Abstract
Purpose
The aim of this study was to investigate the most suitable sperm preparation technique to apply in order to obtain a spermatozoon population with minimal DNA damage during in vitro fertilization procedures. We compared four preparation techniques: direct swim-up (DSU), pellet swim-up (PSU), density gradient (DG), and density gradient followed by swim-up (DG-SU), evaluating the effects of each technique on the DNA damage rate, evaluated by DNA fragmentation index of the spermatozoa obtained.
Methods
In this observational study, 98 semen samples from couples undergoing IVF/ICSI cycles were included. Data were collected between April and November 2014 at the ANDROS Day Surgery Clinic, Palermo, Italy.
Result(s)
The percentages of DNA fragmentation were 18.30 ± 10.8 in raw samples, 6.6 ± 5.7 after DSU, 4.2 ± 3.8 after PSU, 12.9 ± 9.9 after DG, and 3.7 ± 4.0 after DG-SU respectively. Compared to the raw evaluation, all the preparation techniques significantly decreased the total rate of the DNA fragmentation (DSU Z = −8.60, P < 0.008; PSU Z = −8.54, P < 0.008; DG Z = −6.42, P < 0.008, and DG-SU Z = −8.60, P < 0.008, respectively). Comparing them, spermatozoa with intact DNA after PSU and DG-SU were significantly higher than after DSU (Z = −7.12, P < 0.008; Z = −6.59, P < 0.008, respectively) and after DG (Z = −8.41, P < 0.008; Z = −8.60, P < 0.008, respectively). The difference between PSU and DG-SU was not significant (Z = −2.21, P = 0.03).
Conclusion(s)
There are, above all, two techniques of sperm preparation which allow for the recovery of spermatozoa with the lowest DNA fragmentation rate. Furthermore, given low costs and reduced time, we believe that PSU is the best option in the treatment of semen samples during IVF/ICSI.
Keywords: IVF, Sperm preparation techniques, Sperm DNA fragmentation, Sperm chromatin dispersion test
Introduction
Assisted reproductive technologies (ART) involve a variety of procedures for separating spermatozoa from seminal plasma in order to select the most viable gametes capable of fertilizing the eggs. The main aim of these techniques has long been to gain a population of motile spermatozoa with suitable morphology, although these parameters are insufficient to evaluate the viability of a spermatozoon. The processes, such as membrane modifications (which the spermatozoa undergo on entering the female genital tract) can be partially reproduced in an ART laboratory, and consist of separation and migration techniques. The most common sperm-processing protocols used in routine ART laboratories are those of swim-up and density gradient centrifugation.
According to the WHO laboratory manual for the examination and processing of human semen [1], the swim-up method is useful in selecting motile spermatozoa as it is based on the ability of sperm to swim into the culture medium. This method may be performed by layering the culture medium directly over the semen, or layering the culture medium over the pellet, which is obtained after the centrifugation of the sample. Density gradient centrifugation, which can be performed alone or in combination with the swim-up technique, separates the various cell types, thereby providing a selection of spermatozoa with suitable morphology.
Despite the widespread use of these techniques, there exists no consensus, nor are there any guidelines regarding their use, and to date, it has not been established which techniques are more suitable, also considering costs and execution time [2–4]. The limitations of conventional sperm preparation techniques are due to the inability to investigate the functional competence and the genome (nuclear) integrity of the sperm.
Several studies have shown that sperm DNA fragmentation, involving the presence of single- or double-strand DNA damage, is negatively correlated with ART outcomes. Specifically, this damage concerns failed fertilization, impaired embryo development, and an increased risk of pregnancy loss after IUI, IVF, and ICSI [5–12].
Current methods used to establish the degree of sperm DNA fragmentation may be used only as diagnostic tools, meaning that the cells analyzed in the assays cannot be used for in vitro fertilization [13]. Taking this evidence as our starting point, we posited the question as to whether there was a way to distinguish among four preparation techniques: direct swim-up (DSU), pellet swim-up (PSU), density gradient (DG), and density gradient followed by swim-up (DG-SU), also evaluating the effects of each technique on the DNA damage rate of the sperm population obtained. Although all these techniques reduce the presence of damaged spermatozoa [14], we decided to identify which technique might have the greatest capacity to recruit an amount of spermatozoa with a lower DNA fragmentation index (DFI).
Thus, the ultimate goal of our research was to isolate a common criterion in selecting the most eligible sperm preparation technique prior to in vitro fertilization. Accordingly, the aim of this study was to compare the effects of the most commonly used sperm preparation methods in selecting spermatozoa by evaluating DNA fragmentation.
Materials and Methods
Patient inclusion criteria
Semen samples were obtained from patients undergoing IVF/ICSI cycles at the Andros Clinic, Reproductive Medicine Unit, Palermo, Italy, in a period between April and November 2014. Only samples with a sperm concentration ≥10 × 106, progressive motility ≥5 %, total motility ≥35 %, and a volume ≥2 ml were included in the study. All samples were obtained by masturbation. According to the required statistical sample size, a total of 98 semen samples were included in the study.
Sperm preparation
After production, each semen sample was placed in an incubator at 37 °C to liquefy for a maximum period of 1 h. On completion, or within 1 h from production if liquefaction was incomplete, an initial evaluation of semen parameters was performed. A 50-μl sample was drawn in order to ascertain the basal level of DNA fragmentation, and the remaining sample was split into four aliquots and each of them was prepared as follows.
With regard to the direct swim-up technique (DSU), 1 ml of culture medium (Sperm Preparation Medium, Origio, Denmark) was gently stratified above the semen. The tube was inclined at an angle of 45° and incubated for 1 h (37 °C, 6 % CO2). A supernatant was then aspirated and transferred into an empty tube. As regards the pellet swim-up technique (PSU), the semen aliquot was diluted in a 1:2 ratio (1 + 1) with the culture medium (Sperm Preparation Medium, Origio, Denmark) and centrifuged for 10 min at 1500 rpm. Thereafter, the supernatant was discarded and 1 ml of fresh culture medium was layered above the pellet. Again, the tube was incubated for 1 h (37 °C, 6 % CO2) and the supernatant was aspirated and transferred into an empty tube.
With regard to the density gradient technique (DG), the semen sample was gently stratified above the discontinuous density gradients (55 and 80 %, SupraSperm 100, Origio, Denmark) and centrifuged for 20 min at 1500 rpm. Thereafter, the 80 % fraction was aspirated and put into an empty sterile conical tube; at least 3 ml of the culture medium (Sperm Preparation Medium, Origio, Denmark) was added and the tube was centrifuged for 10 min at 1500 rpm. The pellet was then resuspended in 0.5 ml of the same fresh culture medium.
The final technique of sperm preparation (DG-SU) involved the pellet being covered with 1 ml of the abovementioned culture medium, after treatment with the density gradient, to allow the spermatozoa to migrate. The supernatant was then aspirated and transferred into an empty 2-ml Eppendorf tube.
Clinical use
Spermatozoa obtained from one of the aforementioned techniques were used for the insemination of the oocytes, following the empirical criteria usually utilized, irrespective of the results of the DNA fragmentation.
Sperm chromatin dispersion test
In order to assess the degree of sperm DNA fragmentation, the sperm chromatin dispersion (SCD) test (Halosperm® G2 kit, Halotech DNA, S.L., Spain) [15] was utilized. We chose this commercial kit for the following reasons: the test was easy to set up and it did not require access to expensive equipment, which would have been unavailable in the routine laboratory where the study was carried out; this test also bears the CE marking. Moreover, it has been demonstrated that the sperm chromatin dispersion test is an effective tool for assessing sperm DNA fragmentation when compared with other tests [11, 16, 17]. The SCD test and TUNEL assay have the capacity to give similar results in terms of DNA fragmentation, and it has been demonstrated that the SCD test is even more sensitive than the TUNEL assay [18].
Aliquots of 50 μl of semen were gently and rapidly mixed in 100 μl of agarose gel (1 %; low melting point), which had previously been melted in a water bath for 5 min. Thereafter, a drop of 8 μl of each semen-agarose solution was placed onto pre-coated slides and covered with a coverslip. The slides were then refrigerated at 4 °C on a cold surface and left to gel for 5 min. Each coverslip was subsequently removed by gently sliding it off and the slides being incubated with an acid denaturation solution; the latter was used to denature DNA-containing breaks, for 7 min at room temperature. Immediately afterwards, the slides were incubated in a lysis solution for 20 min at room temperature in order to eliminate membranes and proteins. The lysis solution was then washed off with filtered water (Elix ® 5, Millipore) using a disposable pipette for 5 min. After washing, the slides were incubated with 70 % ethanol (2 min), followed by 90 % ethanol (2 min) and left to dry at room temperature. The slides were then stained and left to dry for microscope observation under bright field using a ×40 objective lens. A total of 490 tests were performed (5 tests for each of the 98 semen samples).
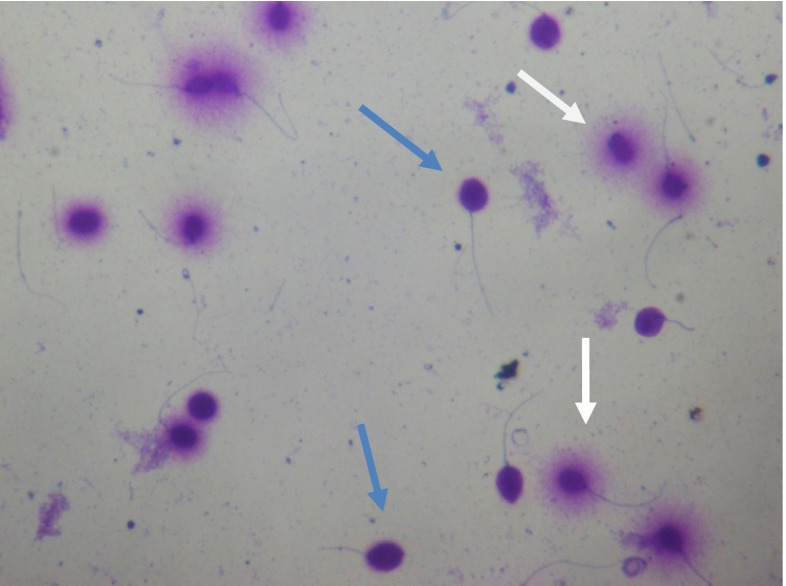
Spermatozoa with large- or medium-sized halos were classified as having intact DNA, whereas those with a small halo, without a halo and degraded ones, were classified as spermatozoa with DNA fragmentation (see Fig. 1). Only sperm cells with intact tails were classified in accordance with the halo size. The DFI was calculated as follows: number of spermatozoa with fragmented DNA divided by the number of spermatozoa analyzed 100×.
Fig. 1.
Spermatozoa after SCD test (magnification ×400). Blue arrows indicate spermatozoa with DNA fragmentation (without any halo); white arrows indicate spermatozoa with intact DNA (with halo)
Statistical analysis and sample size calculation
A preliminary sample size calculation was conducted on a data sample of 25 patients, who had been consecutively recruited. The percentage of DNA fragmentation observed was 15.90 ± 7.8 in a raw semen sample, and a comparison of techniques revealed the largest difference as 9.60, between DG and PSU (1.62 vs 11.22, respectively). Given that the aim of the research was to find at least one significant difference among the four techniques discussed in this study, and assuming P significant at 0.008 with a statistical power of 0.80, the sample size required for the main study was 98 subjects.
An assessment strategy for detecting normality violations was conducted in accordance with the recommendations by Tabachnick and Fidell [19]. Owing to the fact that data were not distributed normally, nonparametric statistics were used, namely the Mann-Whitney and Wilcoxon tests, in order to compare differences between groups in DNA fragmentation. The first test was a statistical test of the difference between the distributions of data, as applied to unmatched groups of cases, by comparing the distributions of the ranks of the scores. The second is a statistical test verifying the relative size of the scores of the same or matched subjects under two experimental conditions, by comparing the distributions for positive and negative differences of the ranks of their absolute values.
The data were initially processed by a square root transformation in order to check for any potential effect of a nuisance factor (subject variability); having achieved a normal distribution, the data were randomized and a complete block design ANOVA was performed thereon. As the primary analysis of the study required multiple comparisons, the Bonferroni correction was applied to protect against the threat of a type 1 error. The resulting P value required for these comparisons to be considered statistically significant was P < 0.008 (0.05/6). The statistical analysis was performed with version 18 PASW software.
Results
The descriptive statistics for the DNA fragmentation of sperm percentages of the different groups are presented in Table 1. Medians, means, and standard deviations are reported in Table 1 as a general indicator of central tendency of data. The resulting percentages of DNA fragmentation were 18.30 ± 10.8 in raw semen samples, 6.6 ± 5.7 after direct swim-up, 4.2 ± 3.8 after pellet swim-up, 12.9 ± 9.9 after density gradient, and 3.7 ± 4.0 after density gradient followed by swim-up. Compared with raw semen, the sperm DNA fragmentation rate was significantly lower after swim-up (Z = −8.60, P < 0.008), pellet swim-up (Z = −8.54, P < 0.008), density gradient (Z = −6.42, P < 0.008), and density gradient followed by swim-up (Z = −8.60, P < 0.008).
Table 1.
Differences in DNA fragmentation among the different sperm preparation techniques
| DNA fragmentation | DNA fragmentation | (Differences vs basal) | Wilcoxon Z | ||
|---|---|---|---|---|---|
| % mean (±SD) | % median | Fold change | Wilcoxon Z | ||
| Basal | 18.3 (±10.75) | 15.95 | |||
| DSU | 6.57 (±5.66) | 5.3 | −2.8 | −8.5961 | |
| PSU | 4.17 (±3.81) | 2.6 | −4.4 | −8.5961 | |
| DG | 12.86 (±9.94) | 10.6 | −1.4 | −6.4221 | |
| DG-SU | 3.74 (±4.04) | 2.62 | −4.9 | −8.5961 | |
| DSU vs PSU | −7.1181 | ||||
| DSU vs DG | −7.7561 | ||||
| DSU vs DG-SU | −6.5581 | ||||
| PSU vs DG | −8.4121 | ||||
| PSU vs DG-SU | −2.2082 | ||||
| DG vs DG-SU | −8.5961 | ||||
DSU direct swim-up, PSU, pellet swim-up, DG density gradient, DG-SU density gradient followed by swim-up
1 P < 0.008, 2 P = N. S.
In pursuing the stated aim of this study, a comparison among the four techniques was performed (Table 1): the results revealed that the percentage of DNA fragmentation in the pellet swim-up and density gradient followed by swim-up methods were significantly lower than in the swim-up method (Z = −7.12, P < 0.008, Z = −6.59, P < 0.008 respectively) and in the density gradient method (Z = −8.41, P < 0.008, Z = −8.60, P < 0.008, respectively). The difference between pellet swim-up and density gradient followed by swim-up was not significant (Z = −2.21, P = 0.03), whereas the percentage of DNA fragmentation in direct swim-up was lower than in density gradient (Z = −6.42, P < 0.008).
These results were confirmed after checking for any potential effect from the nuisance factor (subject variability): regarding the fragmentation percentage, the four sperm preparation techniques remained significantly different to each other (F3,291 = 159.49; P < =0.001). Tukey’s post hoc analysis confirmed the previously reported significant differences among techniques (P < = 0.008), in addition to the lack of any significant differences between the two enhanced techniques: pellet swim-up and density gradient followed by swim-up (=0.230).
Discussion
The increasing demand for assisted reproductive technology has led to necessary improvements in the procedures routinely used in IVF labs worldwide. One of the most subjective areas in the field of IVF work concerns the preparation of semen. Several methods are available and widely used in this field, but there is no consensus as to which method is more suitable than another. Each of these methods, based on the migration or separation of spermatozoa, is useful in selecting suitable morphology and motility cells, but they are not capable of selecting intact genome spermatozoa. To effect the latter, various techniques are available but none of these can be used clinically, since the sperm analyzed is wasted for the test. In combining the use of one of these tests, to investigate sperm DNA integrity, and the use of all the available techniques of sperm preparation, we attempted to ascertain which type of preparation is more suitable, on the basis of the best recovery of spermatozoa with DNA intact.
The suitability of one technique over another is irrefutable as sperm quality negatively influences not only fertilization outcomes, but also embryo development [20]. Moreover, it has been demonstrated that alterations in the paternal genome, such as high levels of fragmentation, may have negative effects on the cleavage speed and embryo morphology, appearing to influence blastocyst formation and implantation rates after IVF and ICSI cycles [21]. Several studies have been carried out in order to identify whether DNA damage may be considered a good predictor of ART outcomes and how it may influence pregnancy rates. In one of the largest ever reported studies investigating the predictive value of a DNA fragmentation assay in the determination of intrauterine insemination (IUI), IVF, and ICSI, the DFI was identified as a new determining factor in ART outcome [22].
Despite the lack of clarity regarding associations between sperm DNA fragmentation and conventional semen parameters, several assays have been developed to assess DNA integrity and, simultaneously, to identify a significant correlation between DNA damage and male infertility. In our study, we decided to use the sperm chromatin dispersion (SCD) test, an assay first described by Fernandez and colleagues in 2003 [23]. This test detects DNA fragmentation, using the principle that sperm with non-fragmented DNA produce a characteristic halo of DNA dispersion after acid denaturation and removal of nuclear proteins. The halo is formed due to relaxed DNA loops still attached to nuclear structures. Despite assessment of the DFI (obtained by using the SCD test) being based on the observation of a few hundred sperm and it being a subjective assessment, this test is a useful assay as it does not require expensive equipment or expertise, and the results are available in a relatively short period of time. In our case, the use of this test enabled us to identify which sperm preparation technique was capable of selecting the most intact DNA from the spermatozoa. A recent study states that the SCD test is even more sensitive than another DNA fragmentation assay (TUNEL, terminal deoxynucleotidyl transferased UTP nick end labeling) in evaluating sperm DNA damage [24].
Moreover, it seems that there might be a correlation between sperm DNA fragmentation and embryo morphology. A study by Tandara and colleagues has shown that halos obtained with the Halosperm represent a prognostic parameter of good embryo quality. In their study, they found a significant correlation between large halos, good morphology embryos, scored on day three of development, and the achievement of a pregnancy [25].
We decided to perform the study including only those samples with a minimum volume of 2 ml, a minimum sperm concentration of 10 million/ml, and a minimum of total motility of 35 %. These inclusion criteria were selected in order to divide each sample into five different aliquots and to obtain a minimum spermatozoon concentration with which to perform the SCD test. Moreover, given that three out of four techniques chosen included swim-up procedures, we excluded sperm samples with total motility of less than 35 %. Out of a total of 490 tests undertaken, we observed that all the sperm preparation techniques were able to decrease the total sperm population with DNA fragmentation, in accordance with the findings of other authors [26]. Specifically, only two methods out of the four used—the pellet swim-up and the density gradient followed by swim-up—exhibited the best results. Mean DFI values obtained after swim-up and density gradient followed by swim-up were significantly lower than those observed in direct swim-up and density gradient.
With regard to the result showing a lower DNA fragmentation rate in direct swim-up compared with density gradient, we assume that the sperm selection obtained by the latter technique is only based on the cells’ molecular weight, which means that the filter action of this procedure might not ensure good selection in terms of fragmentation rate. It is possible that in the direct swim-up, only spermatozoa with good motility and, perhaps, better DNA integrity are selected.
However, our results are inconsistent with the findings presented by Amiri and colleagues [26], which compared the swim-up and density gradient preparations on samples of men undergoing semen analysis. In their study, they found that the DNA-fragmented spermatozoa in the samples prepared with the swim-up method were more numerous than in those processed with the density gradient. We might add that our findings agreed with those of Ghaleno and colleagues [27], which stated that mean DNA fragmentation in samples processed by density gradient centrifugation was found to be higher when compared to conventional swim-up.
Taking these data as a starting point, we believe that despite the two techniques being equivalent in terms of good intact DNA spermatozoa recovery, the density gradient in association with the swim-up is a more expensive procedure, thereby increasing execution time. On the contrary, the conventional pellet swim-up method entails lower costs and is less time-consuming, thereby representing the elective choice in IVF laboratories. One limitation of our study was that we did not correlate sperm DNA fragmentation with any IVF/ICSI outcomes.
In conclusion, given the diminished financial outlay and reduced time requirement, we believe that pellet swim-up is the best option in the treatment of semen samples during IVF/ICSI. As we did not correlate DNA fragmentation rates with any outcomes, we suggest that these findings should be validated with further investigations mainly linked with IVF/ICSI outcomes.
Compliance with ethical standards
Conflict of interest
The authors of this paper declare that they have no conflict of interest which could be perceived as prejudicing the impartiality of the research reported herein.
Funding
This research did not receive any specific grant from any funding agency in the public, commercial, or not-for-profit sector; the funds were provided directly by the ANDROS Day Surgery Clinic, Palermo, Italy.
Footnotes
Capsule Given lower costs and reduced procedure time, we believe that PSU is the best option in the treatment of semen samples during IVF/ICSI.
References
- 1.World Health Organization . WHO laboratory manual for the examination and processing of human semen. 5. Geneva: WHO Press; 2010. [Google Scholar]
- 2.Brandeis VT, Manuel MT. Effects of four methods of sperm preparation on the motile concentration, morphology, and acrosome status of recovered sperm from normal semen samples. J Assist Reprod Genet. 1993;10(6):409–16. doi: 10.1007/BF01228091. [DOI] [PubMed] [Google Scholar]
- 3.Ren SS, Sun GH, Ku CH, Chen DC, Wu GJ. Comparison of four methods for sperm preparation for IUI. Arch Androl. 2004;50(3):139–43. doi: 10.1080/01485010490425566. [DOI] [PubMed] [Google Scholar]
- 4.Barroso G, Chaya M, Bolaños R, Rosado Y, García León F, Ibarrola E. Prognostic value on recovery rates for the application of sperm preparation techniques and their evaluation in sperm function. Ginecol Obstet Mex. 2005;73(5):221–8. [PubMed] [Google Scholar]
- 5.Bungum M, Humaidan P, Spano M, Jepson K, Bungum L, Giwercman A. The predictive value of sperm chromatin structure assay (SCSA) parameters for the outcome of intrauterine insemination, IVF and ICSI. Hum Reprod. 2004;19:1401–8. doi: 10.1093/humrep/deh280. [DOI] [PubMed] [Google Scholar]
- 6.Borini A, Tarozzi N, Bizzaro D, Bonu MA, Fava L, Flamigni C, et al. Sperm DNA fragmentation: paternal effect on early post-implantation embryo development in ART. Hum Reprod. 2006;21:2876–81. doi: 10.1093/humrep/del251. [DOI] [PubMed] [Google Scholar]
- 7.Carrell DT, Liu L, Peterson CM, Jones KP, Hatasaka HH, Erickson L, et al. Sperm DNA fragmentation is increased in couples with unexplained recurrent pregnancy loss. Arch Androl. 2003;49:49–55. doi: 10.1080/01485010290099390. [DOI] [PubMed] [Google Scholar]
- 8.Duran EH, Morshedi M, Taylor S, Oehninger S. Sperm DNA quality predicts intrauterine insemination outcome: a prospective cohort study. Hum Reprod. 2002;17:3122–8. doi: 10.1093/humrep/17.12.3122. [DOI] [PubMed] [Google Scholar]
- 9.Evenson DP, Jost LK, Marshall D, Zinaman MJ, Clegg E, Purvis K, et al. Utility of the sperm chromatin structure assay as a diagnostic and prognostic tool in the human fertility clinic. Hum Reprod. 1999;14:1039–49. doi: 10.1093/humrep/14.4.1039. [DOI] [PubMed] [Google Scholar]
- 10.Seli E, Gardner DK, Schoolcraft WB, Moffatt O, Sakkas D. Extent of nuclear DNA damage in ejaculated spermatozoa impacts on blastocyst development after in vitro fertilization. Fertil Steril. 2004;82:378–83. doi: 10.1016/j.fertnstert.2003.12.039. [DOI] [PubMed] [Google Scholar]
- 11.de la Calle JF V, Muller A, Walschaerts M, Clavere JL, Jimenez C, Wittemer C, et al. Sperm deoxyribonucleic acid fragmentation as assessed by the sperm chromatin dispersion test in assisted reproductive technology programs: results of a large prospective multicenter study. Fertil Steril. 2008;90(5):1792–9. doi: 10.1016/j.fertnstert.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Zini A, Boman JM, Belzile E, Ciampi A. Sperm DNA damage is associated with an increased risk of pregnancy loss after IVF and ICSI: systematic review and meta-analysis. Hum Reprod. 2008;23:2663–8. doi: 10.1093/humrep/den321. [DOI] [PubMed] [Google Scholar]
- 13.Henkel R. Sperm preparation: state-of-the-art-physiological aspects and application of advanced sperm preparation methods. Asian J Androl. 2012;14:260–9. doi: 10.1038/aja.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayaraman V, Upadhya D, Narayan PK, Adiga SK. Sperm processing by swim-up and density gradient effective in elimination of sperm with DNA damage. J Assist Reprod Genet. 2012;29:557–63. doi: 10.1007/s10815-012-9742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez JL, Muriel L, Goyanes V, Segrelles E, Gosàlvez J, Enciso M, et al. Simple determination of human sperm DNA fragmentation with an improved sperm chromatin dispersion test. Fertil Steril. 2005;84:833–42. doi: 10.1016/j.fertnstert.2004.11.089. [DOI] [PubMed] [Google Scholar]
- 16.Chohan KR, Griffin JT, Lafromboise M, De Jonge CJ, Carrell DT. Comparison of chromatin assays for DNA fragmentation evaluation in human sperm. J Androl. 2006;27:53–9. doi: 10.2164/jandrol.05068. [DOI] [PubMed] [Google Scholar]
- 17.Ribas-Maynou J, Garcıa-Peiro A, Fernandez-Encin A, Abad C, Amengual MJ, Prada E, et al. Comprehensive analysis of sperm DNA fragmentation by five different assays: TUNEL assay, SCSA, SCD test and alkaline and neutral Comet assay. Andrology. 2013;1(5):715–22. doi: 10.1111/j.2047-2927.2013.00111.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Qiu Y, Wang K, Wang Q, Tao G, Wang L. Measurement of sperm DNA fragmentation using bright-field microscopy: comparison between sperm chromatin dispersion test and terminal uridine nick end labeling assay. Fertil Steril. 2010;24(3):1027–32. doi: 10.1016/j.fertnstert.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 19.Tabachnick BG, Fidell SL. Using multivariate statistics. 4thed. Needham Heights: Allyn and Bacon; 2001. [Google Scholar]
- 20.Virro MR, Larson-Cook KL, Evenson DP. Sperm chromatin structure assay (SCSA) parameters are related to fertilization, blastocyst development, and ongoing pregnancy in in vitro fertilization and intracytoplasmic sperm injection cycles. Fertil Steril. 2004;81(5):1289–95. doi: 10.1016/j.fertnstert.2003.09.063. [DOI] [PubMed] [Google Scholar]
- 21.Tesarik J, Greco E, Mendoza C. Late, but not early, paternal effect on human embryo development is related to sperm DNA fragmentation. Hum Reprod. 2004;19(3):611–5. doi: 10.1093/humrep/deh127. [DOI] [PubMed] [Google Scholar]
- 22.Bungum M, Humaidan P, Axmon A, Spano M, Bungum L, Erenpreiss J, et al. Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Hum Reprod. 2007;22(1):174–9. doi: 10.1093/humrep/del326. [DOI] [PubMed] [Google Scholar]
- 23.Fernández JL, Muriel L, Rivero MT, Goyanes V, Vazquez R, Alvarez JG. The sperm chromatin dispersion test: a simple method for the determination of sperm DNA fragmentation. J Androl. 2003;24(1):59–66. [PubMed] [Google Scholar]
- 24.Feijo CM, Esteves SC. Diagnostic accuracy of sperm chromatin dispersion test to evaluate sperm deoxyribonucleic acid damage in men with unexplained infertility. Fertil Steril. 2014;101(1):58–63. doi: 10.1016/j.fertnstert.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Tandara M, Bajić A, Tandara L, Bilić-Zulle L, Šunj M, Kozina V, et al. Sperm DNA integrity testing: big halo is a good predictor of embryo quality and pregnancy after conventional IVF. Andrology. 2014;2(5):678–86. doi: 10.1111/j.2047-2927.2014.00234.x. [DOI] [PubMed] [Google Scholar]
- 26.Amiri I, Ghorbani M, Hesmati S. Comparison of the DNA fragmentation and the sperm parameters after processing by the density gradient and the swim up methods. J Clin Diagn Res. 2012;6(9):1451–3. doi: 10.7860/JCDR/2012/4198.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaleno LR, Valojerdi MR, Janzamin E, Chehrazi M, Sharbatoghli M, Yazdi RS. Evaluation of conventional semen parameters, intracellular reactive oxygen species, DNA fragmentation and dysfunction of mitochondrial membrane potential after semen preparation techniques: a flow cytometric study. Arch Gynecol Obstet. 2014;289(1):173–80. doi: 10.1007/s00404-013-2946-1. [DOI] [PubMed] [Google Scholar]