Abstract
Purpose
The aim of this study is to identify potential genes involved in human globozoopsermia.
Methods
Nineteen globozoospermic patients (previously screened for DPY19L2 mutations with no causative mutation) were recruited in this study and screened for mutations in genes implicated in human globozoospermia SPATA16 and PICK1. Using the candidate gene approach and the determination of Spata16 partners by Glutathione S-transferase (GST) pull-down four genes were also selected and screened for mutations.
Results
We identified a novel mutation of SPATA16: deletion of 22.6 Kb encompassing the first coding exon in two unrelated Tunisian patients who presented the same deletion breakpoints. The two patients shared the same haplotype, suggesting a possible ancestral founder effect for this new deletion. Four genes were selected using the candidate gene approach and the GST pull-down (GOPC, PICK1, AGFG1 and IRGC) and were screened for mutation, but no variation was identified.
Conclusions
The present study confirms the pathogenicity of the SPATA16 mutations. The fact that no variation was detected in the coding sequence of AFGF1, GOPC, PICK1 and IRGC does not mean that they are not involved in human globozoospermia. A larger globozoospermic cohort must be studied in order to accelerate the process of identifying new genes involved in such phenotypes. Until sufficient numbers of patients have been screened, AFGF1, GOPC, PICK1 and IRGC should still be considered as candidate genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0715-3) contains supplementary material, which is available to authorized users.
Keywords: Globozoospermia, Linkage analysis, SPATA16, GST pull-down, Male infertility
Introduction
Globozoospermia is a rare (<0.1 % in male infertile patients) and severe teratozoospermia primarily consisting of spermatozoa lacking an acrosome. The acrosome is playing a crucial role during fertilization by allowing spermatozoa to travel through the zona pellucida and to reach the oocyte cytoplasmic membrane. Therefore, patients suffering from a complete form of globozoospermia are infertile. In 2007, we identified spermatogenesis associated 16 (SPATA16) as the first autosomal gene involved in human globozoospermia [1]. We demonstrated that a homozygous mutation (c.848G → A) in SPATA16 led to the production of roundheaded and acrosomeless spermatozoa, by disrupting the splicing site of exon 4 coding for a highly conserved tetratricopeptide repeat domain (TPR domain). The TPR domain is a protein-protein interaction module that acts as an organizing centre for complexes regulating a multitude of biological processes [2]. The encoded protein, specifically expressed in human testis, is localized in the Golgi apparatus and then shifted with Golgi vesicles to the acrosome [3]. Here, we report a new mutation of SPATA16 and we present a study to select genes potentially involved in human globozoospermia using two approaches: (i) the determination of Spata16 partners by Glutathione S-transferase (GST) pull-down and (ii) the candidate gene approach.
Our analysis of 54 globozoospermic patients originating from different countries showed that 36 of them were found mutated for DPY19L2, which represents 67 % of our cohort [4]. However, for the 18 remaining patients, we did not find any causative mutation, confirming that acrosome biogenesis is a complex process that involved many genes. In order to identify genes responsible for globozoospermia in these patients, we initiated our study by screening for SPATA16 and PICK1 mutations, since both have been shown to be implicated in human acrosome biogenesis [1, 5]. Using the candidate gene approach, based on similarity of phenotypes in the null mutant mice and human globozoospermia, i.e. round, spherical and acrosomless sperm [6, 7], we selected two additional genes known as AGFG1 (ArfGAP with FG repeats 1) and GOPC (Golgi-associated PDZ- and coiled-coil motif-containing protein). For instance, in Agfg1-/-mice, proacrosomic vesicles form but fail to fuse [8]. A lack of acrosome formation was observed in Gopc null mutant mice due to the fragmentation of the acrosome in the early stage of spermiogenesis [9]. The same phenotype was also observed in male mice deficient in Pick1, a protein co-localized with GOPC in the Golgi regions [7]. Taking into consideration the mice phenotypes of Agfg1, Gopc, Pick1, knock out as well as the human cases reported implicating SPATA16 and PICK1 [1, 5], we screened the entire coding region and intron/exon boundaries of the four genes in 18 globozoospermic patients for whom no mutation in DPY19L2 was found. In addition, we identified, using the mouse model, Spata16 partners and chosen IRGC as a candidate gene to be screened for mutations. One of our 18 globozoospermic patients showed a homozygous deletion for SPATA16 exon 2, confirmed later during a second parallel study by our team in another Tunisian globozoospermic patient. However, no variation was identified in the selected candidate genes in the 17 remaining patients.
Materials and methods
Patients and controls
A total of 19 globozoospermic patients originating from France, Italy, Tunisia, Turkey, Libya and Morocco were recruited in this study. All the patients were healthy and presented a nonsyndromic infertility. None of them had chromosomal abnormalities and no Y microdeletions were found. The patients have not been exposed to toxic chemicals, radiation or treatment by drugs known to impair fertility. This study was approved by the local Ethical Committee (Comité de protection de la personne, CPP (CPP 09/40-W AC-2008-438 1WDC-2009-I 002)) of Strasbourg University Hospital. For each case analysed, informed written consent was obtained according to the CPP recommendations.
DNA preparation
Genomic DNA from each patient was extracted either from peripheral blood leucocytes using QIAmp DNA Blood Midi Kit (Qiagen, Germany) or from saliva using Oragene DNA Self-Collection Kit (DNAgenotech, Canada).
Linkage analysis
Whole genome scanning using the GeneChip Human mapping 10K 2.0 Xba from Affymetrix was performed on three families of two brothers each (Globo10/11, Globo20/21, Globo23/24). A 250k SNP array was applied on a consanguineous Tunisian globozoospermic patient (Globo65). Single nucleotide polymorphisms (SNP) obtained were analysed with the HomoSNP program that was set to identify homozygous regions of 25 or more consecutive SNPs.
Mutation screening
Mutation screening was performed by polymerase chain reaction (PCR) and Sanger sequencing of the exons and intronic flanking sequences of the SPATA16, PICK1, GOPC, AFGF1 and IRGC genes. PCR primer sequences and amplification conditions are listed in supplementary data. DNA amplicons were purified and double-strand sequencing of each DNA fragment was performed by GATC (Cologne, Germany). Sequence analyses were carried out using an ABI PRISM 3730xl Genetic Analyser (Applied Biosystems).
SNPs genotyping
To establish a haplotype map of the two patients showing the new SPATA16 deletion, we genotyped a total of 61 SNPs around the deletion most of them (51 SNPs) were covered by 5′SP16n7 and 3′SP16n4 primers used to delimit the deletion breakpoints. Besides, additional primers amplifying fragments before and after the deletion were used to genotype the remaining ten SNPs located farther than the breakpoints. Primer sequences and amplification conditions are listed in Table S.1 in supplementary data.
SPATA16 GST pull-down
The fusion protein GST-SPATA16 was constructed by inserting the cDNA encoding the full open reading frame of mouse SPATA16 in the multiple cloning site of the pGex-4T-1.
The murine cDNA was ordered from Biovalley (MMM1013-99826784) and amplified using a forward primer that contained the EcoRI site (5′-GAATTCgattctggcaagagtaggag-3′) and a reverse primer that contained a flag encoding sequence, a stop codon and the XhoI site (5′-caaactgtacagcaaaacgattacaaggacgacgatgacaagtgaccgCTCGAGcgg-3′). The FLAG tag sequence was added to the C terminus of SPATA16 in order to check the complete translated protein. The final construct was verified by sequencing with primers in the plasmid (pGEX3-RP: TCAAGAATTATACACTCCG and pGEX5-FP: AACGTATTGAAGCTATCCC).
The recombinant protein GST-SPATA16 and protein GST were individually expressed in E. coli BL21 cell. The induction by 0.1 mM of IPTG was performed at a cell density of A600 = 0.5 for 3 h at 22 °C and checked by 10 % SDS-PAGE. Cleared lysates were prepared and the soluble fusion proteins were purified by batch procedure incubating the lysate with glutathione sepharose (Amersham Biosciences) for 1 h at 4 °C with gentle shaking. Then the glutathione sepharose was collected by centrifugation and washed five times with lysis buffer (Tris HCl 50 mM pH9, NaCl 500 mM, DTT 2 mM). After boiling, the bound proteins were analysed by 10 % SDS-PAGE followed by western blotting using anti-GST monoclonal antibody (Kangwei, China).
Forty testes were collected from wild-type adult C57BL/6 mice. Proteins were then extracted by adding a lysis buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 0.1 % NP40, 1 mM PMSF, 1 mM DTT and protease inhibitor cocktail) and by mixing for 2 min on ice. The soluble fraction was then collected after centrifugation at 8000 rpm for 10 min at 4 °C.
The pull-down was performed on glutathione sepharose coupled to GST and GST-SPATA16 separately. The soluble fraction of the testes proteins were incubated with the glutathione sepharose at 4 °C with gentle shaking overnight. Then the beads were washed five times with the lysis buffer before analysing by SDS-Page. After running the samples about 2–3 cm into the resolving gel (10 % acrylamide) and stained with Coomassie blue, mass spectrometry was performed at Taplin mass spectrometry (Harvard, USA).
Results
Mutation screening of SPATA16, AGFG1, GOPC and PICK1
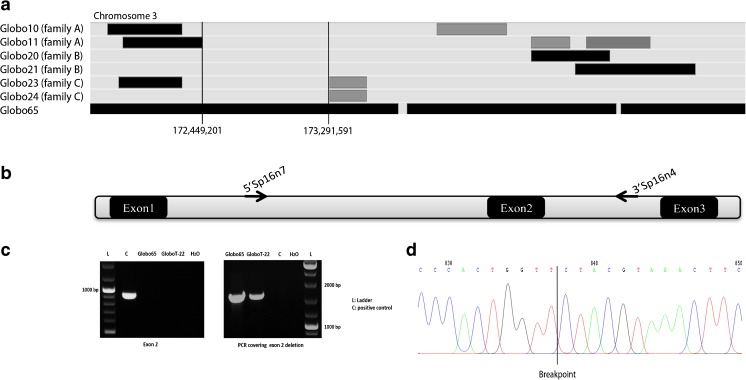
In our previous work, 18 out of 54 patients did not show any mutation for DPY19L2 [4]. Among these patients, three consanguineous families (A, B and C) that each contained two globozoospermic brothers (Globo10/11, Globo20/21 and Globo23/24 respectively) have been analysed by 10K SNP microarrays. Arrays with a higher resolution were not available at this time. A wide genome scan using 250k SNP array was applied later on a consanguineous Tunisian globozoospermic patient (Globo65). A homozygous region was observed at the locus of SPATA16 in Globo65, whereas Globo10, Globo20 and Globo23 were heterozygous (Fig. 1a). Therefore, we performed PCR of all the exons and exon-intron junctions of SPATA16 only on the genomic DNA of Globo65. The patient showed a homozygous deletion of exon 2, since specific amplification was detected in the control but never in the patient. In order to pinpoint the breakpoints within intron 1 and intron 2, subsequent amplifications on both sides of exon 2 were performed.
Fig. 1.
Analysis of SPATA16 gene. a SNP array results of the four infertile patients for the region of chromosome 3. Shared regions of homozygosity are visualized by the HomoSNP software, which displays one patient per line. The areas of homozygosity with 25 or more SNPs are black, whereas homozygosity regions defined by 15–25 consecutive SNPs are grey. Regions of heterozygosity are light grey. Globo65 shows a homozygous region for SPATA16 locus (Loc. Chr3:172, 889,358–173,141,268) which is not shared by the brothers of the three families, Globo10/11, Globo20/21 and Globo23/24. b Schematic representation of exons 1, 2 and 3 of SPATA16. Sequence amplifications on both sides of exon 2 were performed using the primers 5′SPA16n7 at 2500 bp from exon1 and 3′SP16n4 at 2274 bp from exon 2 in order to determine the breakpoints of the deletion. c PCR results of SPATA16 exon 2 and breakpoints. The patients are deleted for exon 2 while the positive control showed amplification. The PCR covering the deletion of exon 2 was performed using the primers 5′SPA16n7 and 3′SP16n4. The deleted patients showed a fragment of 2000 bp while the positive control was homozygous wild type since no fragment was amplified. d Sequence of the PCR covering the deletion in the patients, allowing the identification of the breakpoint
Amplification across the deletion using the primers, 5′SP16n7 as forward primer and 3′SP16n4 as reverse primer, was performed as a control for the deletion (Fig. 1b). Globo65 showed an amplification of 1620 bp, but this was not the case for the fertile control (Fig. 1c). Sequencing the 1620-bp PCR product allowed us to precisely define the breakpoint, which is located at 20,459 bp at 5′ side and at 1547 bp at the 3′ side of the exon 2, and hence to determine the exact size of the deletion which is 22,648 bp (Fig. 1d).
Recently, a second patient (GloboT-22) unrelated to Globo65 showed the same SPATA16 exon 2 deletion and was found to share the same breakpoints described above. To establish the haplotypes for these two patients, we checked different intronic nucleotides reported as single nucleotide polymorphisms (SNPs) in ensembl database (http://www.ensembl.org/index.html) surrounding the exon 2 deletion. In total, 61 SNPs were checked; 34 SNPs before the deletion versus 27 SNPs after the deletion (Table S.2 and S.3, supplementary data). The first SNP genotyped was located at 2426 bp of the breakpoint at 5′ side of the deletion and the last SNP at 5538 bp of the breakpoint at 3′ side. SNPs rs ID, their genomic positions and allelic frequencies were reported in supplementary data (Tables S.2 and S.3). SNPs genotyping revealed the same haplotype for Globo65 and GloboT-22, strongly suggesting the possibility of a founder effect for this new SPATA16 exon 2 deletion. We pursued our study by screening for mutations in SPATA16 and in the three candidate genes AGFG1, GOPC and PICK1 in the 17 remaining patients, but no sequence variation was detected.
Identification of Spata16 testis specific partners and selection of new candidate genes
Since Spata16 contained a highly conserved TPR domain, known to be a scaffold for protein interactions, especially co-chaperone proteins, we wanted to investigate the testis specific partners with which Spata16 interacts. In absence of available antibody against Spata16, we therefore performed GST pull-down using a GST-Spata16 fusion protein. The purified protein was incubated with an extract of wild type mice testes proteins. In order to eliminate the false positive proteins, we analysed the proteins that also interact with GST alone. The two samples were later analysed by 10 % SDS-PAGE and showed specific bands in the GST-Spata16 sample which were not detected in the negative control (supplementary figure 1). We were able to identify the potential partners of Spata16 by mass spectrometry. The proteins shared between the sample and the negative controls were omitted, leading to 35 potential partners (Table 1).
Table 1.
SPATA16 partners. SPATA16 partners were selected after eliminating the proteins identified in the negative control GST protein experiment. Mouse proteins highlighted in dark gray showed an exclusive expression in testes and proteins highlighted in light gray showed a predominant expression in testes with an expression in other tissues

The identified partners could be considered as potential candidate genes involved in human globozoospermia, especially since nine of them are predominantly expressed in the testes (Table 1). Subsequently, we decided to sequence the candidates in the 17 remaining globozoospermic patients who were not mutated for DPY19L2 or SPATA16, hoping to identify new genes.
In order to check if any of these genes could be implicated in human globozoospermia, we combined the linkage analysis of the three consanguineous globozoospermic families and the GST pull-down outcome. Analysis of the three families as one entity, assuming that the same gene is responsible for the phenotype observed in each family, revealed a unique homozygous region located on chromosome 8 (Supplementary data figure 2A). The region of 4.7 Mb contains five genes, but none was identified as a partner of Spata16; hence, they were not prioritized as potential candidate genes and they were not screened for mutations (Supplementary data figure 2A). Examining the families separately, one gene was chosen among the 35 partners: IRGC. In fact, IRGC is chosen because the brothers of family B showed homozygosity state for its locus. However, the amplification and sequencing of its coding sequence did not reveal any nucleotide variation (Supplementary data figure 2B).
Discussion
The genetics of infertility remain largely an unexplored field despite the fact that some genetic causes of male infertility have been identified [1, 4, 10, 11]. In this study, we confirmed the pathogenicity of SPATA16 mutations by identifying a novel deletion of 22.6 Kb encompassing exon 2. The deletion of exon 2 was identified in two unrelated Tunisian patients out of 19 not mutated for DPY19L2. We hypothesized that the absence of exon 2, which is the first coding exon and contains the ATG start codon, lead to the absence of the protein or to the production of a truncated protein. Alternatively, an eventual splice between exon 1 and 3 would lead to the production of an mRNA containing three ATG codons in exon 3. None of these codons are likely to be used since they do not present a Kozac sequence. In addition, the first and third are in frame with each other but are not in frame with the normal frame of SPATA16. In such a case, it is likely that the exon 2 deleted SPATA16 mRNA is eliminated by a nonsense-mediated mRNA decay (NMD) mechanism [12].
The second ATG is in frame with the SPATA16 sequence and, if used, it would produce a truncated protein missing 36 out of 116 amino acids of the highly conserved TPR domain, (representing 31 % of the domain), and thus would probably produce a non-functional protein. Considering the ethical impossibility of getting a testicular biopsy for research purposes from this patient, we have not been able to test any of these hypotheses.
Genotyping SNPs located before and after the deletion breakpoints, we showed that the two patients homozygous for the deletion and originated from two close regions (120Km of distance) shared the same haplotype, pointing to a possible ancestral founder effect for this new SPATA16 mutation.
We also presented two approaches to select genes potentially involved in human globozoospermia: (i) candidate gene approach and (ii) identification of Spata16 partners. Using the candidate gene approach, three genes described from site-directed mutagenesis experiments in mice were of interest: Agfg1, Gopc and Pick1, since mice deficient for these proteins exhibit a phenotype very similar to human globozoospermia [13]. No definitive mutation with a clear link to human globozoospermia was found. We could not confirm in our cohort of patients the missense mutation (G198A) in PICK1 which was proposed as responsible for globozoospermia in humans (Liu et al 2010). Consulting the 1000Genome database, 18 (A) alleles were identified out of 6534 alleles at this position, all in heterozygous state and all were reported in China and Japan (Ghedir et al. unpublished). The transition in question creates a missense substitution (G393R) located in the C-terminal part of the protein. Online available tools (e.g. SIFT and PolyPhen) predict that this variation does not adversely affect protein function. Altogether, we wonder if this is instead an Asian specific polymorphism rather than a mutation causing globozoospermia in humans.
The combined analysis of three consanguineous families and the identification of SPATA16 partners prompted us to consider IRGC as a potential candidate gene. Although, IRGC is exclusively expressed in the testis and was in a homozygous state in one consanguineous family, no mutation was detected. In a future study, IRGC will be screened in the 14 remaining patients who were not the subject of a SNP genome wide screening.
Over all, the results of the present study confirm the pathogenicity of the SPATA16 mutations. The fact that no variation was detected in the coding sequence of AFGF1, GOPC, PICK1 and IRGC does not mean that they are not involved in human globozoospermia. Mutations could also affect the non-coding sequence such as UTRs and promoters, which were not sequenced. Since globozoospermia is a rare teratozoospermia, the main issue remains the lack of sufficient number of patients. A larger globozoospermic cohort must be studied in order to accelerate the process of identifying new genes involved in such phenotypes. Until sufficient numbers of patients have been screened, AFGF1, GOPC, PICK1 and IRGC should still be considered as candidate genes.
In addition to the development of molecular diagnostic test, we believe that the identification and characterization of the genetic bases of male infertility can determine the selection of treatment options and management of such couples [14].
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 18 kb)
(DOCX 24 kb)
(DOCX 23 kb)
(DOCX 27 kb)
(DOCX 17 kb)
GST pull-down. GST-SPATA16 fusion protein (lane 1) and control GST protein (lane 2) were subjected to pull-down reaction in the presence of cell extract derived from adult mice testes. The samples were fractionated in SDS-PAGE and stained with silver. (GIF 75 kb)
Linkage analysis. (A) SNP array results revealing a unique homozygous region shared between the brothers of the three families. The genes located in the region of 4.7 Mb on the chromosome eight are represented in the table below it. (B) SNP array results showing the two brothers of family B sharing a homozygous region for IRGC locus (loc.chr19: 43,716,010 – 43,720,021). (GIF 58 kb)
Acknowledgements
We would like to thank James Turner for his critical reading of the manuscript. We are grateful to the Institute of Genetics and Molecular and Cellular Biology (IGBMC) platforms. The study was funded by Agence Nationale de la Recherche and l’Agence de BioMédecine. This work was supported by the French Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), the Ministère de l’Education Nationale et de l’Enseignement Supérieur et de la Recherche, the University of Strasbourg, the University Hospital of Strasbourg, the Agence Nationale pour la Recherche and the Agence de la BioMédecine.
Compliance with ethical standards
This study was approved by the local Ethical Committee (Comité de protection de la personne, CPP (CPP 09/40 - W AC-2008-438 1W DC-2009-I 002)) of Strasbourg University Hospital. For each case analysed, informed written consent was obtained according to the CPP recommendations.
Footnotes
Capsule In this study, we confirmed the pathogenicity of SPATA16 mutations and we identified potential genes implicated in human globozoospermia.
References
- 1.Dam AH, Koscinski I, Kremer JA, Moutou C, Jaeger AS, Oudakker AR, et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet. 2007;81(4):813–20. doi: 10.1086/521314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krachler AM, Sharma A, Kleanthous C. Self-association of TPR domains: lessons learned from a designed, consensus-based TPR oligomer. Proteins. 2010;78(9):2131–43. doi: 10.1002/prot.22726. [DOI] [PubMed] [Google Scholar]
- 3.Xu M, Xiao J, Chen J, Li J, Yin L, Zhu H, et al. Identification and characterization of a novel human testis-specific Golgi protein, NYD-SP12. Mol Hum Reprod. 2003;9(1):9–17. doi: 10.1093/molehr/gag005. [DOI] [PubMed] [Google Scholar]
- 4.Elinati E, Kuentz P, Redin C, Jaber S, Vanden Meerschaut F, Makarian J, et al. Globozoospermia is mainly due to DPY19L2 deletion via non-allelic homologous recombination involving two recombination hotspots. Hum Mol Genet. 2012;21(16):3695–702. doi: 10.1093/hmg/dds200. [DOI] [PubMed] [Google Scholar]
- 5.Liu G, Shi QW, Lu GX. A newly discovered mutation in PICK1 in a human with globozoospermia. Asian J Androl. 2010;12(4):556–60. doi: 10.1038/aja.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen GL, Ivanov IP, Atkins JF, Campbell B, Carrell DT. Identification of polymorphisms in the Hrb, GOPC, and Csnk2a2 genes in two men with globozoospermia. J Androl. 2006;27(1):11–5. doi: 10.2164/jandrol.05087. [DOI] [PubMed] [Google Scholar]
- 7.Xiao N, Kam C, Shen C, Jin W, Wang J, Lee KM, et al. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J Clin Invest. 2009;119(4):802–12. doi: 10.1172/JCI36230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM. Lack of acrosome formation in Hrb-deficient mice. Science. 2001;294(5546):1531–3. doi: 10.1126/science.1063665. [DOI] [PubMed] [Google Scholar]
- 9.Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, et al. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci U S A. 2002;99(17):11211–6. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghedir H, Ibala-Romdhane S, Okutman O, Viot G, Saad A, Viville S. Identification of a new DPY19L2 mutation and a better definition of DPY19L2 deletion breakpoints leading to globozoospermia. Mol Hum Reprod. 2016;22(1):35–45. doi: 10.1093/molehr/gav061. [DOI] [PubMed] [Google Scholar]
- 11.Okutman O, Muller J, Baert Y, Serdarogullari M, Gultomruk M, Piton A, et al. Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet. 2015;24(19):5581–8. doi: 10.1093/hmg/ddv290. [DOI] [PubMed] [Google Scholar]
- 12.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13(4):246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirrello O, Machev N, Schimdt F, Terriou P, Menezo Y, Viville S. Search for mutations involved in human globozoospermia. Hum Reprod. 2005;20(5):1314–8. doi: 10.1093/humrep/deh799. [DOI] [PubMed] [Google Scholar]
- 14.Kuentz P, Vanden Meerschaut F, Elinati E, Nasr-Esfahani MH, Gurgan T, Iqbal N, et al. Assisted oocyte activation overcomes fertilization failure in globozoospermic patients regardless of the DPY19L2 status. Hum Reprod. 2013;28(4):1054–61. doi: 10.1093/humrep/det005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 18 kb)
(DOCX 24 kb)
(DOCX 23 kb)
(DOCX 27 kb)
(DOCX 17 kb)
GST pull-down. GST-SPATA16 fusion protein (lane 1) and control GST protein (lane 2) were subjected to pull-down reaction in the presence of cell extract derived from adult mice testes. The samples were fractionated in SDS-PAGE and stained with silver. (GIF 75 kb)
Linkage analysis. (A) SNP array results revealing a unique homozygous region shared between the brothers of the three families. The genes located in the region of 4.7 Mb on the chromosome eight are represented in the table below it. (B) SNP array results showing the two brothers of family B sharing a homozygous region for IRGC locus (loc.chr19: 43,716,010 – 43,720,021). (GIF 58 kb)