Abstract
P21 activated kinases-1 (PAK-1)is implicated in various diseases. It is inhibited by thesmall molecule ‘Inhibitor targeting PAK1 activation-3’ (IPA-3), which is highly specific but metabolically unstable. To address this limitation we encapsulatedIPA-3 in sterically stabilized liposomes (SSL). SSL-IPA-3 averaged 139 nm in diameter, polydispersity index (PDI) of 0.05, and a zeta potential of −28.1, neither of which changed over 14 days; however, the PDI increased to 0.139. Analysis of liposomal IPA-3 levels demonstrated good stability, with 70% of IPA-3 remaining after 7 days. SSL-IPA-3 inhibited prostate cancer cell growth in vitro with comparable efficacy to free IPA-3. Excitingly, only a 2 day/week dose of SSL-IPA-3 was needed to inhibit the growth of prostate xenografts in vivo, while a similar dose of free IPA-3 was ineffective. These data demonstrate the development and clinical utility of a novel liposomal formulationfor the treatment of prostate cancer.
The small molecule ‘Inhibitor targeting P21-activated kinase-1 (PAK1) activation-3’ (IPA-3) has potential anti-cancer effects, but is metabolically unstable. We encapsulated IPA-3 in sterically stabilized liposomes (SSL) that averaged 139 nm in diameter, polydispersity index (PDI) of 0.05, and a zeta potential of −28.1, which was stable for over 14 days with 70 % of IPA-3 remaining even after 7 days. A 2 day/week administration of 5 mg/Kg dose of SSL-IPA-3 significantly inhibited the growth of prostate xenografts in vivo as compared to similar dose of free IPA-3, demonstrating the potential benefits of SSL-IPA-3 for the management of prostate cancer.
Keywords: PAK-1, IPA-3, Liposomes, Prostate cancer
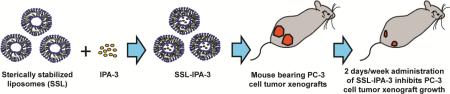
Graphical Abstract
Background
P21 activated kinases (PAKs), which exist in 6 different isoforms, are categorized into group-I and group-II PAKs based on their distinctly different mechanisms of activation and their non-redundant role in the regulation of cellular function1. Group-I PAKs, PAK-1 and PAK-2 in particular, have been implicated in various cancers2. Although PAK-1 is the most expressed PAK isoform in the majority of tissues, the expression level of PAK-1 is below the detectable range in normal prostate tissue and prostatic epithelial cells1. Instead, normal prostate abundantly expresses group-IIPAKs such as PAK-4 and PAK-63. A previous study from ourlaboratory demonstrated that Rac1-mediated cytoskeletal remodeling is essential for prostate cancer invasion4. Since group-I PAKs are the major downstream effectors of Rac1 in mediating cytoskeletal remodeling5, 6, we further investigated the role of PAK-1 in prostate cancer revealing that although not detected in the human prostatic epithelial cells, normal prostate gland and benign prostatic hyperplasia tissues, PAK-1 is abundantly expressed in prostate tumor biopsies and metastasized colonies harbored in the human lung tissues7. Most recently, we demonstrated that PAK-1 is essential for the growth of prostate PC-3 cell tumor xenografts in athymic nude mice as well as transforming growth factor-β (TGFβ)-induced prostate cancer cell epithelial-tomesenchymal transition8. Thus; our studies suggest that PAK-1 is a potential target for prostate cancer therapy.
Among the known inhibitors of PAKs, the most reliable and specific to group-I PAKs is the ‘inhibitor targeting PAK-1 activation-3’(IPA-3), a compound identified through high throughput screening9. A major advantage of IPA-3 is that, unlike other kinase inhibitors, IPA-3 does not interact with and compete for the ATP-binding domain of the kinase10. Instead, itis a non-competitive, allosteric inhibitor of PAK-1 thatinhibits PAK-1 activation,even at higher ATP levels. However, the chemically and metabolically labile nature of IPA-3 becomes a major bottleneck in its use for therapeutic purpose10. Although IPA-3 inhibits prostate cancer cellular function in vitro and tumor growth in vivo, increased frequency (i.e. daily) of drug administration was necessary7, 8. Further, since PAK-1 is necessary for many physiological events11-14, serious toxic side effects of targeting PAK-1 with the free form of IPA-3 cannot be ruled out.This creates a need for a novel method to deliver IPA-3 to prostate cancer cells in vivo to improve the stability and efficacy, to minimize the frequency of drug administration and to reduce any potential side-effects.
Lipid-based nanoparticulate drug carriers, such aslong-circulating sterically-stabilized liposomes (SSL) have the ability to stably encapsulate drugs and facilitate drug delivery15-17. They can alter the pharmacokinetics of the drug, especially compared to free drug and sometimes enhance theirpharmacological activity15. Differences in the half-life and/or tissue and tumor distribution are suggested to be primary drivers for these actions. Additionally, SSL are also suggested to decrease off-targeted toxicity18, 19. Doxil®(Centocor OrthoBiotech, Horsham, PA) is an example of a clinically approved nanoparticle-encapsulating the anti-cancer drug doxorubicin that is essentially a SSL. In addition to their ability to stabilize drugs and enhance their bio-distribution, SSL accumulate passively in solid tumors due to the enhancedpermeability and retention effect mediatedby defects in the vasculature and lack of functionallymphatics20,21. In the current study, we attempted to encapsulate IPA-3 into sterically stabilized liposomes (SSL-IPA-3), characterize the physical properties of these particles, and study the efficacy of these liposomes on prostate cancer cell viability in vitro and growth of PC-3 cell tumor xenografts in athymic nude mice.
Methods
Cell Lines and cell culture
The human prostate cancer cell lines PC-3, LNCaP and DU-145 and the breast cancer cell lines MCF-7 and MDA-231 were purchased from ATCC (Manassas, VA). PC-3, LNCaP, and DU-145 cells were cultured in F12K, RPMI, and EMEM, respectively. MCF-7 and MDA-231cells were cultured in RPMI. All culture media were supplemented with 10% FBS and 1% penicillin/streptomycin antibiotics (ATCC). All the cells were maintained at 37°C in incubators with 5% CO2 and humidified atmosphere.
Chemicals and Reagents
IPA-3 was purchased from Tocris Bioscience (Bristol, United Kingdom). The phospholipids, 1, 2-distearoyl-sn-glycero-3-phosphatidylcholine(DSPC), 1, 2-distearoyl-snglycero-3-phosphatidylethanolamine (DSPE), and 1, 2-distearoyl-sn-glycero-3-phosphoethanolamine–N-[poly (ethyleneglycol) 2000 (DSPE–PEG) were purchased from Avanti Polar Lipids, Inc (Alabaster, Alabama). Cholesterol and MTT [3-(4, 5-dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide] were purchased from Sigma–Aldrich (St. Louis, Missouri). F-12K, EMEM and RPMI cell culture media and their supplements, including antibiotics and fetal bovine serum (FBS), were purchased from ATCC. All other chemicals and solvents were of analytical grade and were obtained from Fisher Scientific (Pittsburgh, PA).
Preparation of Sterically Stabilized IPA-3 Liposomes (SSL-IPA-3)
Liposomes were prepared using the thin lipid hydration method followed by freeze–thaw cycles and a high-pressure extrusion as described previously22, 23.Liposome composition is shown in Table 1. Cholesterol (5 μmol/ml), phospholipids including DSPC (9 μmol/ml) and DSPE–PEG (1 μmol/ml) in chloroform and IPA-3 (4 μmol/ml) in ethanol were added into a round bottom flask, the solvents were then evaporated under vacuum in a water bath at 65°C using a rotary evaporator (BuchiLabortechnik AG, Postlfach, Switzerland). The formed thin film was then hydrated and suspended in ammonium phosphate buffer (250 mM, pH 7.4) or phosphate saline buffer (PBS) to achieve a final lipid concentration of 10 μmol/ml. The formulation then underwent five liquid nitrogen freeze–thaw cycles above the phase transition temperature of the primary lipid, prior to passing five times through a Lipex extruder (Northern Lipids, Inc, Burnaby, BC Canada) at 65°C using double stacked polycarbonate membranes (80 nm, GE Osmonics, Trevose, Pennsylvania). Excess un-encapsulated IPA-3 and lipids were eliminated using dialysis in 10% (w/v) sucrose for at least 20 hours (hrs) with three changings of the dialysis media. The liposome diameter was determined using a dynamic light scattering particle size analyzer (Zetasizer Nano ZS, Malvern Instruments, Enigma Business Park, Grovewood Road, Malvern, Worcestershire, UK).Liposomal size was further confirmed using Tandem Electron Microscopy (TEM) imaging (Supplemental Data Figure 1A). Liposome suspensions were stored at 4°C, protected from light, and used within 24 to 48 hrs of preparation. Empty SSL (made without IPA-3 encapsulation) were also formulated and used as vehicle controls.
Table 1.
Composition of the SSL-IPA-3
| Liposome Component | Concentration |
|---|---|
| 1, 2-distearoyl-sn-glycero-3-phosphocholine (DSPC) | 9 μmol/ml |
| 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG) | 1 μmol/ml |
| Cholesterol | 5 μmol/ml |
| IPA-3 | 4 μmol/ml |
Quantification of IPA-3 in Liposomes
SSL suspensions were collected after an overnight dialysis and IPA-3 was quantified using a spectroscopic method based on its absorbance at 360 nm. Serial dilutions of IPA-3 stock solution were made in acidic ethanol. Absorbance at 360 nm was recorded using a Spectra Max M2 microplate reader (MTX Lab Systems, Inc.Vienna, Virginia 22182 U.S.A.) and plotted against the concentration of IPA-3. Astandard curve (R2> 0.99) was used to determine the concentration of IPA-3 in SSL (Supplemental Data Figure 1B).
Characterization of Size, Zeta Potential and Physical Stability of Liposomal IPA-3
Long circulating liposomes containing IPA-3 were evaluated for physical stability in the storage conditions at 4°C. Drug leakage from liposomes was determined by removing portions of liposome suspension from a pool stored at pH 7.4 and 4°C at various times. IPA-3 liposomes were submitted to dialysis and reanalyzed for IPA-3 content as described above. A significant change in IPA-3 content was used as an indication of liposome stability. Changes in mean diameter and zeta potential of liposomes were also used to assess stability and was monitored overtime using a dynamic light scattering Malvern Zetasizer. Samples were diluted 1 in 10 (v/v) with PBS for the size and zeta potential measurements. As IPA-3 liposomes were to be used for in vivo studies after reconstitution, we assessed IPA-3 stability after reconstitution for increasing time periods, including 3 days, the time corresponding to in vivo dosing. The analysis showed that over 80% of IPA-3 was maintained in SSL after 3 days (Supplemental Figure 2). In fact, good stability appeared to be maintained up to at least 7 days in reconstituted liposomes.
In Vitro Cytotoxicity of Liposomal IPA-3measured using MTT staining
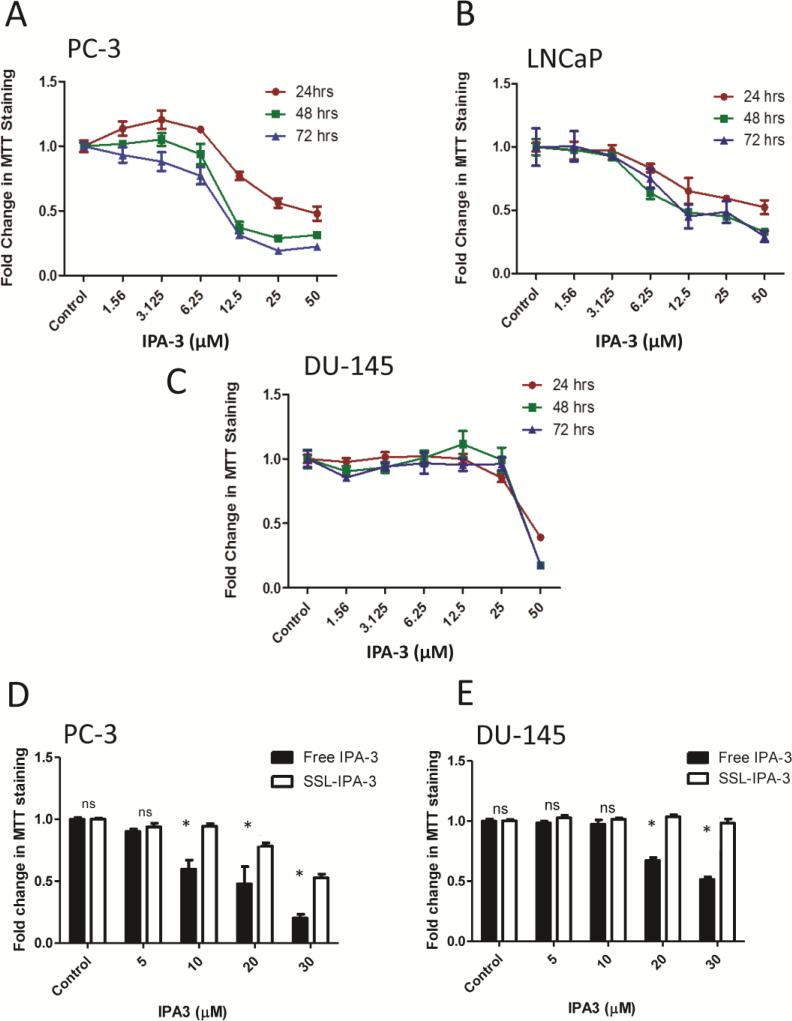
The cytotoxicity of the liposomal IPA-3 as well as the free IPA-3 was determined in three prostate cancer cell lines (PC-3, LNCaP, and DU-145) using the MTT assay24. Free IPA-3 and SSL-IPA-3 were used for comparison. Cells were seeded in 48-well tissue culture plates at 5 × 104 cells/ml and incubated at 37°C in a 5% CO2 incubator for 24 hr. This time was sufficient for the cells to attach and resume their growth. Liposomes were diluted in the different culture media to their final concentrations and experiments were performed in triplicates. Cells were also treated with free IPA-3 (1.56, 3.125, 6.25, 12.5, 25 and 50 μM), DMSO (vehicle for IPA-3) and empty liposomes (vehicles for encapsulated IPA-3; indicated by white bars in the controls of Figure 1D and E) as controls. The cells were incubated for 24, 48, and 72 hrs. MTT was added at each time point, at a final concentration of 0.25 mg/ml and plates were incubated at 37°C. Non-reduced MTT and media were aspirated after 2 hrsand replaced with DMSO to dissolve the MTT formazen crystals. Plates were shaken for 15 min and absorbance was read at 590 nm using a Spectra Max M2 plate reader (BMG Lab Technologies, Inc., Durham, NC). The concentration of IPA-3 resulting in 50% growth inhibition (IC50) was estimated from the concentration-effect curve.
Figure 1.
IPA-3 inhibits prostate cancer cell proliferation. (A-C) Dose- and time-dependent effect of free IPA-3 on MTT staining in human prostate cancer PC-3, LNCaP and DU-145 cells, respectively, 24, 48, and 72 hrs after treatment (n = 3). (D and E)Effect of free IPA-3 and SSL-IPA-3 on MTT staining in human prostate cancer PC-3 and DU-145 cells, respectively, 72 hrs after treatment(n = 3). The white bars in Figure 1D and E for control indicate the effect of empty liposomes. Data are presented as the mean ± SEM of the fold change in MTT staining as function of IPA-3 concentration;*p <0.05; “NS” indicates not significant.
In Vitro Cytotoxicity of Liposomal IPA-3measured usingannexin V and PI staining
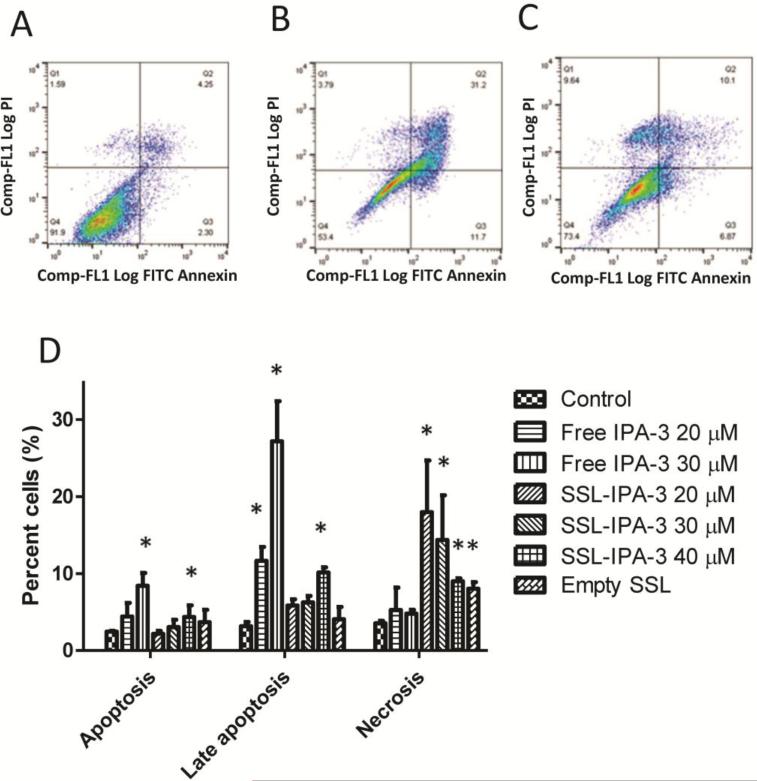
Annexin V and PI staining plus flow cytometry was used to assess NRK cell death as previously described with modifications25. Briefly, cells were seeded and allowed to grow for 24 h prior to addition of free IPA-3, SSL-IPA-3, or empty liposomes. After the indicated time, medium was removed and cells were washed twice with PBS and incubated in binding buffer (10 mMHEPES, 140 mMNaCl, 5 mMKCl, 1 mM MgCl2, 1.8 mM CaCl2, pH 7.4) containing annexin V-FITC (25 mg/ml) and PI (25 mg/ml) for 10 min. Cells were then washed three times with binding buffer, released from monolayers using a rubber policeman, and staining quantified using a Dako Cyan ADP 9 color flow cytometer (Beckman Coulter, Inc., Miami, FL). For each measurement 10,000 events were counted.
Immunoblot Analysis
Proteins from different cell lines were collected in RIPA buffer, which contained protease inhibitor cocktail (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Protein concentrations were determined using the BCA assay. Protein samples of 40 μg were separated on SDS-PAGE gels and transferred to nitrocellulose membranes, which were then blocked in 5% (w/v) nonfat dry milk in Tris-buffered saline–Tween 20 (TBS-T) for 2 hrs. The membranes were then incubated with a rabbit PAK-1 antibody at a dilution of 1:500 in 1% (w/v) BSA TBS-T overnight. Antibodies against GAPDH (Santa Cruz Biotechnology Inc., Santa Cruz, CA) were used at a dilution of 1:200 in 1% (w/v) BSA in TBST for 1 hr. Membranes were then incubated with the appropriate peroxidase-conjugated secondary antibody (Promega, Madison, WI) used at a dilution of 1:2500. The membrane was then washed with TBS-T three times for 10 minutes each and imaged with a Fluorchem SP digital imager (Alpha Innotech, San Leandro, CA, USA). Densitometry was performed using National Institutes of Health Image J software.
In vivo prostate tumor xenograft
All animal procedures listed in this article were performed as per the protocol approved by the Institutional Animal Care and Use Committee at the Charlie Norwood Veterans Affairs Medical Center, Augusta, GA (protocol # 12-06-049). PC-3 cells were grown to 60-70 % confluent in 225-ml flasks. Next, cells were collected and suspended in sterile normal saline. Cell suspension (3 million cells//mouse) was injected subcutaneously (SC) in 6-8-weeks-old male athymicnude mice (Harlan, Indianapolis, IN) (n = 5-6). All the treatments (DMSO, empty liposomes, free IPA-3 (5 mg/Kg), and SSL-IPA-3 (5 mg/kg) were started on day 7 from tumor implantation, and were administered two times a week (i.p.). Tumor diameters were measured with digital calipers on day 7, 14, 21, and 25, and the tumor volume in mm3 were calculated by the modified ellipsoidal formula (Tumor volume = ½[length × width2]). Average size of the tumors prior to the treatment with free IPA-3 and SSL-IPA-3 were 42 mm3. Mice were sacrificed on day 25 and tumors were dissected, weighed, and snap-frozen for further immunohistochemistry analysis.
TUNEL assay
The TUNEL assay for in situ detection of apoptosis was performed using the ApopTag® Fluorescein in situ Apoptosis detection kit (Millipore, MA) as described previously8. Frozen sections taken from prostate tumor xenografts were fixed in 2% paraformaldehyde at 4°C for 30 min. Fixed tissues were then permeabilized in (ethanol: acetic acid [2:1]) and labeled with fluorescein 12-dUTP using terminal deoxynucleotidyl transferase. Nuclei were counterstained with DAPI. Tissue sections were analyzed for apoptotic cells with localized green fluorescence using an inverted fluorescence microscope (Zeiss Axiovert100M, Carl Zeiss, Germany).
Statistical Analysis
All in vitro experiments were repeated at least three times (n = 3) in triplicate. Results are shown as the average of all replicates ± SEM.A two way ANOVA test followed by Bonferroniposttest was used to determine whether the differences between free IPA-3 and SSLIPA-3treatment groups were statistically significant. A two-tailed student'st test was used to compare the differences in PAK-1 expression between different cancer cell lines, and for the direct comparison of the tumor xenograft weights. The significance level (alpha) was set at 0.05 (marked with symbols wherever data are statistically significant).
Results
Characterization of SSL-IPA-3
SSL-IPA-3 (Table 1) had a hydrodynamic particle diameter of approximately 139 nm, and this value did not change significantly over a period of two weeks (Table 2). The polydispersity index (PDI), a measure of the distribution of diameters was maintained at values less than 0.14 for up to 14 days. These PDI values are within the recommended size for medical applications, which is a PDI of less than0.326. Thezeta potential (the overall surface charge of the particles) also did not significantly change during this time. The stability test showed that 70% of IPA-3 was retained by these liposomes after 7 days post- formulation, and 50% was retained after 14 days.The stability tests also showed that a high amount of IPA-3 was retained in SSL after reconstitution for dosing, up to 80% after 3 days, and greater than 60% after 7 days post reconstitution (Supplemental Figure 2).
Table 2.
Physical characterization of SSL-IPA-3
| Mean Diameter (nm) | Polydispersity Index (PDI) | Zeta Potential (mV) | |
|---|---|---|---|
| Day 1 | 139.3 ± 0.115 | 0.050 ± 0.090 | −28.1 ±0.00 |
| Day 7 | 139.4± 2.307 | 0.121 ± 0.021 | −27.1 ±0.451 |
| Day 14 | 137.4 ± 5.726 | 0.139 ± 0.003 | −32.8 ±0.929 |
Anti-tumor activity of IPA-3and SSL-IPA-3in various human prostate cancer cell lines
The anti-tumor activity of freeIPA-3 was initially determined in vitro to verify that this compound inhibited prostate cancer cell growth.Our data showed that freeIPA-3, at concentrations of 5-30 μM, induced concentration- and time-dependent decreases in MTT staining with an IC50 between 10 and 35 μM,depending on the cell line. Interestingly, DU-145 cells were less susceptible to the toxicity of IPA-3 compared to PC-3 and LNCaP cells(Figure 1A-C). This difference was not a result of differences in media composition (Supplemental Figure 3). Treatment of PC-3 cells with SSL-IPA-3 also decreased MTT staining. As expected, SSL-IPA-3 was not as toxic to these cellsas free IPA-3 (Figure 1D-E). Nevertheless, decreases in MTT staining were still detected at doses of 20 μM and higher in PC-3 cells. In contrast, SSL-IPA-3was unable to induce significant decrease in MTT staining in DU-145 cells, even at the maximal encapsulation efficiency of 30 μM IPA-3. These data show that SSL-IPA-3 display anti-cancer activity in vitro.
Annexin V and PI staining were assessed using flow cytometry to further study IPA-3 and SSL-IPA-3 induced death in PC-3 cells (Figure 2). Treatment of cells with empty liposomes did not appreciably increase the percent cells staining positive either for annexin V alone or annexin and PI after 48 hr. In contrast, treatment with free IPA-3 (20-30 μM) significantly increased the percent of cell staining positive for annexin V alone, as well as those staining positive for both annexin V and PI, suggesting the IPA-3 is inducing cell death via apoptosis. The increase in annexin V and PI staining were also time-dependent (data not shown). SSL-IPA-3 also increased the percent cells staining positive for annexin V alone, but appeared to have a greater effect on cells staining positive for both annexin V and PI. Further, SSL-IPA-3 also increased the number of cells staining positive for PI alone, suggesting that the presence of necrosis. The increase in cell staining positive for late apoptosis markers, as well as necrosis, suggest that the encapsulation of IPA-3 in liposomes enhanced cell death and may be more efficacious at killing tumor cell in vivo.
Figure 2.
IPA-3 and SSL-IPA-3 increase annexin V and PI staining. (A-C) Scatter plots demonstrating annexin V (x-axis) and PI (y-axis) staining in control (A), IPA-3 exposed (B) and SSL-IPA-3 exposed (C) PC-3 cells after 48 hr. Annexin V and PI staining were determined using flow cytometry. The quantification of staining is shown in D. Data are presented as the mean ± SEM of the percent of cells in annexin V and PI staining as function of IPA-3 concentration; *p < 0.05.
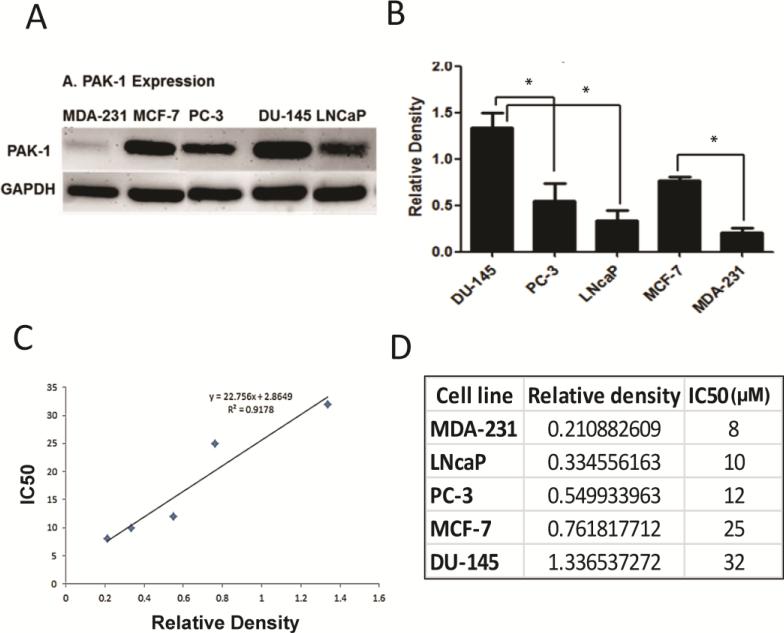
To test whether the differential sensitivity of IPA-3 on DU-145 and PC-3 cells were due to the differences in the activity of PAK-1,PAK-1 protein expression was determined in PC-3, DU-145 and LNCaP cells, and compared to two breast cancer cell lines (MDA-231 and MCF-7). Our results indicated that PAK-1 expression was variable in all cell lines tested,with higher levels being detected in DU-145 cells and the lowest level being detected in MDA-231 cells (Figure 3A-B). Comparison of the protein expression level of PAK-1 to the IC50 of free IPA-3 showed an excellent correlation with an R2 of 0.92 (Figure 3C-D). As expected, cells with the highest levels of PAK-1 expression (DU-145 and MCF-7) had the highest IC50s (~32 and 25 μM, respectively), while cells with the lowest PAK-1 expression (MDA-231 and LNCaP cells) had the lowest IC50s (8 and 10 μM) respectively. These data suggest that differences in the sensitivity of cells to SSL-IPA-3aremediated not by differences in sensitivity to the nanoparticles, but more so by differences in the expression of PAK-1.
Figure 3.
Effect of IPA-3 in various prostate cancer cells relate to its expression levels. (A) Expression of PAK-1 in different prostate and breast cancer cell lines as determined by immunoblot analysis. (B)Histogram showing densitometry analysis of PAK-1 expression in the Western blots (n = 5). (C)Histogram showing correlation between PAK-1 expression and IC50 (μM) of PAK-1 inhibitor IPA-3 (n = 3). (D) Data presenting the linear correlation between PAK-1 expression and IPA-3 toxicity.Data are presented as the mean ± SEM; *p < 0.05.
SSL-IPA-3 inhibits the prostate tumor xenograft growth in vivo
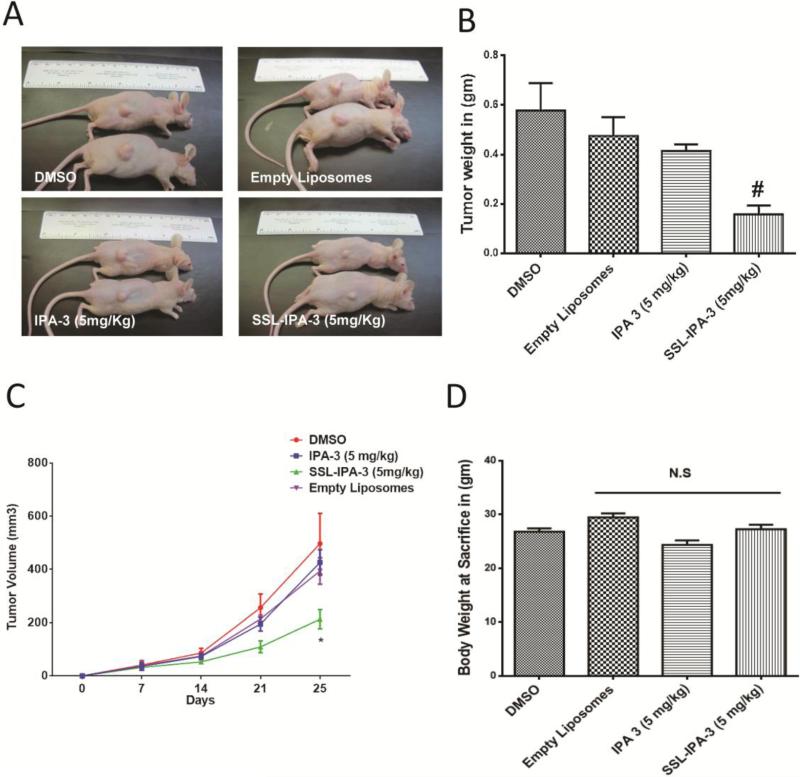
We previously reported that inhibition of PAK-1using IPA-3reduced prostate tumor xenograft growth in athymic nude mice with daily administration8. In the current study, we sought to determine if 2 days/week administration of SSL-IPA-3 can inhibit the PC-3 cell tumor xenograft growth in vivo, compared to the same frequency of free IPA-3. Our results indicated that 2 days/week administration of SSL-IPA-3was indeed sufficient to inhibit the growth ofPC-3 cell tumor xenografts, as comparedtoempty liposomeand free IPA-3 administered groups (Fig 4A-C). In fact, similar doses of free IPA-3 (2 days/week) administration did not have a significant effect on PC-3 tumor xenograft growth. There was no significant effect of empty liposome, free IPA-3 or SSL-IPA-3administration on the body weight of mice even after 25 days (Fig 4D). These data shows the novel finding that SSL-IPA-3 is effective at inhibiting the growth of prostate tumor xenograftsin vivo.
Figure 4.
Sterically stabilizedliposome- encapsulated IPA-3 (SSL-IPA-3) significantly inhibits prostate tumor xenograft growth in vivo. (A)Pictures of athymic nude mice bearing PC-3 cell tumor xenografts and treated with vehicle (DMSO), empty liposomes, IPA-3or liposome encapsulated IPA-3 (SSL-IPA-3). (B)Histogram showing the weight of PC-3 cell tumor xenografts collected from athymic nude mice treated with vehicle (DMSO), empty liposomes, IPA-3or liposome encapsulated IPA-3 on day 25 after tumor implantation and day 18 after the start of treatments (n = 5-6). (C)Histogram showing the volume of PC-3 cell tumor xenografts on day 7, 14, 21 and 25 after implantation in athymic nude mice treated with vehicle (DMSO), empty liposomes, IPA-3or liposome encapsulated IPA-3 as measured using calipers (n = 5-6). (D)Histogram showing the body weight of the PC-3 cell tumor bearing athymic nude mice on day 25 after tumor implantation and day 18 after treatment with vehicle (DMSO), empty liposomes, IPA-3or liposome encapsulated IPA-3. Data are presented as the mean ± SEM; #p < 0.01.
SSL-IPA-3 treatment induces apoptosis in vivo more effectively than free IPA-3
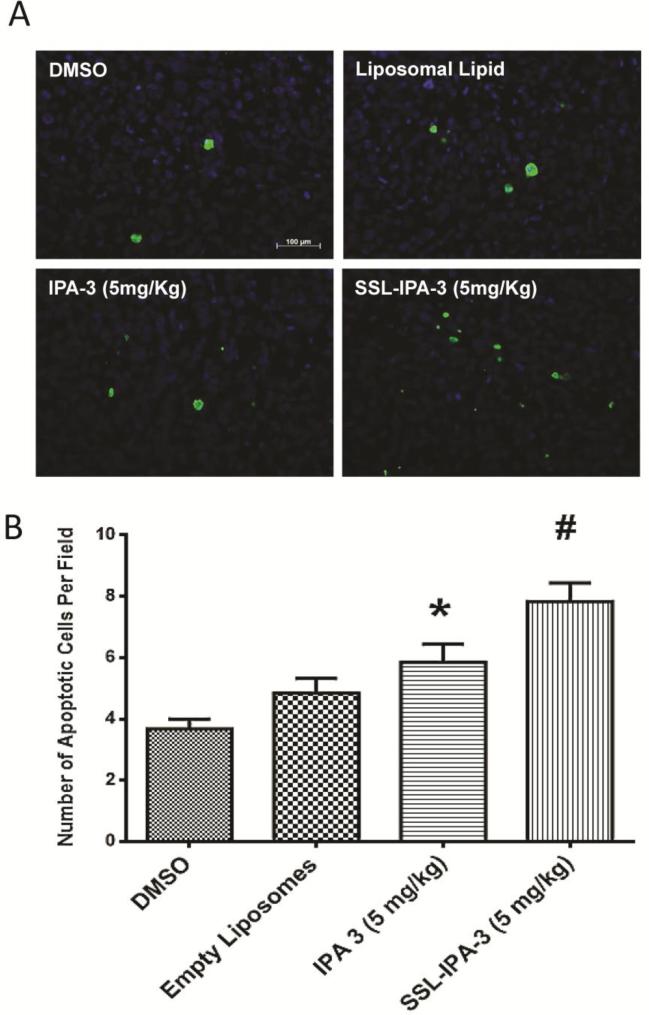
We next determined the effect of SSL-IPA-3 and free IPA-3 on apoptosis in the PC-3 cell prostate tumor xenografts in vivo using TUNEL staining Our analysisindicated that both SSL-IPA-3 andfree IPA-3 treatments resulted in significant increases in the number of apoptotic cells per field compared to the control groups, but, the effect on the apoptosis was more greater in the SSL-IPA-3-treated group, as compared to the free IPA-3-treated group(Fig 5A-B). Thus, SSL-IPA-3'sehanced ability to limit prostate tumor growth in vivo correlates to increased apoptosis.
Figure 5.
Sterically stabilizedliposome- encapsulated IPA-3 induces apoptosis in PC-3 cell tumor xenografts in athymic nude mice. (A)Pictures of PC-3 cell tumor xenograft sections from athymic nude mice treated with vehicle (DMSO), empty liposomes, IPA-3or liposome encapsulated IPA-3 on day 25 after tumor implantation and day 18 after the start of treatments stained with apoptotic marker TUNEL. (B) Histogram showing number of apoptotic (TUNEL positive) cells inPC-3 cell tumor xenograft sections from athymic nude mice treated with vehicle (DMSO), empty liposomes, IPA-3or liposome encapsulated IPA-3 on day 25 after tumor implantation and day 18 after the start of treatments. (n = 4). Data are presented as the mean ± SEM; *p < 0.05; #p < 0.01.
Discussion
Serine–threonine kinases PAKs,PAK-1 in particular, have been associated with a variety of pathological conditions, including cancer1. PAK-1 is regulated by auto-inhibition, and is suggested as good therapeutic target for cancer therapyby using small molecules that may cause conformational changes in the protein disturbing its autoregulation1, 27. Previous studies from our laboratory showed that Rac1, an upstream regulator of PAK-1 promotes prostate cancer cell survival, proliferation and transendothelial-migration4. Although absent in the normal prostate and human benign prostatic hyperplasia tissues, our studies revealed that PAK-1 is expressed in prostate cancer cells, prostate tumors and metastatic colonies in the patient lungs7, and thatPAK-1 is important in the TGFβ-induced epithelial-mesenchymal transition (EMT) in prostate cancer cells8. These studies suggested that targeting PAK-1 may be an effective strategy for prostate cancer therapy.
In a screening process for allosteric inhibitors targeting PAK-1 activation, IPA-3 has been identified as a small molecule inhibitor of PAK-1, and was then proposed as a potential therapeutic candidate for various human pathologies including cancer9. However, IPA-3in its free form has a very short half-lifein vivo because of its rapid metabolism10. Thus, the metabolically labile nature of IPA-3 becomes a major bottleneck in its use for therapeutic purposes. Although the free form of IPA-3 was effective in inhibiting prostate cancer growth in our studies, this required daily administration of the drug8. This demanded a reliable method to specifically deliver IPA-3 to prostate cancer cells in vivothat will improve the stability and efficacy of IPA-3, reduce the frequency of drug administration and avoid any potential side-effects of free IPA-3.
Like in the case of IPA-3, most of the laboratory based research on prostate cancer does not reach the clinic due to various issues relating to the dose, frequency of drug administration, stability of particular drug in vivo and the adverse reactions. The major limitation of many anti-cancer drugs is caused by rapid metabolism and/or toxic side effects. An ideal drug dosage formulation for anti-cancer therapy has to exhibit specific delivery of an effective dose of drug to the target tissue over the required period of time. Tumor specific drug delivery using lipid-based nanoparticulate drug carriers, such as SSL, have been used to encapsulate and release drugs, often with higher efficiency compared to free drug28. This suggested that encapsulation of IPA-3 in SSL may confer stability to IPA-3, lead to its specific delivery to cancer tissues, and hence may be ideal to improve its efficacy in inhibiting prostate cancer growth.
IPA-3 was effectively and stably encapsulated in SSL. Analysis of zeta potential and size indicated that these nanoparticles would be functional for clinical application, and the activity of these formulations were verified in vitro. It is not surprising that SSL-IPA-3 were not as effective as free IPA-3 in vitro and in vivo, as this is commonly seen in many studies, including those reported from our own laboratory16, 22, 29. What was surprising was that the effect of SSL-IPA-3 was shown to be cell type-dependent and mediated by the level of expression of PAK-1. The decreased susceptibility of DU-145 cells to SSL-IPA-3 liposomes indeed mirrored their decreased susceptibility to free IPA-3. These differences were not a result of difference in sensitivity to the formulation used as DU-145 and PC-3 cells were equally sensitive to another anti-cancer drug, doxorubicin, encapsulated in SSL and a modified liposome formulation22, 23. Our in vitrostudies also revealed that free IPA-3 displays greater antitumor activity than SSL-IPA-3. Such an observation is expected as encapsulated IPA-3 in SSL must first be released from the liposomes before acting on its target PAK-1. These data also suggest that the therapeutic efficacy of SSL-IPA-3 may be limited in cancers that express high levels of PAK-1. It is also possible that the differential expression of PAKs in these cancers may also mitigate the toxicity.
Biophysical characterization of SSL-IPA-3 showed excellent stability, as based on the maintenance of zeta potential, size and IPA-3 levels for at least 7 days. Because of its lipophilicity, IPA-3 is probably mostly incorporated in the phospholipid bilayer of the liposomes, which provides a hydrophobic environment. Once trapped, IPA-3may remain in the liposomal bilayer due to their lower affinity for the inner and outer aqueous regions of the liposomes30,31. However, given enough time, IPA-3 may dissociate from the SSL. This may be the cause for the decrease in IPA-3 levels seen in these formulations after 14 days. Nevertheless, our data show that SSL-IPA-3is reasonably stable and therapeutically active, even in biological fluids. Future efforts will be placed on increasing stability for longer time periods.Lyophilization is a promising approach to ensure the extension of shelf-life and longer-term stability of liposomes. Thus, methods such as water replacement and vitrification could also be usedto produce an extended shelf-life32.
The data presented in our study demonstrate the novel finding that 2 days/week administration of SSL-IPA-3is effective in decreasing the growth of human prostate cells in vitro and tumor (PC-3) xenografts in vivo. Further, and more exciting, was that the data showed enhanced efficacy of SSL-IPA-3 compared to free IPA-3, which suggests that the activity of SSL-IPA-3 becomes more effective in an in vivo environment. The increase in efficacy of the SSL-IPA-3, as compared to free IPA-3 is most likely a result of the phenomenon discussed in introduction, including the possibility that the encapsulation of IPA-3 in SSL extends the half-life of IPA-3 and overcomes its rapid metabolism in its free form. The implications of these findings are significant because they suggest decreased dosing, with higher efficacy, which could decrease side-effects and other off-target toxicities associated with anti-cancer agents. Although the experiments in this studyare focused on the activity of SSL-IPA-3on prostate cancer, this targeting strategy may additionally be used to target other cancers such as breast cancer as was shown by our in vitro studies.
In conclusion, we have demonstratedthe formulation and characterization of novel SSL-based nanoparticle formulation that is reasonably stable and shows excellent efficacy both in vitro and in vivo. These data suggest that SSL-IPA-3 isan effective targeting strategy for inhibiting prostate cancer growth. Data in this study also suggest that the activity of IPA-3 is cell dependent and is mediated by the level of expression of PAK-1 in cancer cells.
Supplementary Material
Acknowledgements
The authors would like to thank Mr. Nhat D. Quach for his technical support during liposome preparation.
Financial support: Funds were provided by the National Institutes of Health grant (R01HL103952), UGA-College of Pharmacy, UGA Research Foundation and the American Legion to PRS, National Institutes of Health grant (R01EB0116100) to BSC, and an Achievement Reward for College Scientists Foundation Award to WNM. Research was supported with resources and the use of facilities at the Charlie Norwood VAMC, Augusta, GA. The funders had no role in the study design, data collection, analysis and decision to publish. Preparation of the manuscript and the contents do not represent the views of the Department of Veterans Affairs or the United States Government.
Abbreviations
- PAK-1
P21 activated kinase-1
- SSL
Sterically stabilized liposomes
- IPA-3
Inhibitor targeting PAK1 activation-3
- PDI
polydispersity index
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: Authors have declared that no conflicts of interest exist.
References
- 1.Kichina JV, Goc A, Al-Husein B, Somanath PR, Kandel ES. PAK1 as a therapeutic target. Expert Opin Ther Targets. 2010;14:703–25. doi: 10.1517/14728222.2010.492779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nature reviews Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrantz N, da Silva Correia J, Fowler B, Ge Q, Sun Z, Bokoch GM. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. The Journal of biological chemistry. 2004;279:1922–31. doi: 10.1074/jbc.M311145200. [DOI] [PubMed] [Google Scholar]
- 4.Goc A, Abdalla M, Al-Azayzih A, Somanath PR. Rac1 activation driven by 14-3-3zeta dimerization promotes prostate cancer cell-matrix interactions, motility and transendothelial migration. PLoS One. 2012;7:e40594. doi: 10.1371/journal.pone.0040594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somanath PR, Byzova TV. 14-3-3beta-Rac1-p21 activated kinase signaling regulates Akt1-mediated cytoskeletal organization, lamellipodia formation and fibronectin matrix assembly. Journal of cellular physiology. 2009;218:394–404. doi: 10.1002/jcp.21612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parrini MC, Matsuda M, de Gunzburg J. Spatiotemporal regulation of the Pak1 kinase. Biochemical Society transactions. 2005;33:646–8. doi: 10.1042/BST0330646. [DOI] [PubMed] [Google Scholar]
- 7.Goc A, Al-Azayzih A, Abdalla M, et al. P21 activated kinase-1 (Pak1) promotes prostate tumor growth and microinvasion via inhibition of transforming growth factor beta expression and enhanced matrix metalloproteinase 9 secretion. J Biol Chem. 2013;288:3025–35. doi: 10.1074/jbc.M112.424770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Azayzih A, Gao F, Somanath PR. P21 activated kinase-1 mediates transforming growth factor beta1-induced prostate cancer cell epithelial to mesenchymal transition. Biochim Biophys Acta. 2015;1853:1229–39. doi: 10.1016/j.bbamcr.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deacon SW, Beeser A, Fukui JA, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–31. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudolph J, Crawford JJ, Hoeflich KP, Wang W. Inhibitors of p21-activated kinases (PAKs). J Med Chem. 2015;58:111–29. doi: 10.1021/jm501613q. [DOI] [PubMed] [Google Scholar]
- 11.Ke Y, Wang X, Jin XY, Solaro RJ, Lei M. PAK1 is a novel cardiac protective signaling molecule. Front Med. 2014;8:399–403. doi: 10.1007/s11684-014-0380-9. [DOI] [PubMed] [Google Scholar]
- 12.Koth AP, Oliveira BR, Parfitt GM, Buonocore Jde Q, Barros DM. Participation of group I p21-activated kinases in neuroplasticity. J Physiol Paris. 2014;108:270–7. doi: 10.1016/j.jphysparis.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Taglieri DM, Ushio-Fukai M, Monasky MM. P21-activated kinase in inflammatory and cardiovascular disease. Cellular signalling. 2014;26:2060–9. doi: 10.1016/j.cellsig.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Tsui H, Bolton EL, et al. Novel insights into mechanisms for Pak1-mediated regulation of cardiac Ca(2+) homeostasis. Frontiers in physiology. 2015;6:76. doi: 10.3389/fphys.2015.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muggia FM. Liposomal encapsulated anthracyclines: new therapeutic horizons. Current Oncol. 2001;3:156–62. doi: 10.1007/s11912-001-0016-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhu G, Mock JN, Aljuffali I, Cummings BS, Arnold RD. Secretory phospholipase A responsive liposomes. J Pharm Sci. 2011;100:3146–59. doi: 10.1002/jps.22530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marra M, Salzano G, Leonetti C, et al. New self-assembly nanoparticles and stealth liposomes for the delivery of zoledronic acid: a comparative study. Biotechnology advances. 2012;30:302–9. doi: 10.1016/j.biotechadv.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Lasic DD, Martin FJ, Gabizon A, Huang SK, Papahadjopoulos D. Sterically stabilized liposomes: a hypothesis on the molecular origin of the extended circulation times. Biochimica et biophysica acta. 1991;1070:187–92. doi: 10.1016/0005-2736(91)90162-2. [DOI] [PubMed] [Google Scholar]
- 19.Sharma US, Sharma A, Chau RI, Straubinger RM. Liposome-mediated therapy of intracranial brain tumors in a rat model. Pharm Res. 1997;14:992–8. doi: 10.1023/a:1012136925030. [DOI] [PubMed] [Google Scholar]
- 20.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–84. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 21.Yuan F, Leunig M, Huang SK, Berk DA, Papahadjopoulos D, Jain RK. Microvascular permeability and interstitial penetration of sterically stabilized (stealth) liposomes in a human tumor xenograft. Cancer research. 1994;54:3352–6. [PubMed] [Google Scholar]
- 22.Mock JN, Costyn LJ, Wilding SL, Arnold RD, Cummings BS. Evidence for distinct mechanisms of uptake and antitumor activity of secretory phospholipase A2 responsive liposome in prostate cancer. Integr Biol (Camb) 2013;5:172–82. doi: 10.1039/c2ib20108a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu G, Mock JN, Aljuffali I, Cummings BS, Arnold RD. Secretory phospholipase A(2) responsive liposomes. J Pharm Sci. 2011;100:3146–59. doi: 10.1002/jps.22530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer. 1987;56:279–85. doi: 10.1038/bjc.1987.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummings BS, Schnellmann RG. Cisplatin-induced renal cell apoptosis: caspase 3-dependent and -independent pathways. The Journal of pharmacology and experimental therapeutics. 2002;302:8–17. doi: 10.1124/jpet.302.1.8. [DOI] [PubMed] [Google Scholar]
- 26.De La Vega JC, Elischer P, Schneider T, Hafeli UO. Uniform polymer microspheres: monodispersity criteria, methods of formation and applications. Nanomedicine (London, England) 2013;8:265–85. doi: 10.2217/nnm.12.210. [DOI] [PubMed] [Google Scholar]
- 27.Cheetham GM. Novel protein kinases and molecular mechanisms of autoinhibition. Curr Opin Struct Biol. 2004;14:700–5. doi: 10.1016/j.sbi.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Gabizon A. Liposomes as a drug delivery system in cancer chemotherapy. Horiz Biochem Biophys. 1989;9:185–211. [PubMed] [Google Scholar]
- 29.Quach ND, Mock JN, Scholpa NE, et al. Role of the phospholipase A2 receptor in liposome drug delivery in prostate cancer cells. Mol Pharm. 2014;11:3443–51. doi: 10.1021/mp500174p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen TM. Liposomal drug formulations. Rationale for development and what we can expect for the future. Drugs. 1998;56:747–56. doi: 10.2165/00003495-199856050-00001. [DOI] [PubMed] [Google Scholar]
- 31.Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. International journal of nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Han D, Cai C, Tang X. An overview of liposome lyophilization and its future potential. Journal of controlled release : official journal of the Controlled Release Society. 2010;142:299–311. doi: 10.1016/j.jconrel.2009.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.