Abstract
Introduction
We tested the relationship between genotype, gene expression and suicidal behavior and MDD in live subjects and postmortem samples for three genes, associated with the hypothalamic-pituitary-adrenal axis, suicidal behavior and major depressive disorder (MDD); FK506 binding protein 5 (FKBP5), Spindle and kinetochore-associated protein 2 (SKA2) and Glucocorticoid Receptor (NR3C1).
Materials and Methods
Single-nucleotide polymorphisms (SNPs) and haplotypes were tested for association with suicidal behavior and MDD in a live (N=277) and a postmortem sample (N=209). RNA-seq was used to examine gene and isoform-level brain expression postmortem (Brodmann Area 9) (N=59). Expression quantitative trait loci (eQTL) relationships were examined using a public database (UK Brain Expression Consortium).
Results
We identified a haplotype within the FKBP5 gene, present in 47% of the live subjects, that was associated with increased risk of suicide attempt (OR=1.58, t=6.03, p=0.014). Six SNPs on this gene, three SNPs on SKA2 and one near NR3C1 showed before-adjustment association with attempted suicide, and two SNPs of SKA2 with suicide death, but none stayed significant after adjustment for multiple testing. Only the SKA2 SNPs were related to expression in the prefrontal cortex. One NR3C1 transcript had lower expression in suicide relative to non-suicide sudden death cases (b=-0.48, SE=0.12, t=-4.02, adjusted p=0.004).
Conclusion
We have identified an association of FKBP5 haplotype with risk of suicide attempt and found an association between suicide and altered NR3C1 gene expression in the prefrontal cortex. Our findings further implicate hypothalamic pituitary axis dysfunction in suicidal behavior.
Keywords: suicide/self harm; depression, genetics, mood disorders, biological markers
Introduction
Major depressive disorder (MDD)1-5 and suicidal behavior6-10 are associated with HPA axis over-activity and with excessive cortisol response to stress. One cause of excessive cortisol release is impaired glucocorticoid receptor (GR) feedback inhibition11. This can be caused by less expression of the gene NR3C1 (nuclear receptor subfamily 3, group C, member 1), which encodes for GR, due to genetic variation12; 13 or DNA methylation14-16 resulting in less gene expression. Another potential cause is less expression of GR-related chaperone proteins such as FKBP5 (FK506 binding protein 5, FKBP5), a gene located on human chromosome 6 (chromosome 6p21.31) and encoding the 51kDa protein FKBP517; 18 or SKA219; 20, involved in the transport of GR into the nucleus. FKBP5 can reduce GR signaling by promoting the nuclear translocation of inactive β- isomer of GR21. SKA2 (spindle and KT associated 2), also known as FAM33A (family with sequence similarity 33, member A), located on 17q22, is a recently described member of the SKA complex, which is essential for proper chromosomal segregation22. A previous study found that overexpression of SKA2 resulted in modest enhancement of GR transactivation, while knockdown of SKA2 markedly inhibited GR transactivation20.
Evidence exists linking GR, and these three genes in particular, to suicidal behavior23; 24 and MDD25-28. However, no study to-date to our knowledge has combined the examination of these three genes from a gene variant and gene expression perspective to determine which of these mechanisms or combination of these mechanisms is associated with suicidal behavior and MDD. We have therefore conducted such a study, which had three parts. First, we examined and studied associations between genotype and suicidal behavior in a population of DSM-IV diagnosed major depressive disorder (MDD) suicide attempters, MDD nonattempters and healthy volunteers, and in a postmortem sample of subjects with and without a diagnosis of MDD who died by suicide, compared to sudden death, nonpsychiatric controls, in order to evaluate the effect of genotype on MDD and suicidal behavior separately. Second, we compared the expression of these three genes in prefrontal cortex postmortem in a second, overlapping postmortem sample consisting of three groups: MDD subjects who died from causes other than suicide; MDD suicides; and non-psychiatric, not suicide controls. Third, using results from a pre-existing publically available Brain eQTL Almanac (http://www.braineac.org/, UK Brain Expression Consortium, UKBEC), we examined each of the aforementioned SNPs to determine whether presence of a given variant affected the transcriptome of the respective gene. Such an effect would lend support to the hypothesis that genotype associations with suicidal behavior or MDD are mediated through altered gene expression.
MATERIALS AND METHODS
Participants
Data for three samples of subjects was used in this study. Two separate samples were genotyped, one live and one postmortem. RNA-seq data were obtained for a third subsample of 59 postmortem subjects, partially overlapping with the genotyped postmortem sample. All subjects were of European ancestry and were screened for genetic ancestry based on genotype , as described below.
Descriptive statistics for the three subsamples are included in Table 1.
Table1.
Description of live and postmortem subjects.
| Sample group | Group | Psychiatric Diagnosis (N) | Age(years) | Sex (N) | ||||
|---|---|---|---|---|---|---|---|---|
| MDD | Other diagnosis1 |
No diagnosis |
Mean | SD | M | F | ||
| Genotype Sample |
Live subjects | |||||||
| Suicide attempter |
87 | 0 | 0 | 38.67 | 11.24 | 30 | 57 | |
| Non-attempter | 133 | 0 | 57 | 41.19 | 13.73 | 73 | 117 | |
| Total | 220 | 0 | 57 | 40.4 | 13.03 | 103 | 174 | |
| Postmortem | ||||||||
| Suicide death | 75 | 46 | 0 | 50.24 | 18.96 | 80 | 41 | |
| Non-suicide death |
10 | 11 | 67 | 50.85 | 17.46 | 66 | 22 | |
| Total | 85 | 57 | 67 | 50.5 | 18.31 | 146 | 63 | |
| RNA Sample2 |
Postmortem | |||||||
| Suicide death | 21 | 0 | 0 | 52.19 | 21.73 | 13 | 8 | |
| Non-suicide death |
9 | 0 | 29 | 46.92 | 20.78 | 29 | 9 | |
| Total | 30 | 0 | 29 | 48.80 | 21.08 | 42 | 17 | |
The Other Diagnosis category includes subjects with diagnosis other than MDD, including bipolar disorder, depression NOS, schizophrenia, schizoaffective disorder, delusional disorder, psychotic disorder, adjustment disorder, organic mood syndrome, senile dementia NOS, conduct disorder, pathological gambling and substance abuse/dependence
This sample overlaps with the postmortem subjects from the Genotype Sample above, sharing n=38 subjects.
Genotyped Live Subsample
The live subsample consisted of 277 unrelated individuals: 87 MDD suicide attempters (31%), 133 subjects (48%) with MDD diagnosis but no suicide attempt and 57 healthy volunteers (21%), recruited in the New York and Pittsburgh area. Participants gave written informed consent as required by the Institutional Review Board of New York State Psychiatric Institute (NYSPI). Psychiatric diagnosis was established using the Structured Clinical Interview for DSM IV (SCID-I). History of past suicide attempts was recorded on the Columbia Suicide History Form29; the definition of suicide attempts was: a self-injurious act that has at least partial intent to end one’s life. A group with MDD and no history of a suicide attempt was used as a psychiatric control group. Unrelated healthy volunteers were recruited through advertising. They were assessed by psychiatrists or clinical psychologists and evaluated using the SCID-NP to rule out axis I diagnoses, cluster B personality disorder, substance use disorder and lifetime history of a suicide attempt. Exclusion criteria included mental retardation, dementia and acute psychosis.
Genotyped Postmortem Subsample
All postmortem subjects, suicides and non-suicides, died suddenly. This subsample consisted of 209 subjects: 121 died of suicide and 88 were non-suicides. Cause of death for postmortem subjects, excluding suicide, was determined by the coroner or medical examiner. Suicide as a cause of death was determined by the research team using Columbia Classification Algorithm for Suicide Assessment and was in agreement with the medical examiner’s determination in all cases30. Diagnosis of major psychiatric disorder was determined using the SCID I31 and our validated psychological autopsy method as previously described32. All of the suicides and 24% of the non-suicide deaths had a psychiatric diagnosis (some subjects had more than one). In particular, 62% of the suicides and 11% of the non-suicide deaths had a lifetime MDD diagnosis. Biological samples were obtained from the Medical Examiner’s Office in accordance with local regulations and protocols approved by the applicable Institutional Review Boards. Brain samples were dissected from Brodmann Area 9 as previously reported33. All brains were free of gross neuropathology and had negative brain toxicology for psychoactive and neurotoxic drugs.
Postmortem sample for RNA sequencing
A second subsample of postmortem subjects, with RNA sequencing data available, consisted of N=21 suicides with MDD diagnosis, 9 sudden death victims with MDD, and 29 sudden death victims without psychiatric diagnosis. Some of the postmortem subjects with RNAseq data (n=38 out of 59) also had been included in the genotyped sample. Procedures for brain collection and psychological autopsy, psychiatric diagnosis, inclusion and exclusion criteria are as describe above for the genotyped postmortem sample.
Genotyping and RNA sequencing methods
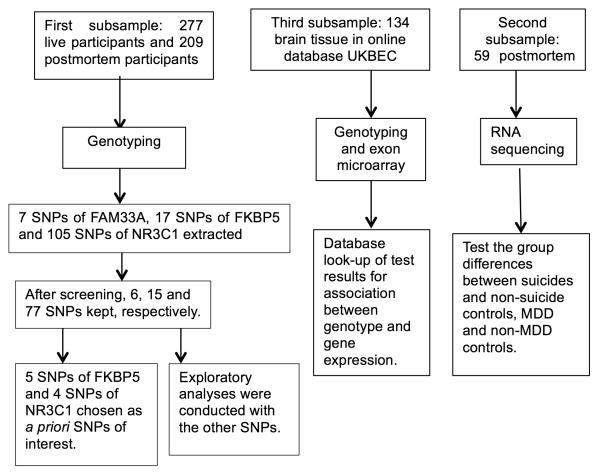
The flowchart for genotyping and RNA sequencing of the samples is in Figure 1. The live sample and the genotyped postmortem subsample were part of a larger genome-wide association (GWAS) study and genotyping methods have been previously reported34 using the HumanOmni-1-Quad Beadchip (1,014,770 SNPs, Illumina, San Diego, CA, USA). In that study, Multidimensional Scaling Analysis and comparison to a HapMap Phase 3 population were used to exclude individuals of non-European ancestry, defined as outliers more than 3 trimmed standard deviations away from the sample average. The GWAS did not find any association with suicide attempt or completed suicide. In the current study, for the New York subsample of the original subjects, we extracted SNP genotype from the following genes: SKA2, FKBP5 and NR3C1. Hardy–Weinberg Equilibrium test, minor allele frequency (MAF) were calculated for all SNPs. Haploblocks of SNPs were established using PLINK v. 1.07. Six SNPs from SKA2, fifteen from FKBP5 and seventy-seven from NR3C1 were included in the analyses after the exclusion of the SNPs not in Hardy-Weinberg equilibrium in the control sample or with MAF less than 10%. Five SNPs of FKBP5 (rs3800373, rs9296158, rs3777747, rs4713902 and rs9470080)35-37 and four SNPs of NR3C1 (rs6196, rs33388, rs33389 and rs10052957)17; 38; 39 that were previously reported to be associated with suicidal behavior or MDD were chosen as of primary interest. Analyses of the remaining 89 SNPs were exploratory. Location, genotype, Hardy–Weinberg Equilibrium chi-square values, and frequency distribution for all SNPs are in Supplementary Table S1.
Figure 1.
Flowchart of the three steps in the analysis.
Details of the RNA sequencing methodology have been previously reported40. Briefly, RNA was extracted from dorsal prefrontal cortex (BA9) using previously established protocols33 and conducted according to the guidelines recommended by the NIH Roadmap Epigenomics Mapping Consortium (REMC).
Paired-end, strand specific sequencing for total RNA was performed on Illumina HiSeq 2500 with 100 bp read lengths. For each sample, raw RNA-seq reads were aligned and mapped to the human reference genome and assembled using Tophat v2.0.9.; these assemblies were then merged together to provide a uniform basis for calculating gene and transcript expression in each sample using Cufflinks v2.2.0 software41. Cuffquant was then used to quantify gene and isoform-level expression values, and the output CXB files were then input to Cuffnorm v2.2.1. which produced normalized expression values for each sample.
Statistical Analysis
The data analysis plan consisted of three steps: 1. Testing the association between genotype and phenotype, 2. Testing the association between gene expression and phenotype, and 3. Corroborating the functional link between genotype and gene expression for the SNPs found to be associated with suicidal behavior or MDD.
Part 1
We tested the association between genotypes in the 3 genes of interest and suicidal behavior, and, separately, with MDD, on both individual SNPs and haplotypes on multiple markers. We also tested the association between gene variants and suicide attempt separately in MDD subjects only, to separate the effect of diagnosis and suicidal behavior. One non-attempter MDD subject with a co-morbid diagnosis of psychotic disorder was removed from this analysis. As the sample size for postmortem subjects with MDD who did not die of suicide was small, the respective analysis for the association with suicide within MDD subjects could not be conducted. For live subjects, two SNP-wise logistic regressions were performed in R (http://www.r-project.org/) for each SNP that passed the screening, with genotype as the predictor, indicator variable for suicidal behavior history (yes/no) or MDD as response. Age and sex were included as covariates in the model. The SNP-wise significance levels were not adjusted for multiple testing for those SNPs previously cited in the literature (see Table 2 for the list); for those SNPs not previously cited, the Benjamini-Hochberg adjustment for multiple testing was performed separately by outcome variable for a total of k=89 tests by outcome variable. This same analytic strategy was used in postmortem subjects. Due to limited sample size in the postmortem sample, haplotype analysis was performed in live subjects only. We used PLINK v. 1.07, to find haploblocks of SNPs within the same gene that were in Linkage Disequilibrium, and then to test association with suicide attempt history and MDD in separate models for each haploblock in logistic regression models adjusted for age.
Table 2.
Significance levels for SNPs previously reported to be associated with suicidal behavior or MDD in live and postmortem subjects (uncorrected p-values). All models were adjusted by age and sex.
| Y=attempted suicide/suicide death | Y=MDD | ||||||
|---|---|---|---|---|---|---|---|
| postmortem | live | live | live | ||||
| Gene | SNP | suicide death (n=121) vs non-suicide (n=88) |
suicide attempter (n=87) vs non-attempter (n=190) |
MDD attempter (n=87) vs MDD non-attempter (n=132) |
MDD (n=219) vs healthy volunteer (n=56) |
||
| p-value | p-value | risk genotype |
p-value | risk genotype |
p-value | ||
| FKBP5 | rs3800373 | 0.628 | 0.062 | 0.188 | 0.179 | ||
| rs9296158 | 0.525 | 0.038 | GG | 0.102 | 0.292 | ||
| rs3777747 | 0.533 | 0.017 | GG | 0.018 | GG | 0.749 | |
| rs4713902 | 0.799 | 0.031 | GG | 0.041 | GG | 0.535 | |
| rs9470080 | 0.602 | 0.154 | 0.240 | 0.450 | |||
| NR3C1 | rs6196 | 0.257 | 0.621 | 0.576 | 0.982 | ||
| rs33388 | 0.443 | 0.860 | 0.824 | 0.426 | |||
| rs33389 | 0.173 | 0.286 | 0.286 | 0.968 | |||
| rs10052957 | 0.153 | 0.958 | 0.911 | 0.732 | |||
Part 2
The second step tested for group differences in the expression of the three genes in the postmortem sample only. Significance levels were computed for two comparisons: for suicides vs. those who died from other causes; and for subjects with and without an MDD diagnosis. Since the distribution of gene expression was skewed with several outliers for each isoform, robust regression models were fit using the function lmrob from the library robustbase in R, adjusting for age, sex and RIN score. Two subjects were missing RIN scores due to technical issues (one NPC and one DSUI) and were excluded from analyses that covaried for RIN score. The brain pH values for the three groups were very consistent (DSUI: 6.41 ±0.30, DC: 6.41 ±0.32, NPC: 6.49 ±0.33), and there were no significant differences in the post-mortem interval (PMI) of the three groups (DS: 16.08 ±7.00,: DC:15.06 ± 4.38, NPC: 13.21 ± 4.61).
Transcripts were screened before being entered into the group comparisons, and those with expression levels that were not significantly different from 0, or those for which the robust regression model failed to converge, were excluded. For the remaining transcripts, significance levels were adjusted for multiple testing using the Benjamini-Hochberg method for controlling the False Discovery Rate, separately by outcome, using k=19 as the number of tests for each.
Part 3
Since our sample of sudden death subjects with both genotype and gene expression data was too small to allow for an association analysis,, p-values were extracted for the association between genotype and gene expression from the public online dataset UKBEC (http://www.braineac.org/). This dataset comprises data from 134 brains from individuals free of neurodegenerative disorders and up to twelve brain regions per brain. We examined the extracted significance levels corresponding to expression quantitative trait loci (eQTL) analysis for the association of candidate SNPs (corresponding to associations with uncorrected p<0.05 with suicide or MDD in our project) with expression in the frontal cortex of all the exon-level probesets from the same gene. We selected the minimal p-value for each SNP from all pairwise associations with probesets on the gene. We did not adjust for multiple testing, since this is a database look-up instead of an independent analysis.
Results
Part 1. Association between genotype and suicidal behavior and/or MDD
After screening the SNPs based on minor allele frequency and the Hardy-Weinberg Equilibrium test, there were 15 SNPs in FKBP5, 77 SNP in NR3C1, and 6 SNPs in SKA2 genes. Of the five SNPs in FKBP5 and four SNPs in NR3C1 previously reported in the literature,, SNP-wise logistic regressions with genotype, age and sex as predictors and suicide attempt as outcome, indicated that three SNPs in FKBP5 had significance levels below 0.05 for an association with attempted suicide in both the full live sample including healthy volunteers, and the sample restricted to depressed subjects (see Table 2). There was no evidence for an association between these genotypes and suicide in the postmortem sample or MDD diagnosis subsample in the live group.
Results from exploratory analyses of SNPs not previously reported in the suicide literature are provided in Supplementary Table S3, and the results with uncorrected p<0.05 are displayed in Table 3. No significant results were found after adjustment for multiple testing using the Benjamini-Hochberg method. Three SNPs of FKBP5, three SNPs of SKA2 and one SNP of NR3C1 showed evidence for association at the uncorrected p<0.05 level with attempted suicide, and two SNPs of SKA2 showed association with suicide death (all uncorrected p-values<0.05, see Table 3), but all these findings lost significance after multiple testing adjustment.
Table 3.
Association between genotype on SNPs not previously reported on and suicidal behavior or MDD in live and postmortem subjects (only SNPs with uncorrected p<0.05 are listed). All models were adjusted by age and sex.
| Y=attempted suicide/suicide death | Y=MDD | |||||||
|---|---|---|---|---|---|---|---|---|
| postmortem | live | live | live | |||||
| Genes | SNPs | suicide death(n=121)vs sudden death(n=88) |
suicide attempter (n=87) vs non-attempter (n=190) |
MDD attempter (n=87) vs MDD non-attempter (n=132) |
MDD(n=219) vs healthy volunteer n=56) |
|||
| p-value | risk genotype |
p-value | risk genotype |
p-value | risk genotype |
p-value | ||
| FKBP5 | rs7757037 | 0.718 | 0.033 | AA | 0.044 | AA | 0.471 | |
| rs737054 | 0.726 | 0.049 | AA | 0.084 | 0.293 | |||
| rs9380529 | 0.053 | 0.011 | GG | 0.011 | GG | 0.657 | ||
| SKA2 | rs8082544 | 0.036 | AG | 0.638 | 0.929 | 0.278 | ||
| rs12945875 | 0.636 | 0.306 | 0.024 | AA | 0.358 | |||
| rs9911583 | 0.543 | 0.014 | GG | 0.008 | GG | 0.665 | ||
| rs8067682 | 0.488 | 0.081 | 0.043 | AA | 0.597 | |||
| rs7502947 | 0.013 | AG | 0.401 | 0.754 | 0.229 | |||
| NR3C1 | rs9324924 | 0.295 | 0.078 | 0.037 | AC | 0.402 | ||
Haplotype analysis
Analysis of LD values indicated the existence of 2 haploblocks on the FKBP5, 13 on the NR3C1, and 2 on the SKA2. Haplotype analysis using logistic regression, testing the association with suicide attempt risk adjusted for age, yielded one significant association for a haploblock on FKBP5 (omnibus test p= 0.0368). Subjects with haplotype AA (frequency=47%), on the SNPs rs3800373 and rs7757037 had higher odds of suicide attempt than others (OR=1.58, t=6.03, p=0.014), and those with haplotype CG (frequency=28%) had lower odds (OR=0.63, t=4.89, p=0.027). None of the haplotypes were associated with MDD. No significant haplotype associations were found for the NR3C1 or SKA2 haploblocks.
Part 2. FKBP5, SKA2 and NR3C1 brain expression in suicide and MDD postmortem
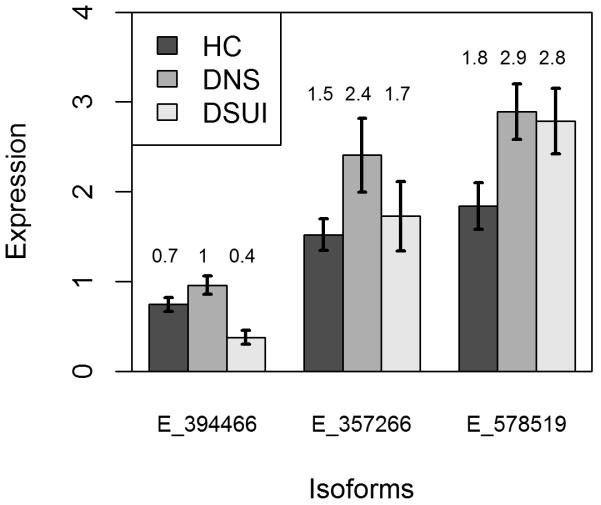
Summary statistics for the brain expression levels of all the transcripts that passed the screening phase (i.e. had average expression levels significantly above 0) are presented in Table 4 for non-psychiatric non-suicide controls (NPC, N=28), depressed non-suicides (DNS, N=9) and depressed suicides (DSUI, N=20). Robust regression models showed that one protein coding transcript of NR3C1 and one protein coding transcript of FKBP5 have lower expression in suicide vs. non-suicide (age, sex and RIN-adjusted statistics: NR3C1-006: B=−0.48; SE=0.12, t=−4.02, p<0.001; FKBP5-001: B=−0.71, SE=0.32, t=−2.18, p=0.034, see Table 4 and Figure 2). After using the Benjamini-Hochberg method for controlling the False Discovery Rate, the association for NR3C1-006 remained significant (adjusted p=0.004). A SKA2 transcript involved in nonsense mediated decay showed higher expression in MDD vs. non-MDD subjects (SKA2-005: B=1.06; SE=0.42; t=2.55; p=0.014). There were no group differences in the total (gene-level) expression levels of NR3C1, FKBP5 and SKA2 (see Table 4). All analyses were adjusted for age, sex and RIN score, and all the significant results stayed significant after adjusting for postmortem interval (PMI).
Table 4.
Gene expression summary statistics for three groups of subjects (NPC=non-psychiatric controls, DNS=depressed non-suicides, DSUI=depressed suicides). Significance levels are reported for two tests comparing MDD and non-MDD subjects (DSUI+DNS vs. NPC), and suicide and non-suicide (DSUI vs. DNS+HC) subjects.
| Gene | Transcript ID | Name | Biotype | Length(bp) | Protein | NPC(n=29) | DNS(n=9) | DSUI(n=21) | MDD: non-MDD |
suicide: non-suicide |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | Q | M | Q | M | Q | p-value | p-value | ||||||
| NR3C1 | XLOC_NR3C1 | Total | 1.893E+01 | 4.072E+00 | 1.956E+01 | 7.019E+00 | 1.834E+01 | 5.372E+00 | 0.942 | 0.838 | |||
| ENST00000504572 | NR3C1-010 | protein coding | 3389 | 778aa | 2.821E−01 | 8.384E−02 | 2.415E−01 | 1.651E−01 | 2.189E−01 | 1.885E−01 | 0.270 | 0.206 | |
| ENST00000394464 | NR3C1-002 | protein coding | 6795 | 777aa | 1.528E+01 | 4.569E+00 | 1.471E+01 | 3.583E+00 | 1.515E+01 | 5.153E+00 | 0.117 | 0.208 | |
| ENST00000343796 | NR3C1-201 | protein coding | 7286 | 777aa | 2.907E−01 | 2.066E−01 | 1.653E−01 | 2.831E−01 | 2.052E−01 | 3.319E−01 | 0.215 | 0.751 | |
| ENST00000394466 | NR3C1-006 | protein coding | 3245 | 778aa | 7.848E−01 | 5.700E−01 | 9.833E−01 | 6.217E−01 | 4.246E−01 | 6.456E−01 | 0.128 | <0.001 | |
| ENST00000231509 | NR3C1-001 | protein coding | 3556 | 778aa | 2.507E−04 | 4.405E−03 | 1.848E−04 | 5.918E−03 | 2.975E−03 | 5.456E−01 | 0.095 | 0.065 | |
| ENST00000503201 | NR3C1-007 | protein coding | 2594 | 777aa | 1.298E+00 | 9.094E−01 | 6.722E−01 | 1.379E+00 | 1.321E+00 | 1.240E+00 | 0.589 | 0.722 | |
| ENST00000505058 | NR3C1-012 | processed transcript | 491 | no protein | 3.421E−01 | 1.891E−01 | 3.259E−01 | 2.901E−01 | 2.773E−01 | 2.925E−01 | 0.358 | 0.289 | |
| FKBP5 | XLOC_FKBP5 | Total | 1.913E+00 | 2.097E+00 | 3.160E+00 | 2.810E+00 | 3.907E+00 | 3.576E+00 | 0.205 | 0.276 | |||
| ENST00000357266 | FKBP5-001 | protein coding | 3772 | 457aa | 1.589E+00 | 1.597E+00 | 2.571E+00 | 2.157E+00 | 9.831E−01 | 1.917E+00 | 0.961 | 0.034 | |
| ENST00000542713 | FKBP5-203 | protein coding | 7384 | 268aa | 1.935E−01 | 4.183E−01 | 4.979E−01 | 8.417E−01 | 1.304E−01 | 2.848E−01 | 0.772 | 0.111 | |
| SKA2 | XLOC_SKA2 | Total | 1.032E+01 | 1.930E+00 | 1.115E+01 | 1.481E+00 | 1.131E+01 | 4.993E+00 | 0.720 | 0.700 | |||
| ENST00000578519 | SKA2-005 | Nonsense mediated decay | 894 | 61aa | 1.811E+00 | 2.647E+00 | 3.207E+00 | 1.774E+00 | 3.057E+00 | 2.510E+00 | 0.014 | 0.087 | |
| ENST00000437036 | SKA2-002 | protein coding | 625 | 75aa | 3.451E−01 | 9.310E−01 | 2.115E−01 | 4.825E−01 | 5.057E−01 | 5.394E−01 | 0.317 | 0.673 | |
| ENST00000578105 | SKA2-009 | protein coding | 689 | 92aa | 5.313E−01 | 8.867E−01 | 3.431E−01 | 4.041E−01 | 4.545E−01 | 8.985E−01 | 0.550 | 0.641 | |
| ENST00000583927 | SKA2-007 | processed transcript | 533 | no protein | 1.367E+00 | 8.962E−01 | 9.805E−01 | 6.434E−01 | 1.368E+00 | 7.110E−01 | 0.889 | 0.267 | |
| ENST00000581068 | SKA2-008 | protein coding | 595 | 62aa | 1.289E+00 | 8.004E−01 | 1.803E+00 | 1.155E+00 | 1.432E+00 | 1.271E+00 | 0.694 | 0.913 | |
| ENST00000330137 | SKA2-001 | protein coding | 2798 | 121aa | 3.129E+00 | 2.832E+00 | 2.173E+00 | 2.535E+00 | 2.915E+00 | 3.281E+00 | 0.087 | 0.780 | |
| ENST00000580541 | SKA2-003 | protein coding | 689 | 46aa | 5.188E−01 | 7.740E−01 | 4.666E−01 | 6.348E−01 | 3.605E−01 | 7.086E−01 | 0.510 | 0.447 | |
Adjusted by age, sex and RIN score.
Figure 2.
Group comparison of the expression of transcripts ENST00000394466 (NR3C1-006) of NR3C1, ENST00000357266 (FKBP5-001) of FKBP5 and ENST00000578519 (SKA2-005) of SKA2. Error bars represent standard deviations. Expression is in units of normalized Fragments Per Kilobase of transcript per Million mapped reads (FPKM). Only the NR3C1-006 transcript shows significant difference after adjustment for multiple testing.
Part 3. Corroboration of association between genotype and gene expression
Only the SNPs with uncorrected significance levels below 0.05 for the association with suicide and/or MDD were examined for an association with gene expression (i.e. quantitative trait loci (eQTL) relationship) using a public online dataset (UKBEC, http://www.braineac.org/). P-values for eQTL analyses were extracted for the prefrontal cortex (pFCTX) in order to keep results consistent with RNA-seq expression results above (Brodmann Area 9) (see Supplementary Table S3). Out of all exon-specific probesets from the same gene, the one with lowest (uncorrected) p-value is reported (see Table 5).. In the SKA2 gene, genotype AG of rs8082544, genotype GG of rs9911583, genotype AA of rs8067682 and genotype AG of rs7502947 showed lower expression of some exon-specific probesets for the same gene in dorsolateral prefrontal cortex (uncorrected p-values=0.02, 0.01, 0.01, 0.03, respectively). No SNPs in FKBP5 and NR3C1 showed association with gene expression (Table 5).
Table 5.
Association between genotype of our candidate risk SNPs and expression of exon-specific probesets for the same gene in the frontal cortex in UKBEC (n=134). (Only the minimal p-value for each SNP is listed).
| Gene | SNP | exprID | tID | chromosome | p-value |
|---|---|---|---|---|---|
| FKBP5 | rs9296158 | 2951640 | 2951567 | chr6 | 0.099 |
| rs3777747 | 2951601 | 2951567 | chr6 | 0.079 | |
| rs4713902 | 2951648 | 2951567 | chr6 | 0.160 | |
| rs7757037 | 2951644 | 2951567 | chr6 | 0.190 | |
| rs737054 | 2951648 | 2951567 | chr6 | 0.160 | |
| rs9380529 | 2951601 | 2951567 | chr6 | 0.060 | |
| SKA2 | rs8082544 | 3764760 | 3764738 | chr17 | 0.019 |
| rs12945875 | 3764761 | 3764738 | chr17 | 0.140 | |
| rs9911583 | 3764740 | 3764738 | chr17 | 0.006 | |
| rs8067682 | t3764738 | 3764738 | chr17 | 0.008 | |
| rs7502947 | 3764760 | 3764738 | chr17 | 0.025 | |
| NR3C1 | rs9324924 | 2879385 | 2879312 | chr5 | 0.071 |
tID:The tID is the transcript cluster ID from Affymetrix.
exprID:The exprID is the Probe Set ID from Affymetrix.
Discussion
In this study we identified associations of suicide attempt with gene haplotypes of FKBP5, suicide with altered NR3C1 gene expression in prefrontal cortex, and found a possible association between SKA2 expression in prefrontal cortex and MDD but not suicidal behavior. FKBP5 and SKA2 are chaperone proteins that affect the functioning of the glucocorticoid receptor (GR), which mediates negative feedback regulation of the HPA axis42. Lower expression of GR or either FKBP5 or SKA2 potentially results in less GR function and excessive cortisol release under stress due to impaired feedback inhibition of glucocorticoids via GR. Altered function of the HPA-axis is associated with suicidal behavior23; 24 and MDD43, and DNA methylation reducing GR expression may mediate the relationship of childhood adversity on suicide24. In this study, we hypothesized that polymorphisms in GR-related genes that favor higher levels of HPA activity in response to stress will be associated with suicidal behavior or MDD. This association could occur through a structural gene variant or epigenetic effect that could alter gene expression. Although for none of the three genes could we identify a definite pathway from gene polymorphism through altered gene expression to association with suicidal behavior, we believe our results strengthen the evidence for an association between suicidal behavior and MDD and GR-related genes. Future studies that combine deep sequencing and gene expression of genes associated with GR can both verify and extend our findings.
Association between genotype and suicidal behavior or MDD
We found that a haploblock involving SNPs rs3800373 and rs7757037 was associated with risk of suicide attempt. This result is partially consistent with a previous study35 performed in an African American sample. Other studies reported risk haplotypes for suicidal behavior on the FKBP5 genes in a Japanese17; and in a Polish bipolar sample44. The latter study also reported that the risk haplotype identified for the suicidal phenotype was not associated with a melancholic bipolar phenotype, which parallels our finding of no association between the haplotype and MDD diagnosis.
None of the SNPs on FKBP5 showed an association with either suicidal behavior or MDD in the postmortem sample, which is not consistent with some studies44-46 in which these SNPs were associated with depressive symptoms (but not MDD). Discrepancy between suicidal behavior-related findings in our live and postmortem subjects could be explained by demographic, clinical and biological differences between suicides (mostly males, greater suicide intent, substantial serotonin system deficits and HPA axis abnormalities) and nonfatal suicide attempters who vary greatly in terms of lethality of behavior, which is associated with serotonin system deficits and HPA axis abnormalities8; 47.
Suicide and MDD-related differences in gene expression
Most multi-exon genes undergo alternative splicing, which greatly increases the functional diversity of protein coding species, and isoforms of the same gene may have different, even opposing, functions. We used next generation RNA sequencing of RNA extracted from dorsal prefrontal cortex (Brodmann Area 9) to examine both gene-level and isoform-level expression differences. Previous studies of the relationship between FKBP5, SKA2 and NR3C1 brain gene expression in samples with suicidal behavior or MDD have focused on total gene expression. Although we did not find group differences in the total expression of the three genes, we found lower expression of a protein-coding transcript of NR3C1 in the prefrontal cortex (Brodmann Area 9) of suicides compared to sudden death controls, that survived adjustment for multiple testing. These isoforms may contribute to the risk of suicide through reduced expression and poorer GR feedback leading to excessive HPA response under stress. Lower NR3C1 (GR) gene expression is reported in the amygdala of suicides compared with controls48, and lower NR3C1 expression in suicides with a history of childhood adversity24, while no expression differences in NR3C1 were reported between MDD and controls49.
Association between genotype and gene expression
Using a database look-up, four SNPs on SKA2 showed an association with expression in the dorsolateral prefrontal on exon-specific probesets of the same gene. However, in part 2 of this study, we did not find any SKA2 isoforms with altered expression in suicide (although we found altered expression of a SKA2 isoform in MDD). Thus, although the SNPs modulate SKA2 expression, the evidence for association between genotype and suicide risk, and suicide risk and expression, is missing. SNPs in the FKBP5 haploblock that showed associations with suicide, did not appear to be associated with altered gene expression, perhaps because either these are not real associations or the SNPs confer risk via epigenetic or other mechanisms.
Conclusions
We provide further evidence for the involvement of NR3C1 and FKBP5 in suicidal behavior through gene-phenotype associations and differential mRNA transcript expression in suicide. Modest sample size and unaccounted-for phenotypic heterogeneity may have resulted in limited power for some analyses. Future work in larger samples combining genetics (in interaction with environment such as childhood trauma), epigenetics and functional genomics is needed to determine the link between HPA axis-related gene variation, brain gene expression, and risk for suicidal behavior and MDD.
Supplementary Material
Acknowledgments
This research was funded by NIMH 5R01MH082041, R01MH40210 and P50MH62185, and a Paul Janssen Translational Neuroscience Postdoctoral Fellowship (SPP). J. Mann, M. Oquendo and A. Burke receive royalties from the Research Foundation for Mental Hygiene for commercial use of the Columbia Suicide Severity Rating Scale, which was not used in this study. M. Oquendo received financial compensation from Pfizer for the safety evaluation of a clinical facility, unrelated to this study. She has received unrestricted educational grants and/or lecture fees from Astra-Zeneca, Bristol Myers Squibb, Eli Lilly, Janssen, Otsuko, Pfizer, Sanofi-Aventis, and Shire. Her family owns stock in Bristol Myers Squibb. H. Galfalvy’s family owns stock in Illumina, Inc.
Footnotes
Disclosures
H. Yin, S.P. Pantazatos and Y. Huang do not have any conflict of interest to report.
References
- 1.Baek JH, Kang ES, Fava M, et al. Thyroid stimulating hormone and serum, plasma, and platelet brain-derived neurotrophic factor during a 3-month follow-up in patients with major depressive disorder. J Affect Disord. 2014;169:112–7. doi: 10.1016/j.jad.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Hardeveld F, Spijker J, Vreeburg SA, et al. Increased cortisol awakening response was associated with time to recurrence of major depressive disorder. Psychoneuroendocrinology. 2014;50:62–71. doi: 10.1016/j.psyneuen.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Coryell W, Young E, Carroll B. Hyperactivity of the hypothalamic-pituitary-adrenal axis and mortality in major depressive disorder. Psychiatry Res. 2006;142(1):99–104. doi: 10.1016/j.psychres.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Rubinow DR, Post RM, Pickar D, et al. Relationship between urinary free cortisol and CSF opioid binding activity in depressed patients and normal volunteers. Psychiatry Res. 1981;5(1):87–93. doi: 10.1016/0165-1781(81)90064-0. [DOI] [PubMed] [Google Scholar]
- 5.Slotkin TA, Hays JC, Nemeroff CB, Carroll BJ. Dexamethasone suppression test identifies a subset of elderly depressed patients with reduced platelet serotonin transport and resistance to imipramine inhibition of transport. Depress Anxiety. 1997;6(1):19–25. doi: 10.1002/(sici)1520-6394(1997)6:1<19::aid-da3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Braquehais MD, Picouto MD, Casas M, Sher L. Hypothalamic-pituitary-adrenal axis dysfunction as a neurobiological correlate of emotion dysregulation in adolescent suicide. World J Pediatr. 2012;8(3):197–206. doi: 10.1007/s12519-012-0358-0. [DOI] [PubMed] [Google Scholar]
- 7.Ghaziuddin N, King CA, Welch K, Ghaziuddin M. Depressed suicidal adolescent males have an altered cortisol response to a pharmacological challenge. Asian J Psychiatr. 2014;7(1):28–33. doi: 10.1016/j.ajp.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann JJ, Currier D, Stanley B, et al. Can biological tests assist prediction of suicide in mood disorders? Int J Neuropsychopharmacol. 2006;9(4):465–74. doi: 10.1017/S1461145705005687. [DOI] [PubMed] [Google Scholar]
- 9.Pfeffer CR, Stokes P, Shindledecker R. Suicidal behavior and hypothalamic-pituitary-adrenocortical axis indices in child psychiatric inpatients. Biol Psychiatry. 1991;29(9):909–17. doi: 10.1016/0006-3223(91)90057-s. [DOI] [PubMed] [Google Scholar]
- 10.Traskman-Bendz L, Ekman R, Regnell G, Ohman R. HPA-related CSF neuropeptides in suicide attempters. Eur Neuropsychopharmacol. 1992;2(2):99–106. doi: 10.1016/0924-977x(92)90018-4. [DOI] [PubMed] [Google Scholar]
- 11.Juruena MF. Early-life stress and HPA axis trigger recurrent adulthood depression. Epilepsy Behav. 2014;38C:148–159. doi: 10.1016/j.yebeh.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Kumsta R, Moser D, Streit F, et al. Characterization of a glucocorticoid receptor gene (GR, NR3C1) promoter polymorphism reveals functionality and extends a haplotype with putative clinical relevance. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):476–82. doi: 10.1002/ajmg.b.30837. [DOI] [PubMed] [Google Scholar]
- 13.Leszczynska-Rodziewicz A, Szczepankiewicz A, Pawlak J, et al. Association, haplotype, and gene-gene interactions of the HPA axis genes with suicidal behaviour in affective disorders. ScientificWorldJournal. 2013;2013:207361. doi: 10.1155/2013/207361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Na KS, Chang HS, Won E, et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One. 2014;9(1):e85425. doi: 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perroud N, Paoloni-Giacobino A, Prada P, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiger H, Labonte B, Groleau P, et al. Methylation of the glucocorticoid receptor gene promoter in bulimic women: associations with borderline personality disorder, suicidality, and exposure to childhood abuse. Int J Eat Disord. 2013;46(3):246–55. doi: 10.1002/eat.22113. [DOI] [PubMed] [Google Scholar]
- 17.Supriyanto I, Sasada T, Fukutake M, et al. Association of FKBP5 gene haplotypes with completed suicide in the Japanese population. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(1):252–6. doi: 10.1016/j.pnpbp.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Tatro ET, Everall IP, Kaul M, Achim CL. Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain Res. 2009;1286:1–12. doi: 10.1016/j.brainres.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guintivano J, Brown T, Newcomer A, et al. Identification and Replication of a Combined Epigenetic and Genetic Biomarker Predicting Suicide and Suicidal Behaviors. Am J Psychiatry. 2014 doi: 10.1176/appi.ajp.2014.14010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice L, Waters CE, Eccles J, et al. Identification and functional analysis of SKA2 interaction with the glucocorticoid receptor. J Endocrinol. 2008;198(3):499–509. doi: 10.1677/JOE-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Clark AF, Yorio T. FK506-binding protein 51 regulates nuclear transport of the glucocorticoid receptor beta and glucocorticoid responsiveness. Invest Ophthalmol Vis Sci. 2008;49(3):1037–47. doi: 10.1167/iovs.07-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi W, Gerster K, Alajez NM, et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71(8):2926–37. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 23.Mann JJ, Arango VA, Avenevoli S, et al. Candidate endophenotypes for genetic studies of suicidal behavior. Biol Psychiatry. 2009;65(7):556–63. doi: 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGowan PO, Sasaki A, D'Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pariante CM, Miller AH. Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol Psychiatry. 2001;49(5):391–404. doi: 10.1016/s0006-3223(00)01088-x. [DOI] [PubMed] [Google Scholar]
- 26.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–95. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 27.van West D, Van Den Eede F, Del-Favero J, et al. Glucocorticoid receptor gene-based SNP analysis in patients with recurrent major depression. Neuropsychopharmacology. 2006;31(3):620–7. doi: 10.1038/sj.npp.1300898. [DOI] [PubMed] [Google Scholar]
- 28.van Rossum EF, Binder EB, Majer M, et al. Polymorphisms of the glucocorticoid receptor gene and major depression. Biol Psychiatry. 2006;59(8):681–8. doi: 10.1016/j.biopsych.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Oquendo MA, Placidi GP, Malone KM, et al. Positron emission tomography of regional brain metabolic responses to a serotonergic challenge and lethality of suicide attempts in major depression. Arch Gen Psychiatry. 2003;60(1):14–22. doi: 10.1001/archpsyc.60.1.14. [DOI] [PubMed] [Google Scholar]
- 30.Posner K, Oquendo MA, Gould M, et al. Columbia Classification Algorithm of Suicide Assessment (C-CASA): classification of suicidal events in the FDA's pediatric suicidal risk analysis of antidepressants. Am J Psychiatry. 2007;164(7):1035–43. doi: 10.1176/appi.ajp.164.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spitzer RL, Davies M, Barkley RA. The DSM-III-R field trial of disruptive behavior disorders. J Am Acad Child Adolesc Psychiatry. 1990;29(5):690–7. doi: 10.1097/00004583-199009000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Kelly TM, Mann JJ. Validity of DSM-III-R diagnosis by psychological autopsy: a comparison with clinician ante-mortem diagnosis. Acta Psychiatr Scand. 1996;94(5):337–43. doi: 10.1111/j.1600-0447.1996.tb09869.x. [DOI] [PubMed] [Google Scholar]
- 33.Sibille E, Arango V, Galfalvy HC, et al. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29(2):351–61. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- 34.Galfalvy H, Haghighi F, Hodgkinson C, et al. A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2015 doi: 10.1002/ajmg.b.32330. [DOI] [PubMed] [Google Scholar]
- 35.Roy A, Gorodetsky E, Yuan Q, et al. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology. 2010;35(8):1674–83. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy A, Hodgkinson CA, Deluca V, et al. Two HPA axis genes, CRHBP and FKBP5, interact with childhood trauma to increase the risk for suicidal behavior. J Psychiatr Res. 2012;46(1):72–9. doi: 10.1016/j.jpsychires.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatro ET, Nguyen TB, Bousman CA, et al. Correlation of major depressive disorder symptoms with FKBP5 but not FKBP4 expression in human immunodeficiency virus-infected individuals. J Neurovirol. 2010;16(5):399–404. doi: 10.3109/13550284.2010.504248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnamurthy P, Romagni P, Torvik S, et al. Glucocorticoid receptor gene polymorphisms in premenopausal women with major depression. Horm Metab Res. 2008;40(3):194–8. doi: 10.1055/s-2007-1004541. [DOI] [PubMed] [Google Scholar]
- 39.Szczepankiewicz A, Leszczynska-Rodziewicz A, Pawlak J, et al. Glucocorticoid receptor polymorphism is associated with major depression and predominance of depression in the course of bipolar disorder. J Affect Disord. 2011;134(1-3):138–44. doi: 10.1016/j.jad.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 40.Pantazatos SP, Andrews SJ, Dunning-Broadbent J, et al. Isoform-level brain expression profiling of the spermidine/spermine N1-Acetyltransferase1 (SAT1) gene in major depression and suicide. Neurobiol Dis. 2015;79:123–34. doi: 10.1016/j.nbd.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell GM, Henley DE, Leendertz J, et al. Rapid glucocorticoid receptor-mediated inhibition of hypothalamic-pituitary-adrenal ultradian activity in healthy males. J Neurosci. 2010;30(17):6106–15. doi: 10.1523/JNEUROSCI.5332-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasserman D, Wasserman J, Sokolowski M. Genetics of HPA-axis, depression and suicidality. Eur Psychiatry. 2010;25(5):278–80. doi: 10.1016/j.eurpsy.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Szczepankiewicz A, Leszczynska-Rodziewicz A, Pawlak J, et al. FKBP5 polymorphism is associated with major depression but not with bipolar disorder. J Affect Disord. 2014;164:33–7. doi: 10.1016/j.jad.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Shinozaki G, Jowsey S, Amer H, et al. Relationship between FKBP5 polymorphisms and depression symptoms among kidney transplant recipients. Depress Anxiety. 2011;28(12):1111–8. doi: 10.1002/da.20879. [DOI] [PubMed] [Google Scholar]
- 46.Velders FP, Kuningas M, Kumari M, et al. Genetics of cortisol secretion and depressive symptoms: a candidate gene and genome wide association approach. Psychoneuroendocrinology. 2011;36(7):1053–61. doi: 10.1016/j.psyneuen.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan GM, Oquendo MA, Milak M, et al. Positron emission tomography quantification of serotonin(1A) receptor binding in suicide attempters with major depressive disorder. JAMA Psychiatry. 2015;72(2):169–78. doi: 10.1001/jamapsychiatry.2014.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Ortiz JM, Garcia-Gutierrez MS, Navarrete F, et al. Gene and protein alterations of FKBP5 and glucocorticoid receptor in the amygdala of suicide victims. Psychoneuroendocrinology. 2013;38(8):1251–8. doi: 10.1016/j.psyneuen.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Iacob E, Light KC, Tadler SC, et al. Dysregulation of leukocyte gene expression in women with medication-refractory depression versus healthy non-depressed controls. BMC Psychiatry. 2013;13:273. doi: 10.1186/1471-244X-13-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.