBPs with developmentally regulated expression patterns (Yaniv and Yisraeli, 2002). IGF2BP3 is of particular interest because it is undetectable in most adult tissue but strongly expressed in embryos and in diverse tumor types (Mueller-Pillasch et al., 1999, Kobel et al., 2009, Schaeffer et al., 2010, Findeis-Hosey and Xu, 2011). For example, it is up-regulated in 90% of pancreatic ductal adenocarcinomas, suggesting that it may have a role in initiation or progression of cancer (Schaeffer et al., 2010). Several groups proposed that elevated IGF2BP3 expression is prognostic for malignancy in PDAC, colorectal, ovarian cancers and B-acute lymphocytic leukemia (B-ALL) and negatively influences patient survival (Suvasini et al., 2011, Stoskus et al., 2011, Schaeffer et al., 2010, Lochhead et al., 2012, Kobel et al., 2009, Bell et al., 2013). Indeed, Taniuchi et al demonstrated aberrant IGF2BP3 expression in pancreatic tumors promotes metastasis in xenograft assays in nude mice (Taniuchi et al., 2014). It is likely that this role of IGF2BP3 in cancer metastasis mirrors a normal function of IGF2BP3-mediated cell migration during embryogenesis (Mueller et al., 2003, Mueller-Pillasch et al., 1999).
The evidence for IGF2BP3 acting as a bona fide pathoprotein continues to mount (Wagner et al., 2003, Palanichamy et al. 2016) yet several fundamental questions remain unanswered. For example, IGF2BP3 regulates cytoplasmic steps of posttranscriptional gene expression but the molecular mechanisms are unknown (Nielsen et al., 1999, Vikesaa et al., 2006, Jonson et al., 2014, Gu et al., 2012). Another challenge is elucidating the RNA binding specificity of IGF2BP3 in cancer cells. Recent studies identified transcripts associated with exogenous or endogenous IGF2BP3 using PAR-CLIP and RIP-seq (Hafner et al., 2010, Taniuchi et al., 2014). Here, we use a combination of genomic approaches to discover cancer-related IGF2BP3 mRNA targets. These data also suggest a mechanism for IGF2BP3-dependent gene regulation. We find that IGF2BP3 modulates the association of target transcripts with the RISC complex. Taken together, our results reveal a malignancy associated RNA network regulated by IGF2BP3.
RESULTS
Identification of IGF2BP3 mRNA targets in PDAC cells
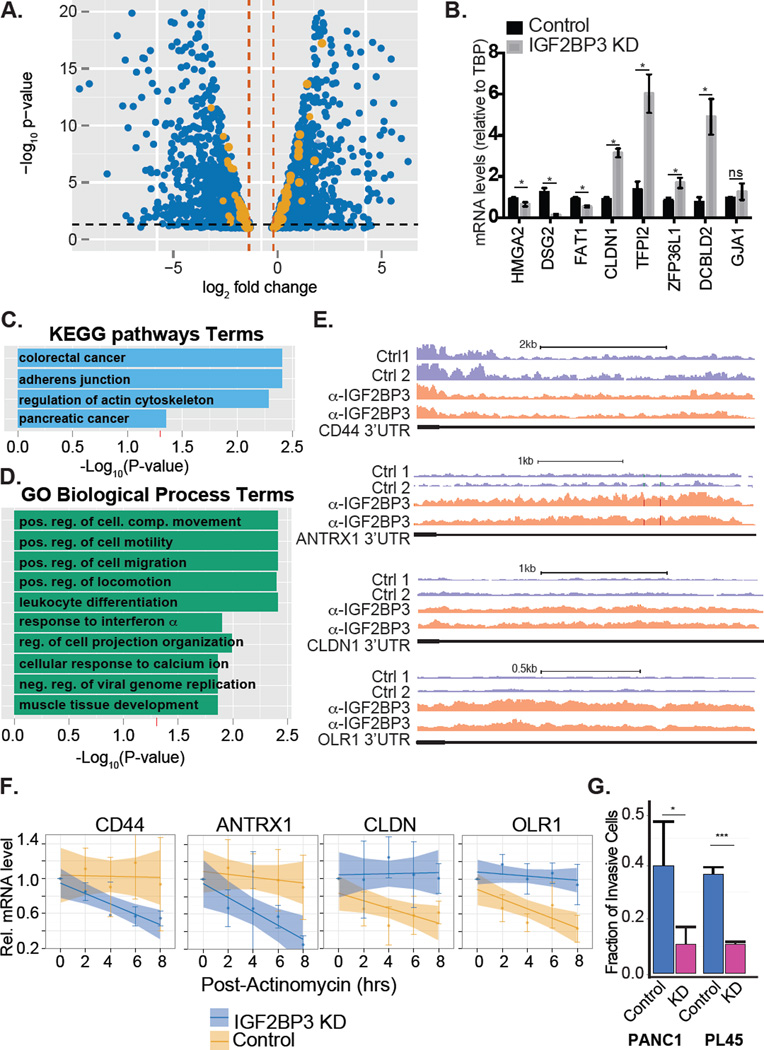
To characterize the mRNA targets of IGF2BP3 we performed RIP-seq in two PDAC cell lines. We identified 2,223 and 1,718 transcripts enriched in the anti-IGF2BP3 immunoprecipitate relative to the control from PL45 and PANC1 cells, respectively (Supplemental Figure 1 and Supplemental Table 1). A significant fraction of the PANC1 target transcripts (1,069, ~63%, Supplemental Figure 1D) were also identified in the PL45 RIP dataset (odds-ratio = 14.2, P< 1.50e-195; Fisher's exact test). This common set of transcripts encodes proteins over-represented for functions in focal adhesion junctions, cell migration, and regulation of the actin cytoskeleton (See Supplemental Table 2). Included in this common set are CD44, HMGA2, EIF4EBP2, ARF6 and ARHGEF4, previously characterized as IGF2BP3 targets (Vikesaa et al. 2006; Jonson et al. 2014; Taniuchi et al. 2014; Mizutani et al. 2015)
To identify IGF2BP3 regulatory targets we analyzed steady state mRNA levels in IGF2BP3-depleted and control PDAC cells (PL45 and PANC1 cell lines) using high-throughput RNA sequencing (RNA-Seq). DEseq analysis revealed 2,795 differentially expressed PANC1 genes (Figure 1A and Supplemental Table 3). We validated several candidate genes from PANC1 cells by RT-qPCR. Of the 10 transcripts selected for validation, 9 agreed with the RNA-Seq data (Figure 1B). In PL45 cells, DEseq identified 137 differentially expressed transcripts (Supplementary Table 3). We obtained similar results using Affymetrix exon arrays (Figure 1A and Supplemental Table 4). Of the 137 differentially expressed genes in PL45 cells, 53 were also identified in the PANC1 experiment (P<0.0001, Fishers Exact Test). Comparison of control PANC1 and PL45 revealed thousands of differentially expressed genes, suggesting that these cell models are quite different (Supplemental Table 5 and Ryu et al., 2002) To identify direct IGF2BP3 regulatory targets, we focused on differentially expressed transcripts that are reproducibly associated with IGF2BP3 in RIP-Seq assays (Figure 1A, orange dots). This approach revealed 164 transcripts encoding proteins enriched for functions in cancer pathways, cellular migration/motility, and regulation of the actin cytoskeleton (Figure 1C and 1D, adjusted P < 0.05, Supplemental Table 2).
Figure 1. IGF2BP3 regulates transcripts associated with cancer pathways.
A. Volcano plot depicting differentially expressed genes from both PANC1 and PL45 (blue dots) and those mRNAs bound by IGF2BP3 (orange dots). B. Bar graph showing relative quantification of mRNA levels in IGF2BP3-depleted or control PANC1 cells. C. KEGG pathway enrichment analysis of differentially expressed-IGF2BP3 bound mRNAs (orange dots from panel A). D. Gene ontology enrichment analysis of differentially expressed-IGF2BP3 bound mRNAs. E. Genome Browser snapshots of read coverage from replicate RIP-seq experiments from PL45 cells for CD44 and CLDN1 3’UTR (top and bottom panel, respectively). F. Relative quantification RT-qPCR analysis of CD44 and CLDN1 mRNA stability in control or IGF2BP3-depleted PL45 cells. G. Bar graph quantifying the invasiveness of control or IGF2BP3-depleted PANC1 or PL45 cells through matrigel filters relative to control filters. The bars represent an average of three independent experiments. Error bars correspond to standard deviation. Statistical significance estimated using unpaired T-test, *** P<0.001, * P<0.05.
We investigated how IGF2BP3 affects steady state mRNA levels using the PL45 cell model. In PL45 cells as previously described for HeLa cells (Vikessa et al. 2006), IGF2BP3-depletion correlates with reduced mRNA levels of the bona fide target CD44 (Supplemental Table 1 and 3). Previous studies determined that IGF2BP3 stabilizes CD44 mRNA (Vikessa et al. 2006). We used actinomycin D to inhibit transcription and measure the decay rate of CD44 and three additional targets, ANTRX1, CLDN1 and OLR1 mRNAs. As expected we confirmed that CD44 mRNA is less stable in IGF2BP3-depleted cells. Likewise, ANTRX1 mRNA also has a shorter half-life in IGF2BP3-depleted cells relative to control cells. By contrast, decay of CLDN1 and OLR1 mRNA is attenuated when IGF2BP3 is depleted from PL45 cells (Figure 1E and 1F).
IGF2BP3 modulates the focal adhesion junction and promotes PDAC cell invasiveness
To determine how IGF2BP3 expression influences cancer cell biology, we analyzed focal adhesion junctions and cell migration in control or IGF2BP3-depleted PDAC cells (Supplemental Figure 2). Genes from both of these pathways are enriched in the IGF2BP3-RNA interaction network (Figure 1C,D and Supplemental Table 2). We stained focal adhesion complexes with antibody recognizing focal adhesion kinase (FAK). Although FAK staining localizes to the periphery of both control and IGF2BP3-depleted cell lines, their size difference was significant in IGF2BP3-depleted cells relative to control (Supplemental Figure 2D and E, P<0.0020, Student’s T-test). To determine whether depletion of IGF2BP3 affects the invasive behavior of PDAC cells we performed in vitro invasion assays. IGF2BP3-depleted cells exhibited significantly reduced invasiveness as compared to control cells (Student’s t-test, P<0.01 for both lines, Figure 1G and supplementary Figure 2C). Taken together these data show that IGF2BP3 alters the focal adhesion junction and promotes PDAC invasiveness in vitro.
A single nucleotide resolution map of IGF2BP3-RNA interactions
We used iCLIP to determine in situ binding specificity of IGF2BP3 in PL45 and PANC1 cells. Anti-IGF2BP3 antibodies precipitated nuclease-sensitive protein-RNA complexes from whole cell extracts of UV-irradiated cells (Supplemental Figure 3A–C). We obtained three libraries from two replicate experiments from PL45 and a single PANC1 assay. We identified 244 and 124 mRNA targets that directly crosslinked with IGF2BP3 in PL45 and PANC1 cells, respectively (Supplementary Table 6). Unique reads from the IGF2BP3 iCLIP libraries map with high frequency to exonic sequences. This distribution differs from another RBP, hnRNPA1, which crosslinks to intronic sequences (Figure 2A). A background set of simulated crosslinking sites map primarily to intergenic regions (Figure 2A). We annotated IGF2BP3, hnRNPA1 and simulated binding sites using the Piranha peak caller software (Uren et al., 2012). Both IGF2BP3 and hnRNPA1 peaks occur with highest frequency in 3’UTRs of target transcripts (Figure 2B). By contrast, simulated data exhibit a larger proportion of peaks in first and internal exons.
FIGURE 2. Global Single nucleotide-resolution IGF2BP3-RNA interaction map.
A. Bar graph showing genomic distribution of IGF2BP3 from PDAC cell lines, hnRNPA1 (HEK293 cells) and simulated (hg19 background) crosslinking sites. The bars represent the average of replicate experiments (N=2 from PL45; N=1 from PANC1), respectively. Standard deviations are indicated by error bars. B. Distribution of peaks called from IGF2BP3-, hnRNPA1- and simulated crosslinking sites within mRNA. C. Meta-analysis of IGF2BP3 and hnRNPA1 iCLIP crosslink sites relative to the stop codon. Relative crosslinking density (normalized count) is shown −/+ SEM. D. IGF2BP3 crosslinking density from PDAC cell lines mapped relative to annotated miRNA target sites. hnRNPA1 crosslinking sites from HEK293 cells are included as a control. E. Over represented pentamers found near IGF2BP3 crosslinking sites in PDAC cells.
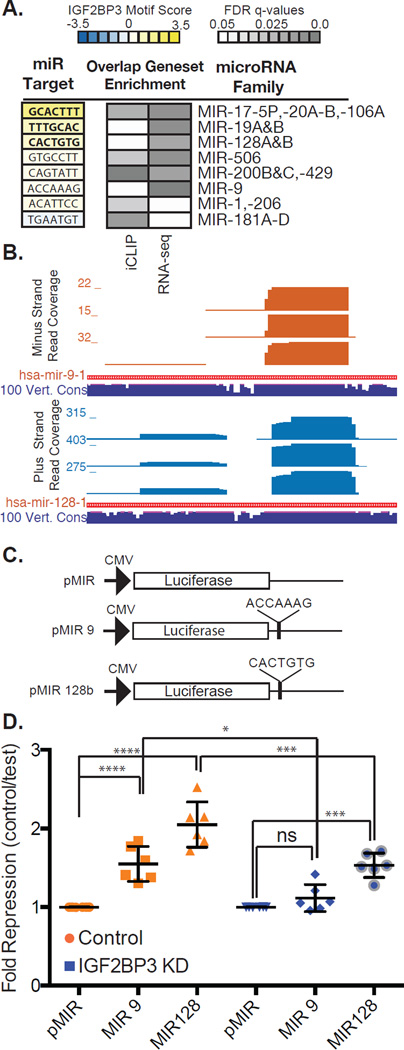
IGF2BP3 binding sites occur frequently within 3’UTRs. We hypothesized that cis regulatory signals associated with 3’UTR overlap IGF2BP3 binding sites. To test this hypothesis, we refined the IGF2BP3-RNA interaction map by isolating the 5'end of each read. These positions correspond to the protein-RNA crosslink site. We determined crosslinking frequency relative to cis-acting RNA elements within 3'UTRs (Figure 2C and D). IGF2BP3 and hnRNPA1 crosslinking density relative to stop codons is similar (Figure 3C). By contrast, crosslinking density for IGF2BP3, but not hnRNPA1, peaked within a 25 bp window centered on predicted miRNA target sequences (Figure 2D). These data suggest that IGF2BP3 binding is specifically enriched over predicted miRNA target sites. Next, we used IGF2BP3 crosslink sites to identify over-represented motifs using a pentamer-clustering approach (See Methods). Two over-represented motifs occur within a 10 nt window of each crosslinking site. One motif resembles a previously defined IGF2BP3 consensus binding sequence (Figure 2E, lower panel and Supplemental Table 7; (Hafner et al., 2010)). Our results are also consistent with a bipartite motif described for IGF2BP1 (Chao et al., 2010). ~50–70% of the PL45 and PANC1 iCLIP targets significantly overlapped the RIP-Seq data (Fisher’s Exact test P < 2.74 e-19 and 1.0 e-90, respectively; Supplemental Figure 3F, J and K). These cross-validated IGF2BP3 targets encode proteins with functions in cellular adhesion, migration, and remodeling of the extracellular matrix (Supplemental Figure 3G).
Figure 3. IGF2BP3 modulates expression of miRNA-regulated transcripts.
A. Examples of miRNA families with significant sequence similarity between their target site and the IGF2BP3 consensus motif (left panel). Enrichment of miRNA target sites in 3’UTRs identified by iCLIP and by RNA-Seq analysis of control or IGF2BP3-depleted PANC1 cells. B. UCSC genome browser screen shots showing read coverage across miRNA-9 and -128 loci (upper and lower panel, respectively). Histogram shows number of reads at each position. Sequence conservation across 100 vertebrate genomes (navy blue track). C. Schematic diagram of luciferase reporter plasmids. D. Scatter plot showing normalized luciferase reporter expression (fold repression) in control cells (orange) and IGF2BP3-depleted cells (blue).
IGF2BP3-RNA crosslinks overlap miRNA targets sites in 3'UTRs. We determined if IGF2BP3 shares sequence specificity with miRNAs. We scored 221 miRNA seed-target site pairings (Lewis et al., 2005) by their similarity to IGF2BP3-bound pentamers (see Methods). miRNAs score significantly higher if their target site crosslinks to IGF2BP3 than isolated sites (Figure 3A and Supplemental Table 8; P<0.00153 Wilcoxon rank-sum test). Specific miRNA target sites are over represented in both the iCLIP data and within the 3’UTRs of differentially expressed transcripts in PANC1 cells (Figure 3A, grey boxes, Supplemental Figure 3H–I, Supplemental Tables 9 and 10). These include miR-1, -9, 17, -20A/B, -19A/B, -106, -128A/B, -181, -200B/C, -429, and -506, amongst others. Small RNA sequencing confirmed the expression of this subset of miRNA in PANC1 cells (Figure 3B and Supplemental Table 11).
RBPs can stabilize mRNA by competing with miRNAs for common regulatory motifs (Jens and Rajewsky, 2015). To test this hypothesis we re-examined IGF2BP3-dependent changes in steady state mRNA levels. Transcripts targeted by both IGF2BP3 and miRNAs tended toward a reduction in steady state mRNA levels. By contrast, transcripts containing only miR-target sites were largely unaffected by IGF2BP3-depletion (Supplemental Figure 3I). These data suggest that IGF2BP3 may attenuate miRNA-mediated mRNA decay. To further determine if IGF2BP3 antagonizes specific miRNAs, we generated luciferase mRNA reporters containing exact matches to miR-9 and -128 in their 3'UTR (Figure 3C). Both miRNAs target a sequence predicted to bind IGF2BP3. If the attenuation model is correct we predict IGF2BP3-depletion would result in a significant decrease in luciferase reporter expression. In wild type cells, both miR-9 and -128 reporters showed significant repression relative to the control reporter construct (Figure 3D, P<0.0022). By contrast, repression of miR-9 and -128 luciferase reporters were significantly attenuated in IGF2BP3-depleted cells (Figure 3D, compare repression of miR-9 and -128 in IGF2BP3-depeleted or control cells, P<0.0022).
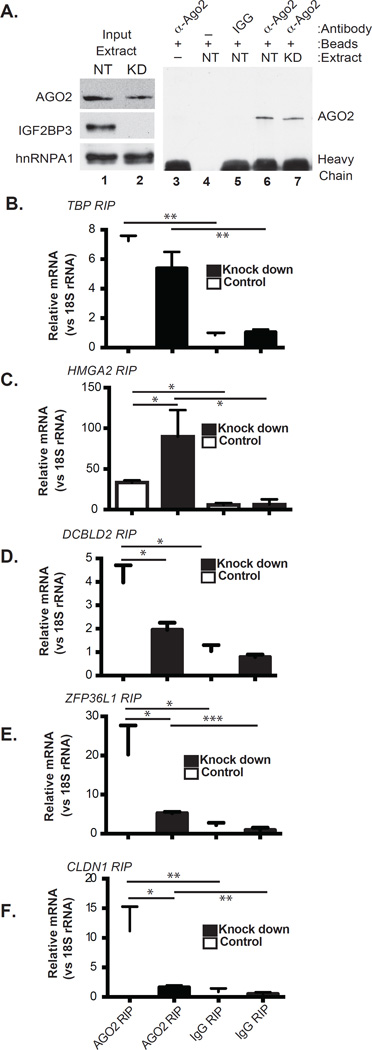
IGF2BP3 promotes Ago2-mRNA interactions
The data presented above suggest that IGF2BP3 may function in concert with the RNA induced silencing complex in PDAC cells. We tested this hypothesis by immunoprecipitating Argonaute 2 (Ago2), a RISC-component (Gerstein et al., 2010), from control or IGF2BP3-depleted PANC1 cells and further quantified co-purified mRNA by RT-qPCR. Western blotting confirmed precipitation of Ago2 from both control and IGF2BP3-depleted cells (Figure 4A). TBP and HMGA2 mRNAs served as controls for this experiment. TBP mRNA was not detected in any of the IGF2BP3 protein-RNA interaction assays (iCLIP or RIP) and its steady-state mRNA levels are not affected by IGF2BP3 depletion (Supplemental Figure 4). By contrast, HMGA2 is a bona fide IGF2BP3-regulatory target. IGF2BP3 stabilizes HMGA2 mRNA by antagonizing its association with RISC (Jonson et al., 2014). Besides HMGA2, we selected several other IGF2BP3-bound transcripts for analysis including ZFP36L1, DCBLD2 and CLDN1. IGF2BP3-depletion correlates with increased ZFP36L1, DCBLD2 and CLDN1 mRNA levels and decreased HMGA2 mRNA (Supplemental Figure 4). All the mRNAs co-precipitate with Ago2 from both control and knock down cells (Figure 4, compare Ago2-RIP to IgG). TBP mRNA binding to Ago2 was independent of IGF2BP3 protein levels (Figure 4B). As expected, HMGA2 mRNA coprecipitation with Ago2 increased in IGF2BP3-depleted cells (Figure 4C). However, Ago2-association with ZFP36L1, DCBLD2 and CLDN1 mRNAs diminished. (Figure 4D–F, P< 0.0161, 0.0135 and 0.0144, respectively). These data show that IGF2BP3 is a bimodal modulator of RISC-mRNA association.
FIGURE 4. IGF2BP3 alters mRNA association with RISC.
A. Western blot of Ago2 immunoprecipitation from control (NT) or IGF2BP3-depleted (KD) PANC1 cells. (B–F) RT-qPCR analysis of RNA precipitated with α-Ago2 or nonspecific rabbit IgG (Ago2 RIP or IgG RIP, respectively). Relative quantification for each transcript was performed using 18S rRNA as a reference gene. Statistical significance was estimated for each comparison using an unpaired T-test (* P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001).
Discussion
In this study, we identify a set of IGF2BP3 mRNA targets enriched in cancer-related pathways including focal adhesions, adherens junctions, actin-cytoskeleton and cell migration. The IGF2BP3 RNA regulon reported here is strikingly similar to the homologous drosophila protein, IMP (dIMP) (Hansen et al., 2015). Our study provides evidence that IGF2BP3 and miRNAs converge on the 3’UTRs of target transcripts to coordinately up- and down-regulate programs of gene expression that are associated with malignancy and invasiveness. The results further show that this dual activity of IGF2BP3 likely operates through its ability to protect transcripts from – or enhance – miRNA-mediated post-transcriptional gene silencing.
The expression of IGF2BP3 correlates with malignancy. Elucidating direct IGF2BP3-mRNA targets is a major challenge for the field (Bell et al., 2013). Earlier studies identified thousands of IGF2BP3 targets using highly sensitive assays with low specificity (Taniuchi et al., 2014, Hafner et al., 2010). Here, we delineate a refined set of mRNA targets by integrating different genomic approaches. A similar strategy significantly reduced the mRNA target space for HuR and hnRNP H1(Uren et al., 2016, Mukherjee et al., 2011). This integrated approach to elucidating IGF2BP3 target transcripts was also applied to human B-ALL cells (Palanichamy et al. 2016). We identified a regulon of similar size (216 transcripts in B-ALL cells vs 164 in PDAC cells), but with very different classes of enriched transcripts such as cell cycle regulation and blood cell differentiation (Palanichamy et al. 2016). Further a significant fraction (~65%) of the B-ALL targets are down regulated upon IGF2BP3 depletion. These data suggest a role for IGF2BP3 in mRNA stabilization in B-ALL cells. It will be interesting to determine how IGF2BP3 activities are defined in different cell types.
miRNA modulation is a major function of RBPs (Ho et al., 2013, Kundu et al., 2012, Kedde et al., 2007, Jafarifar et al., 2011, Bhattacharyya et al., 2006, Jing et al., 2005, Xue et al., 2013). IGF2BP3 promotes mRNA stability, as described for CD44 and HMGA2 (Vikesaa et al., 2006, Jonson et al., 2014). One mechanism suggested by Jønson et al., and partially supported by our iCLIP data, is that in certain contexts miRNAs and IGF2BP3 compete for common binding sites within 3’UTRs. The sequence similarity between the consensus IGF2BP3 RNA recognition elements and target sites of several different miRNA families support this hypothesis (Figure 3) as does the concomitant decrease in expression of mRNAs that are targeted by both regulatory systems (Supplemental Figure 3I). However, our miRNA reporters and Ago2 RIP experiments revealed an unexpected role for IGF2BP3 in promoting the association of mRNAs with Ago2 (Figures 3D and 4). The mechanism we propose here is consistent with a recent finding that IGF2BP3 destabilizes the EIF4EBP2 mRNA in HeLa cells (Mizutani et al., 2015).
Our results demonstrate that IGF2BP3 is a bi-modal regulator of target mRNA stability. It remains to be determined how the dual roles of IGF2BP3 in mRNA stability are regulated and if IGF2BP3 has different functions in different types of cancer cells. Other RBPs, including HuR, PTB, MOV10 and FMRP function as bi-modal modulators of RISC function (Kenny et al., 2014, Kim et al., 2009, Kundu et al., 2012, Mukherjee et al., 2011, Young et al., 2012, Xue et al., 2013). Given that several of the miRNA families presented in Figure 3C play roles in cancer cell biology it is possible that the modulation of RISC by IGF2BP3 contributes to PDAC pathogenesis.
Experimental Procedures
Cell lines and constructs
We maintained PDAC cell lines, PL45 and PANC1, according to the recommended conditions (ATCC Manassas, VA). We created independent clones of IGF2BP3-depleted or control cell lines by transfecting PDAC cells with plasmids that express IGF2BP3-targeting short hairpin RNA or a non-targeting control (Santa Cruz Biotechnologies, Santa Cruz, CA) with Lipofectamine 2000 (Life Technologies) and selecting with puromycin (1 ug/mL).
Western blot and antibodies
We performed Western blotting as previously described (Sterne-Weiler et al., 2011). We probed nitrocellulose filters with the following antibodies against: IGF2BP3 (clone D7, Santa Cruz Biotechnology), GAPDH (Santa Cruz Biotechnology, clone FL335), hnRNPA1 (clone 4B10, Santa Cruz Biotechnology) and anti-4EBP (Santa Cruz Biotechnology, Clone D-10).
Immunofluorescence microscopy
Cells grown on acid washed, fibronectin-coated coverslips were fixed in 4% paraformaldehyde in PBS for 15 min, washed in PBS for 10 min and 0.15M glycine in PBS for 10 min on a shaker. Cells were permeabilized in 0.02% Triton in PBS for 10 min with a subsequent wash in PBS for another 10 min. Coverslips were blocked using 10% serum (to secondary host animal) in PBS and incubated with primary antibody diluted in 5% serum in a humidified chamber for 12 hours at 4°C. anti-FAK (C-20, Santa Cruz Biotech), anti-IGF2BP3 (N-19, Santa Cruz Biotech). After 3 washes in PBS for 15 min each, the samples were incubated with secondary antibody and a nuclear stain diluted in 5% serum for 45 min in a humidified chamber at room temperature. All samples were mounted onto slides with Fluoromount-G (SouthernBiotech) and allowed to dry overnight. Slides were imaged on Keyence Biorevo BZ-9000 Digital Widefield Microscope.
Invasion Assay
In vitro invasion assay used Transwell inserts with 8.0-µm pore size (BDbiosciences). Each transwell insert (matrigel coated or control) received 5 × 10^5 PDAC cells suspended in serum-free medium. The Transwell inserts were then placed into 6-well plates containing medium supplemented with 10% FBS. After 72 hours, we removed cells from the upper surface of the Transwell inserts and fixed/stained cells on the lower surface using the Quik-Diff kit and DAPI (IMEB, San Marcos, CA). We counted DAPI stained nuclei from four fields of each well using an inverted microscope and ImageJ software. We performed each experiment in triplicate.
Gene expression profiling of PDAC cells by RNA-Seq
We isolated RNA from cytosolic fractions of IGF2BP3-depleted or control PANC1 and PL45 cells using TRI-Reagent LS (Sigma). Illumina compatible sequencing libraries were prepared using the NEXTflex™ Rapid Directional qRNA-Seq™ Kit (BIOO Scientific, Austin, TX). We analyzed each condition in duplicate using the HiSeq2500 platform. MiRNA expression was profiled in PANC1 cells using the NEXTflex Small RNA sequencing kit v2 (BIOO Scientific, Austin TX). MiRNA expression was analyzed in triplicate. Mapping statistics can be found in Supplemental Table 12.
RIP-Seq assay
RNA immunoprecipitation with anti-IGF2BP3 (RN009P, MBL Inc.) or normal rabbit IgG (AB-105-C, R&D) was performed with PL45 and PANC1 cells as previously described (Uren et al., 2016). Each RIP and control IP was performed in duplicate (PL45) or triplicate (PANC1). Mapping statistics can be found in Supplemental Table 12.
iCLIP assay
iCLIP was performed as previously described from PL45 and PANC1 cells (König J et al., 2010). Two replicate experiments were performed in PL45 cells and one experiment was performed in PANC1. The three data sets were pooled for analysis. Mapping statistics can be found in Supplemental Table 12.
cDNA synthesis and RT–qPCR
cDNA was synthesized from RNA purified from cytosolic extracts and RIP samples using the High-Capacity cDNA reverse transcriptase kit from ABI Scientific. qPCR was perfomed using a Roche Lightcycler 480 (Roche Diagnostics), Titanium Taq (Clontech) and SYBR green dye. Based on melting point analysis primers corresponding to TBP1, CLDN1, ZFP36L1, DCBDL2, FAT1, DSG2, TFPI2, GJA1, HMGA2, IGF2BP3 and 18S rRNA generated a single specific amplicon. Primer sequences can be found in the Supplementary text. For normalization, TBP was used as a reference gene for comparisons in cytoplasmic RNA whereas 18S rRNA was used in the RIP experiments. To compare TBP levels between cells, the geometric mean of three stable genes (CFL1, RHOA and EGFR) was used as a reference. Relative expression levels were determined using the ΔΔCT method with the Roche Lightcycler Analysis Software Package version 1.5 (Roche Diagnostics). All experiments use a minimum of three technical replicates per sample and at least three biological replicates per analysis. Statistical significance was determined by comparing the mean normalized ratios of each mRNA using a non-parametric Mann-Whitney U test (Prism6, GraphPad).
Ago2 RNA immunoprecipitation
To immunoprecipitate Ago2 mouse monoclonal antibody 9E8.2 (Millipore) was tethered to Dynal protein A beads using a rabbit anti-mouse IgG bridging antibody (Jackson ImmunoResearch, Fcγ Fragment Specific). Mouse IgG (Pierce Biotechnology) was used as a negative control for RIP assays. Beads were washed three times in lysis buffer (25 mM Tris-HCl at pH 8.0, 150 mM NaCl, 2 mM MgCl2, 0.5% NP-40, and 5 mM DTT) to remove the unbound antibody. Whole cell extracts were prepared from IGF2BP3-depleted or control PANC1 cells by lysing cells on ice for 10 min in 1 mL of fresh lysis buffer with protease inhibitors (Complete Protease Inhibitor Cocktail Tablets, EDTA-free, Roche Applied Science) and RNasin (1/1000 dilution; Applied Biosystems), followed by sonication and centrifugation at 10,000g for 10 minutes. Following the washes in lysis buffer (as described in Supplemental experimental procedures), beads were treated with 5U DNase 1 (Promega) for 10 minutes at 37°C. An aliquot from each IP was used for western blot analysis and RNA was purified from the remainder using TRIzol LS (Invitrogen). Total RNA from cell lysates was isolated using the same procedure, and was also subjected to DNA digestion as described above prior to cDNA synthesis.
iCLIP-, RIP-, mRNA and miRNA-seq data analysis pipeline
Bioinformatic analyses are described in the supplementary experimental procedures.
Luciferase Reporter Assays
Luciferase reporters (pMIR, Life Scientific) containing sequences with perfect complementarity to miR-9 and -128 in their 3’UTR were transfected into control or IGF2BP3-depleted PANC1 cells using Lipofectamine 2000 (Life Scientific). Renilla luciferase reporter plasmid (Promega) was cotransfected as a control for transfection efficiency. Renilla and firefly luciferase activity was assayed 24 hours post-transfection using the Dual-Glo system (Promega).
Supplementary Material
Acknowledgments
This work was funded through grants from the NIH GM109146 (to JRS), HG007336 (MT) and HG006015 (ADS and LOP), Santa Cruz Cancer Benefit Group (to JRS), University of California Cancer Research Coordinating Committee (to JRS) and CIRM TG2-00157 (to HE), the CIRM Major Facilities grant FA1-00617-1 and the CIRM Shared Stem Cell Facilities grant CL1-00506-1.2. TSW was part supported in part by an operating grant from the Canadian Institutes of Health Research and a CH Best Foundation Postdoctoral Fellowship. We thank The Genome Sequencing and Analysis Facility at the University of Texas at Austin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
RNA-seq data sets are available in the GEO SRA (Accession number GSE79147).
Author Contributions
HE, JMH, TSW and JRS: Experimental design, performed experiments, analyzed data, prepared figures, prepared manuscript.
DJC, MD, JMD, AJW, OF: Performed experiments, analyzed data.
PU, SK: Analyzed data.
FJ, SCB and MQ: Performed experiments.
LH, AS, LOFP and MMT: Designed experiments, prepared manuscript.
BB: Edited manuscript.
Reference
- Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M, Huttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014;34:44–54. doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeis-Hosey JJ, Xu H. The use of insulin like-growth factor II messenger RNA binding protein-3 in diagnostic pathology. Hum Pathol. 2011;42:303–314. doi: 10.1016/j.humpath.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;18:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Katz Z, Wu B, Park HY, Li D, Lin S, Wells AL, Singer RH. Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J Cell Sci. 2012;125:81–91. doi: 10.1242/jcs.086132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HT, Rasmussen SH, Adolph SK, Plass M, Krogh A, Sanford J, Nielsen FC, Christiansen J. Drosophila Imp iCLIP identifies an RNA assemblage co-ordinating F-actin formation. Genome Biol. 2015;16:123. doi: 10.1186/s13059-015-0687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JJ, Robb GB, Tai SC, Turgeon PJ, Mawji IA, Man HS, Marsden PA. Active stabilization of human endothelial nitric oxide synthase mRNA by hnRNP E1 protects against antisense RNA and microRNAs. Mol Cell Biol. 2013;33:2029–2046. doi: 10.1128/MCB.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CArich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–1334. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Jonson L, Christiansen J, Hansen TV, Vikesa J, Yamamoto Y, Nielsen FC. IMP3 RNP Safe Houses Prevent miRNA-Directed HMGA2 mRNA Decay in Cancer and Development. Cell Rep. 2014;7:539–551. doi: 10.1016/j.celrep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, Le Sage C, Nagel R, Voorhoeve PM, Van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, Ceman S. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014;9:1729–1741. doi: 10.1016/j.celrep.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel M, Xu H, Bourne PA, Spaulding BO, Shih Ie M, Mao TL, Soslow RA, Ewanowich CA, Kalloger SE, Mehl E, et al. IGF2BP3 (IMP3) expression is a marker of unfavorable prognosis in ovarian carcinoma of clear cell subtype. Mod Pathol. 2009;22:469–475. doi: 10.1038/modpathol.2008.206. [DOI] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lochhead P, Imamura Y, Morikawa T, Kuchiba A, Yamauchi M, Liao X, Qian ZR, Nishihara R, Wu K, Meyerhardt JA, et al. Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur J Cancer. 2012;48:3405–3413. doi: 10.1016/j.ejca.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani R, Imamachi N, Suzuki Y, Yoshida H, Tochigi N, Oonishi T, Suzuki Y, Akimitsu N. Oncofetal protein IGF2BP3 facilitates the activity of protooncogene protein eIF4E through the destabilization of EIF4E-BP2 mRNA. Oncogene. 2015 doi: 10.1038/onc.2015.410. [DOI] [PubMed] [Google Scholar]
- Mueller F, Bommer M, Lacher U, Ruhland C, Stagge V, Adler G, Gress TM, Seufferlein T. KOC is a novel molecular indicator of malignancy. Br J Cancer. 2003;88:699–701. doi: 10.1038/sj.bjc.6600790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Pillasch F, Pohl B, Wilda M, Lacher U, Beil M, Wallrapp C, Hameister H, Knochel W, Adler G, Gress TM. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech Dev. 1999;88:95–99. doi: 10.1016/s0925-4773(99)00160-4. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, Kern SE. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–826. [PubMed] [Google Scholar]
- Palanichamy K, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, Katzman S, Toloue M, Yan W, Basso G, Pigazzi M, Sanford JR, Rao DS. Hematopoietic progenitor proliferation is promoted by the RNA-binding protein IGF2BP3. Journal of Clinical Investigation. 2016;126:1495–1511. doi: 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer DF, Owen DR, Lim HJ, Buczkowski AK, Chung SW, Scudamore CH, Huntsman DG, Ng SS, Owen DA. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer. 2010;10:59. doi: 10.1186/1471-2407-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne-Weiler T, Howard J, Mort M, Cooper DN, Sanford JR. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011;21:1563–1571. doi: 10.1101/gr.118638.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoskus M, Gineikiene E, Valceckiene V, Valatkaite B, Pileckyte R, Griskevicius L. Identification of characteristic IGF2BP expression patterns in distinct BALL entities. Blood Cells Mol Dis. 2011;46:321–326. doi: 10.1016/j.bcmd.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Suvasini R, Shruti B, Thota B, Shinde SV, Friedmann-Morvinski D, Nawaz Z, Prasanna KV, Thennarasu K, Hegde AS, Arivazhagan A, et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J Biol Chem. 2011;286:25882–2590. doi: 10.1074/jbc.M110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K, Furihata M, Hanazaki K, Saito M, Saibara T. IGF2BP3-mediated translation in cell protrusions promotes cell invasiveness and metastasis of pancreatic cancer. Oncotarget. 2014;5:6832–6845. doi: 10.18632/oncotarget.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Bahrami-Samani E, De Araujo PR, Vogel C, Qiao M, Burns SC, Smith AD, Penalva LO. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol. 2016:0. doi: 10.1080/15476286.2015.1138030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikesaa J, Hansen TV, Jonson L, Borup R, Wewer UM, Christiansen J, Nielsen FC. RNA-binding IMPs promote cell adhesion and invadopodia formation. Embo J. 2006;25:1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Kunsch S, Duerschmied D, Beil M, Adler G, Mueller F, Gress TM. Transgenic overexpression of the oncofetal RNA binding protein KOC leads to remodeling of the exocrine pancreas. Gastroenterology. 2003;124:1901–1914. doi: 10.1016/s0016-5085(03)00402-5. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287:49–54. doi: 10.1016/s0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.