Abstract
A promising recent development in the study of brain function is the dynamic analysis of resting-state functional MRI scans, which can enhance understanding of normal cognition and alterations that result from brain disorders. One widely used method of capturing the dynamics of functional connectivity is sliding window correlation (SWC). However, in the absence of a “gold standard” for comparison, evaluating the performance of the SWC in typical resting-state data is challenging. This study uses simulated networks (SNs) with known transitions to examine the effects of parameters such as window length, window offset, window type, noise, filtering, and sampling rate on the SWC performance. The SWC time course was calculated for all node pairs of each SN and then clustered using the k-means algorithm to determine how resulting brain states match known configurations and transitions in the SNs. The outcomes show that the detection of state transitions and durations in the SWC is most strongly influenced by the window length and offset, followed by noise and filtering parameters. The effect of the image sampling rate was relatively insignificant. Tapered windows provide less sensitivity to state transitions than rectangular windows, which could be the result of the sharp transitions in the SNs. Overall, the SWC gave poor estimates of correlation for each brain state. Clustering based on the SWC time course did not reliably reflect the underlying state transitions unless the window length was comparable to the state duration, highlighting the need for new adaptive window analysis techniques.
Keywords: Resting-state functional MRI, Functional connectivity, Sliding window correlation, Network dynamics, k-Means, States
Introduction
Resting-state functional MRI (rsfMRI) has had much success as a tool for the study of normal and disordered brain function (Rombouts et al., 2005; Sorg et al., 2007; Zang et al., 2007; Xia et al., 2013). Initially, rsfMRI analysis assumed networks in the resting-brain were stationary over the whole scan length (typically ranging from six to ten minutes), but more recently methods that examine the network connectivity as a function of time have been applied. Several studies have reported that the connectivity of these networks changes over the course of the scan (within a few seconds) and reveal a number of functional connectivity (FC) states in the brain, which can be sensitive to changes related to neurological disorders (Sakoğlu et al., 2010; Leonardi et al., 2013a, 2013b; Damaraju et al., 2014; Li et al., 2014; Ou et al., 2015). These dynamics are also linked to changes in human behavior (Kucyi et al., 2013; Thompson et al., 2013a, 2013b; Jia et al., 2014; Sadaghiani et al., 2015).
Sliding window correlation (SWC) is the simplest and most commonly used method for dynamic FC analysis and most of the dynamic FC studies use it at some point (Schulz and Huston, 2002; Chang and Glover, 2010; Kiviniemi et al., 2011; Handwerker et al., 2012; Chang et al., 2013; Hutchison et al., 2013a, 2013b; Keilholz et al., 2013; Thompson et al., 2013a, 2013b; Wilson et al., 2015). It should be noted that in this study dynamic FC refers to the dynamics of resting-state networks only and not the dynamics because of any environmental input or task. In the SWC, a temporal window of a certain size and shape is selected, and the correlation coefficient between two signals of interest within that window is computed. Afterwards the window is shifted (slided) by some offset, and the process is repeated for the entire scan length. Despite the popularity of the SWC, results are strongly dependent on window length (Sakoğlu et al., 2010; Hutchison et al., 2013a, 2013b; Keilholz et al., 2013; Wilson et al., 2015) and the ideal values for this and other parameters for the dynamic FC analysis remain unknown. A nice but simplified examination of the relationship between the minimum window length and the frequency components of the signals has been presented (Leonardi and Van De Ville, 2015). Another study used windows of different sizes on resting state and sleep data and reported that short epochs can be used effectively for dynamic FC analysis (Wilson et al., 2015). However, no study has convincingly identified the best window length for dynamic FC analysis. Furthermore, since these brain networks change states at random times, using the same window over the entire rsfMRI scan may not be the optimum method to capture the true dynamic configurations of these networks. The effect of window length, offset, and other parameters has not been systematically examined in realistic data, and a recent study that looked at the effect of window length on the correlation between the BOLD signal and simultaneously-acquired local field potentials found that the optimal window length is somewhat ambiguous (Thompson et al., 2013a, 2013b).
After the SWC is performed pairwise for the brain areas of interest, clustering is often used to find the number of ‘states’ that occur over the length of the scan, and the times at which transitions occur (Hutchison et al., 2013a, 2013b; Allen et al., 2014; Damaraju et al., 2014; Shakil et al., 2014). The most commonly used method for clustering SWC results is based on the k-means algorithm (Hutchison et al., 2013a, 2013b; Allen et al., 2014; Damaraju et al., 2014; Shakil et al., 2014). The accuracy of the clustering depends on the clustering algorithm and the ability of the SWC to resolve transitions of interest, emphasizing the need to evaluate the SWC parameters.
The biggest obstacle in identifying the best approach to the SWC and clustering for dynamic FC analysis is that there is no ‘ground truth (GT)’ in standard rsfMRI data, since the actual network dynamics, number of states, and state transitions are all unknown. This study circumvents this problem by using simulated networks (SNs) with known transition points created from real rsfMRI data. We evaluate the SWC algorithm and the effects of window size, window shift, window type, noise, filtering, and sampling or repetition time (TR) on the SWC results, and on the correct identification of state transitions and durations obtained from these results using k-means clustering. As expected, window size and offset had a substantial impact on the accuracy of the results, followed by the impact of noise and filtering, while TR had a very small impact. Tapered windows resulted in poorer state identification than rectangular windows due to the abrupt sharp state transitions present in the SNs. These findings motivate further work on methods that can dynamically adapt the length of the window during the analysis or the formulation of an algorithm which can more accurately detect the state transition points.
Material and methods
Data and preprocessing
We used rsfMRI scans of nine healthy human subjects (four females, ages: 21–57 years, downloaded from Nathan Klein Institute's Enhanced Rockland dataset of 1000 Functional Conectome Project (http://www.nitrc.org/projects/fcon_1000/). The scans were done on SIEMENS MAGNETOM TrioTim syngo MR B17 scanner. The scanning parameters were: TR = 645 ms, voxel size = 3 mm isotropic, duration = 10 min, TE = 30 ms, slices = 40, multi-band accel, factor = 4, and time points = 900. Each scan contained 900 volumes and the initial 10 volumes of each scan were discarded to compensate for transient scanner instability. All preprocessing was done in statistical parametric mapping (SPM 12, http://www.fil.ion.ucl.ac.uk/spm/). Preprocessing included motion correction, coregistration of the functional images with the anatomical image, segmentation, normalization, and smoothing. Default parameter values from SPM12 were used during preprocessing but smoothing was done using a Gaussian kernel of size 8 and for normalization a voxel size of 3 × 3 × 3 was chosen. The images were coregistered to the AAL atlas (Tzourio-Mazoyer et al., 2002) using nearest neighbor interpolation without any warping.
After preprocessing, five functional networks (dorsal DMN, ventral DMN, anterior-salience, visuospatial, and sensorimotor) were extracted using the masks from the Stanford FIND (http://findlab.stanford.edu/home.html) lab (Shirer et al., 2012) for all subjects.
Region-of-interest (ROI) time series
For each subject, seven, non-overlapping, three-dimensional, regions-of-interest (ROIs) consisting of 3 × 3 × 3 voxels were chosen from each of the abovementioned five networks (dorsal DMN, ventral DMN, anterior-salience, visuospatial, and sensorimotor). The anatomical location of the ROIs in the five networks (taken from Supplementary data of Shirer et al. (2012)) is given in Supplementary Fig. 1. Maps of the five functional networks (taken from Supplementary data of Shirer et al. (2012)) along with the locations of the ROIs selected for the current study (arrows) are given in Supplementary Fig. 2. Each ROI time series was formed by extracting the intensities of the voxels in the ROI and then computing their mean at each time point. In order to observe the dependency of the analysis on the location of ROIs, we later performed the analysis on a second, entirely different sets of ROIs (shown in Supplementary Fig. 3). These ROIs were used to create simulated networks (SNs) as described in the next section. The averaged time series of each ROI was extracted and bandpass filtered (0.016–0.08 Hz, order 20 FIR) before the formation of the SNs. As expected, the ROIs that came from the same network were highly correlated, which was confirmed by computing the pairwise stationary correlations (Supplementary Fig. 4).
Sliding window correlation of actual resting-state networks
The main goal of this study was to analyze the performance of the SWC with variable parameters using SNs with known timing formed from real rsfMRI data. However, before starting this analysis we computed the pairwise SWC of the time series of the five actual networks (dorsal DMN, ventral DMN, anterior-salience, visuospatial, and sensorimotor) using the same window sizes as the ones used for the SNs (discussed in detail in the Simulated networks and sliding window correlation section). The purpose was to compare the SWC of the actual data with the results of previous studies (Hutchison et al., 2013a, 2013b; Keilholz et al., 2013; Wilson et al., 2015), and to determine how the abrupt intensity changes (outliers) introduced in our SNs due to state transitions (explained in the Simulated networks and sliding window correlation section) might influence results of the SWC.
Simulated networks and sliding window correlation
To form a SN, seven ROIs from one of the abovementioned rsfMRI networks were used. A portion of the time courses for these ROIs was taken and used as the time courses for the seven nodes of the SN until the first state transition point t1. At t1, a portion of the time courses from the seven ROIs of a different network was added to the SN to create a new state lasting from t1 to t2. This process was repeated until the desired length of 890 time points was obtained. For example, if we chose the nodes from ventral DMN from t1 to t2, then the nodes from t2 to t3 were from another network e.g. sensorimotor network of the same subject, and this process continued till we reached the last interval from tn – 1 to tn. Formation of the SNs in such a manner incorporated real rsfMRI data but gave us control over the time at which the SNs changed states (switched from one resting-state functional network to another) since we chose the transition times t1 to tn. It should be noted here that our SNs were formed from five resting-state networks but some of them had more than five transitions which means the data from the same resting-state network would be taken more than once in formation of these SNs. However, apart from one SN (QPeriodicSN explained later in this section) there is no repetition of data. For example, if the data from ventral DMN is taken for the durations tx – 1 to tx and ty – 1 to ty (x and y are integers) for a SN then it would be from two entirely different non-overlapping intervals of the ventral DMN. This step insured that no two parts of a SN has exactly the same correlation values between the node pairs (except for QPeriodicSN).
The configuration of a SN between any two consecutive transition points is referred to as one ‘state’ of that SN since it was formed by data from one FC network (state) of original resting-state scans. All the states of a SN (except QPeriodicSN) have different correlation values between the node pairs but more than one of these states may be from the same resting-state network. By changing the transition points we were able to form five different SNs. Three of these SNs were formed with state transitions at equal intervals (state transition at every 50 TRs for 50SN, at every 100 TRs for 100SN, and at every 200 TRs for 200SN, TR = 645 ms). Real resting-state networks are unlikely to change configurations at equal intervals but the analysis with these SNs allowed us to explore the best choice of the sliding window parameters for the ideal case of regularly changing networks. However, we also formed two SNs where the state transitions occurred at random intervals to evaluate the performance of the SWC in more realistic scenario. In the first randomly changing network, we used quasi periodicity by repeating three states at random points during the formation of the SN (QPeriodicSN). The second randomly changing network (RandSN) had completely random state change points with state durations from 20 TRs (≈13 s) to 130 TRs (≈84 s).The formation of these five SNs allowed us to examine the sensitivity of the SWC to regular states of different lengths, and randomly changing states with or without quasi periodicity. The state durations for the SNs ranged from ≈13 s to ≈129 s, while the window lengths used were ≈17, 33, 65 and 129 s (25 TRs, 50 TRs, 100 TRs, and 200 TRs, TR = 645 ms). All of these window lengths were within the sizes commonly reported in the literature that vary from 8 to 240 s (Chang and Glover, 2010; Kiviniemi et al., 2011; Handwerker et al., 2012; Chang et al., 2013; Keilholz et al., 2013; Thompson et al., 2013a, 2013b; Kucyi and Davis, 2014; Wilson et al., 2015).
In their papers (Leonardi and Van De Ville, 2015; Shakil et al., 2015), explored the relationship of frequency components in the correlating signals and window length. (Leonardi and Van De Ville, 2015) established the mathematical relationship between the window length and the minimum frequency (fmin) of the correlating time series. By using simple sinusoids (Leonardi and Van De Ville, 2015) reported that the minimum window length should at least be equal to 1/fmin in order to avoid spurious fluctuations arising due to the SWC algorithm itself. Using the same guidelines (Shakil et al., 2015) explored the effect of two frequencies on the SWC results. Since the frequency components of the signals may influence the results of the SWC, we plotted log10 (power spectrum) vs. log10 (frequency) of the actual (ROI) and SN time series in Supplementary Figs. 5(a) and (b) respectively. The frequency characteristics are similar, however, due to abrupt transitions in the SNs, the power spectrum changes in the SNs are less smooth. It should be mentioned here that there is no one to one correspondence between the plots in (a) and (b). The plots in (a) are for original rsfMRI networks, while those in (b) are for SNs that were formed by combination of time series in (a).
Overall five SNs were formed from each subject's data: Three with equal duration states and two with randomly changing states. The equal duration SNs had 18 (50SN), 9 (100SN), and 5 (200SN) distinct states of FC. QPeriodicSN had a total of 15 FC states, of which three were repeated at random time points, while RandSN had 10 distinct FC states changing at random times. We computed SWC for all of these SNs for different window lengths and other parameters to determine the best combination of parameters for SWC as a dynamic FC analysis method. Detailed results from the SNs created from one randomly selected subject's data along with the mean results of all SNs created from all nine subjects are reported in this study.
Ground truth
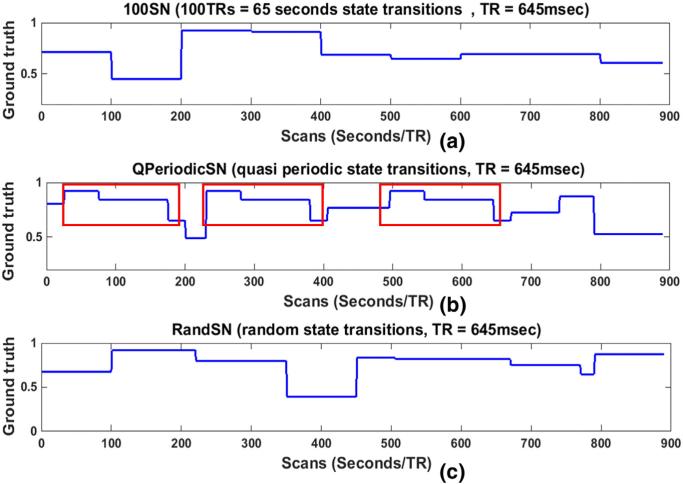
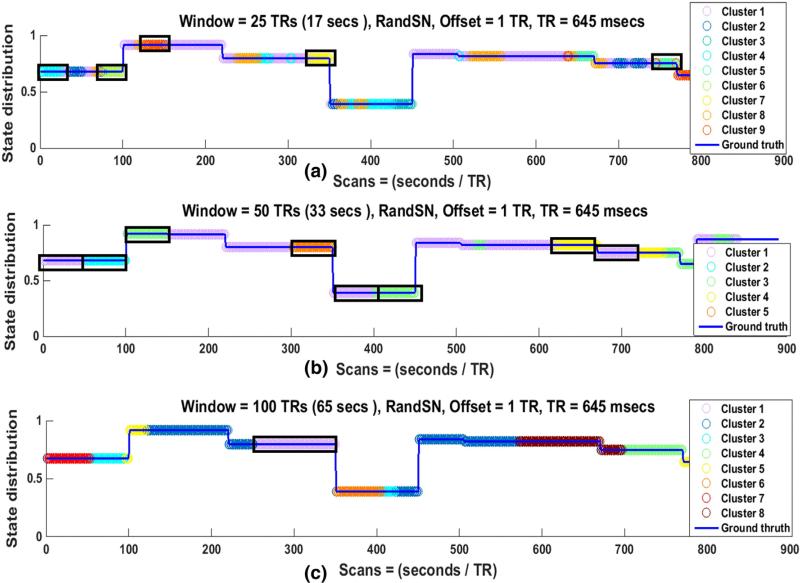
State changes in the SNs occurred at times t1, t2, t3 ..., tn – 1, and tn, when ROIs were taken from a different resting-state network. This resulted in a change of pairwise correlations between all of the nodes in a SN at each transition point. To examine the influence of various SWC parameters on its sensitivity to network transitions, actual correlations between the nodes of the SNs were computed for the durations for the which the state remained the same. The actual correlation of a node pair for any state was correlation coefficient of the node pair time series for the duration of the state. This correlation value was the same for the whole state duration and can be replicated. For example, if the correlation coefficient between two time series was 0.5 for the state between t1 and t2 then 0.5 would be replicated from t1 to t2 for representation purposes. The computation of correlation in this manner is the same as the computation of stationary correlation with length of the correlating time series equal to the state duration instead of the whole scan length as in the case of stationary correlation. The mean of the resulting actual correlations of an SN was called the “ground truth (GT)” for that SN. Because there were n = 7 nodes in each SN, overall there were , distinct correlations for each state, and the GT was the mean of these correlations. Fig. 1 shows the GT for 100SN in (a), QPeriodicSN in (b), and RandSN in (c). The GTs for the other two SNs (50SN and 200SN) are shown in Supplementary Fig. 6. Discontinuities in Fig. 1 (and Supplementary Fig. 6) are the state change points, which will be referred as “state transition points” in our study, and the duration between two consecutive discontinuities corresponds to one state of the SN. It is also important to note that the GTs are not the actual correlation time series between the node pairs, but are their means plotted to clearly show the state transitions and durations.
Fig. 1.
Ground truths (GTs) for (a) 100SN, (b) QPeriodicSN, and (c) RandSN. 100SN has state transitions at every 100 TRs (65 s), QPeriodicSN has quasi periodic states (red rectangles) repeated at random times, and RandSN has all the states transitions at random times. Each SN has seven nodes and ground truth for any SN is the mean of actual correlations of all of its node pairs.
Inter-subject variability of the SWC
We computed the actual correlations of all SN node pairs in all subjects and their mean was taken as GTs (Fig. 1 and Supplementary Fig. 6). In order to observe the deviation of the SWC results from actual correlations, we computed the correlations of the actual correlations and SWCs for all node pairs for all the windows in each subject. We then calculated the means of these correlations for each subject (over all node pairs) to observe the inter-subject variability of these correlations. Afterwards, we computed the overall mean for all the nine subjects to show the overall trend of correlation changes for each window.
Sliding window correlation parameters
Six parameters were varied to explore their influence on the sensitivity of the SWC and subsequent clustering. For all parameter values, the SWC was calculated pairwise for all nodes of each SN.
-
a)
Window length: Gonzalez-Castillo et al. (2014) reported that resting-state networks exhibit long-term stability and that the similarity of within-subject whole brain connectivity is a function of window duration. In various dynamic FC studies based on the SWC, the window length ranges from 8 to 240 s (Chang and Glover, 2010; Kiviniemi et al., 2011; Keilholz et al., 2013; Allen et al., 2014). We examined windows of 25, 50, 100, and 200 TRs (≈17, 33, 65 and 129 s). All the window sizes were within the limits reported in the literature (Chang and Glover, 2010; Kiviniemi et al., 2011; Keilholz et al., 2013; Allen et al., 2014; Kucyi and Davis, 2014; Wilson et al., 2015).
-
b)
Signal-to-noise ratio (SNR): The signal-to-noise ratio (SNR) of rsfMRI data depends on a number of factors and can include structured noise due to motion or physiological cycles (Kruger and Glover, 2001; Greve et al., 2013; Bright and Murphy, 2015). In order to explicitly examine the influence of random noise on the identification of states using the SWC, additive white Gaussian noise (AWGN) (using awgn() in Matlab) was added to the time series of each SN at levels of 10 dB and 20 dB. We also examined the effect of reduced SNR by selecting random single voxels from random slices as nodes of our SNs instead of 3 × 3 × 3 voxels ROIs.
-
c)
Window offset: Typical window offsets used in previous studies range from 1 to 20 s (which covers the range from a single TR step to 50% of the window length) (Chang et al., 2013; Keilholz et al., 2013; Kucyi and Davis, 2014; Shakil et al., 2014). The majority of this study uses a window offset of one TR (645 ms), but the effects of offsets equal to one-fourth or one-half of the window lengths were also explored.
-
d)
Window type: Typically rectangular windows are used for the SWC analysis (Chang et al., 2013; Keilholz et al., 2013; Kucyi and Davis, 2014; Shakil et al., 2014). The majority of the work in this study also utilized a rectangular window but the impact of using tapered (Hamming, Hanning) windows were also examined.
-
e)
Filtering: Typical rsfMRI studies bandpass filter (0.01–0.08 Hz or 0.01–0.1 Hz) the time series in order to reduce the effects of noise. Two recent studies (Leonardi and Van De Ville, 2015; Shakil et al., 2015) have shown that the frequency components of the signal and the window size interact to impact final SWC results. Both the studies showed that the spurious correlations of the SWC results are reduced when the size of the window is greater than 1/fmin (fmin = minimum frequency). Since both of these studies reported a relationship between the minimum frequency of the correlating signals and the shortest recommended window length, we explored this relationship by varying the minimum frequency content of the rsfMRI signal. For this purpose we used highpass filtered (>0.016 Hz, >0.05 Hz and >0.08 Hz) time series to form SNs, corresponding to minimum window lengths of 62.5, 20, and 12.5 s respectively.
-
f)
Repetition time (TR): Most simulations used SNs formed from scans acquired with a TR of 645 ms, but analysis was also performed on scans with 1400 ms TR to determine how a lower sampling rate influences SWC results and their state distributions.
Clustering
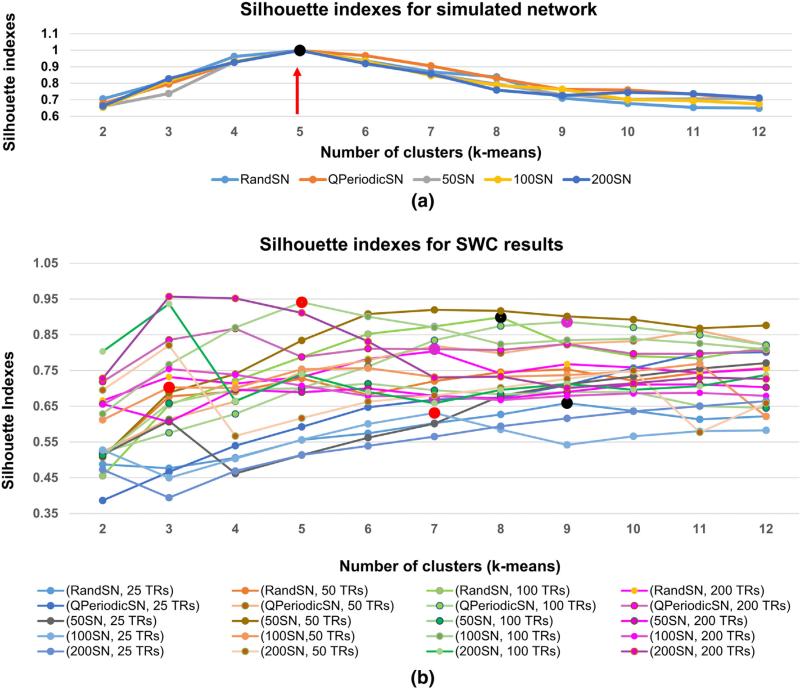
One of the major goals of this study was to determine how well the clustering based on the SWC performs at identifying network state durations and transitions. In previous dynamic FC studies, k-means clustering was used on SWC results to extract states of FC (Leonardi et al., 2013a, 2013b; Allen et al., 2014; Damaraju et al., 2014; Shakil et al., 2014). We also applied k-means clustering to the SWC results to determine whether state transitions and durations were identified correctly. For this purpose, the SWC results were grouped into clusters called “Cstates” and the results were examined to determine how well the Cstates coincided with the known state transitions and durations shown in the GTs (Fig. 1 and Supplementary Fig. 6). In order to evaluate the performance of k-means clustering alone for state identification, we clustered the raw time courses for the seven nodes of each SN in addition to clustering the SWC results. Silhouette criteria was used to determine the best number of clusters for each data set. The number of clusters was varied from two to twelve and the overall mean of silhouettes was computed (Rousseeuw, 1987). We selected the number of clusters (raw SNs) and Cstates (SWC results) for each case when this mean became constant or started to decrease.
In general, the number of Cstates for all windows and all SNs was less than or equal to the actual number of states in the SN (10 for RandSN, 15 for QPeriodicSN, 18 for 50SN, 9 for 100SN, and 5 for 200SN), resulting in the assignment of more than one state to the same Cstate. To determine the similarity of states assigned to the same Cstate, the Euclidean distance between them was computed.
Results
Sliding window correlation of actual ROI time series
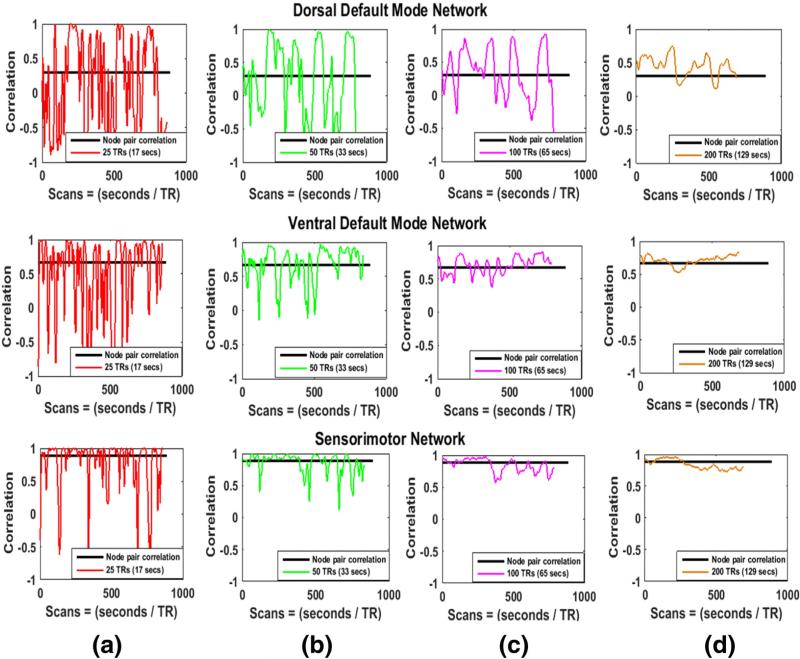
We computed pairwise SWC of the actual ROI time series of the five functional networks (dorsal DMN, ventral DMN, anterior-salience, visuospatial, and sensorimotor) for all windows. Fig. 2 shows the results of the pairwise SWC along with the stationary correlations (black horizontal lines). The node pairs are selected at random from three of the five networks (dorsal DMN, ventral DMN, and sensorimotor network). It can be observed that for all cases the SWC varies around the actual correlation but the extent of variability is largest for the smallest window (25 TRs, red in (a)) and decreases as the window size increases towards right. The same plots (overlaid on each other) are shown for three other pairs of randomly selected nodes in Supplementary Fig. 7. All the plots of these figures show a similar trend and are in accordance with the results reported in Hutchison et al. (2013a, 2013b), Keilholz et al. (2013), and Wilson et al. (2015).
Fig. 2.
Stationary and sliding window correlations of three node pairs (selected at random) from three different real resting-state networks. Stationary correlations of the node pairs are shown by black horizontal lines. The size of the window increases from left to right. The SWC fluctuates around the stationary correlation and these fluctuations are largest for smallest window in (a) and decrease as the window size increases towards the right.
Effects of window length
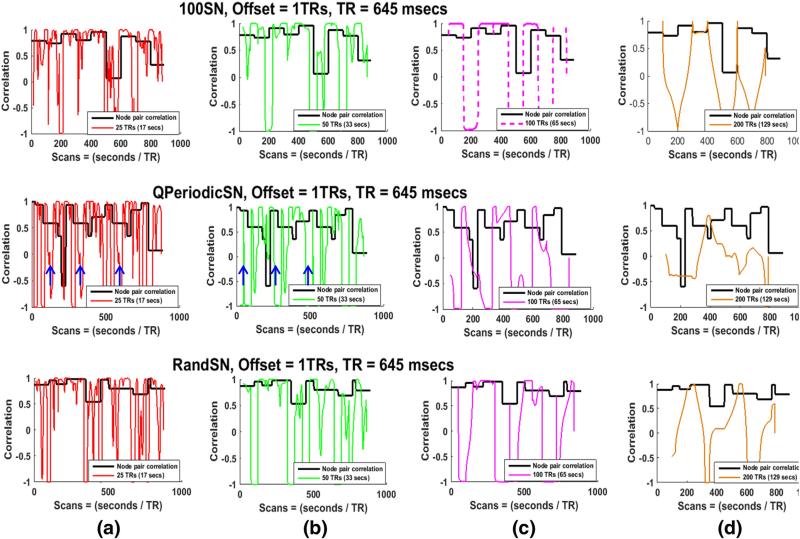
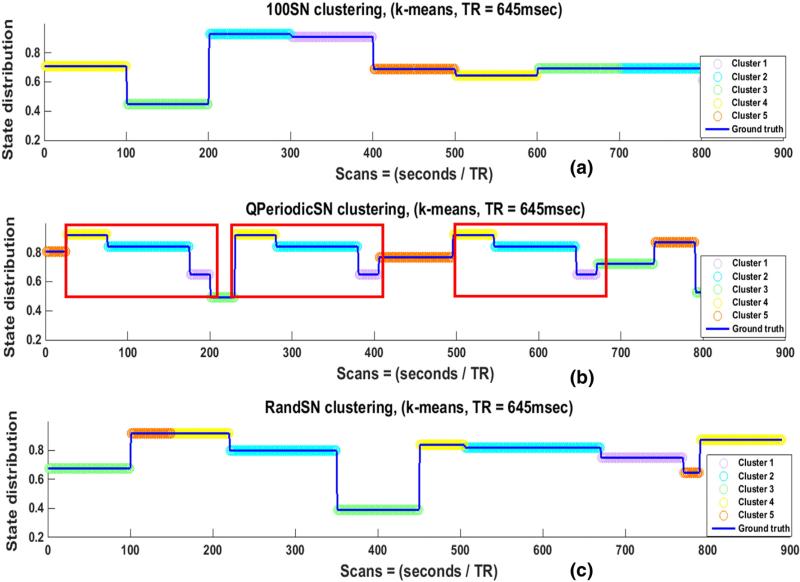
Four different window sizes (25, 50, 100, and 200 TRs, or 17, 33, 65 and 129 s respectively) were employed to determine the effect of window length on the sensitivity to actual correlations, state transitions, and state durations in the SNs. For this portion of the study, the window offset was kept constant at one TR (645 ms), the window shape was rectangular, and all time courses were bandpass filtered (0.016–0.08 Hz). The SWC for all pairs of nodes for all SNs exhibited variability over time. Greater variability was observed when shorter windows were used, in accordance with previous studies (Hutchison et al., 2013a, 2013b; Keilholz et al., 2013). Fig. 3 shows the actual correlations (black lines) and the SWC for one node pair in 100SN (row 1), QPeriodicSN (row 2), and RandSN (row 3) for all window lengths. Actual correlations and SWCs for 50SN and 100SN for the same node pair are plotted in Supplementary Fig. 8. Results were similar for all node pairs and all subjects. A few observations can be made:
-
a)
None of the SWC time courses accurately capture the different states of correlation for the SNs. The amount of variability in the SWC time course is typically higher than the variation across different states of correlation. Large changes in the SWC sometimes correspond to transitions between states due to the abrupt changes that occur at the transition points, but often exist in the absence of state transitions (especially in smaller windows). SWC results vary widely between −1 and 1 for almost all windows, though these variations are more rapid for smaller windows (red and green) compared to larger ones (pink and brown).
-
b)
Smaller window sizes can capture short-lived variations in correlation more accurately than longer windows, as can be seen from the rapidly fluctuating plots of the 25 TRs window (red) and 50 TRs window (green), as compared to the longer windows (pink and brown). However, a portion of these fluctuations may arise since smaller windows also create more high-amplitude variations and spurious fluctuations (Leonardi and Van De Ville, 2015; Shakil et al., 2015) that do not correspond to state transitions.
-
c)
For the SNs with state transitions at equal intervals, the SWC is sensitive to the state transitions if the window size is equal to the state duration, although the actual correlation values are not always captured. 100SN is shown as an example in Fig. 3 (row 1, (c)) and similar results were obtained for 50SN and 200SN (shown in Supplementary Fig. 8 for the same node pair). We plotted the SWC values at the center of each window (Fig. 3) but shifting the values to the start of the window reveals that the SWC changes at the transition points when the actual correlation (state of the SN) is changing sharply (Supplementary Fig. 9). These results show the inability of the SWC to deal with abrupt amplitude changes (outliers) in the intensity levels of correlating signals (Lindquist et al., 2014).
-
d)
For QPeriodicSN a quasi-periodic pattern was observed in the SWC results for smaller windows (blue arrows in Fig. 3, row 2 (a) and (b)) but this pattern was lost for larger windows.
-
e)
For the SN with randomly changing state transitions (Fig. 3, row 3), no window size consistently identifies transitions at the state change points.
Fig. 3.
Actual correlations and SWCs for one node pair in 100SN (row 1), QPeriodicSN (row 2), and RandSN (row 3). The SWC is plotted for windows of 25 TRs, 50 TRs, 100 TRs, and 200 TRs ((≈17, 33, 65 and 129 s). The window offset is one TR and filtered (0.016–0.08 Hz) time series are used. The actual correlation between the node pair is plotted in black. Smaller windows (red, green) result in more variable correlation time series. The large amount of variability indicates that smaller windows could capture short-lived correlations but some of these variations may be due to the spurious fluctuations introduced due to the small size of the window. Smaller windows are also able to capture the quasi periodic pattern of QPeriodicSN indicated by the blue arrows but this pattern is lost with increasing window size. The transitions in the SWC results occur at an interval of 65 s (100 TRs) for 100SN when the state duration and window size are the same (pink dashed) which shows that SWC is good in identifying the transition of states when window size and state duration are well matched.
Inter-subject variability
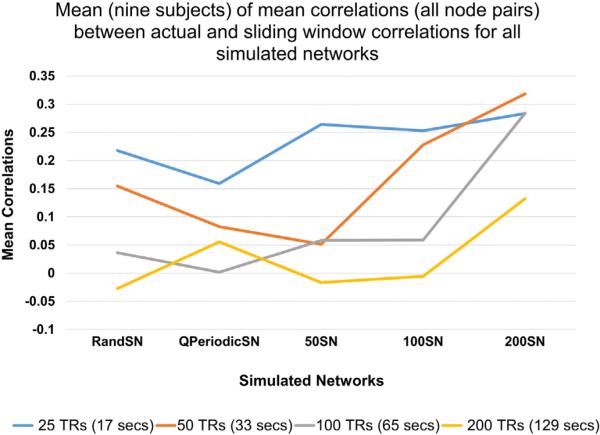
Stationary correlation between the actual correlation and the SWC of all node pairs for all windows was also computed. Means of these correlations for each subject are shown in Fig. 4. The SWC was not strongly correlated with the actual correlation since there were sharp changes at points where state transitions occurred. However, the smaller windows gave better correlation values regardless of the SN. This may be because smaller windows capture dynamics of correlation at smaller scales compared to larger windows. It can also be observed that regardless of the window size, the means were highest for 200SN, in which the states were stable for longest amount of time. We also plotted the mean correlations of all subjects individually in Supplementary Fig. 10. In most of the cases, the means were highest for the smallest window size (blue) and decreased as window size increased. Furthermore, similar to Fig. 4, the mean correlation for almost all the windows was highest for 200SN.
Fig. 4.
Mean of mean correlations (of all node pairs) over all subjects. The overall mean is not very high because the SWC is very sensitive to abrupt transitions between the states. However, smaller windows give SWC results that are more similar to the actual correlation values compared to larger windows.
Silhouette indexes
Fig. 5(a) shows the silhouette indices for k-means clustering on the raw signal for five SNs for the subject selected for detailed analysis in this study, and (b) shows the silhouette indices for the SWC results of the same subject. For the raw SNs, the silhouette indices are highest for five clusters in all cases (shown by black dot and red arrow) and the results of clustering raw 100SN, QPeriodicSN, and RandSN with five clusters are shown in Fig. 6. Silhouette indexes for the raw SNs for the other eight subjects are plotted in Supplementary Fig. 11 and as expected the highest silhouette value was obtained for five clusters (shown by red dots and pointed to by green arrows).
Fig. 5.
Silhouette indexes for raw SNs in (a) and SWC results in (b) for one subject. (a) The highest silhouette value (black dot and red arrow) corresponding to the best number of clusters for all the raw SNs were five since the SNs were formed from five resting-state networks. (b) The silhouette values for SWC results were different for different windows. Red dots show the silhouette indexes for 100SN (25 TRs, 50 TRs, and 100 TRs) windows. Pink dots are for QPeriodicSN and black are for RandSN for the same window sizes. The cluster (Cstate) distribution of the SWC results for these SNs are plotted in Figs. 7–9.
Fig. 6.
Clustering of raw SNs (100SN in (a), QPeriodicSN in (b), and RandSN in (c)) using k-means. The best number of clusters was identified using Silhouette criteria. Discontinuities in the blue lines indicate state transition points, while the colors of the overlaid circles represent the Cstate at each time point that was assigned by the clustering algorithm (k-means). When the circles between two adjacent state transition points remain the same color, it indicates that the state is correctly identified to be in single Cstate and a color change at state transition points indicates that the state transitions are correctly identified. The division of the raw signals gives perfect state transitions and state durations. The repeated states of QPeriodicSN (red rectangles) are assigned to the same Cstates, as expected.
The SWC changes the cluster assignment, resulting in different values of the highest silhouette for different combinations of SNs and window length, as shown in Fig. 5(b) for the subject selected for detailed analysis in this study. Large red dots are the selected silhouette indexes for 100SN (25 TRs, 50 TRs, 100 TRs windows) and the results of corresponding clustering are shown in Fig. 7. Similarly, pink dots are the selected silhouettes for QPeriodicSN (25 TRs, 50 TRs, 100 TRs windows) and black dots are for RandSN (25 TRs, 50 TRs, 100 TRs windows) with the corresponding clustering plotted in Figs. 7 and 8 respectively. Silhouette indices became constant before the number of clusters reached twelve. Silhouette indices for the SWC results of the other eight subjects are plotted in Supplementary Fig. 12. We can observe from the concentration of the color distribution in the plots that the silhouette indexes have similar trends for a given window size in almost all subjects. The mean number of Cstates varied from three to seven, with an overall mean of five for all the SNs and all windows.
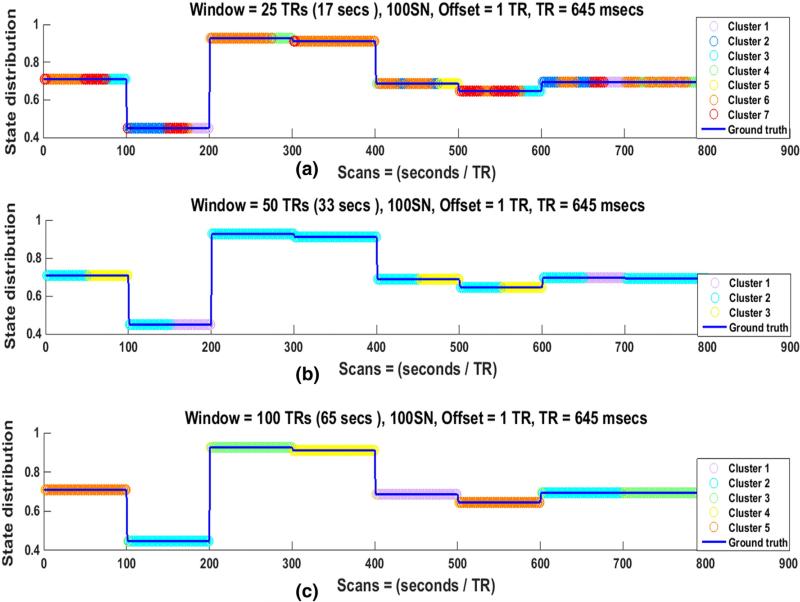
Fig. 7.
State distribution of SWC results for 100SN for (a) 25 TRs, (b) 50 TRs, and (c) 100 TRs windows in one subject. The best number of clusters (Cstates) in each case was identified by Silhouette criteria. Discontinuities in the blue lines indicate state transition points, while the colors of the overlaid circles represent the Cstate at each time point that was assigned by the clustering algorithm (k-means). When the circles between two adjacent state transition points remain the same color, it indicates that the state is correctly identified to be in single Cstate and a color change at transition points indicates that the state transitions are correctly identified. (a) State distribution for a 25 TR window seven clusters or Cstates. Here state transitions are identified accurately but the number of transitions is much greater than the number of actual state transitions. State durations are not identified correctly in this case. (b) State distribution for a 50 TR window (three clusters or Cstates). Here state transitions are identified accurately but again the number of transitions is greater than the actual number of state transitions. State durations are not identified correctly, with most actual states split evenly into two Cstates. (c) State distribution when the window size is equal to the state duration of 100 TRs. The state transitions are correctly identified as seen by the change of color at the discontinuities. Furthermore, the state durations are also identified reliably as shown by consistent assignment of each actual state to a single Cstate.
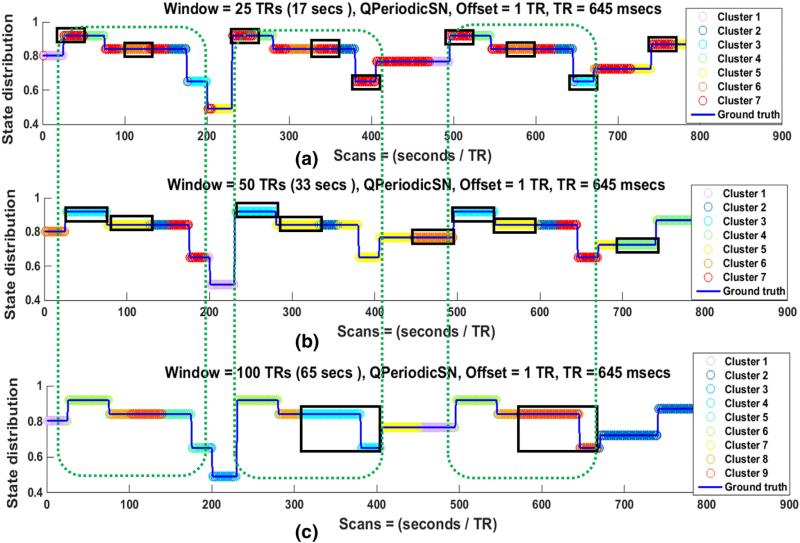
Fig. 8.
State distribution of QPeriodicSN for (a) 25 TRs, (b) 50 TRs, and (c) 100 TRs windows in one subject. Discontinuities in the blue lines indicate that state transition points and the colors of the overlaid circles represent the Cstate at each time point that was assigned by the clustering algorithm (k-means). When the circles between two adjacent state transition points remain the same color, it indicates that the state is correctly identified to be in single Cstate, while a color change at transition points indicates that the state transitions are correctly identified. The 25TRs window is able to capture the state transitions accurately than the larger windows in (a) but the number of overall state transitions in this case is also larger than for the 50TRs or 100TRs windows in (b) and (c). The quasi periodic pattern of the SN is identified in 25 TRs and 50 TRs windows as shown by the same color distributions inside the green dotted rectangles. Furthermore, the state identification is dominated by the size of the window since most of the state durations are almost equal to the window size as shown by black rectangles.
The Silhouette criteria for raw SNs identified five as the best number of clusters for all subjects, which was an expected result since the raw SNs were originally extracted from five resting-state networks. As a consequence all the raw SNs were divided into five clusters (Cstates) as shown in Fig. 6 and Supplementary Fig. 13.
Dependence of state identification on window length
We clustered the actual (raw) SNs in five Cstates based on Silhouette criteria to determine whether the k-means algorithm would be able to identify the state transitions and durations in raw data. The results of this clustering for 100SN, QPeriodicSN, and RandSN are given in Fig. 6. The blue lines in the plots show the GTs of the SNs with state transition points at the discontinuities. Circular colored markers on these lines indicate the Cstate in which the SN resides at that particular time. If all circles between two state transition points are in the same color, then the SN remained in one Cstate during that time. A change of marker color at a state transition point indicates correct detection of the state transition. Good state transition and duration identification implies that the color should change at the state transition points but should remain the same between any two adjacent state transition points. It can be observed from the figure that k-means perfectly identified the state transitions and durations for the raw SNs. Results for 50SN and 200SN are shown in Supplementary Fig. 13.
To examine how the window length of SWC impacts the identification of ‘brain states’, the SWC results were divided into clusters (Cstates) using the k-means algorithm. Figs. 7–9 show the clustering results for different SNs and different window lengths.
Fig. 9.
State distribution of RandSN for (a) 25 TRs, (b) 50 TRs, and (c) 100 TRs windows in one subject. Discontinuities in the blue lines indicate state transition points, while the colors of the overlaid circles represent the Cstate at each time point that was assigned by the clustering algorithm (k-means). When the circles between two adjacent state transition points remain the same color, it indicates that the state is correctly identified to be in single Cstate, while a color change at transition points indicates that the state transitions are correctly identified. None of the window correctly identifies the state transitions and durations. For smaller windows ((a) and (b)), identified state durations are mostly equal to the size of the window.
For 100SN, it can be seen in Fig. 7(a) that the Cstate changes at almost every discontinuity, indicating that a state transition has been correctly identified even when the window size is only about one-fourth of the state duration. Similar findings were observed for windows of 50 TRs and 100 TRs ((b) and (c)). These results show that the state transitions can be correctly identified when the window size is proportional to the state duration (one-fourth, one-half, or equal) for the SNs that change state at equal intervals. However, especially for shorter windows, it is common for a single actual state to be assigned to multiple Cstates, artificially increasing the number of observed transitions. Results for other regularly changing SNs (50SN, 200SN) for the same subject are shown in Supplementary Figs. 14 and 15 respectively. Similar observations were made for all SNs in all the subjects.
In 100SN the changes in Cstate consistently occurred at known state transitions for all window lengths, but Cstates changed between true transitions for window lengths that were smaller than the state duration (25 and 50 TRs in Fig. 7). The time period of a state duration is assigned to more than one Cstate (typically two for the 50 TR window and three or more for the 25 TR window), while for the 100 TR window it is typically assigned to a single Cstate. These findings were similar for other regular networks and indicate the failure of the SWC to correctly identify the state for the whole of its duration if the window size is not at least as long as the state duration interval. Reducing the window size to one-half of the state duration resulted in each state typically being divided into two Cstates, as can be seen from Fig. 7(b). Similar results can be observed for 50SN and 200SN for the same subject in Supplementary Figs. 14 and 15 respectively. These observations were consistent across all window sizes and in all the regularly changing SNs of all the subjects. The number of correct state transitions and state duration assignments for all SNs were assessed by computing the mean for all subjects in Tables 1 and 2. These results are explained further in the next section.
Table 1.
Mean percentages of full state identifications for all SNs along with the mean percentages of the closest states that are assigned to the same cluster (Cstate, given in parentheses) for all nine subjects. Results are shown for bandpass filtered data with a window offset of one TR. The second row contains the SN type along with the total number of states (parentheses). A state was considered fully identified if it remained in the same Cstate for more than 98% of its total duration. For a specific window size, the maximum full state identification occurs for the SN with state durations equal to that window size (50 TRs window for 50SN, 100 TRs window for 100SN, and 200 TRs window for 200SN), shown in (row 4, column 4), (row 5, column 5), and (row 6, column 6). None of the windows provided good full state identification for the QPeriodicSN and RandSN. The number of actual state transitions (15 for QPeriodicSN, 10 for RandSN, 18 for 50SN, 9 for 100SN, and 5 for 200SN) was greater than the number of Cstates (mean varying between three and seven), which means that in the case of full state identifications, more than one state would be assigned to the same Cstate. The numbers in the parenthesis next to the full state identification percentages show how many of the states, which were closest to each other, are assigned to a single Cstate. Only 55% of the states assigned to the same Cstate are closest to each other compared with the other states for 50SN (50 TRs window in row 4, column 4). This percentage becomes better with an increase in the size of the window for equal duration networks (row 5, column 5 & row 6, column 6). No specific pattern is observed for the full state identification of QPeriodicSN and RandSN except that in both SNs the identification became better with the increase in the size of the window.
| Mean percentages of full state identification (mean percentages of closest states) for nine subjects for filtered (0.016–0.08 Hz) simulated networks | |||||
|---|---|---|---|---|---|
| RandSN (10) | QPeriodicSN (15) | 50SN (18) | 100SN (9) | 200SN (5) | |
| 25 TRs (17 s) window | 21 (97) | 9 (94) | 13 (58) | 11 (40.7) | 0 (0) |
| 50 TRs (33 s) window | 50 (64) | 58 (97) | 99 (55) | 21 (56) | 25 (56) |
| 100 TRs (65 s) window | 30 (50) | 50 (75) | 42 (66) | 100 (85) | 15 (44) |
| 200 TRs (129 s) window | 69 (59) | 65 (55) | 44 (73) | 39 (82) | 100 (97) |
Table 2.
Total state transitions along with the mean percentages of correct state transitions (with respect to total detected state transitions and actual state transitions given in the parentheses) are shown for all window sizes and SNs. The second row contains the SN type along with the total number of state transitions for each one in the parenthesis. The total number of transitions is dependent on the window size as well as the actual number of states in a SN. QPeriodicSN and 50SN have the most state transitions (14 and 17) and all the windows show the maximum number of transitions for these SNs (columns 3 & 4) but the largest of them occurs for smallest window of 25 TRs (row 3, column 3 and 4). Mean percentages of correct state transitions with respect to the total and actual state transitions are largest for an equal interval SN when the window size is equal to the state durations ((row 4, column 4), (row 5, column 5), and (row 6, column 6)). Furthermore, the state transitions are fairly well identified if the window size is less the state durations.
| Mean total state transitions (mean((correct state transitions/total state transitions) × 100), mean ((correct state transitions/actual state transitions) × 100)) for nine subjects for filtered (0.016–0.08 Hz) simulated networks | |||||
|---|---|---|---|---|---|
| RandSN (9) | QPeriodicSN (14) | 50SN (17) | 100SN (8) | 200SN (4) | |
| 25 TRs (17 s) window | 39 (16, 68) | 39 (32, 90) | 44 (28, 73) | 33 (19, 81) | 42 (6, 64) |
| 50 TRs (33 s) window | 15 (37, 63) | 25 (40, 70) | 25 (59, 86) | 13 (45, 74) | 9.6 (27, 64) |
| 100 TRs (65 s) window | 13 (17, 25) | 18 (27, 33) | 15 (12, 10) | 12 (57, 86) | 5.8 (44, 64) |
| 200 TRs (129 s) window | 4.3 (15, 7) | 7.1 (3, 2) | 10 (0, 0) | 6.8 (8, 6.9) | 6 (48, 72) |
Since real resting-state networks in the brain would not be expected to exhibit regularly spaced state transitions, we also explored the effect of window size on state transitions and state durations in QPeriodicSN and RandSN. QPeriodicSN was formed based on the identification of quasi periodic patterns of FC networks in rats and humans (Majeed et al., 2011). Majeed et al. reported that the same patterns of FC dynamics occur at random times during the scan. We presented similar quasi periodicity by repeating three states (shown by red rectangles in Fig. 1) at random times in QPeriodicSN. Fig. 8 shows the state distribution of QPeriodicSN for 25 TRs, 50 TRs, and 100 TRs windows. It can be observed from the color changes at the state transition points that the 25 TRs window was able to identify state transitions better than the larger windows (50 TRs and 100 TRs). Smaller windows (25 TRs and 50 TRs) also identified the quasi periodicity of the SN as shown by almost identical Cstates assignments inside green dotted rectangles. However, most of the actual states were assigned to multiple Cstates for all windows, indicating that the state durations are not correctly identified. An interesting observation is that it appears that the state durations are dominated more by the size of the window than by the actual transitions of the SN, as can be seen by the width of the black rectangles that are almost equal to window sizes.
Fig. 9 illustrates the state distribution of the SWC results of RandSN for 25 TRs, 50 TRs, and 100 TRs windows. The results shown here are similar to the results of QPeriodicSN, in which each state is assigned to more than one Cstate, and state assignment is dominated by the window size, especially for smaller windows (25 TRs and 50 TRs). Figs. 8 and 9 clearly identify the inefficiency of the SWC in recognizing the state transitions and durations for randomly changing networks.
Correct state duration and transition identification
Clustering based on the SWC performed well for identifying state transitions and durations for certain combinations of SNs and window sizes but not for other combinations. In order to quantitatively analyze the SWC results, mean percentages of full state identifications and number of correct state transitions for each SN and each window size were calculated for all nine subjects. The Cstate transitions were defined by a change of cluster assignment at any time, and the identification was said to be correct if the change occurred at a state transition point. For example, if there are total ‘m’ state transitions in a SN and ‘n’ of these are coinciding with the Cstate transitions then the correct transition percentage would be .
A state was defined as fully identified if it remained in the same Cstate for more than 98% of its duration. Because the number of Cstates was less than or equal to number of states in the SNs (15 for QPeriodicSN, 10 for RandSN, 18 for 50SN, 9 for 100SN, and 5 for 200SN), even in the case of successful full state identification, more than one of the states would be assigned to the same Cstate. To examine how well the Cstates reflected the underlying patterns of correlation, the distance between the states assigned to a single Cstate was calculated and compared across Cstates. The mean percentages of full state identification and the mean percentages of closest states assigned to the same Cstate are given in Table 1.
As expected from Figs. 7–9, poor full state identification was achieved with small window sizes. For a 25 TRs window the percentage of full state durations assigned to the same Cstates varied from 0% to 21%. This is not surprising given the large amount of variability observed in the SWC obtained with short windows. States were identified well (>99%) when the SN had transitions at equal intervals, and the window size was equal to the interval between transitions. The mean percentages of the closest states assigned to a Cstate was also high for 100SN and 200SN, but not for 50SN (50 TRs window) though the mean percentage of full state identification was high (99%) for this case too. This low percentage may be due to sensitivity of the window to small correlation changes and presence of spurious fluctuations.
Table 2 gives the total number of state transitions along with the mean percentages of correct state transitions with respect to both the total detected state transitions and the actual state transitions (in the parenthesis) for all windows sizes and SNs. The total number of transitions was dependent on the window size as well as the actual number of states in a SN. QPeriodicSN and 50SN had largest number of state transitions (17 and 14), and in general all the windows showed the largest number of transitions for these SNs (columns 3 & 4), but the largest (44) of them occurred for smallest window of 25 TRs. For any SN, the 25 TRs window captured the highest number of state transitions, some of which may be due to spurious fluctuations. The correct state transition percentages with respect to total and actual state transitions were maximum for a SN ((59, 86) for 50SN, (57, 86) for 100SN, and (48, 72) for 200SN) when the window size was equal to the state duration in regularly changing SNs. These percentages were reasonably high when the window size was smaller than or equal to the state duration but dropped when window size became larger than the state duration. For both QPeriodicSN and RandSN, in the absence of any regularity, correct identification was observed for smaller windows and decreased with the increase in the length of the window. However, these percentages were higher for QPeriodicSN as a result of quasi periodic patterns.
Effects of signal-to-noise ratio (SNR)
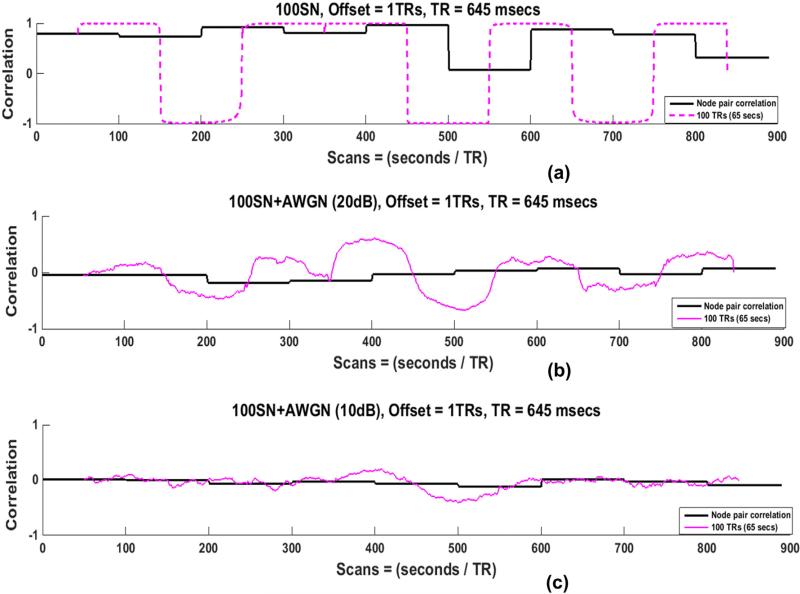
Various types and levels of noise are present in actual resting-state scans. Filtering the data minimizes some of the noise sources but cannot eliminate them completely. In order to examine the influence of noise, we added random additive white Gaussian noise (AWGN) at levels of 10 dB and 20 dB to all SNs before performing the SWC. The SWC results of one node pair in 100SN for the 100 TRs window are shown in Fig. 10, since this combination of state and window length performed best for the noise-free case. The addition of noise reduced the actual correlation and lessened the sharp state transitions in the SWC results. Higher levels of noise had stronger effects.
Fig. 10.
Actual correlation and SWC of one node pair (100SN for 100 TRs window) before addition of the noise in (a) and after addition of additive white Gaussian noise (AWGN) with an SNR of 20 dB in (b) and 10 dB in (c). Noise reduces the sharp transitions of the SWC results. The identification of transition points becomes more difficult for small SNR (c) compared with large SNR (b).
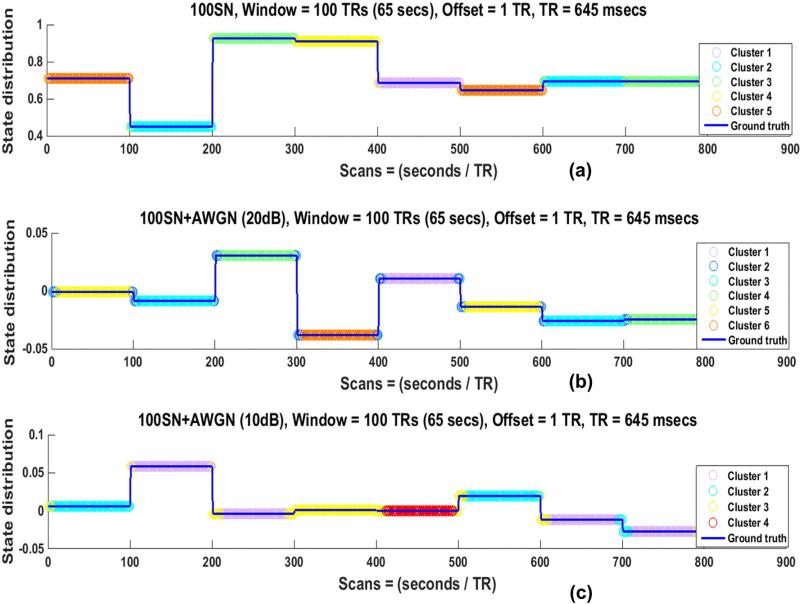
Fig. 11 shows the state distribution of the noisy 100SN for 100 TRs windows. As expected from the result of the SWC for a node pair, the state transitions are lost after the addition of noise as can be observed from the lack of a color change at the discontinuities in (b) and (c). Similar results were observed for other regularly changing SNs (50SN for 50TRs window and 200SN for 200 TRs window).
Fig. 11.
State distribution of 100SN for 100 TRs window before addition of the noise in (a) and after addition of additive white Gaussian noise (AWGN) with an SNR of 20 dB in (b) and 10 dB in (c). Addition of noise resulted in poorer identification of state transitions.
We also examined a more typical reduction in SNR for rs-fMRI by taking a single voxel as a node of our SNs instead of 3 × 3 × 3 voxel ROIs. All analysis was performed on seven of the nine subjects for four SNs (RandSN, 50SN, 100SN, and 200SN) and mean percentages of full and closest state identifications are shown in Supplementary Table 1. The results indicate that higher SNR improves the estimation of SWC, state durations, and transitions.
Effects of window offset
Up to this point all of the results were shown for a window offset of one TR (645 ms). Since some previous studies have used larger offsets (Chang and Glover, 2010; Chang et al., 2013; Kucyi and Davis, 2014; Wilson et al., 2015), we also explored the effect of larger offsets on the SWC and the resulting Cstates. It was observed earlier in this study (Fig. 7(c)) that for a window of size 100 TRs applied to 100SN, the SWC transitioned at the point of the actual state change. When the same 100 TRs window is applied to 100SN but with a window offset of one-fourth (25 TRs) or one-half (50 TRs) the window length, an additional transition occurring at approximately 25 TRs (50 TRs) after the transition at the state change point is introduced into the SWC time course (Supplementary Figs. 16(b) and (c), blue oval markers show one of these additional transitions in each of these figures).
We also plotted state distributions for 100SN with a 100 TRs window for an offset of one TR, an offset of 25 TRs, and an offset of 50 TRs (Supplementary Fig. 17). State durations were successfully identified for an offset of one TR, but not for an offset of 25 TRs and 50 TRs. Almost all of the actual states were separated into two Cstates in both cases, one of which was equal to the duration of the offset (black ovals). Results were consistent across all windows and all SNs.
Effects of window type
A rectangular window was used for the majority of our simulations, similar to most of the previous dynamic FC studies (Keilholz et al., 2013; Shakil et al., 2014; Leonardi and Van De Ville, 2015), but we also explored the effects of window type on the SWC results. Hamming and Hanning windows of the same lengths as the rectangular windows (25 TRs, 50 TRs, 100 TRs, and 200 TRs) were used for the SWC computations. The state distributions for a Hamming window for 50SN, 100SN, and 200SN and windows lengths of 50 TRs, 100 TRs, and 200 TRs respectively are shown in Supplementary Fig. 18. Results for the Hanning window were similar. The Cstates are not localized to true correlation states and state transitions are poorly identified. The results for both tapered windows were worse than those obtained for rectangular windows, possibly because the SNs in our model have sharp discontinuities that are tracked better by rectangular windows.
Effects of filtering
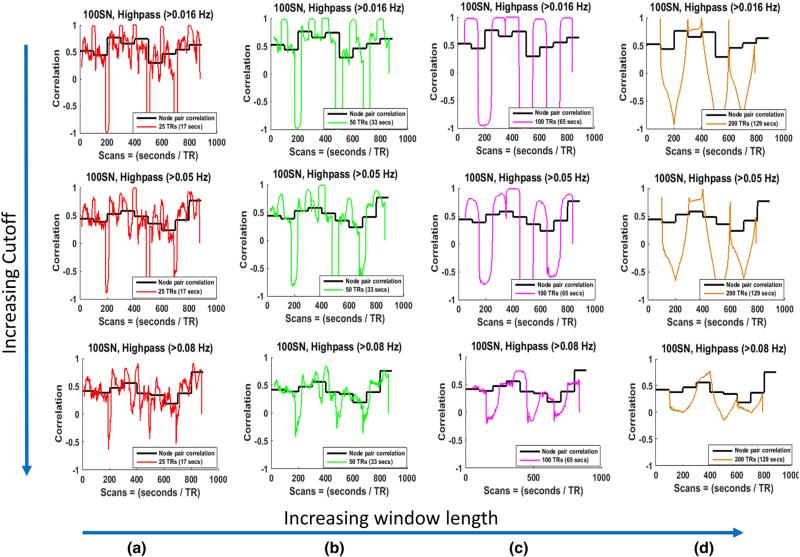
Recent studies showed that the SWC for two sinusoids approaches the steady state value only if the window size ≥1/fmin, where fmin is the minimum frequency of the correlating time series (Leonardi and Van De Ville, 2015; Shakil et al., 2015). To determine if matching the lowest frequency in the time course of the signals to the window size impacts the calculation of the SWC and corresponding clustering in more complex data, we performed the same analysis on the SNs created using highpass filtered (>0.016 Hz, >0.05 Hz, and >0.08 Hz) time series. Based on the findings of (Leonardi and Van De Ville, 2015; Shakil et al., 2015), windows of size greater than or equal to 62.5 s (for fmin = 0.016 Hz), 20 s (for fmin = 0.05 Hz), and 12.5 s (for fmin = 0.08 Hz) would result in the SWC approaching the steady state values for our highpass filtered SNs. The SWC between the node pairs were computed and compared with the actual correlations for these highpass filtered SNs. The results for one node pair for 100SN are given in Fig. 12. We observed decrease in the SWC values for almost all of our windows (17, 33, 65, 129 s) when fmin was 0.08 Hz (row 3), but this reduction was more noticeable for larger windows and did not follow the steady state correlations shown by black lines. Furthermore, increasing the highpass cutoff also resulted in the loss of sharp transitions at the state change point even when the window size was the same as the state duration, which can be observed in row 3 (c). The state transitions and duration detections were not strongly influenced by the lower cutoff values of the ideal case (100SN with 100 TRs window) as shown in Supplementary Fig. 19. However, there was some error in the detection of state transitions (Supplementary Fig. 19(c)). This deterioration may be the result of the SWC's inability to detect the state transitions for higher cutoffs as was observed in Fig. 12 (row 3 (c)). Since most of the power in the low frequency BOLD fluctuations lie in the very low frequencies (<0.1 Hz), the filters with higher cutoffs discard much of the information. Similar results were observed in all the subjects.
Fig. 12.
Actual and sliding window correlations of one node pair for 100SN for different highpass cutoffs. The cutoffs are increasing along the rows, while the window sizes are increasing along the columns. (a) Results for 25 TRs (17 s) window. The window size is larger than minimum window size (1/fmin = 12.5 s) to suppress the spurious correlations for row 3 only. Not much difference in the SWC values are observed from first two rows for the same window size. (b) Results for 50 TRs (33 s) window. The window size is larger than minimum window size (1/fmin = 20 s, and 12.5 s) to suppress the spurious correlations for row 2, and 3. Significant reduction in the SWC values are observed for cutoff > 0.08 Hz. (c) and (d) follow the same trend as (b) for windows of sizes 100 TRs (65 s), and 200 TRs (129 s) respectively. In (c) the sharp transitions observe for a cutoff of >0.01 and >0.05 Hz (row 2 and 3) are not present for cutoff > 0.08 Hz (row 3).
Effects of the change in the repetition time
The simulations described up to this point were performed using high temporal resolution data acquired using a multiband sequence. Most rsfMRI studies, however, are performed with longer TRs (1–3 s), which results in a lower sampling rate and provide fewer points in any given window. To examine the effect of this reduction in samples, the same analysis was performed using data from scans with 1400 ms TR for two subjects for low SNR SNs formed by single voxels as nodes. RandSN changed states at random times, while 50SN, 100SN, and 200SN changed states after every 24 TRs, 47 TRs, and 93 TRs, respectively giving the states almost the same durations in seconds as that of the SNs formed with 645 ms TR scans. Window sizes of 13, 24, 47, and 93 TRs (≈19, 34, 66, 131 s) were used, which were almost equal (in seconds) to the sizes of the windows used for the 645 ms TR scans. The results of the analysis were similar to the results for the 645 ms TR scans, illustrating that the outcome of the SWC depends more strongly on the size of the window (in seconds) used than on the TR of the scans. We did not perform this analysis on higher SNR ROI based SNs because the results were so similar for the voxel-based SN case.
Effects of regions-of-interest selection
In order to determine if the results were dependent upon the ROIs selected, we selected entirely different sets of ROIs for all the five resting-state networks as shown in Supplementary Fig. 3. Afterwards, we formed the SNs with these ROIs and performed the analysis on new SNs. The mean percentages of full state identification for these SNs are shown in Supplementary Table 2. Even though the means differ from the previous SNs (Table 2), the trend remained the same with highest percentages obtained for a window when the size of the window was equal to the intervals of the SN (red entries in the Supplementary Table 2).
Discussion
Dynamic analysis of resting state FC holds the potential to provide new insights into the organization and interplay of network activity in the brain. Already it has been shown that dynamic analysis can improve sensitivity to changes that occur in psychiatric disorders (Sakoğlu et al., 2010; Jones et al., 2012; Damaraju et al., 2014; Li et al., 2014). One of the major challenges in furthering the field of dynamic FC is a lack of gold standards that can be used to evaluate how well analysis techniques reflect the actual network interactions in a system where the timing, duration, and composition of states are all unknown.
Previous studies have taken two approaches to the problem, either linking network dynamics to measurable behavioral outputs (Thompson et al. 2013, Kucyi and Davis, 2014) or tying them to simultaneous measures of electrical activity made with other modalities (Tagliazucchi et al., 2012; Chang et al., 2013; Pan et al., 2013; Thompson et al., 2013a, 2013b). These studies impart confidence that dynamic analysis methods reflect at least some of the underlying neural dynamics. However, they do not readily adapt to a systematic investigation of analysis techniques and parameters.
SWC is one of the simplest and most widely used approaches to dynamic functional analysis, and is sometimes followed by clustering to identify common states of correlation patterns throughout the brain (Allen et al., 2014; Shakil et al., 2014). Most dynamic functional connectivity studies use SWC but the method's performance depends on the choice of parameters like window length, offset etc. In this study, we formed SNs from real rsfMRI data in order to obtain SWC results similar to those of real FC networks but with controlled state transition times.
Two recent studies (Hindriks et al., 2015; Shine et al., 2015) analyzed the performance of the SWC for dynamic analysis and identified shortcomings of the method. (Hindriks et al., 2015) reported that the mere presence of fluctuations in the SWC time series cannot be taken as the evidence of dynamic functional connectivity and that it is important to select an appropriate null hypothesis when performing the dynamic FC analysis and (Shine et al., 2015) compared their proposed method with the SWC. Our approach complements these studies since similar to Shine et al. (2015), we observed a significant influence of a change in signal amplitude on the SWC results for all windows (Fig. 3, Supplementary Figs. 8 and 9). We also observed spurious fluctuations in the results of the SWC for actual rsfMRI networks (Fig. 2) and for the SNs (Fig. 3, Supplementary Figs. 8 and 9), supporting the findings of Hindriks et al. (2015) that the mere presence of fluctuations in the SWC results cannot be taken as evidence of dynamic FC. These similarities in the results validate the use of our SNs for evaluating the impact of parameter choice on the performance of SWC.
SWC has successfully identified changes linked to electrophysiology, behavior or disorders in previous studies (Tagliazucchi et al., 2012; Chang et al., 2013; Thompson et al., 2013a, 2013b; Damaraju et al., 2014; Li et al., 2014), despite its relatively poor performance in this study. As mentioned in the Sharp transitions section, our simulated networks are only approximations of actual brain states, which may not exhibit the same sharp state transitions. However, there are several reasons why SWC could perform adequately in previous studies. First, it is important to note that the “ground truth” in these empirical studies is unknown, preventing an examination of the accuracy of the SWC measurements. Instead, BOLD SWC is compared to other measures of neural activity, behavioral output, or across populations. In studies that relate SWC of BOLD to sliding window measures of neural activity (Tagliazucchi et al., 2012; Chang et al., 2013; Thompson et al., 2013a, 2013b), the sliding windows might affect both signals in a similar manner, preserving relationships between them. In studies that look at transitions and states across groups (Damaraju et al., 2014; Li et al., 2014), a similar effect may be preserving the differences between groups even if the actual correlation is poorly estimated. For comparison to behavior, ROIs are often large and may increase the SNR enough to improve SWC performance (Thompson et al., 2013a, 2013b).
The effects of different parameter choices for the SWC and clustering were examined using these model networks. While the SNs were created using data from actual resting state networks, several implicit assumptions may affect their similarity to actual networks in the brain:
-
1)
All nodes were assumed to be connected at all times. The SNs were spatially invariant, meaning that nodes did not join or leave the network at any time.
-
2)
Abrupt transitions occurred between consecutive states. In practice, the characteristics of ‘state’ transitions in the brain are not known, but it is likely that smoother transitions over the course of seconds occur alongside or instead of abrupt transitions.
-
3)
The average duration of a real brain ‘state’ is not known. For our study it was modeled as 13 to 129 s, ranging from time scales on the order of a cohesive thought to more long-term changes (in vigilance, for example) that occur on the scale of minutes. It is likely that these time scales are expressed simultaneously in the actual brain.
These assumptions are excellent targets for future extensions of this work and more complex model networks may prove more realistic for examining the accuracy of dynamic analysis techniques. Nevertheless, this simple model provides some guidance about the impact of parameter choice on the SWC.
Sharp transitions
Despite the fact that the data in our SNs was from rsfMRI scans, our SNs had sharp state transitions as shown in Fig. 1 and Supplementary Fig. 4, which is different from the gradual changes expected in real resting-state networks. These sharp state transitions were due to abrupt changes in the signal intensities at the state transition points (Lindquist et al., 2014) that acted as outliers, especially for smaller windows (25 TRs, 50 TRs) in which the averaging in the SWC includes a smaller number of points. Although the transitions in our SNs may not reflect the actual state transitions of rsfMRI networks, they clearly identify the important issue of SWC's very high sensitivity to even one large short-lived intensity change which may happen due to some form of noise. Similar to the results of actual rsfMRI networks (Fig. 2., Supplementary Fig. 7) and previous studies (Hutchison et al., 2013a, 2013b; Keilholz et al., 2013; Wilson et al., 2015) the SWC fluctuated between negative and positive values even when the actual correlation was positive for the SNs. Furthermore, similar to Wilson et al. (2015) we observed that despite the large variations for smaller windows, the pattern and timings of these correlation variations were the same. For example, in Fig. 3, the −1 correlations appear almost at the same points for all the windows, though other lesser variabilities are smoothed out by an increase in the size of the window, as expected. However, our results differ from those of Wilson et al. (2015) in the sense that the correlations vary between extreme values of −1 and 1 regardless of the actual correlation values between two state transition points. A closer inspection of these results show that these variations are mostly accompanied by abrupt signal changes at state transition points. Once the SWC value changed at the state transition point, it remained the same in most of the cases till the next abrupt signal change (at the next state transition point) especially for longer windows (pink and brown). This may be because the data between two consecutive state transition points was from the same resting-state networks so would not have any abrupt changes in signal. For smaller windows (red and green) the window size was less than 1/fmin which may have introduced spurious fluctuations (Leonardi and Van De Ville, 2015; Shakil et al., 2015). These results emphasized the vulnerability of the SWC as an efficient and effective dynamic analysis method for FC. While sharp transitions are troublesome for SWC, the same sharp transitions in our SNs in some sense provided a “best case scenario” for clustering, with distinct boundaries between states that resulted in efficient clustering performance by k-means, with perfect identification of state transitions and durations in the raw SNs.
Window length
As expected, window length had a substantial effect on the amount of variability captured by the SWC and was the single most important determinant of the overall accuracy of the technique. Smaller windows (17 and 33 s) had more variability in the SWC results regardless of the SN. Some of these fluctuations maybe the result of spurious fluctuations arising from the SWC algorithm itself based on the findings of recent studies (Leonardi and Van De Ville, 2015; Shakil et al., 2015). In these studies it was reported that the minimum window length to avoid the spurious fluctuations arising due to the SWC itself should be at least equal to 1/fmin, where fmin is the minimum frequency in the simplified correlating signal. In our study the fmin is 0.016 Hz that corresponds to a minimum window length of 62.5 s and window lengths less than this (17 and 33 s) would give rise to spurious fluctuations. Regardless of window length, we found the overall correspondence between the actual correlation and the SWC to be rather poor. SWC results were more divergent from the actual correlation values at the time of state transitions illustrating that even a single outlier or abrupt change can have significant influence on the SWC output (Lindquist et al., 2014). This effect was more pronounced for smaller windows, since the averaging of SWC includes a smaller number of samples, and a large change in the value of even one sample would greatly influence the averaging. Furthermore, smaller windows would introduce spurious fluctuations also (Leonardi and Van De Ville, 2015; Shakil et al., 2015). Window sizes that were well matched to the duration of the states provided fairly good sensitivity to state transitions but still gave poor estimates of the actual correlation values. Perhaps because of the sensitivity to transitions, clustering after the SWC provided surprisingly good state identification for window lengths well matched to state durations, despite the poor estimation of correlation values. To some degree, the clustering was able to salvage information from the relatively poor accuracy of the SWC time courses. However, this was only true for certain situations where window lengths aligned well with state durations. It was also noted that in some situations, the duration of Cstates appears to be dominated by the window length rather than by the actual underlying state durations. Surprisingly, the clustering of raw SNs resulted in perfect state transition and duration identifications, giving far better performance than when SWC was used. These ideal results may be due to the sharp state transitions of our SNs which would provide distinct grouping to the clustering algorithm. However, these results also suggest that the application of the SWC is in general detrimental to the identification of brain states, and that clustering based on raw data provides a cleaner estimate of the patterns of underlying activity. If windowing methods are used, the results of these simulations highlight the need for development of adaptive techniques to maximize sensitivity to states of different durations.
Inter -subject variability was influenced by the size of the window more than the type of the SN. The smaller windows could capture the short-lived variabilities of the node pair correlations much better than the larger windows as is evident from Fig. 4 and Supplementary Fig. 7. However, the largest correlation was present for 200SN in almost all the subjects for all windows because the states were stable for the longest duration (200 TRs). This result suggests that the SWC could perform well when the underlying network is changing very slowly.
Signal-to-noise ratio (SNR)
Additive white Gaussian noise (AWGN) deteriorated the results of both SWC and k-means clustering since it reduced the identification of sharp state transitions. This AWGN (20 dB and 10 dB SNRs) was added to the whole length of time series in SNs. This resulted in less sharp state transitions and the SWC result being closer to actual correlation value influencing the state assignments of the SWC results even for best case scenarios (50SN with 50TRs, 100SN with 100 TRs windows, and 200SN with 200 TRs window). The noise that we added was random, but actual noise is rsfMRI often arises from physiological sources (heart beat at ≈1 Hz, respiration at ≈0.3 Hz) and head motion (Kruger and Glover, 2001; Greve et al., 2013; Bright and Murphy, 2015) and is spatially structured. These types of noise may introduce structured deterioration in the SWC results which may introduce smooth but large variability in the correlation resulting in poor performance of the SWC in estimating the actual correlation. Influence of these structured noises is reduced during the preprocessing by filtering (heart and respiration) and regressing (motion parameters), however, they cannot be eliminated completely. In addition, the different filtering ranges are used (0.01–0.08 or 0.01–0.1 Hz usually) which may also influence the final output as discussed in the Time domain filtering section and reported by Leonardi and Van De Ville (2015) and Shakil et al. (2015).
Number and size of ROIs
For actual FC networks the number of ROIs is different for different networks. For example, the atlas used in this study (from FIND lab, http://findlab.stanford.edu/home.html) has a total of 90 functional ROIs and out of these the number of ROIs for different FC networks vary from two (primary and higher visual networks) to twelve (post salience network). However, another expanded atlas by the same lab covering more gray matter contains 499 functional ROIs. The selection of seven nodes (ROIs) for our SNs was random but this number had no influence on the results of the SWC between the node pairs and affected the results of clustering only in that fewer nodes gave poorer clustering results. We performed an additional analysis by reducing the number of nodes to three and clustered the SWC using k-means. In that analysis for most of the cases, the best Silhouette mean was obtained for two clusters only, which would result in assignment of different states to the same Cstates and deteriorate the clustering results. Increasing the number of nodes should improve the state assignments as each additional node provides more information that can be used to distinguish between states.
Sizes of the ROIs in actual FC networks range from a few voxels to hundreds of voxels. In this study we found that the size of the ROI influences the results of the SWC because the SNR depends on the size of the ROI. Reducing the size of the ROI to a single voxel deteriorated the correct state identifications (Supplementary Table 1) compared to larger ROIs (Table 1). As a result, it may be deduced that the SWC and k-means combination would provide better estimates of actual correlations and state distributions if bigger ROIs were used that are similar to ROIs of real FC networks.
Window offset
An offset of a single TR proved best for detecting both state changes and durations. Increasing the window offset reduced the sensitivity of the SWC towards the correlation changes, as well as its ability to detect state transitions and durations. The larger the offset, the worse the results became. A closer inspection of the results showed that this deterioration may be due to shifts in the SWC results due to uncertainty in the times which best reflect the average in the window, and to the introduction of new transitions at intervals equal to the offset. To some extent, this may be related to the sharp transitions between networks used for this study and could well be mitigated for networks with more gradual transitions.
Time domain filtering
Typical rsfMRI scans use a bandpass filter to minimize contributions from noise (typically 0.01–0.1 Hz or 0.01–0.08 Hz). Following the findings of Leonardi and Van De Ville (2015) and Shakil et al. (2015), we applied highpass filters with different frequency cutoffs to the data. We observed that the highpass filtered SNs provide results similar to bandpass filtered networks when the highpass cutoff was the same as the that of the bandpass lower cutoff. As we increased the highpass cutoff to higher values (>0.05 Hz and >0.08 Hz) magnitudes of the SWC results for windows larger than 1/fmin were reduced, but the ability of the SWC to follow actual correlation values did not improve though the sharp state transitions of the SWC results were lost. The ability of the clustering algorithm to identify state transitions deteriorated, particularly for larger windows. This suggests that in our model, other sources of variability dominate the signal rather than the instability introduced by a mismatch in frequency and window length. One reason for the differences observed from Leonardi and Van De Ville (2015) and Shakil et al. (2015) may be that in our case, correlations were computed for the duration of the states, rather than for the whole scan time, making the sample length shorter. In some sense, the finding that the inclusion of lower frequencies does not greatly impact the performance of the SWC is encouraging. The combined requirements of highpass filtering to match the window length and low pass filtering to limit contributions from noise would place rather stringent limits on the range of windows that could be used to examine the SWC dynamics. The unfortunate corollary of this finding is that since the inclusion of lower frequencies does not greatly impact the SWC, the highpass filter does not appreciably improve the SWC performance, leaving it far from ideal as an analysis technique.
Repetition time (TR)
TR did not substantially affect the results of the SWC as far as sensitivity towards correlation changes and state identification were concerned. While shorter TRs provide more samples which should theoretically improve the estimation of correlation values, longer TRs often provide stronger signal-to-noise ratios that could offset this effect. As many rsfMRI studies routinely use TRs of 1–3 s, it is encouraging to see that the reduced sampling rate can have a minimal effect on the SWC and clustering.
Considerations for future SWC studies
The SWC gives a mediocre estimation of the actual underlying correlation even in the best of circumstances for our SNs, as shown in Fig. 3. However, with appropriate window lengths it can detect transition points fairly well, at least in networks with abrupt or sharp transitions. Clustering based on these window lengths results in reasonably accurate state identification. The results from our model suggest that the use of a window length equal to or longer than the expected state duration will provide the most accurate state characterizations. This requirement will of course need to be balanced with the necessity of using windows short enough to provide an estimation of changes over time. Shorter windows also identified greater proportions of actual transitions between states, though this is counterweighted by the number of false transitions that occur. An offset of a single TR and a rectangular window provided the most accurate results, although this may be due in part to the abrupt changes in the model network.
Finally, the results of the SWC applied to QPeriodicSN and RandSN (arguably the most realistic models for the types of transitions expected in rsfMRI) highlight the way that window size impacts the resulting states. A large proportion of the states identified were approximately equal in length to the sliding window, regardless of the length of the actual state duration. This may be an important point to consider during the interpretation of the SWC-based clustering on real rsfMRI data.
A surprising and novel aspect of this study was perfect state transition and duration identification obtained using k-means clustering on the raw SN signal. These ideal results may be partly due to abrupt transitions of our SNs and will require further examination. However, they suggest that researchers may wish to consider using clustering-based approaches on the raw signal to identify the states of FC, rather than the SWC time courses whenever possible.
Future directions
This study evaluated the performance of the SWC as a suitable method for capturing network dynamics with rsfMRI. After examining the effects of various parameters on the dynamic analysis results of the SWC, we observed that no set of parameters were suitable for optimum performance of the SWC under conditions that are likely to exist in the human brain (e.g., random lengths of state durations). Windows that matched the intervals at which brain states changed provided the most accurate information, but this situation is unlikely to be found in rsfMRI.
The clear outcome of this study is the need to develop algorithms that can adaptively detect state transitions of different durations. This is a clear target for future research and may be addressed by the use of multiscale approaches (e.g., wavelet-based analysis) that can simultaneously examine multiple time scales. An ideal analysis method would identify each state based on quantitative relationships between areas, along with transition points between states. The challenge of identifying an optimum number of ‘brain states’ is another target for future efforts.
Any algorithm that aims to address these issues will need to be tested against modeled network dynamics, but even the best performance on a model network will not guarantee success in identifying transitions of interest in actual rsfMRI data. Validation with multimodal data and behavioral outputs will be necessary to conclusively establish a link between the output of the algorithm and activity within the brain. Despite the large amount of work, remaining, dynamic rsfMRI has the potential to have a profound impact on our understanding of the brain.
Supplementary Material
Acknowledgments
This research was supported by NIH R01NS078095. The authors would like to thank their lab members Mr. Joshua Grooms and Mr. Jacob Billings for being helpful by discussing various aspects of this study. The authors would also like to thank their lab members Mr. Anzar Abbas and Mr. Hyun Koo Chung for proofreading the paper.
Abbreviations
- rsfMRI
resting-state functional magnetic resonance imaging
- FC
functional connectivity
- SWC
sliding window correlation
- SN
simulated network
- GT
ground truth
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2016.02.074.
References
- Allen E, Damaraju E, Plis S, Erhardt E, Eichele T, Calhoun V. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex. 2014;24(3):663–676. doi: 10.1093/cercor/bhs352. http://dx.doi.org/10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, Murphy K. Is fMRI “noise” really noise? Resting state nuisance regressors remove variance with network structure. NeuroImage. 2015;114(1):158–169. doi: 10.1016/j.neuroimage.2015.03.070. http://dx.doi.org/10.1016/j.neuroimage.2015.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover G. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50(1):81–89. doi: 10.1016/j.neuroimage.2009.12.011. http://dx.doi.org/10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Liu Z, Chen M, Liu X, Duyn J. EEG correlates of time-varying BOLD functional connectivity. NeuroImage. 2013;15(72):227–236. doi: 10.1016/j.neuroimage.2013.01.049. http://dx.doi.org/10.1016/j.neuroimage.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E, Allena E, Belger A, Ford J, McEwen S, Mathalon D, Mueller B, Pearlson G, Potkin S, Preda A, Turner J, Vaidya J, van Erp T, Calhoun V. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. http://dx.doi.org/10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Handwerker D, Robinson M, Hoy C, Buchanan L, Saad Z, Bandettini P. The spatial structure of resting state connectivity stability on the scale of minutes. Front. Neurosci. 2014 doi: 10.3389/fnins.2014.00138. http://dx.doi.org/10.3389/fnins.2014.00138. [DOI] [PMC free article] [PubMed]
- Greve D, Brown G, Mueller B, Glover G, Liu T. A survey of the sources of noise in fMRI. Psychometrika. 2013;78(3):396–416. doi: 10.1007/s11336-012-9294-0. http://dx.doi.org/10.1007/s11336-013-9344-2. [DOI] [PubMed] [Google Scholar]
- Handwerker D, Roopchansingh V, Gonzalez-Castillo J, Bandettini P. Periodic changes in fMRI connectivity. NeuroImage. 2012;63(3):1712–1719. doi: 10.1016/j.neuroimage.2012.06.078. http://dx.doi.org/10.1016/j.neuroimage.2012.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriks R, Adhikari M, Murayama Y, Ganzetti M, Mantini D, Logothetis N, Deco G. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? NeuroImage. 2015;127:242–256. doi: 10.1016/j.neuroimage.2015.11.055. http://dx.doi.org/10.1016/j.neuroimage.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison R, Womelsdorf T, Allen E, Bandettini P, Calhoun V, Corbetta M, Della Penna S, Duyn J, Glover G, Gonzalez-Castillo J, Handwerker D, Keilholz S, Kiviniemi V, Leopold D, de Pasquale F, Sporns O, Walter M, Chang C. Dynamic functional connectivity: promise, issues, and interpretations. NeuroImage. 2013a;(80):360–378. doi: 10.1016/j.neuroimage.2013.05.079. http://dx.doi.org/10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed]
- Hutchison R, Womelsdorf T, Joseph G, Everling S, Menon R. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum. Brain Mapp. 2013b;34(9):2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Hu X, Deshpande G. Behavioral relevance of the dynamics of the functional brain connectome. Brain Connect. 2014;4(9):741–759. doi: 10.1089/brain.2014.0300. http://dx.doi.org/10.1089/brain.2014.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Vemuri P, Murphy M, Gunter J, Senjem M, Machulda M, Przybelski S, Gregg B, Kantarci K, Knopman D, Boeve B, Petersen R, Jack CJ. Nonstationarity in the “resting brain's” modular architecture. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0039731. http://dx.doi.org/10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilholz S, Magnuson M, Pan W-J, Willis M, Thompson G. Dynamic properties of functional connectivity in the rodent. Brain Connect. 2013;3(1):31–40. doi: 10.1089/brain.2012.0115. http://dx.doi.org/10.1089/brain.2012.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Vire T, Remes J, Elseoud A, Starck T, Tervonen O, Nikkinen J. A sliding time-window ICA reveals spatial variability of the default mode network in time. Brain Connect. 2011;1(4):339–347. doi: 10.1089/brain.2011.0036. http://dx.doi.org/10.1089/brain.2011.0036. [DOI] [PubMed] [Google Scholar]
- Kruger G, Glover G. Physiological noise in oxygenation-sensitive magnetic resonance imaging. Magn. Reson. Med. 2001;46:631–637. doi: 10.1002/mrm.1240. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Davis K. Dynamic functional connectivity of the default mode network tracks. NeuroImage. 2014;100:471–480. doi: 10.1016/j.neuroimage.2014.06.044. http://dx.doi.org/10.1016/j.neuroimage.2014.06.044. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Salomons T, Davis K. Mind wandering away from pain dynamically engages antinociceptive and default mode brain networks. Proc. Natl. Acad. Sci. U. S. A. 2013;110(46):18692–18697. doi: 10.1073/pnas.1312902110. http://dx.doi.org/10.1073/pnas.1312902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N, Van De Ville D. On spurious and real fluctuations of dynamic functional connectivity during rest. NeuroImage. 2015;104:430–436. doi: 10.1016/j.neuroimage.2014.09.007. http://dx.doi.org/10.1016/j.neuroimage.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Leonardi N, Richiardi J, Van De Ville D. Functional Connectivity EIGENNETWORKS Reveal Different Brain Dynamics in Multiple Sclerosis Patients. ISBI; San Francisco: 2013a. [Google Scholar]
- Leonardi N, Richiardi J, Gschwind M, Simioni S, Annoni J, Schluep M, Vuilleumier P, Van De Ville D. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. NeuroImage. 2013b;83:937–950. doi: 10.1016/j.neuroimage.2013.07.019. http://dx.doi.org/10.1016/j.neuroimage.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu D, Jiang X, Jin C, Zhang X, Guo L, Zhang J, Hu X, Li L, Liu T. Dynamic functional connectomics signatures for characterization and differentiation of PTSD patients. Hum. Brain Mapp. 2014;35(4):1761–1778. doi: 10.1002/hbm.22290. http://dx.doi.org/10.1002/hbm.22290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M, Xu Y, Nebel M, Caffo B. Evaluating dynamic bivariate correlations in resting-state fMRI: a comparison study and a new approach. NeuroImage. 2014;101:531–546. doi: 10.1016/j.neuroimage.2014.06.052. http://dx.doi.org/10.1016/j.neuroimage.2014.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W, Magnuson M, Hasenkamp W, Schwarb H, Schumacher E, Barsalou L, Keilholz S. Spatiotemporal dynamics of low frequency BOLD fluctuations in rats and humans. NeuroImage. 2011;54(2):1140–1150. doi: 10.1016/j.neuroimage.2010.08.030. http://dx.doi.org/10.1016/j.neuroimage.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Xie L, Jin C, Li X, Zhu D, Jiang R, Chen Y, Zhang J, Li L, Liu T. Characterizing and differentiating brain state dynamics via hidden Markov models. Brain Topogr. 2015;28(5):666–679. doi: 10.1007/s10548-014-0406-2. http://dx.doi.org/10.1007/s10548-014-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W-J, Thompson G, Magnuson M, Jaeger D, Keilholz S. Infraslow LFP correlates to resting-state fMRI BOLD signals. NeuroImage. 2013;74 doi: 10.1016/j.neuroimage.2013.02.035. http://dx.doi.org/10.1016/j.neuroimage.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts S, Barkhof F, Goekoop R, Stam C, Scheltens P. Altered resting state networks in mild cognitive impairment and mild Alzheimer's disease: an fMRI study. Hum. Brain Mapp. 2005;26(4):231–239. doi: 10.1002/hbm.20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. http://dx.doi.org/10.1016/0377-0427(87)90125-7. [Google Scholar]
- Sadaghiani S, Poline J, Kleinschmidt A, D'Esposito M. Ongoing dynamics in large-scale functional connectivity predict perception. Proc. Natl. Acad. Sci. U. S. A. 2015;112(27):8463–8468. doi: 10.1073/pnas.1420687112. http://dx.doi.org/10.1073/pnas.1420687112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoğlu Ü, Pearlson G, Kiehl K, Wang Y, Michael A, Calhoun V. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. MAGMA. 2010;23(5–6):351–366. doi: 10.1007/s10334-010-0197-8. http://dx.doi.org/10.1007/s10334-010-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D, Huston J. The sliding window correlation procedure for detecting hidden correlations: existence of behavioral subgroups illustrated with aged rats. J. Neurosci. Methods. 2002;121(2):129–137. doi: 10.1016/s0165-0270(02)00224-8. http://dx.doi.org/10.1016/S0165-0270(02)00224-8. [DOI] [PubMed] [Google Scholar]
- Shakil S, Keilholz S, Lee C-H. IEEE Conference on BioInformatics and Bioengineering. Maryland: 2015. On frequency dependencies of sliding window correlation. [Google Scholar]
- Shakil S, Magnuson M, Keilholz S, Lee C-H. IEEE Eng Med Biol Soc. Chicago: 2014. Cluster-based Analysis for Characterizing Dynamic Functional Connectivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Koyejo O, Bell P, Gorgolewski K, Gilat M, Poldrack R. Estimation of dynamic functional connectivity using multiplication of temporal derivatives. NeuroImage. 2015;122:399–407. doi: 10.1016/j.neuroimage.2015.07.064. http://dx.doi.org/10.1016/j.neuroimage.2015.07.064. [DOI] [PubMed] [Google Scholar]
- Shirer W, Ryali S, Rykhlevskaia E, Menon V, Greicius M. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb. Cortex. 2012;22(1):158–165. doi: 10.1093/cercor/bhr099. http://dx.doi.org/10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Muhlau M, Calhoun V, Eichele T, Laer L, Drzezga A, Forstl H, Kurz A, Zimmer C, Wohlschlager A. Selective changes of resting-state networks in individuals at risk for Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104(47):18760–18765. doi: 10.1073/pnas.0708803104. http://dx.doi.org/10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, von Wegner F, Morzelewski A, Brodbeck V, Laufs H. Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Front. Hum. Neurosci. 2012;28(6):339. doi: 10.3389/fnhum.2012.00339. http://dx.doi.org/10.3389/fnhum.2012.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G, Magnuson M, Merritt M, Schwarb H, Pan W-J, McKinley A, Tripp L, Schumacher E, Keilholz S. Short time windows of correlation between large scale functional brain networks predict vigilance intra-individually and inter-individually. Hum. Brain Mapp. 2013a;34(12):3280–3298. doi: 10.1002/hbm.22140. http://dx.doi.org/10.1002/hbm.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G, Merritt M, Pan W-J, Magnuson M, Grooms J, Jaeger D, Keilholz S. Neural correlates of time-varying functional connectivity in the rat. NeuroImage. 2013b;83:826–836. doi: 10.1016/j.neuroimage.2013.07.036. http://dx.doi.org/10.1016/j.neuroimage.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuorImage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Wilson R, Mayhew S, Rollings D, Goldstone A, Przezdzik I, Arvanitis T, Bagshaw A. Influence of epoch length on measurement of dynamic functional connectivity in wakefulness and behavioural validation in sleep. NeuroImage. 2015;112:169–179. doi: 10.1016/j.neuroimage.2015.02.061. [DOI] [PubMed] [Google Scholar]
- Xia W, Wang S, Sun Z, Bai F, Zhou Y, Yang Y, Wang P, Huang Y, Yuan Y. Altered baseline brain activity in type 2 diabetes: a resting-state fMRI study. Psychoneuroendocrinology. 2013;38(11):2493–2501. doi: 10.1016/j.psyneuen.2013.05.012. http://dx.doi.org/10.1016/j.psyneuen.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Zang YF, He Y, Zhu CZ, Cao QJ, Sui MQ, Liang M, Tian LX, Jiang TZ, Wang YF. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007;29(2):83–91. doi: 10.1016/j.braindev.2006.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.