Abstract
Autophagy is an essential cellular survival mechanism that is required for adaptive lymphocyte development; however, its role in innate lymphoid cell (ILC) development remains unknown. Furthermore, the conditions that promote lymphocyte autophagy during homeostasis are poorly understood. Here we demonstrate that Atg5, an essential component of the autophagy machinery, is required for the development of mature natural killer (NK) cells and group 1, 2, and 3 innate lymphoid cells (ILC). While inducible ablation of Atg5 was dispensable for the homeostasis of lymphocyte precursors and mature lymphocytes in lymphoreplete mice, we found that autophagy is induced in both adaptive and innate lymphocytes during homeostatic proliferation in lymphopenic hosts to promote their survival by limiting cell-intrinsic apoptosis. Induction of autophagy through metformin treatment following homeostatic proliferation increased lymphocyte numbers through an Atg5-dependent mechanism. These findings highlight the essential role for autophagy in ILC development and lymphocyte survival during lymphopenia.
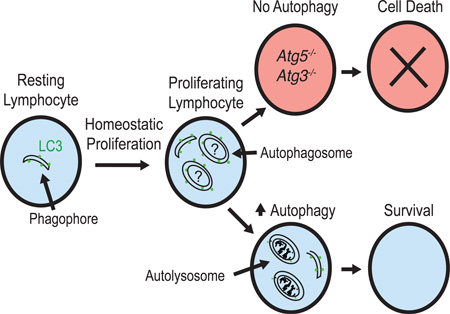
Graphical abstract
eTOC Blurb
Autophagy is an essential cellular survival mechanism induced during periods of stress. O’Sullivan et al. demonstrate that the essential autophagy gene Atg5 is required for the development and survival of all ILC lineages by limiting cell-intrinsic apoptosis.
Introduction
Apoptosis and autophagy are evolutionarily conserved pathways that often elicit contrasting cellular outcomes in response to cellular stress (Marino et al., 2014; Reed and Kroemer, 2000). Autophagy is a process in which cytosolic contents are engulfed into double-membrane vacuoles, or autophagosomes, and delivered to the lysosome for degradation (Levine et al., 2011; Marino et al., 2014). Whereas apoptosis executes cell death programs during periods of metabolic starvation or stress, autophagy can serve as an essential cellular survival mechanism by maintaining energy homeostasis through its self-catabolic activity (Levine et al., 2011; Marino et al., 2014).
Mice with conditional deletion of the gene encoding the essential autophagy machinery component Atg7 in the hematopoietic system exhibit a drastic loss of self-renewing hematopoietic stem cells (HSCs) (Mortensen et al., 2011), which utilize Atg12-dependent autophagy during cellular stress for their survival (Warr et al., 2013). In the immune system, conditional gene deletion of Atg7, Atg5, or Atg3 during early T cell development (Jia and He, 2011; Pua et al., 2007; Pua et al., 2009), Atg7 or Atg5 in the hematopoietic system (Miller et al., 2008; Mortensen et al., 2010), and Atg5 or Atg7 during invariant natural killer T (iNKT) cell development (Pei et al., 2015; Salio et al., 2014) results in diminished numbers of mature peripheral T, B, and iNKT cells that display increased apoptosis and dysfunctional organelle homeostasis, suggesting that autophagy is continuously utilized as a maintenance mechanism to promote adaptive lymphocyte development and homeostasis. Although these studies underpin the pro-survival role of autophagy during HSC and adaptive lymphocyte development and homeostasis, deletion of Atg7 in the hematopoietic system, or conditional deletion of Atg7 in the myeloid compartment does not negatively impact the development of innate immune cells such as macrophages, dendritic cells, or neutrophils (Bhattacharya et al., 2015; Mortensen et al., 2010), challenging the hypothesized role of autophagy as a constitutive pro-survival intracellular quality control mechanism in all leukocytes.
The family of innate lymphocytes consist of mature NK cells (mNK), group 1 ILCs (ILC1s), group 2 ILCs (ILC2s), and group 3 ILCs (ILC3s) including lymphoid-tissue inducer (LTi) cells (Artis and Spits, 2015). Although ILCs do not undergo RAG-dependent somatic rearrangement of antigen receptors (Spits et al., 2013), they share similar cytokine signaling and transcription factor requirements for their development, as well as functional attributes, with their adaptive T helper cell counterparts (Artis and Spits, 2015; De Obaldia and Bhandoola, 2015; Sun and Lanier, 2011; Verykokakis et al., 2014). Because it is not known whether autophagy is induced or necessary for the development of the ILC lineage, we investigated the impact of Atg5 in regulating the development and homeostasis of innate lymphocyte populations using genetic ablation of Atg5 at distinct developmental checkpoints.
Results and Discussion
Atg5 is required for ILC development
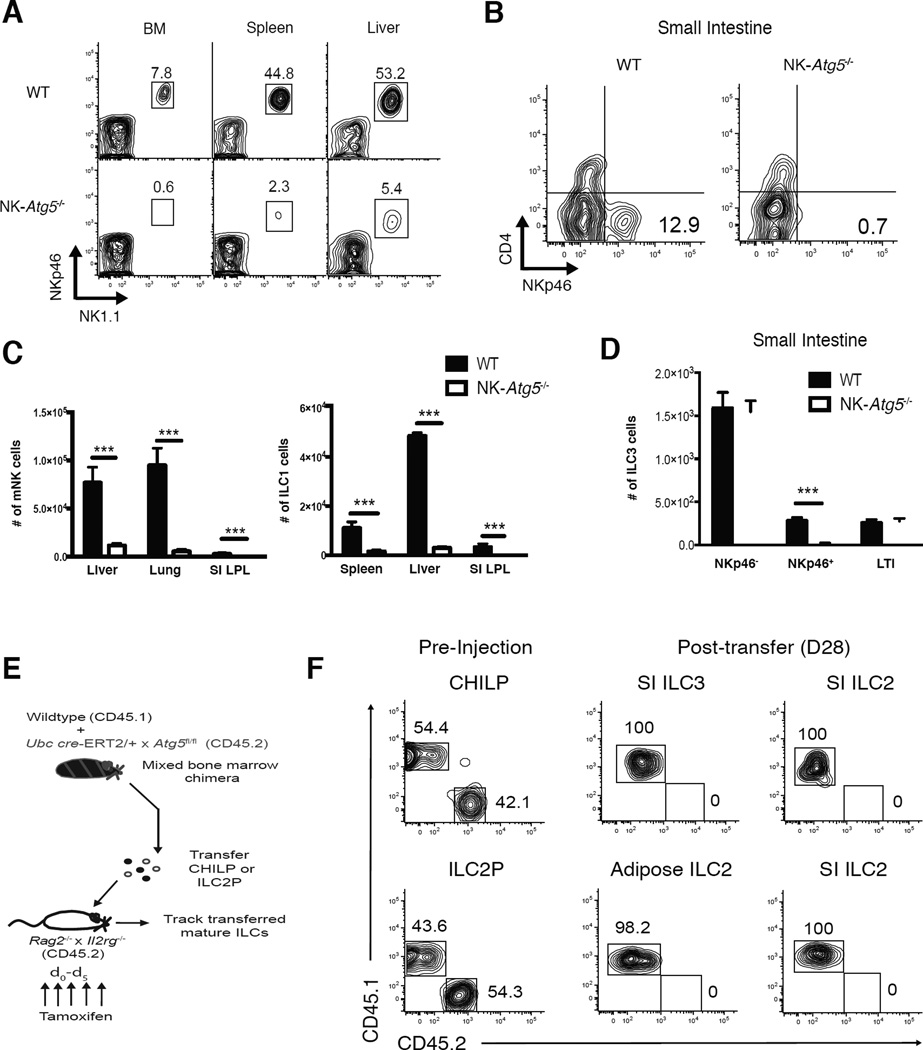
In order to demonstrate the physiological importance of autophagy in group 1 and certain group 3 ILCs, we generated mice with NKp46+ cell-specific deletion of the essential autophagy machinery component Atg5 (NKp46iCre × Atg5fl/fl), hereafter referred to as “NK-Atg5−/− mice”. Analysis of the peripheral organs of NK-Atg5−/− mice revealed a pronounced reduction in the frequency (Fig. 1A,B) and absolute numbers (Fig. 1C,D) of mature NK (mNK), ILC1, and NKp46+ ILC3 cells compared to iCre-negative Atg5fl/fl littermate mice (called ‘WT’ in this figure). Peripheral T and B cells were found at normal numbers in NK-Atg5−/− mice (data not shown). We found a marked reduction in the percentage (Fig. S1A) and number (Fig. S1B) of both iNK (NKp46+DX5− NK1.1+ Eomes+) and NKp46+DX5− NK1.1+ Eomes− cells in the bone marrow of NK-Atg5−/− mice compared to littermate controls, whereas the numbers of “refined” NK progenitor (rNKP) cells were unaffected, consistent with the lack of NKp46 surface expression on these early precursor cells (Narni-Mancinelli et al., 2011). Because of the drastically reduced peripheral mNK cell numbers, NK-Atg5−/− mice quickly succumbed to mouse cytomegalovirus (MCMV) infection (Fig. S1C), and displayed higher viral titers when compared to littermate controls (Fig. S1D). Together these results demonstrate the critical role for Atg5 in the development of NKp46+ ILCs in the bone marrow and periphery.
Figure 1. Atg5 is essential for innate lymphocyte development.
(A) Representative flow cytometric plots of lineage marker negative (Lin−) NKp46+NK1.1+ cells and (B) Lin−NK1.1−CD127+CD90.2+Rorγt+ (ILC3s) in indicated peripheral organs of NKp46iCre × Atg5fl/fl mice (“NK-Atg5−/−”) or littermate Atg5fl/fl controls (called “WT”) (C) Absolute numbers of mature NK cells (mNK; Lin−NK1.1+NKp46+DX5+Eomes+), ILC1 (Lin−NK1.1+NKp46+DX5−Eomes−), and (D) ILC3s in indicated peripheral organs of WT or littermate NK-Atg5−/− mice (E,F) WT mice (CD45.1 × CD45.2) were lethally irradiated and reconstituted with an equal number of bone marrow cells from WT (CD45.1) and UbcCre-ERT2 × Atg5fl/fl (CD45.2) mice. (E) Schematic of experiment. (F) Both WT (CD45.1) and UbcCre-ERT2 × Atg5fl/fl (CD45.2) CHILP (Lin−NK1.1−DX5−FLT3−CD127+α4β7+CD25−) and ILC2P (Lin−NK1.1−DX5−FLT3−CD127+α4β7+CD25+) cells were adoptively transferred into sub-lethally irradiated Rag2−/− × Il2rg −/− hosts and injected i.p. with 1mg of tamoxifen daily for 5 days. After 28 days, the SI and adipose tissue were harvested from recipient mice and analyzed for mature ILC2 (Lin−NK1.1−DX5−CD127+CD90.2+GATA3+KLRG1+) and ILC3 cells. (Lin− refers to TCRβ−CD19−CD3ε−Ly6G−TER119−TCRγδ− cells). Data are representative of two independent experiments, with n=5 per time point. Samples were compared using an unpaired, two-tailed Student’s t test, and data presented as the mean ± s.e.m. (***p < 0.001). See also Figure S1.
After observing a near complete loss of NKp46+ ILCs in NK-Atg5−/− mice, we investigated whether Atg5 may also regulate the development or homeostasis of additional ILC populations. Mature ILC2s and ILC3s can be derived from a common helper ILC progenitor (CHILP) in the bone marrow (Constantinides et al., 2014; Klose et al., 2014; Yang et al., 2015), whereas mature ILC2 cells can be exclusively derived from a ILC2 progenitor (ILC2P) population in the bone marrow upon adoptive transfer into lymphopenic hosts (Hoyler et al., 2012). To test if Atg5-deficiency also impacted the development of mature ILC2 and ILC3s, specifically from their respective committed progenitors, we generated mice with inducible deletion of Atg5 (UbcCre-ERT2 × Atg5fl/fl) upon administration of 4-hydroxytamoxifen. We then generated mixed bone marrow chimeric (mBMC) mice by co-adoptively transferring equal numbers of WT (CD45.1) and UbcCre-ERT2 × Atg5fl/fl (CD45.2) mouse bone marrow cells into lethally-irradiated WT (CD45.1 × CD45.2) hosts (hereafter referred to as WT:i-Atg5 mBMC) (Fig. 1E). 8 weeks following bone marrow transplantation, bone marrow from WT:i-Atg5 mBMC was harvested and CHILP or ILC2P populations were sorted to high purity (Fig. S1E) and adoptively transferred into irradiated Rag2−/− × Il2rg−/− hosts (Fig. 1E). At the time of sorting, reconstitution by donor populations in WT:i-Atg5 mBMC was comparable (Fig. 1F and data not shown). Following tamoxifen treatment of recipient mice, we observed that adoptively transferred Atg5-deficient CHILPs failed to generate mature ILC2 and ILC3s in the small intestine, while Atg5-deficient ILC2Ps failed to generate mature ILC2s in peripheral organs (Fig. 1F). In contrast, mature WT ILC2 and ILC3 populations (CD45.1) were observed in peripheral organs of recipient mice 4 weeks after transfer (Fig. 1F), highlighting the importance of Atg5 in both ILC2 and ILC3 development in vivo.
Lymphocytes induce autophagy in lymphopenic hosts
Recent studies have shown that autophagy is induced in antigen-specific T, B, and NK cells following clonal proliferation in response to viral infection to mediate cell survival (Chen et al., 2014; O'Sullivan et al., 2015; Puleston et al., 2014; Xu et al., 2014). In mice devoid of mature lymphocytes, adoptively transferred T and NK cells rapidly proliferate and persist long-term in peripheral organs in a process known as homeostatic proliferation (Jamieson et al., 2004; Prlic et al., 2003; Ranson et al., 2003; Sun et al., 2011). Similar to splenic NK cells, we observed that liver ILC1s could also undergo homeostatic proliferation and persist for at least a month following transfer into Rag2−/− × Il2rg−/− hosts (Fig. S2A–C). Since immature adaptive and innate lymphocytes undergo proliferation during development to generate a mature compartment in the periphery (Hoyler et al., 2012; Kim et al., 2002; Koch and Radtke, 2011; Yang et al., 2015), we hypothesized that autophagy could also be induced following homeostatic proliferation during lymphopenia.
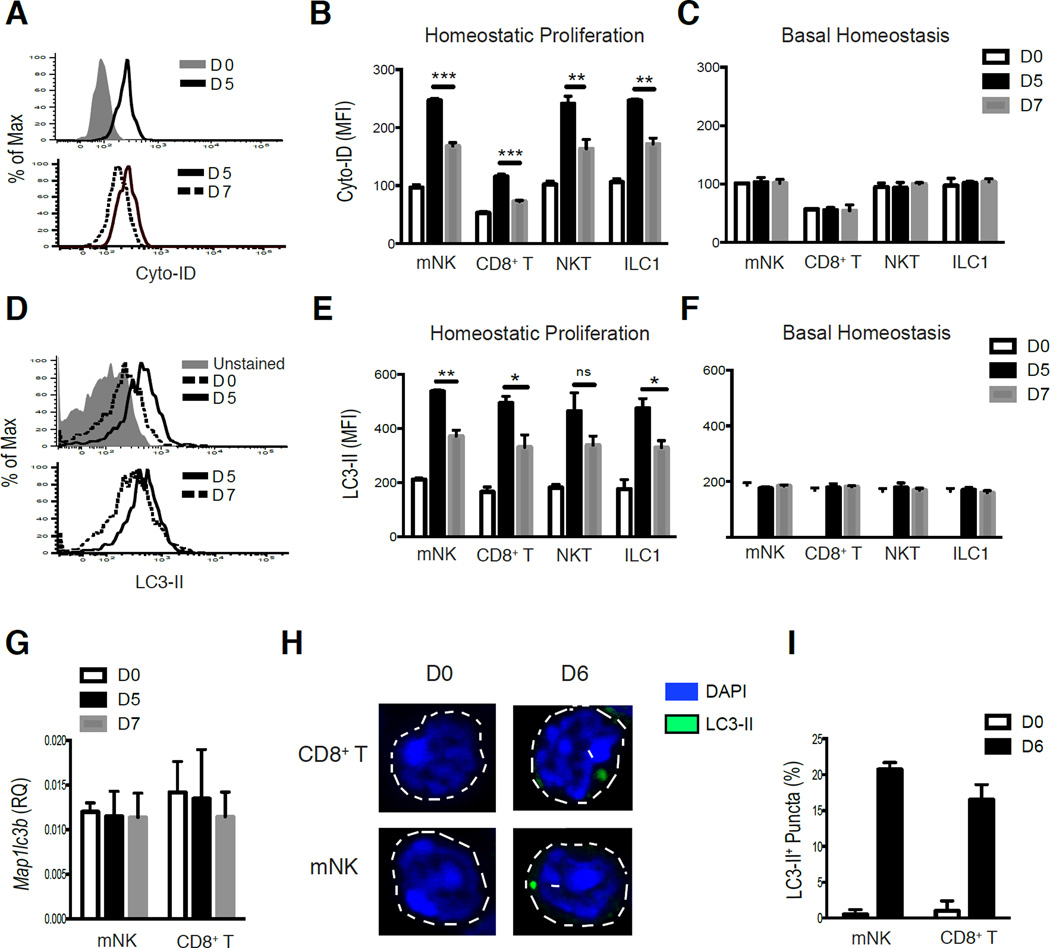
To investigate whether mature adaptive and innate lymphocytes induce autophagy during homeostatic expansion, we utilized Cyto-ID staining (which labels both autophagosomes and autolysosomes) (O'Sullivan et al., 2015; Puleston et al., 2014) in parallel with LC3-GFP transgenic mice (Mizushima et al., 2004) to assess autophagic activity by measuring the degradation of autophagosomes and LC3-II by flow cytometric analysis (LC3-II is the lipidated form of LC3-I that is selectively incorporated into the elongation membrane of early autophagosomes and subsequently degraded following lysosomal fusion with autophagosomes) (Eng et al., 2010; Klionsky et al., 2016). We found that Cyto-ID and LC3-II staining were both increased in adoptively transferred lymphocytes on d5 following transfer into Rag2−/− × Il2rg−/− hosts (homeostatic proliferation) compared to resting lymphocytes (Figures 2A,B and 2D,E), whereas adoptively transferred lymphocytes recovered from WT lymphoreplete hosts (basal homeostasis) did not display increased staining of LC3-II or Cyto-ID (Fig. 2C,F). Accumulation of LC3-II and autophagosomes/autolysosomes during the first five days would be consistent with either increased production of LC3 protein, induction of autophagy, or the inhibition of autophagosome degradation by lysosomes. However, we did not observe an increase in Map1lcb (LC3) transcripts on d5 following homeostatic proliferation in sorted mNK or CD8+ T cells compared to resting cells (Fig. 2G), suggesting that increased LC3 protein is not due to elevated transcription in lymphocytes during early homeostatic proliferation. In contrast, we observed that adaptive and innate lymphocytes collectively displayed a reduction in Cyto-ID staining (Fig. 2A,B) with a concomitant loss of LC3-II fluorescence (Fig. 2D,E) between day 5 and 7 after transfer into Rag2−/− × Il2rg−/−, but not WT hosts (Fig. 2C,F), indicating that lymphocytes may induce autophagy following homeostatic proliferation but not during basal homeostasis. In support of this hypothesis, adoptively transferred LC3-GFP transgenic lymphocytes did not alter the amounts of LC3 transcripts at these time points (Fig. 2G), and we observed the presence (Fig. 2H) and higher frequency (Fig. 2I) of LC3-II puncta by confocal microscopy in lymphocyte populations following homeostatic proliferation compared to resting cells. Together, these results suggest that lymphocytes induce autophagy following homeostatic proliferation.
Figure 2. Autophagy is induced in lymphocytes following homeostatic expansion.
(A,B) WT (CD45.1) liver lymphocytes were adoptively transferred into Rag2−/− × Il2rg−/− (homeostatic proliferation) (A) Representative histograms and (B) quantification of Cyto-ID mean fluorescence intensity (MFI) in adoptively transferred NK, ILC1, NKT, and CD8+ T cells harvested from livers of recipient mice on day 5 and 7 after transfer. (C) WT (CD45.1) liver lymphocytes were adoptively transferred into WT (CD45.1 × CD45.2) recipients (basal homeostasis) and quantified for Cyto-ID staining on day 5 and 7 after transfer from recipient livers. (D,E) LC3-GFP (CD45.2) transgenic liver lymphocytes were adoptively transferred into Rag2−/− × Il2rg−/− recipients, and livers were harvested on either day 5 or 7 post-transfer. (D) Representative histograms and (E) quantification of LC3-II MFI for indicated adoptively transferred cell subsets. (F) LC3-GFP transgenic liver lymphocytes were adoptively transferred into WT (CD45.1 × CD45.2) recipients and quantified for LC3-II staining for each indicated cell subset on day 5 and 7 after transfer from recipient livers (G–I) LC3-GFP transgenic splenocytes were adoptively transferred into Rag2−/− × Il2rg−/− recipients. (G) mNK and CD8+ T cells were sorted at the indicated time points from recipient mice, and analyzed for Map1lc3b (LC3) mRNA levels by qRT-PCR (n=3 for each time point) (H) Confocal microscopy (original magnification, 63×) of sort-purified mNK or CD8+ T cells and (I) quantification of LC3-II+ puncta on either day 0 or 6 post-transfer (presented as the frequency of cells with more than one LC3-II+ punctum). Data are representative of two independent experiments, with n=3–4 per time point. Samples were compared using an unpaired, two-tailed Student’s t test, and data presented as the mean ± s.e.m. (*p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant).
Autophagy is required for lymphocyte survival following homeostatic proliferation, but dispensable in lymphoreplete mice
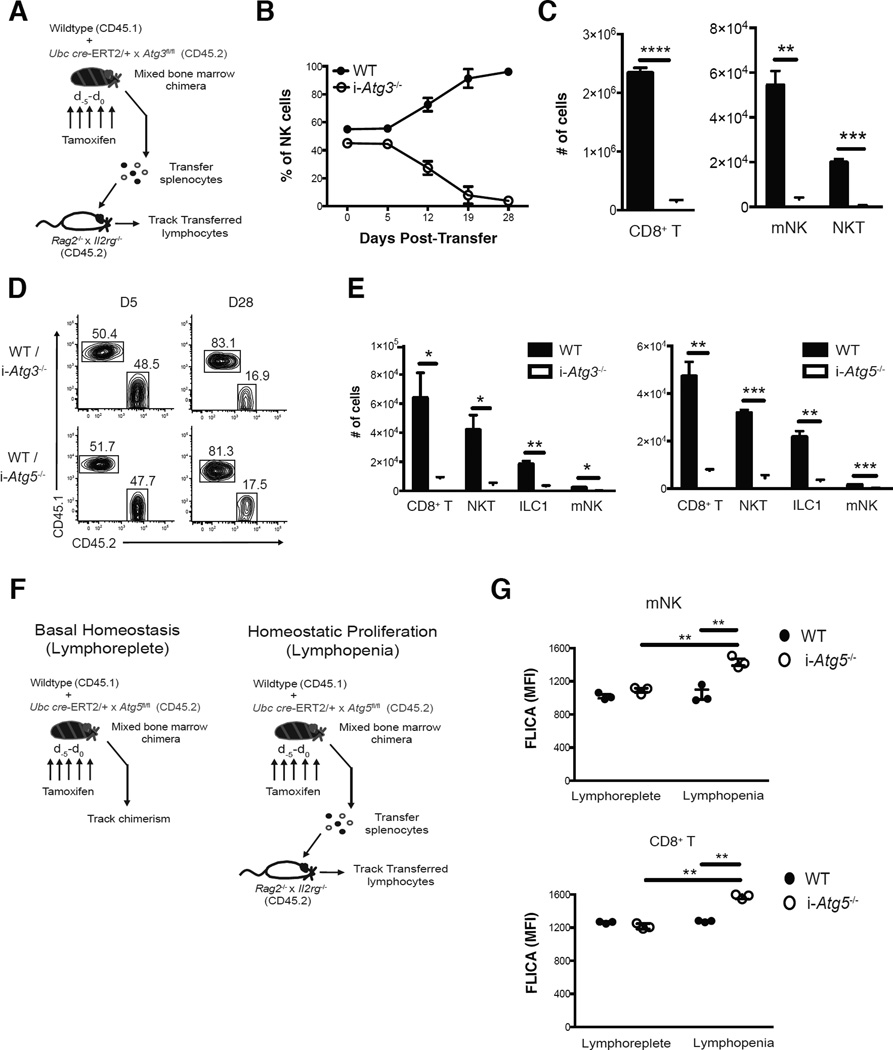
To study the functional relevance of increased autophagic activity in lymphocytes during lymphopenia without the confounding developmental defects, we utilized mice with either inducible deletion of the essential autophagosome machinery component Atg5 (UbcCre-ERT2 × Atg5fl/fl) or Atg3 (UbcCre-ERT2 × Atg3fl/fl) (Jia and He, 2011). WT (CD45.1) and UbcCre-ERT2 × Atg3fl/fl (CD45.2) mBMC mice were generated and treated with tamoxifen for 5 days prior to adoptive transfer of splenocytes into Rag2−/− × Il2rg−/− hosts (Fig. 3A). Whereas inducible deletion of Atg3 (i-Atg3−/−) resulted in similar expansion of adoptively transferred i-Atg3−/− and WT NK and CD8+ T cells after transfer, we observed a drastic reduction in the percentage of i-Atg3−/− NK and CD8+ T cells during later time points (Figures 3B and S2D), with a significant loss in the absolute numbers of indicated i-Atg3−/− lymphocytes in peripheral organs compared to WT controls on day 28 after transfer (Figures 3C and S2E). Inducible deletion of either Atg3 or Atg5 resulted in significant reductions in percentage (Fig. 3D) and absolute number (Fig. 3E) of liver lymphocyte lineages on day 28 after adoptive transfer. Furthermore, the persistence of both i-Atg5−/− ILC3 and ILC2s was greatly diminished compared to WT controls following either interleukin (IL)-7 or IL-33-induced proliferation in WT:i-Atg5 mBMC mice, respectively, although cytokine treatment resulted in similar expansion of these populations (Fig. S3A–I).
Figure 3. Autophagy is critical for lymphocyte survival following homeostatic proliferation.
(A–C) WT:i-Atg3 mixed bone marrow chimeras were injected i.p. with 1mg of tamoxifen (i-Atg3−/−) daily for 5 days and splenocytes were adoptively transferred into Rag2−/− × Il2rg−/− hosts. (B) Percentages of adoptively transferred WT or i-Atg3−/− NK cells in the blood at indicated time points post-transfer and (C) absolute numbers of indicated lymphocytes from the spleen of recipients on d28 post-transfer. (D,E) WT:i-Atg5 or WT:i-Atg3 mixed bone marrow chimeras were injected i.p. with 1mg of tamoxifen (i-Atg3−/−, or i-Atg5−/−) daily for 5 days and liver lymphocytes were adoptively transferred into Rag2−/− × Il2rg−/− hosts. (D) Representative plots show adoptively transferred ILC1 populations on day 5 and day 28 post-transfer from the liver and (E) absolute numbers of indicated lymphocyte populations harvested from the liver on day 28 post-transfer. (F) Schematic of experiment. Briefly, WT:i-Atg5 mixed bone marrow chimeras were injected i.p. with 1mg of tamoxifen (i-Atg5−/−) daily for 5 days and lymphocyte chimerism was subsequently measured in the blood (basal homeostasis) or splenocytes were adoptively transferred into Rag2−/− × Il2rg−/− hosts (homeostatic proliferation). (G) Quantification of FLICA MFI of indicated lymphocytes in the blood on d9 for each cohort. Data are representative of two independent experiments, with n=3–4 per cohort. Samples were compared using an unpaired, two-tailed Student’s t test, and data presented as the mean ± s.e.m. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). See also Figures S2,S3, and S4.
Only mature innate and adaptive lymphocytes require autophagy following homeostatic or cytokine-induced proliferation, because we did not observe a loss of Atg5-deficient lymphocyte precursors in the bone marrow (Fig. S4A) or mature lymphocytes in the periphery (Fig. S4B,C) in tamoxifen treated WT:i-Atg5 mBMC mice after 3 weeks of basal homeostasis. Furthermore, these results were not due to decreased proliferation of Atg5-deficient lymphocytes, because similar Ki67 staining was observed between adoptively transferred i-Atg5−/− and WT mNK and CD8+ T cells during homeostatic proliferation and both ILC2/ILC3 populations during cytokine-induced proliferation (Fig. S4D,E and data not shown). Instead, the observed survival defect in Atg5-deficient lymphocytes was likely due to enhanced apoptosis following homeostatic expansion, as i-Atg5−/− mNK and CD8+ T cells both displayed increased FLICA staining on d9 following homeostatic proliferation compared to co-transferred WT controls that was not observed in lymphoreplete hosts (Fig. 3F,G). Together, these data indicate that autophagy is essential for the survival of lymphocytes by shielding them against apoptosis following homeostatic proliferation, but not during basal homeostasis in lymphoreplete mice.
Induction of autophagy by metformin enhances lymphocyte survival following homeostatic proliferation
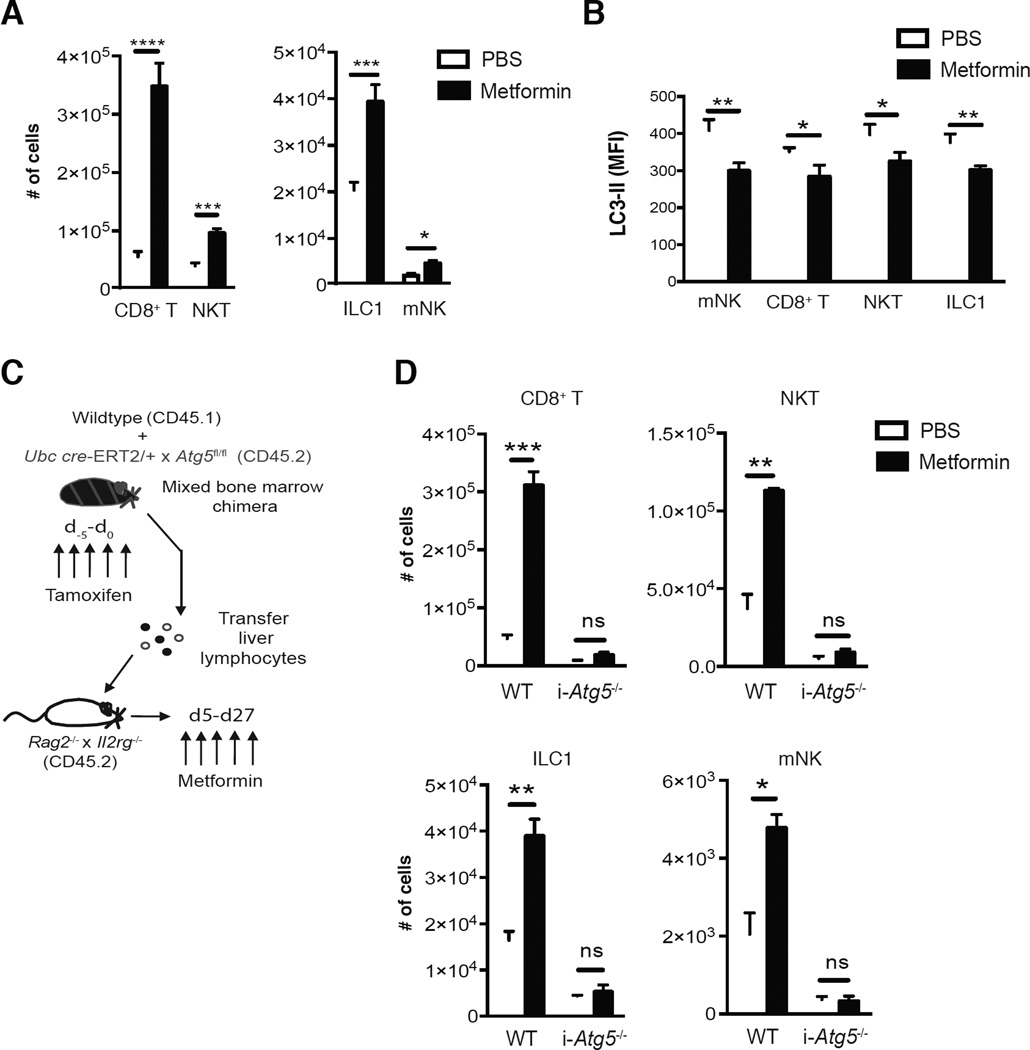
In order to address whether augmenting autophagic activity in lymphocytes by pharmacological means could promote their survival during lymphopenia, we adoptively transferred lymphocytes from LC3-GFP mice into Rag2−/− × Il2rg−/− hosts and treated recipient mice with metformin, an AMPK activator that can induce autophagy through inhibition of mTOR (Kim et al., 2011). Treatment of recipient mice with metformin dramatically increased the absolute numbers of CD8+ T cells, NKT cells, mNK cells, and ILC1 on day 28 after transfer (Fig. 4A). Metformin administration increased autophagic activity in these transferred lymphocytes following homeostatic proliferation, as we observed a reduction in LC3-II in indicated lymphocytes from metformin treated mice compared to PBS treated controls (Fig. 4B). To test whether metformin treatment enhanced survival of lymphocytes in an autophagy-dependent mechanism, lymphocytes from tamoxifen-treated WT:i-Atg5−/− mBMC were transferred into Rag2−/− × Il2rg−/− hosts and metformin was administered daily from day 5 to 28 after transfer (Fig. 4C). Indeed, the increase in these lymphocytes from metformin-treated mice was Atg5-dependent, as i-Atg5−/− lymphocytes cells failed to demonstrate enhanced survival (Fig. 4D). These results support the hypothesis that increasing autophagic activity through metformin treatment is sufficient to enhance both adaptive and innate lymphocyte survival following lymphopenia-driven expansion.
Figure 4. Induction of autophagy through metformin treatment enhances lymphocyte survival following homeostatic proliferation in an Atg5-dependent manner.
(A,B) LC3-GFP transgenic liver lymphocytes were adoptively transferred Rag2−/− × Il2rg−/− (CD45.2) recipients and either treated with 200µg of metformin or control PBS i.p. daily from day 5 to day 27 post-transfer. (A) Absolute numbers on day 28 post-transfer and (B) LC3-II MFI of indicated lymphocytes on day 9 post-transfer from harvested recipient livers of each indicated cohort. (C,D) WT:i-Atg5 mixed bone marrow chimeras were injected i.p. with 1mg of tamoxifen (i-Atg5−/−) daily for 5 days and liver lymphocytes were adoptively transferred into Rag2−/− × Il2rg−/− hosts that either received 200µg of metformin or control PBS daily from day 5 to day 27 post-transfer. (D) Absolute numbers of indicated lymphocytes on day 28 post-transfer from livers of each indicated cohort. Data are representative of two independent experiments, with n=3–4 per cohort. Samples were compared using an unpaired, two-tailed Student’s t test, and data presented as the mean ± s.e.m. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
In summary, our study demonstrates a critical role for the essential autophagosome machinery component Atg5 in the development of all innate lymphoid cell subsets. Although lymphocytes may block the degradation of autophagsomes early during homeostatic expansion and then transiently relive this inhibition later during homeostatic proliferation, our evidence suggests that autophagy is induced during this process, and that autophagy is necessary and sufficient for both adaptive and innate lymphocyte survival following lymphopenia-driven proliferation by limiting cell-intrinsic apoptosis. These results provide a general molecular mechanism for how lymphocytes are able to persist long-term in lymphopenic hosts, and support the hypothesis that autophagy is necessary for lymphocyte survival following proliferation during ontogeny. Although autophagy has been implicated in preventing mature T cell apoptosis by promoting organelle turnover in vitro (Jia and He, 2011), our results indicate that autophagy is not induced in mature lymphocytes during basal homeostasis to limit apoptosis, because Atg5 is dispensable for the survival of mature lymphocytes and bone marrow precursors during basal homeostasis in lymphoreplete mice. These results suggest that autophagy is not used continuously as a survival mechanism in mature lymphocytes, and is only utilized to inhibit apoptosis following periods of cellular differentiation or stress, such as clonal proliferation in response to viral infection (Chen et al., 2014; O'Sullivan et al., 2015; Puleston et al., 2014; Xu et al., 2014), cytokine deprivation (Warr et al., 2013), or lymphopenia and cytokine-driven expansion.
How autophagy enables lymphocytes to evade apoptosis during development and following homeostatic proliferation remains unknown. Atg3-deficient T cells, Atg7 and Atg5-deficient iNKT cells, and Atg7-deficient HSCs all displayed higher mitochondrial mass than littermate controls (Jia and He, 2011; Mortensen et al., 2011; Pei et al., 2015; Salio et al., 2014), suggesting that removal of damaged mitochondria through mitophagy may promote the survival of lymphocytes during development. Although we have previously demonstrated that antigen-specific NK cells rapidly remove dysfunctional mitochondria through BNIP3 and BNIP3L-mediated mitophagy following MCMV infection (O'Sullivan et al., 2015), Bnip3 is not required for T, NKT, or mNK survival during development or following homeostatic expansion (T.E.O. and J.C.S., unpublished observations). Furthermore, lymphocytes undergoing homeostatic expansion do not reduce their mitochondrial content 28 days after transfer into lymphopenic hosts (T.E.O. and J.C.S., unpublished observations). Together, these observations suggest that mitophagy may be dispensable for lymphocyte development and survival during homeostasis. Future work will be necessary to elucidate the molecular mechanisms by which autophagy regulates lymphocyte survival during lymphopenia and whether these findings will have clinical significance during immune reconstitution following bone marrow transplantation.
METHODS
Mice
Mice were bred at Memorial Sloan Kettering Cancer Center in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC). The following strains were used in this study: C57BL/6 (CD45.2), B6.SJL (CD45.1), Rag2−/− × Il2rg−/−, Nkp46iCre (Narni-Mancinelli et al., 2011), UbERT2-Cre × Atg3fl/fl (Jia and He, 2011), LC3-GFP transgenic (Mizushima et al., 2004), Atg5fl/fl, UbERT2-Cre. Mixed bone marrow chimeric (mBMC) mice were generated as previously described (Sun et al., 2009). Experiments were conducted using age- and gender-matched mice in accordance with approved institutional protocols.
Virus infection
MCMV (Smith strain) was passaged serially through BALB/c hosts two times, and viral stocks were prepared by using a dounce homogenizer to dissociate the salivary glands of infected mice 3 weeks after infection. Experimental mice were infected by intraperitoneal (i.p.) injection with 2.5 × 104 PFU.
Adoptive transfer of mature and progenitor innate lymphocytes
In adoptive transfer experiments, splenocytes or liver lymphocytes from indicated mBMC mice, or liver lymphocytes from LC3-GFP transgenic mice were injected intravenously (i.v.) into Rag2−/− × Il2rg−/− mice. In certain experiments, lymphocyte populations were pre-incubated with Cell Trace Violet (Life Technologies) according to the manufacturers instructions, and analyzed on day 5 after transfer. In ILC progenitor adoptive transfer experiments, bone marrow from indicated mBMC mice was stained with surface antibodies and ~1,000 CHILP or ~ 3,000 ILC2P populations were sorted to >95% purity and adoptively transferred into individual irradiated Rag2−/− × Il2rg−/− recipients. Mature ILC populations were harvested from these mice on day 28 after transfer. In some experiments, Rag2−/− × Il2rg−/− recipient mice or mBMC mice were injected i.p. with 1 mg/day of hydroxytamoxifen (4-OHT) dissolved in corn oil (Sigma), or corn oil alone as a control for five consecutive days. In certain adoptive transfer experiments, recipient mice were injected i.p. with PBS or 200 µg/day metformin (Sigma) from day 5 to 9, or 5 to 27 after transfer and tamoxifen treatment. In certain experiments, mBMC mice were injected i.p. with 1 mg/day of tamoxifen or control corn oil for five consecutive days and then received 3 i.p injections of either 15µg anti-IL-7 complex (clone M25, Bio × Cell)/ 2.5µg rhIL-7 (eBioscience) every other day to expand ILC3s (Abt et al., 2015), or 500ng rmIL-33 (Biolegend) every other day to expand ILC2s (Molofsky et al., 2015).
Flow cytometry and cell sorting
Cell surface staining was performed using the following fluorophore-conjugated antibodies (BD Biosciences, eBioscience, BioLegend, Tonbo, and R&D Systems): NK1.1 (PK136), CD11b (M1/70), CD19 (ID3), CD27 (LG.3A10), CD49b/DX5 (DX5), KLRG1 (2F1), NKp46 (29A1.4), CD45.1 (A20), CD45.2 (104), CD8α (53-6.7), TCRβ (H57-597), Eomes (Dan11mag), FLT3 (A2F10), a4B7 (DATK32), CD127 (A7R34), CD3e (17A2), TCRγδ (GL3), Ly6G (1A8), CD25 (PC61), GATA3 (TWAJ), Rorγt (B2D), CD122 (TM-B1), CD90.2 (30-H12), TER119 (TER-119). Intranuclear staining was performed with the Foxp3 transcription factor staining buffer set (eBioscience). Pan-caspase staining was carried out using the carboxyfluorescein FLICA in vitro poly caspase kit (Immunochemistry Technologies). Liver lymphocytes were harvested and stained with cell surface antibodies, and then incubated with 1:400 Cyto-ID autophagy detection reagent (Enzo Life Sciences) for 30 min at 37°C to measure autophagosomes. LC3-GFP transgenic liver lymphocytes or splenocytes were harvested and stained with cell surface antibodies, and then permeabilized using a selective permeabilization reagent (FlowCellect Autophagy LC3 kit, Millipore) to remove cytosolic LC3-I. Autophagosome-associated LC3-II was then stained using an anti-GFP Alexa Fluor 488 antibody (Invitrogen). Flow cytometry and cell sorting were performed on the LSR II and Aria II cytometers (BD Biosciences), respectively. Data were analyzed with FlowJo software (Tree Star).
Cellular imaging
LC3-GFP transgenic mNK or CD8+ T cells were sort purified and then permeabilized using a selective permeabilization reagent (FlowCellect Autophagy LC3 kit, Millipore) to remove cytosolic LC3-I. Autophagosome-associated LC3-II was then stained using an anti-GFP Alexa Fluor 488 antibody (Invitrogen). Cells were then analyzed on a Leica SP8 confocal microscope at the MSKCC Molecular Cytology Core. Images were acquired at a magnification of 63×. Spot-counting analysis was used for evaluation of LC3-II puncta+ cells in each cohort.
Quantitative realtime PCR
Sorted NK or CD8+ T cells were lysed in Tri-Reagent (Ambion). RNA purification and cDNA synthesis were carried out with the Qiagen RNeasy kit (with on-column DNase I treatment), and MuLV reverse transcriptase and oligo(dT)16 primers (Applied Biosystems). iQ Sybr Green SuperMix (BioRad) was used for qRT-PCR. Data were normalized to that for Actb and expressed as relative target abundance via the ΔΔCt method, where Ct (threshold cycle) is the cycle number at which the amplification curve intersects the threshold value. Map1lc3b forward primer: TTATAGAGCGATACAAGGGGGAG, Map1lc3b reverse primer: CGCCGTCTGATTATCTTGATGAG. Actb forward primer: TGCGTGACATCAAAGAGAAG, Actb reverse primer: CGGATGTCAACGTCACACTT.
Statistical analyses
For graphs, data are shown as mean ± s.e.m. and, unless otherwise indicated, statistical differences were evaluated using a two-tailed unpaired Student's t-test, assuming equal sample variance. P < 0.05 was considered significant. Graphs were produced and statistical analyses were performed using GraphPad Prism.
Supplementary Material
Highlights.
Atg5 is required for innate lymphocyte development
Adaptive and innate lymphocytes induce autophagy during homeostatic expansion
Autophagy is essential for lymphocyte survival during lymphopenia
Metformin increases autophagy to enhance lymphocyte survival in lymphopenic hosts
Acknowledgments
We thank members of the Sun lab for technical support and experimental assistance, insightful comments, and helpful discussions. You-Wen He, Eric Vivier, Liang Deng, and Noboru Mizushima provided mice, reagents, and expertise critical to this study. T.E.O. was supported by the American Cancer Society. M.R. was supported by a fellowship from the DAAD (Germany). T.L.G. is supported by a F31 award from the National Institute of Allergy and Infectious Diseases (AI114019). G.W.D. was supported by a grant from the National Institutes of Health (HL59888). J.C.S. was supported by the Ludwig Center for Cancer Immunotherapy, the Cancer Research Institute, and grants from the National Institutes of Health (AI100874 and P30CA008748). The MSKCC Cytology core is supported by core grant (P30 CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no financial conflicts of interest.
Author Contributions
T.E.O. and J.C.S. designed the study; M.O. provided critical discussion and interpretation of results; G.W.D. provided mice; T.E.O. and C.D.G. performed cell sorting; M.R., O.W., and T.L.G. aided with tissue processing; T.E.O. performed all additional experiments; T.E.O. and J.C.S. wrote the manuscript.
References
- Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, Ling L, Leiner I, Pamer EG. Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell host & microbe. 2015;18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Wei Q, Shin JN, Abdel Fattah E, Bonilla DL, Xiang Q, Eissa NT. Autophagy Is Required for Neutrophil-Mediated Inflammation. Cell reports. 2015;12:1731–1739. doi: 10.1016/j.celrep.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Chen M, Hong MJ, Sun H, Wang L, Shi X, Gilbert BE, Corry DB, Kheradmand F, Wang J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nature medicine. 2014;20:503–510. doi: 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annual review of immunology. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- Eng KE, Panas MD, Karlsson Hedestam GB, McInerney GM. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 2010;6:634–641. doi: 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson AM, Isnard P, Dorfman JR, Coles MC, Raulet DH. Turnover and proliferation of NK cells in steady state and lymphopenic conditions. Journal of immunology. 2004;172:864–870. doi: 10.4049/jimmunol.172.2.864. [DOI] [PubMed] [Google Scholar]
- Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. Journal of immunology. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, Yokoyama WM. In vivo developmental stages in murine natural killer cell maturation. Nature immunology. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Koch U, Radtke F. Mechanisms of T cell development and transformation. Annual review of cell and developmental biology. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nature reviews. Molecular cell biology. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, Virgin HWt. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular biology of the cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Van Gool F, Liang HE, Van Dyken SJ, Nussbaum JC, Lee J, Bluestone JA, Locksley RM. Interleukin-33 and Interferon-gamma Counter-Regulate Group 2 Innate Lymphoid Cell Activation during Immune Perturbation. Immunity. 2015;43:161–174. doi: 10.1016/j.immuni.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of experimental medicine. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity. 2015;43:331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei B, Zhao M, Miller BC, Vela JL, Bruinsma MW, Virgin HW, Kronenberg M. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. Journal of immunology. 2015;194:5872–5884. doi: 10.4049/jimmunol.1402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic M, Blazar BR, Farrar MA, Jameson SC. In vivo survival and homeostatic proliferation of natural killer cells. The Journal of experimental medicine. 2003;197:967–976. doi: 10.1084/jem.20021847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. The Journal of experimental medicine. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. Journal of immunology. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, Cerundolo V, Townsend AR, Klenerman P, Simon AK. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife. 2014;3 doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson T, Vosshenrich CA, Corcuff E, Richard O, Muller W, Di Santo JP. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood. 2003;101:4887–4893. doi: 10.1182/blood-2002-11-3392. [DOI] [PubMed] [Google Scholar]
- Reed JC, Kroemer G. Mechanisms of mitochondrial membrane permeabilization. Cell death and differentiation. 2000;7:1145. doi: 10.1038/sj.cdd.4400777. [DOI] [PubMed] [Google Scholar]
- Salio M, Puleston DJ, Mathan TS, Shepherd D, Stranks AJ, Adamopoulou E, Veerapen N, Besra GS, Hollander GA, Simon AK, Cerundolo V. Essential role for autophagy during invariant NKT cell development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5678–E5687. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews. Immunology. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Bezman NA, Lanier LL. Homeostatic proliferation generates long-lived natural killer cells that respond against viral infection. The Journal of experimental medicine. 2011;208:357–368. doi: 10.1084/jem.20100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nature reviews. Immunology. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M, Zook EC, Kee BL. ID'ing innate and innate-like lymphoid cells. Immunological reviews. 2014;261:177–197. doi: 10.1111/imr.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nature immunology. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, Wang H, Wang X, Yu S, Zhou X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nature immunology. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.