Abstract
A major step towards an HIV-1 vaccine is an immunogen capable of inducing neutralizing antibodies. Envelope glycoprotein (Env). mimetics, such as the NFL and SOSIP designs, generate native-like, well-ordered trimers and elicit tier 2 homologous neutralization (SOSIPs). We reasoned that the display of well-ordered trimers by high-density, particulate array would increase B cell activation compared to soluble trimers. Here, we present the design of liposomal nanoparticles displaying well-ordered Env spike trimers on their surface. Biophysical analysis, cryo- and negative-stain electron microscopy and binding analysis with a panel of broadly neutralizing antibodies confirm high-density, well-ordered trimer particulate array. The Env trimer-conjugated liposomes were superior to soluble trimers in activating B cells ex vivo and germinal center B cells in vivo. In addition, the trimer-conjugated liposomes elicited modest tier 2 homologous neutralizing antibodies. The trimer-conjugated liposomes represent a promising initial lead towards the development of more effective HIV vaccine immunogens.
eTOC Blurb
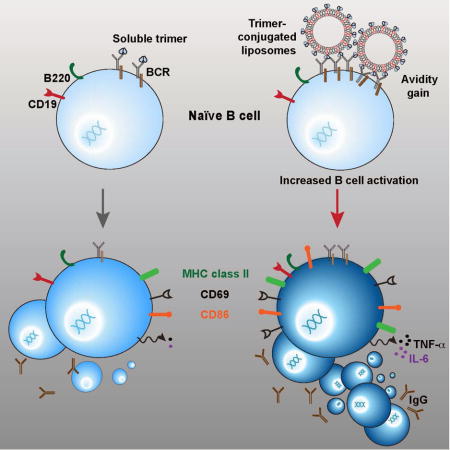
Ingale et al array well-ordered HIV envelope glycoprotein (Env) trimers at high density on the surface of synthetic liposomes. The trimer-liposomes are homogenous, and well recognized by broadly neutralizing antibodies against HIV-1. High-density particulate trimer array more efficiently activates Env-specific B cells ex vivo, enhancing the generation of germinal center B cells in vivo.
Introduction
A broadly effective HIV-1 vaccine will likely require the elicitation of broadly effective neutralizing antibodies. Although broadly neutralizing antibodies (bNAbs) arise sporadically following chronic HIV infection such bNAbs are exceedingly difficult to elicit by vaccination(Burton et al., 2004). One roadblock to the elicitation of neutralizing antibodies by vaccination has been the development of an immunogen that stably presents a gp120-gp41 envelope glycoprotein (Env) trimer in its native state, as it is presented on the virus surface. With the recent developments of SOSIP and NFL trimeric Env platforms, it is now possible to express and purify soluble, native-like and well-ordered Env trimers that are structurally and antigenically well characterized(Guenaga et al., 2015; Julien et al., 2013; Pancera et al., 2014; Sharma et al., 2015). To date, these trimeric mimetics of the HIV-1 native spike elicit homologous “tier 2” neutralization to a few selected strains of undetermined long-term durability (Sanders et al., 2015) and most potently in small animals. Although this is a promising first step, the next step is to quantitatively or qualitatively increase B cell activation elicited by the well-ordered trimers, toward the longer-term goals of increased somatic hypermutation and more durable antibody responses leading to trimer-dependent elicitation of broader neutralizing activity.
An impediment to elicit neutralizing antibodies by HIV itself or HIV-based virus-like particles (VLP) may be the low levels of natural Env incorporation, resulting in sparse numbers of spikes per VLP(Deml et al., 1997). Here, we generated Ni+2-bearing, fully synthetic single bilayer liposomes arrayed with the newly developed, well-ordered HIV-1 soluble trimer mimetics possessing three C-terminal His-tags per trimer. This design allows the variable incorporation of Ni+2-lipids to generate a series of JRFL trimer-conjugated liposomes displaying different levels of either SOSIP or NFL well-ordered(Kovacs et al., 2014; Sanders et al., 2013; Sharma et al., 2015) trimers arrayed on the liposomal surface. This means of high-density particulate display as well blocks access to the non-neutralizing, non-glycosylated underside of the trimers. This solvent accessible surface, usually occluded by the viral lipid bilayer, was recently shown to be highly immunogenic using soluble BG505 SOSIP trimers(Hu et al., 2015) in mice, however no “tier 2” autologous neutralizing antibodies were detected as they are in rabbits(Sanders et al., 2015). HIV gp140 trimers conjugated to interbilayer-cross-linked multilamellar vesicles (ICMVs) generate high titer binding responses in mice(Pejawar-Gaddy et al., 2014) and biotin-labeled gp120 onto avidin-containing liposomes stimulate anti-gp120 B cells in vitro(Ota et al., 2013), however well-ordered Env trimers arrayed at high-density particulate array have not been assessed in an animal model capable of generating tier 2 autologous neutralization.
In our current study, we showed by biochemical and biophysical analysis, cryo-electron microscopy (EM) and negative staining-EM that the predominantly single bilayer liposomes, when optimized, present the well-ordered trimers with high-density, multi-valent array. On the liposome surface, these well-ordered trimers retained qualities of a closed native trimer and were stable for several months at 4°C. We demonstrated that this high-density array better activated B cells ex vivo compared to strain-matched soluble trimers and that the liposome-conjugated trimers more efficiently generated germinal center B cells compared to soluble trimers in a statistically significant manner. Compared to the soluble trimers, there was a trend for the liposome-conjugated trimers to more efficiently elicit binding antibodies to native-like trimers and modest tier 2 (JRFL) homologous neutralizing titers. The clinical efficacy of human papillomavirus (HPV) L1 virus-like particles (VLPs) to provide long-lasting protection against a virus that enters by mucosal routes, while, in contrast, the HPV L1 monomer is not protective suggests that particulate display of ordered HIV trimers might hasten development toward a more effective HIV-1 vaccine(Caldeira Jdo et al., 2010; Safaeian et al., 2013)’(Schiller and Chackerian, 2014; Schiller and Lowy, 2015). Combining the well-ordered trimers with particulate high-density display presents a scalable platform to enhance B cell responses to HIV-1 Env and potentially to envelope glycoproteins from other viruses that are relevant vaccine targets.
Results
Well-ordered His-tagged trimers for liposomal array
The development of well-ordered trimers by two independent platforms, SOSIP and NFL, present the opportunity to assess the multivalent array of such trimers on the surface of liposomal nanoparticles to study their impact on antigenicity and immunogenicity. These well-ordered trimers are well characterized by negative stain followed by EM at both the level of 2D classifications and 3D reconstructions, including the JRFL strain-derived trimer prototypes used here(Guenaga et al., 2015; Sharma et al., 2015). To contrast the ordered appearance of the JRFL SOSIP and NFL trimers with the previously described disordered foldon trimers(Ringe et al., 2015; Tran et al., 2014) we performed negative staining followed by EM at the level of resolution amenable for analysis of liposomes. As shown in Figure 1A (left panel), the previously described JRFL-based gp140-foldons displayed an amorphous mixture of oligomeric states. In contrast, the more faithful mimetics of the HIV-1 Env spike, JRFL SOSIP and NFL trimers, presented a relatively well-ordered appearance. Depending upon the random orientation of the trimers on the carbon-coated grid, the soluble spikes displayed 3-fold symmetry typified by a “propeller-like” appearance by negative staining-EM, at the level of resolution analyzed here (Figure 1A, center and right panels). With the well-ordered trimers in hand, each containing C-terminal His6-tags on each protomeric subunit, we sought to array these spike mimetics on a repetitive, nanoparticle platform to assess potential improvements in B cell activation and immunogenicity. We generated liposomes by standard procedures comprised of a mixture of 60% 1,2-distearoyl-sn-glycero-3-phosphocholine (DGPC) and 40% cholesterol (Avanti Polar Lipids; see Methods). We used a relatively high concentration of cholesterol to increase liposomal membrane stability and integrity in vivo(Arsov and Quaroni, 2007). In brief, polar lipids in chloroform were dried onto glass, re-suspended in aqueous buffer, sonicated in the buffer and extruded through filters to generate approximately 100 nm diameter nanoparticles. To produce His-binding liposomes, we incorporated 1,2-dioleoyl-sn-glycero-3-((N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl) (nickel salt) (DGS-NTA-Ni) into the lipid mixture at levels of 1 to 4%, substituting for the cholesterol lipid component. We reasoned that the NTA-Ni would randomly disperse in the lipid bilayer, and that approximately 50% of the time, the polar head groups would align on the outside of the lipid bilayer to be available for conjugation with the His-tags present on the C-terminus of the ordered HIV-1 trimers (see schematic, Figure 1B). We could also incorporate TLR ligands into the liposomes by including them in the chloroform mixture prior to drying the lipids on to the glass surface (see Methods).
Figure 1. HIV trimers and their particulate display.
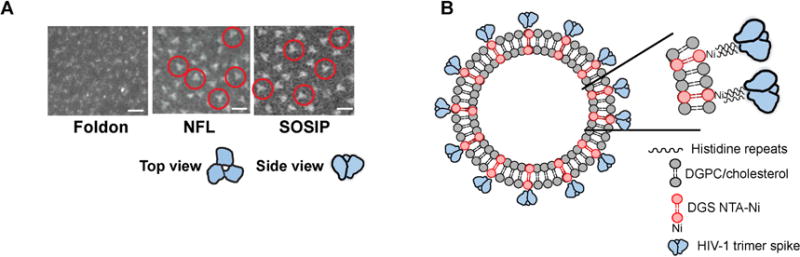
(A) Negative stain EM micrographs of JRFL gp140-foldon oligomers, JRFL NFL2P, and JRFL SOSIP trimers. Scale bars = 20nm. (B) Schematic representation of liposomes displaying HIV-1 trimers. Zoomed field depicts binding of the 6-histidine repeats (His6 tag) present as a fusion on the C-terminus of each protomer of each trimer to the Ni+2 chelated at the hydrophilic head group of the DGS-NTA(Ni) polar lipid.
Analysis of well-ordered trimers coupled to liposomes by SDS-PAGE and EM
Following incubation of the well-ordered NFL and SOSIP His-tagged timers with the Ni-bearing liposomes, containing 1, 2 and 4% DGS-NTA(-Ni) lipids, respectively, we performed size exclusion chromatography (SEC) to separate free trimers from liposomes. Next, we analyzed the liposomes by SDS-PAGE and detected increasing trimer bands with increasing levels of Ni+2 incorporated into the liposomes following staining of the gel by Commassie blue solution (Figure 2). We subjected the 4% DGS-NTA(Ni) liposomes to dynamic light scattering (DLS) to assess liposomal diameter and uniformity before and after conjugation with the well-ordered NFL or SOSIP trimers. As shown in Figure 2B, the non-conjugated liposomes displayed an average diameter of 152 nm while the average diameter of trimer-conjugated liposome was 172 nm. To further assess liposome uniformity, diameters and to visually determine state of trimers arrayed on the surface of the liposomes, we performed cryo-EM of both the JRFL SOSIP- and JRFL NFL-conjugated 4% DGS-NTA(Ni) liposomes (Figure 2 and Figure S1A). Using the cryo-EM images, we measured the liposome diameters to be from 75 nm to 250 nm (see Figure S1B), consistent with the DLS data. Using the measuring tools, we determined the lipid bilayer to be ~5 nm in width with individual trimers spaced at ~12–14 nm apart on the liposomes. As detected by the cryo-EM analysis, the majority of liposomes possessed a single lipid bilayer and virtually all nanoparticles displayed a high-density array of the SOSIP or NFL trimers, respectively. An apparent double lipid bilayer was visible on a small percentage of liposomes and, for even fewer, multiple layered lipid bilayers were observed (Figure S1A), all possessing trimers on their outermost surface. The liposome interior appeared to contain only buffer as no other density was observed in the enclosed volumes. In addition, no other materials were observed in the sample, indicating that the sample was free of any adventitious agents.
Figure 2. Characterization of JRFL SOSIP-conjugated liposomes.
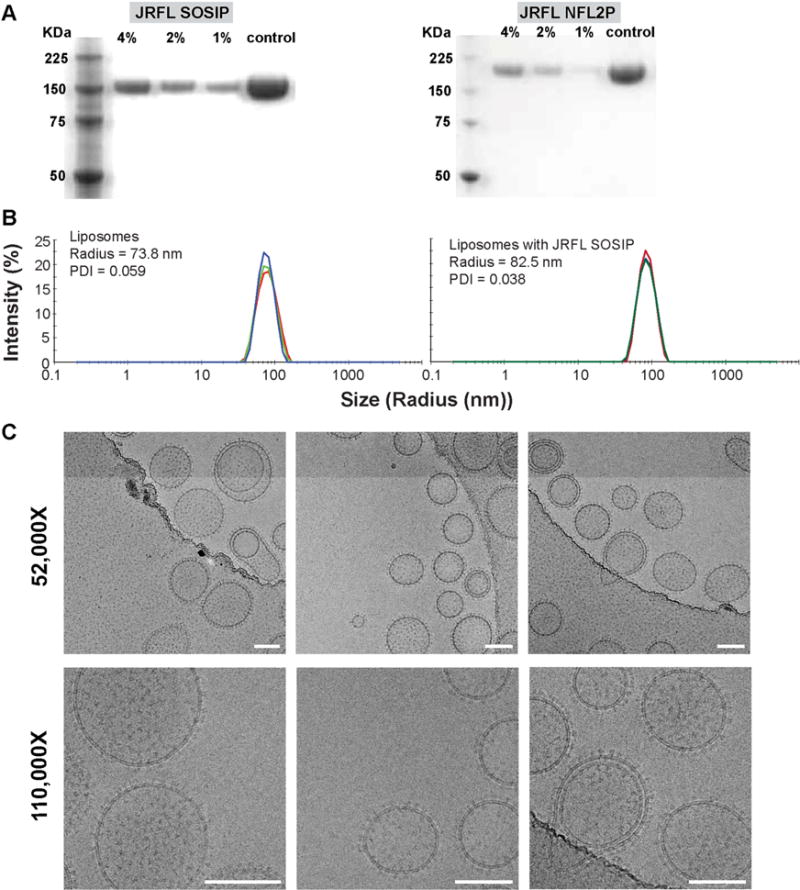
(A) Reducing SDS PAGE of 4%, 2% and 1% Ni DGS-NTA(Ni) JRFL SOSIP and JRFL NFL trimer-conjugated liposomes. JRFL SOSIP and JRFL NFL2P soluble trimeric glycoproteins are included as controls. (B) Dynamic light scattering (DLS) of the 4% DGS-NTA(Ni) liposomes and JRFL SOSIP-conjugated liposomes was performed using a using Zetasizer Nano instrument to measure particle size and the polydispersity index. (C) Cryo-EM images of 4% Ni JRFL SOSIP liposomes at 52,000 and 110,000× magnification. Scale bar = 100 nm. See also Figure S1.
To increase contrast and resolution of the trimers arrayed on the liposomal surface, we performed negative staining followed by EM at selected magnifications of the trimer-conjugated liposomes. As seen in Figure 3, with 1% Ni-lipid formulation, the conjugated well-ordered trimers were detected as a “ring” visible around the circumference of the liposomes, observable by EM in what appeared to be two dimensions. In the case of the 2% Ni-containing liposomes, beside trimers ringing the circumference of individual nanoparticles, trimers could be detected on the surface of the liposomes encircled by the lipid bilayer. For the 4% Ni-containing liposomes, we observed densely packed and evenly spaced trimers arrayed on the surface of the liposomes by the negative staining-EM. The patterns of trimer array were very similar for both the JRFL SOSIP- and JRFL NFL Env-containing liposomes. Because the trimers were relatively well resolved, we next quantitated the approximate number of trimers per liposomal field by constructing a grid to aid manual counting (see Figure 3B). The total number of trimers per visible field was in the range of 300 spikes per liposome, likely an underestimate since not all surfaces of the liposome are observable in these EM images. Using iTEM (EMSIS, GMbH) measuring tools, we determined the relatively uniformly arrayed trimers to be spaced approximately 14–15 nm apart, center-to-center, on the surface of the nanoparticles (see Figure 3C and D). In addition, we constructed a square area of approximately 12 nm per side that would encompass each trimer, and adjacent unoccupied surface area, to circumscribe an area of ~144 nm2. We calculated the surface area of a spherical liposome with a radius of ~75 nm to be 70,865 nm2 and by simple division would yield ~492 trimers per liposome, bracketing an estimated range between 300 and 500 trimers per particle.
Figure 3. Incorporation of different amounts of DGS-NTA(Ni) into the liposomes to increase JRFL trimer density on the liposomal surface.
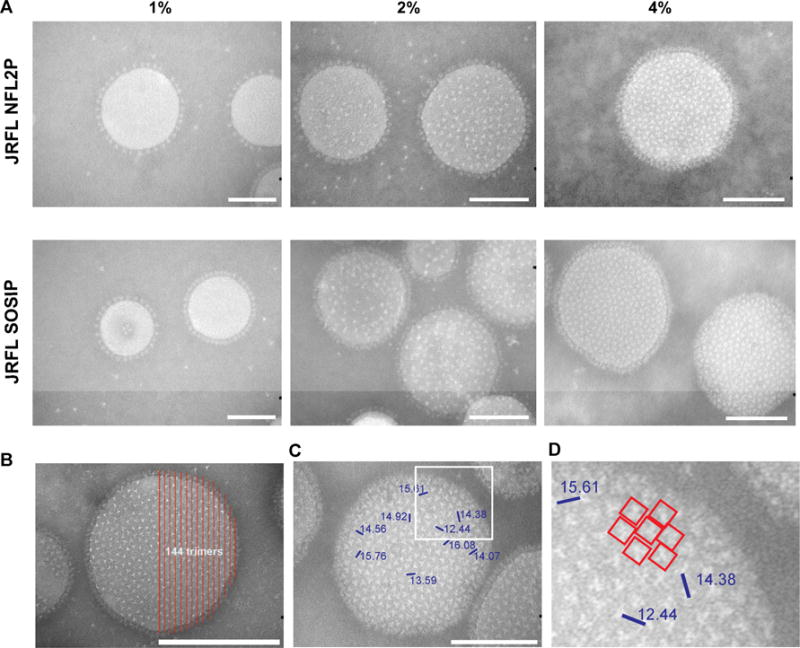
Negative stain EM images of DGS-NTA(Ni) liposomes made with 1%, 2% and 4% DGS-NTA(Ni) and conjugated with either JRFL NFL or JRFL SOSIP trimers. All images are at 18,000 × magnification. Scale bar = 100 nm. (B) Representative negative stain image of 4% JRFL SOSIP-conjugated liposomes with a counting grid (red lines) to manually determine the approximate number of trimers visible in half the area of the trimer-liposome image. (C) Measurement of distances (nm) between selected trimers as demarked by blue bars, center to center. Numbers indicate the distance between the two adjacent trimers. (D) Zoomed image of the white square area from panel C. See also Figure S2.
To confirm that trimer conjugation to the liposomes was Ni-dependent, DGPC liposomes without any Ni-lipid were generated and the JRFL SOSIP trimers were added to assess interaction. Most trimeric glycoprotein remained dissociated from the liposomal fraction by SEC and EM further confirmed that no trimers were associated with the Ni-lacking liposomes (Figure S2A).
Binding analysis of the liposome bound trimers by Biolayer Light Interferometry (BLI) and EM
We next assessed if the well-ordered trimers maintained quaternary packing on the surface of the liposomes by probing the trimers with selected bNAbs and mAbs using BLI. Accordingly, we captured the trimer-conjugated liposomes on the Octet sensor surface by wheat germ agglutinin (WGA), which recognizes and binds to carbohydrates that are abundant as N-glycans located on the trimeric spike surface. In this format, due to the dense array of the trimers on the liposomal surface, there will be avidity effects in regards to the bivalent IgGs as analytes, therefore we used the BLI binding analysis not to derive actual affinities, but to qualitatively assess relative avidities to confirm that once the trimers were conjugated to the liposomes, they displayed the same binding pattern as assessed previously by more quantitative binding kinetics(Guenaga et al., 2015; Sharma et al., 2015). Relative avidity was determined by the maximal response value detected for each antibody at the completion of the association phase. Using the WGA capture of the trimer-conjugated lipoosomes, we did attempt binding by selected Fabs to obtain affinities, but due to the smaller mass of the Fab relative to the liposomes, we could not detect reliable signals by this approach, so we proceeded with the qualitative binding assessments using bivalent IgG.
To begin the avidity analysis, we assessed recognition by the CD4 binding site-directed bNAb, VRC01(Wu et al., 2011) and the glycan-dependent 2G12, to determine overall levels of liposome-conjugated trimers, since these mAbs can recognize trimeric or monomeric forms of HIV Env (Figure 4A). The VRC01 and 2G12 bNAbs efficiently recognized the trimers conjugated to the liposomes, consistent with previous binding studies, and the EM analysis presented here,as did the other CD4bs-directed bNAbs PGV04(Falkowska et al., 2014), CH103(Liao et al., 2013) and b12(Burton et al., 1994). We then utilized the trimer-specific, V2-directed bNAb, PGT145(McLellan et al., 2011), to confirm that, following conjugation, the quaternary variable region cap of the trimers remained intact. As expected, the trimer-specific PGT145 bNAb efficiently recognized the trimers arrayed on the liposomal surface as did other trimer-preferring bNAbs such as VRC03(Li et al., 2012; Tran et al., 2012), VRC06(Li et al., 2012), PGDM1400(Sok et al., 2014) and PG16(Pejchal et al., 2010) (Figure 4A). In contrast, the V3 loop specific mAb, 447-52D(Stanfield et al., 2004), did not bind to the JRFL NFL or JRFL SOSIP trimers on the liposomes, indicating that the V3 region of these trimers is not accessible when arrayed on the liposomal surface. These results contrast with the BLI binding data shown here (Figure S3A), and previously published data(Sharma et al., 2015) that were generated with the JRFL ordered trimers in solution and 447-52D on the sensor surface. In this configuration, the well-ordered JRFL trimers were well recognized by the V3-directed mAb. To confirm this result in the same binding format used for the liposomes, we assessed 447-52D recognition of the JRFL SOSIP and JRFL NFL trimers when they were captured on the WGA sensors. In this context, 447-52D recognized the trimers, indicating occlusion of the V3 region occurs only when the trimers are arrayed on the liposomal surface. The non-neutralizing mAb, F105(Posner et al., 1993), does not efficiently recognize the soluble JRFL NFL or SOSIP trimers(Guenaga et al., 2015; Sharma et al., 2015) (Figure 4A and S3B), but does efficiently recognize disordered trimers, such as foldon(Guenaga et al., 2015; Sharma et al., 2015). Here, F105 did not recognize the liposome-conjugated trimers as assessed by BLI, indicating the maintenance of a well-ordered trimeric state following liposomal conjugation (Figure 4A and Figure S3C). Since the previous affinities using selected bNAbs were not determined by WGA capture but by His-tag capture of the JRFL trimers(Guenaga et al., 2015; Sharma et al., 2015), we performed a comparative binding analysis with most of the antibodies used to probe the trimer-conjugated liposomes to confirm the relative rank order of the antibody avidities to the trimers and the apparent occlusion of V3 on the trimer-conjugated liposomes (Figure 4A).
Figure 4. Binding of HIV-1 antibodies to JRFL SOSIP trimer-conjugated liposomes and soluble JRFL SOSIP trimer assessed by Bio-Layer Interferometry using Octet and negative stain EM.
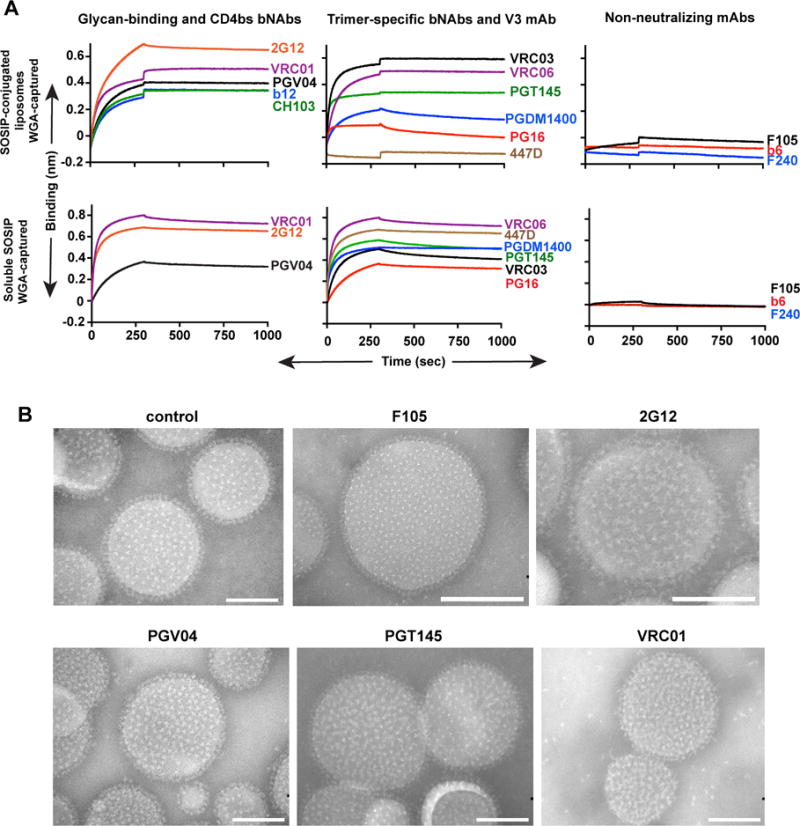
(A) JRFL SOSIP trimer conjugated to 4% DGS-NTA(Ni) liposomes (equivalent to 75 nmoles of phospholipids) or JRFL SOSIP trimers (10 mg/ml) were immobilized on WGA-captured streptavidin sensors and 20 mg/ml monoclonal antibodies (IgGs) were used as analyte. (B) 2% DGS-NTA(Ni) liposomes conjugated to JRFL SOSIP were incubated with 10 molar excess of respective IgG mAbs at 37°C for 30 min, stained with phospho-tungstate, viewed by EM and images were obtained with a CCD camera. All images are at 180,000× magnification. Scale bar = 100 nm. See also Figure S3.
We then probed both JRFL SOSIP and NFL trimers arrayed on the 2% DGS-NTA(Ni)-lipid-containing liposomes with a subset of Env-directed antibodies, but now qualitatively analyzed by negative stain EM, to determine relative accessibility of the neutralizing and non-neutralizing epitopes once the trimers are coupled to the solid phase. Consistent with the binding analysis, when the Env trimer-conjugated liposomes were incubated with non-neutralizing F105 mAb, in excess, there was no detectable binding to the ordered trimers compared to the unliganded control trimer-conjugated liposomes (Figure 4B and Figure S3D). In stark contrast, when the glycan-directed bNAb, 2G12(Trkola et al., 1996) was incubated with the trimer-conjugated liposomes, the propeller-like pattern of the well-ordered trimers was noticeably perturbed by this full IgG. Similarly, but with less disruption of trimer symmetry, PGV04 and PGT145 binding to the trimers could be observed following incubation and negative stain EM (Figure 4B and Figure S3D).
Stability of trimer-conjugated liposomes at selected temperatures by EM
We next sought to determine the stability of the trimer-conjugated liposomes in aqueous buffer at both 4°C and 37°C. Accordingly, we stored the liposomes at 4°C for an extended period of time and assessed overall conformation by negative stain EM. Both the liposomes and the trimers arrayed on the surface of these nanoparticles were very stable for up to 4 months at 4°C (Figure S2B). We performed the same analysis of trimer-bound liposomes stored at 37°C and detected more of an impact on both the liposomes and trimers on the surface of the liposomes. In brief, approximately 50% of trimers were lost from the liposomal surface over a period of 7 days when stored at 37°C (Figure S2C).
Trimer-conjugated liposomes are more efficient at activating B cells than soluble trimers
Next, we determined if the well-ordered trimers on the liposomes were able to activate B cells following BCR engagement. For this purpose, we used B cells isolated by negative selection from the previously described mature b12-expressing knock-in mice(Ota et al., 2013). The B cells were over 99% pure as characterized by flow cytometry using the B cell markers CD19 and B220 (Figure S4). The B cells expressing matched b12 H and L chains, possessing either IgM or IgD transmembrane regions, were activated for 18–20 hours with JRFL SOSIP as soluble protein or liposomal preparation and stained with fluorescent antibodies specific for the B cell activation markers CD69, CD86 and MHC class II. The B cell activation markers CD86, CD69, and MHC II were upregulated in a dose-dependent manner in both formulations. However, increases in the levels of CD69 and CD86 were significantly greater following incubation with the trimer-conjugated liposomes compared to the soluble trimers (Figure 5A), indicating that the multivalent array of the HIV-1 trimers was more effective for the induction of BCR signaling and activation. As shown in Figure 5B, in addition to the increased MFI values associated with liposomal activation, a clear shift in CD69 levels present on the cell-surface was observed in a greater percentage of B cells incubated with the trimer-conjugated liposomes compared to those incubated with soluble trimers. Next, we assessed the expression of proinflammatory cytokines following overnight incubation of the B cells with the two trimer-types. We assessed proinflammatory cytokines since they are often produced at higher quantities benefitting ease of detection and WEHI B cells were shown previously to express TNF-α(Canfield et al., 2005). By ELISA, the levels of TNF-α and IL-6 levels were significantly increased in the culture medium of the B cells incubated with the trimer-conjugated liposomes compared to the soluble trimers (***P=0.0001 and **P=0.008 respectively). These data are consistent with the benefit of multi-valent trimer particulate array to enhance B cell activation. We performed additional experiments using blank liposomes (4% DGS-NTA(Ni) liposomes lacking trimers) as negative controls compared to the JRFL SOSIP-conjugated liposomes. The induction of cell-surface activation markers and the levels of cytokines induced by the blank liposomes was negligible compared to the trimer-conjugated liposomes, confirming that that the B cells were specifically activated by the trimers arrayed on the surface of the liposomes (Figure 5D). Since the liposomes used for these experiments were generated without MPLA and R848, we confirmed that their integrity was similar to liposomes formulated with these TLR agonists by negative stain EM (Figure S2E).
Figure 5. Activation of primary B cells by soluble JRFL SOSIP trimers and JRFL SOSIP trimer-conjugated liposomes.
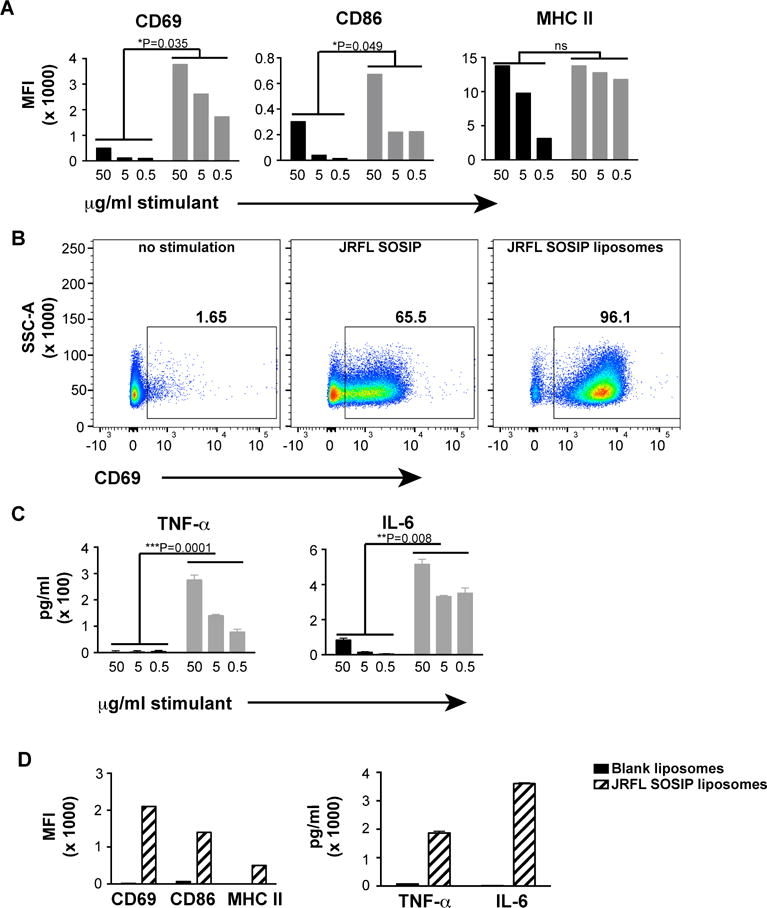
B cells from b12 mature knock-in mice were negatively selected from splenocytes and induced by overnight incubation with either soluble JRFL SOSIP trimers or 4% liposomes conjugated with JRFL SOSIP trimers. The cell-surface activation markers and the cytokines secreted by the activated cells were analyzed by cell-surface staining or ELISA. (A) FACS staining of cell-surface activation markers plotted as MFI values. Soluble JRFL SOSIP (black); JRFL SOSIP conjugated to liposomes (grey bars). (B) Frequency of CD69+ cells upon activation by 50 μg/ml of soluble trimers or JRFL SOSIP trimer-conjugated liposomes. (C) TNF-α and IL-6 levels present in the supernatants of the B-cells upon overnight activation by soluble JRFL SOSIP trimers (black bars) or JRFL trimer-conjugated liposomes (grey bars) B cells were assessed. (D) MFI values of cell surface activation markers and levels of cytokines produced by B cells upon activation by 50 μg/ml of JRFL SOSIP liposomes or similar dilution of blank liposomes without any trimers on the surface. Statistical comparisons between groups are performed by paired t-test. See also Figure S4.
Germinal center (GC) B cells are more efficiently activated by the trimer-conjugated liposomes compared to soluble trimers
GCs are formed in secondary lymphoid organs such as lymph nodes (LNs) and spleen, where activated B cells proliferate and undergo immunoglobulin isotype class switching and somatic hypermutation(Victora and Nussenzweig, 2012). To assess the capacity of the multivalent trimer liposomal array in formation of GCs relative to the soluble trimers, we separated C57BL/6 mice into 3 groups (5 mice per group) and inoculated with PBS (naïve), soluble JRFL SOSIP trimers in ISCOMATRIX™ adjuvant (CSL, Australia), and JRFL SOSIP trimer-containing liposomes in ISCOMATRIX™ adjuvant. We confirmed that the ISCOMATRIX™ adjuvant did not affect the trimers by EM (Figure S2D). Fourteen days following inoculation, we observed that the draining LNs were larger in the protein/adjuvant-inoculated mice as compared to the naïve mice. We isolated single cells from the LNs of the 5 mice in each of the three groups and performed flow cytometry analysis of CD19+ B cells positive for the GC marker, GL7. The flow cytometry analysis indicated a higher percentage of B cells positive for GL7 in mice inoculated with the JRFL SOSIP-conjugated liposomes compared to mice inoculated with the soluble JRFL SOSIP trimers. Specifically, we observed that the soluble trimers displayed a significantly increased percentage of GL19+GL7+ B cells compared to naïve mice analyzed similarly (**P=0.0012) as did the trimer-conjugated liposome mice (**P=0.0015). More importantly, the Env trimers arrayed on the surface of the liposomes elicited a statistically significant increase in the percentage of CD19+GL7+ B cells compared to the levels elicited by the soluble trimers (*P=0.0152, Figure 6A and 6B), indicating more efficient GC formation was induced by particulate, multivalent trimer array. To ensure that the increase in GL7+ B cells was elicited by the trimers conjugated to liposomal surface and was not due to non-specific activation from the liposomes themselves or inadvertent acquisition of a contaminant during processing (ie, endotoxin), we performed additional control experiments with blank liposomes (4% DGS-NTA(Ni) liposomes lacking trimers) or JRFL SOSIP-conjugated liposomes. Fourteen days post inoculation, the percentage of CD19+GL7+ cells present in LNs derived from individual mice were analyzed. Mice immunized with the JRFL SOSIP-conjugated liposomes possessed significantly higher GL7+ B cells compared to mice immunized with the blank liposomes (***P=0.0004; Figure 6C).
Figure 6. Immunization with JRFL SOSIP trimer-conjugated liposomes induced enhanced germinal center (GC) formation.
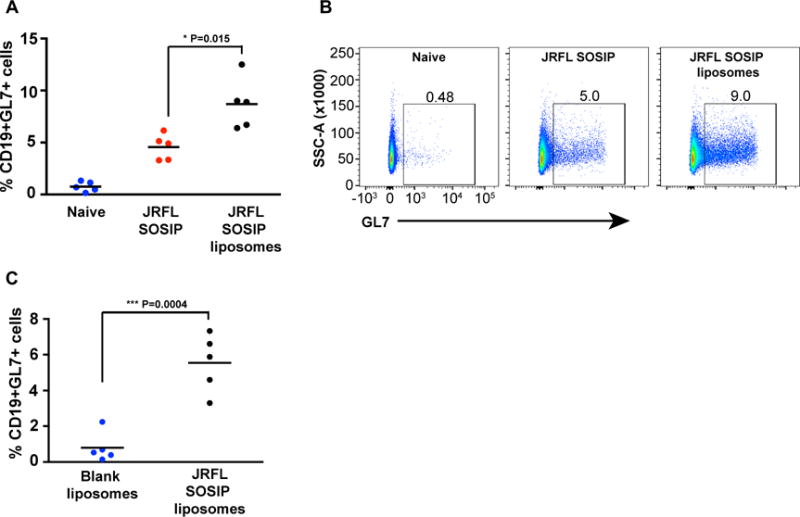
(A) Three groups of five C57Bl/6 mice were subcutaneously administered PBS, soluble JRFL SOSIP trimers or JRFL SOSIP trimer-conjugated liposomes. After 14 days, lymph node B cells were analysed for the activation marker, GL7. The percentage of CD19+ GL7+ cells are enumerated. (B) Representative flow cytometry scatter plots from each group of mice shown in (A) were gated over CD19+ cells from lymph nodes. (C) Two groups of five C57Bl/6 mice were subcutaneously administered either blank liposomes or JRFL SOSIP trimer-conjugated liposomes. After 14 days, the lymph nodes are processed and the percentages of CD19+ GL7+ cells are enumerated. P values were calculated with a two-tailed unpaired t test.
Binding and neutralizing antibodies elicited by trimer liposomal array compared to soluble trimers
Given the promising antigenic profile of the well-ordered trimers and the ability of the liposome array of the spike mimetics to activate B cells more efficiently both ex vivo and in vivo compared to the soluble spikes, we next tested the trimer-conjugated liposomes formulated in adjuvant for immunogenicity in a pilot rabbit study. We also sought to determine if inclusion of innate-response-activating TLR agonists into the liposomes would augment antibody responses. Three groups of four rabbits each were immunized with 25 μg of protein either as soluble protein trimer in adjuvant or arrayed on the surface of 4% DGS-NTA(Ni) liposomes containing TLR ligands either with or without adjuvant. Control animals were immunized with blank liposomes containing TLR ligands with adjuvant. Prior to inoculation, the trimer protein concentrations on the liposomes were assessed by protein dye to confirm and quantify the Env content per volume of liposome (see Methods).
After 3 immunizations, IgG titers were elicited against JRFL SOSIP as measured by ELISA with JRFL SOSIP captured on the plate by the anti-His mAb (see Figure 7B). The soluble trimers in adjuvant elicited relatively low, but detectable, binding titers to the His-captured SOSIP immunogen. The trimer-conjugated liposomes with the incorporated TLR agonists, but not formulated in the exogenous adjuvant, elicited very little IgG antibody response, indicating that the TLR ligands contributed little to activate the adaptive immune response to Env in vivo. In contrast, the trimer-conjugated liposomes (+TLR ligands) but formulated in exogenous adjuvant, elicited much higher native-trimer binding titers compared to both trimer:liposomes(+TLR ligands) lacking exogenous adjuvant and the soluble trimers formulated in exogenous adjuvant. Taken together, these comparative data sets demonstrated that the liposomal presentation of the trimers rendered them more immunogenic than soluble trimers in the presence of exogenous adjuvant. Blank liposomes in adjuvant, as expected, elicited no detectable trimer binding antibodies in the serum.
Figure 7. Immunogenicity of the JRFL SOSIP trimer-conjugated liposomes.
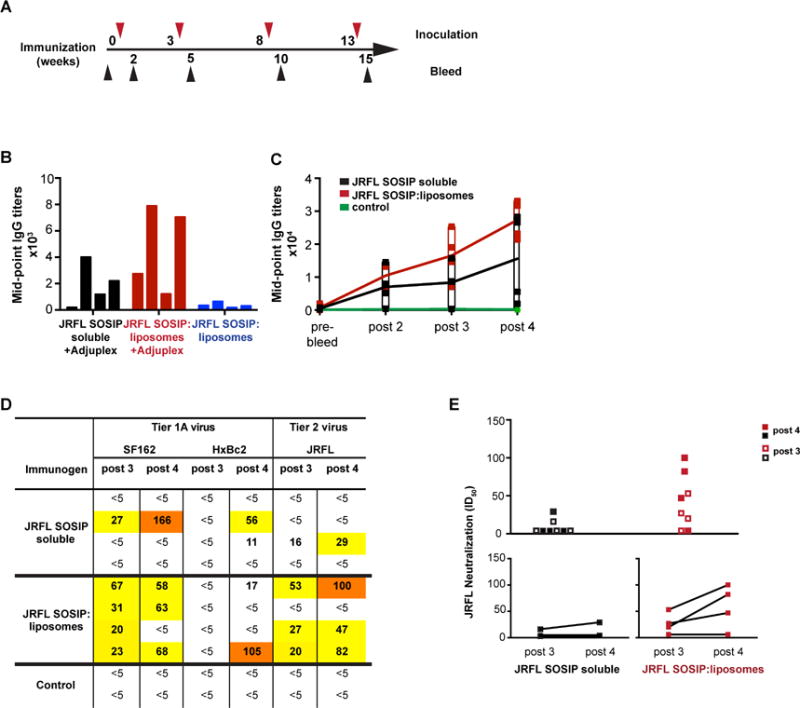
(A) Timeline of inoculations and bleeds. Bleeds were collected 2 weeks after each injection. (B) Mid-point IgG titers of individual rabbits immunized 3 times with 25 ug JRFL SOSIP protein as soluble trimers or conjugated to liposomes in the presence or absence of exogenous adjuvant Adjuplex were determined. Sera were collected 2 weeks after the 3rd injection and were analyzed by ELISA with JRFL SOSIP trimers captured on ELISA plate via the C-terminal His6tag. (C) Mid-point IgG titers of rabbits immunized 4 times with JRFL SOSIP soluble protein or JRFL SOSIP conjugated to liposomes. ELISA plates were coated with anti-His monoclonal antibody to capture JRFL SOSIP trimers via the C-terminal His6-tag. (D) Neutralization ID50 values of SF162, HxBc2, and JRFL viruses by antisera following the third and fourth inoculations. Control animals were inoculated with blank liposomes in Adjuplex. (E) Combined JRFL neutralization ID50 values elicited by the soluble trimers compared to the trimer-conjugated liposomes are plotted after third and fourth inoculations. Lower panel shows boosts in ID50 values after the fourth inoculation for both group of rabbits. See also Figure S5.
Due to the initial results indicating that the trimer-conjugated liposomes in adjuvant were more immunogenic than the soluble trimers in adjuvant, we performed additional boosts at 5 week intervals and assessed binding titers in a longitudinal manner. We observed that there was a trend for the liposomal trimers to elicit higher binding titers to the native spike mimetics compared to the soluble trimers over the course of immunization (Figure 7C). Together these data indicated that the trimer-conjugated liposomes were more immunogenic, and that the trimer integrity was maintained in vivo when present on the liposome surface. To further evaluate the quality of the antibody response, we assessed the avidity of the sera to JRFL SOSIP captured on plate by anti-His antibody. The animals immunized with trimer-conjugated liposomes possessed antibodies displaying higher avidity than those receiving soluble trimers, however, the increased avidity was not statistically significant (Figure S5A).
Next, to determine the quality of the neutralizing response, we performed HIV pseudovirus neutralization assays(Li et al., 2005). We used sera isolated from individual animals immunized with each trimer-type 14 days following each inoculation with either soluble or trimer-conjugated liposomes. Following both the 3rd and 4th immunization, we detected modest autologous tier 2-like JRFL neutralizing titers from 3 of 4 rabbits receiving the trimer-conjugated liposomes. Only one animal immunized with the soluble trimers displayed weak neutralizing activity at these time points. Although there was a trend for increased neutralization titers elicited by the trimer-conjugated liposomes compared to the soluble trimers, and in more animals per group, as well as boosting of trimer-conjugated liposomes compared to the soluble trimers, these differences were not statistically significant in this small pilot study (Figure 7D and E).
Discussion
In this study, we made use of the new well-ordered Env trimers recently designed in our laboratory to create a high-density multi-variant array of these recombinant glycoprotein HIV spike mimetics on the surface of fully synthetic liposomes. We generated synthetic nickel containing liposomes by incorporating lipids bearing Ni+2 at the polar head group into the lipid bilayer, to capture the C-terminally His6-tagged trimers. We demonstrated by both negative staining and cryo-EM that both the NFL and SOSIP trimers could be arrayed at high-density, were stable, and displayed a favorable antigenic profile following coupling to the surface of the fully synthetic liposomes. We showed that the well-ordered, trimer-conjugated liposomes more efficiently activated B cells ex vivo compared to soluble trimers and better generated GC B cells in vivo in wild-type mice in a statistically significant manner. The trimer-conjugated liposomes displayed a trend to elicit both native binding antibodies and neutralizing antibodies against the autologous HIV-1 tier 2 strain that is more resistant to vaccine-elicited neutralizing antibodies.
These data indicate that we have developed a flexible platform, in which potentially any His-tagged Env trimer can be arrayed at high density. We could array literally hundreds of trimers per particle, perhaps overcoming immune evasion mechanisms that HIV has evolved naturally to reduce activation of host B cells. The non-glycosylated bottom surface of the trimers is blocked by capture using the His tag on the C-terminus of the trimers on the liposomal surface. This non-glycosylated surface was recently shown to be highly immunogenic, potentially diverting neutralizing responses, rendering the SOSIP immunogens incapable of eliciting neutralizing titers in mice(Hu et al., 2015). The high density and uniform array indicates that all three of the His6 tags bound to nickel on the liposome, although we do not have direct evidence for this possibility. Array of the trimers on the liposomes may stabilize more distal regions of Env as the V3 is partially exposed on JRFL Env (more so than most other tier 2 viruses), however, on the JRFL trimer-conjugated liposomes, we could not detect any V3 binding, even with IgG avidity in play. The well-ordered trimers also exhibit enhanced stabilization on the liposomes, as at 4°C they remain tightly trimeric and arrayed at high density over a period of multiple months.
In terms of B cell activation, the multivalent trimer array on the surface of the liposomes seems to impart advantages in terms of B cell activation both ex vivo and in vivo. We used mouse B cells expressing the bNAb b12, and showed that there is enhanced activation of these B cells, presumably via avidity gained by the high-density multivalent array. Although the increased secretion of IL-6 from the purified B cells following induction by the trimer-conjugated liposomes was consistent with known cytokine profiles, that TNF alpha secretion was also increased was somewhat surprising as this cytokine is not usually secreted by naïve murine B cells, although it has been reported to be secreted by human B cells (Plzakova et al., 2014)and WEHI B cells(Canfield et al., 2005). This may indicate super-antigen effects by trimer multi-valent array or most likely, an altered cytokine profile in these engineered B cells. Less likely, TNF-a secretion is indirect and occurs by dose-dependent activation of a small percentage of myeloid cells in the greater than 99% pure B cell population.
In terms of immunogenicity in vivo, in a pilot rabbit study, the trimer-conjugated liposomes displayed a trend to induce increased binding titers to the native-like trimers compared to the soluble trimers. The enhanced binding titers were detectable beyond a boost or two, an initial indication of longevity of the B cell response. This scenario is reminiscent of the arrayed L1 protein of the highly successful HPV vaccine, which imparts long-lived antibody secreting B cells in humans(Schiller and Chackerian, 2014; Schiller and Lowy, 2015). It will be very interesting to determine if the Env-conjugated liposomes can accomplish prolonged antibody secretion in non-human primates on the pre-clinical pathway toward a vaccine candidate using this type of system. Inclusion of TLR agonists into the liposomal formulation did not render the trimers very immunogenic, indicating that at least for the levels of TLR agonist incorporated here, and the subcutaneous route of administration, these ligands contributed little to the adaptive immune response to the trimers. The data shown here indicates a clear trend of benefit of multivalent array over soluble trimers in vivo in the presence of exogenous adjuvant. However, due to the small numbers of animals per group in this initial study, statistical significance was not achieved. Neutralizing capacity, although not robust in our study, again displayed a trend to be preferentially induced by the trimer-conjugated liposomes, but again did not achieve statistical significance. Why we don’t elicit tier 2 autologous neutralizing responses approaching the levels reported by Sanders et al(Sanders et al., 2015) is unclear, but the JRFL strain used here is generally resistant to vaccine-induced antibodies, as apparently is not the case for BG505 when SOSIPs are inoculated into rabbits. In fact, for BG505, monomeric gp120 can efficiently elicit autologous tier 2 neutralization in rabbits, whereas JRFL gp120 does not elicit autologous tier 2 neutralizing antibodies(Beddows et al., 2007). Further investigation will be needed to determine trimer-virus pairing in regards to the elicitation of autologous tier 2 neutralizing antibodies, as well as the elicitation of neutralizing antibodies in different animal models using matched well-ordered trimer immunogens.
Due to the flexibility of the liposomal system described here, expansion to incorporate well-ordered trimers from other clades onto the liposomal surface is possible either as a diverse array of trimers on independent particles or with different trimers from the same subtype or from different clades arrayed on the same liposome. Such array may have advantages to enhance responses to conserved and common B cell epitopes and neutralizing determinants. Using the approach described here, we demonstrate proof-of-concept using Ni-dependent capture of well-ordered HIV trimers as immunogens. For clinical applications of this approach, we may need to use other divalent cationic lipids possessing cobalt or zinc due to potential nickel inflammatory issues or it might be beneficial to use available maleimide-conjugated lipids to capture trimers possessing a free C-terminal cysteine per protomer of each trimer to reduce any concerns about release from the liposomal surface in vivo.
In summary, we present here initial analysis of multivalent array of ordered HIV trimers conjugated to liposomes in vitro, improved over our previous Env:proteoliposome studies(Grundner et al., 2002), and their potential advantages as an improved immunogenic platform to more efficiently activate B cell responses ex vivo and in vivo.
Experimental Procedures
Expression and purification of recombinant HIV-1 Trimer proteins
JRFL SOSIP and JRFL NFL trimers were expressed in serum-free medium by transient transfection of HEK293F cells (Invitrogen) with plasmid DNA as described previously(Sharma et al., 2015). In brief, the secreted proteins were purified by galanthus lectin affinity chromatography followed by size exclusion chromatography (SEC). Next, the trimer peak was subjected to negative selection by the non-neutralizing monoclonal antibody, F105 to retain disordered trimers on the column. The flow through from the F105 column, containing the well-ordered trimers, was resolved by a second SEC column to isolate a homogenous fraction of well-ordered trimers.
Liposome preparation, protein conjugation and lipid and protein quantitation
For DLS, Octet, EM, GC analysis, and immunization, the liposomes were composed of a molar ratio of 50:36:4:5:5 of DGPC, cholesterol, DGS-NTA(Ni), Monophosphoryl lipid A (MPLA) (synthetic) PHAD™ (Avanti Polar Lipids), and R848 (InvivoGen). For ex-vivo studies, liposomes were composed of a molar ratio of 60:36:4 of 1,2-distearoyl-sn-glycero-3-phosphocholine (DGPC) (Avanti Polar Lipids), cholesterol (Sigma Lifescience), and [(5-amino-1-carboxypentyl) imino di acetic acid) succinyl] (nickel salt) (DGS-NTA(Ni)) (Avanti Polar Lipids). To form the liposomes, the above constituents were mixed in the appropriate ratios in chloroform, incubated over glass and chloroform was evaporated in the presence of gaseous nitrogen. The resulting lipid film was dried further O/N in a desiccator. The lipids were hydrated in PBS for 2 hours at 37°C with constant shaking followed by vigorous sonication for 30 seconds. Next, the liposomes were extruded for a minimum of 15 times through 1 μm, 0.8 μm, 0.4 μm, 0.2 μm, and 0.1 μm filters using a hand-held mini-extrusion device (Avanti Polar Lipids) at room temperature (RT). For conjugation of protein to the liposomes, 2.2mg of trimer protein was added to 500 μl of liposomes and incubated at RT for 2 hours. The unbound protein was removed from the liposomes by passing the liposome mixture through Superdex 100 column. The liposome fractions were collected, pooled and stored at 4°C.
JRFL SOSIP and JRFL NFL trimers conjugated on the liposomes were quantitated using a standard curve generated by either soluble JRFL SOSIP or JRFL NFL trimers, respectively, using Advanced Protein Assay Reagent (Cytoskeleon) according to manufacturer’s instructions. Phosphorous in liposomes was estimated by a colorimetric assay reported earlier. Briefly, phosphorous (Sigma) standard curve was generated and used to determine the amounts of phosphorous in the liposome samples. First, the organic samples were digested to inorganic phosphate by heating the samples at 215°C for 25 min, followed by addition of hydrogen peroxide and continued heating for additional 30 min. Next, ammonium molybdate and ascorbic acid were added sequentially and again the samples were heated at 100°C for 7 min. The absorbance at 820 nm was determined for both the standard and the experimental samples.
Electron Microscopy
For negative stain EM, liposomes were applied for 3 minutes onto a glow discharged carbon-coated 400-Cu mesh grids (Electron Microscopy Sciences, Hatfield, PA). Excess sample was removed and the grids were immediately placed on a droplet of 2% phosphotungstic acid solution (pH 6.9) for 2 minutes. Excess stain was removed and the grids were allowed to dry thoroughly. Grids were examined on a Philips CM100 electron microscope (FEI, Hillsbrough OR) at 80 kV, and images were acquired with a Megaview III charge-coupled device (CCD) camera (Olympus Soft Imaging Solutions Germany). For cryo EM, samples were preserved undiluted in vitrified ice supported by holey carbon films on 400-mesh copper grids. Samples were prepared by applying 3 μl drop of sample suspension to a clean grid blotting away excess with filter paper and immediately proceeded with vitrification in liquid ethane. Grids were stored under liquid nitrogen until transferred to the electron microscope for imaging. Electron microscopy was performed using an FEI Tecnai T12 electron microscope operating at 120keV equipped with an FEI Eagle 4k × 4k CCD camera. Vitreous ice grids were transferred into the electron microscope using a cryostage that maintains the grids at a temperature below −170°C.
Bio-Layer Light Interferometry (BLI)
Binding interactions between monoclonal antibodies (mAbs) and the trimers conjugated to the liposome surface or soluble trimers were examined by BLI using an Octet RED system (ForteBio). Biotinylated Wheat Germ Agglutinin (WGA) (Vector Laboratories) was captured on Streptavidin biosensors (ForteBio) at 50 μg/ml in PBS-B (PBS with 0.1%BSA) for 180 sec followed by wash for 180 sec in PBS-B. Next, the liposomes conjugated with trimers (10 μg/ml) were loaded onto the WGA sensors for 30 min followed by wash for 60 min. The biosensors were immersed in PBS-B aliquoted in 96 well plates to generate a baseline. Next, the biosensors were immersed in a separate 96 well plate filled with PBS-B containing the mAbs (20 μg/ml) for 300 sec to allow association of the immobilized trimers with antibody. Association was followed by dissociation in PBS-B for 30 min. A constant temperature of 30°C was maintained inside the instrument during all reactions. A reference sensor was generated during each experimental run to ensure that there was minimal level of non-specific binding of mAbs to the WGA sensors. Binding avidity of each IgG to the trimer-conjugated liposomes was rank ordered as determined by the maximal binding signal at the end of the association phase at 300 sec.
B-cell Activation Assay
B-cells from spleen and lymph nodes were purified by negative selection using MACS® separation (Miltenyi Biotec) that uses monoclonal antibodies specific for T cells and monocytes conjugated to the paramagnetic bead to retain all cells but B cells on the solid phase. Cells were plated as 1×105 cells/well in a 96 well plate and stimulated with 50 μg/ml, 5 μg/ml and 0.5 μg/ml of soluble JRFL SOSIP trimers, JRFL SOSIP trimers conjugated to the 4% DGS-NTA(Ni) liposomes or 4% DGS-NTA(Ni) liposomes for 18–20 hours. The supernatants were stored at −20 °C for TNF-α and IL-6 ELISAs. Next, the cells were stained with fluorescent antibodies specific for CD86, CD69, and MHC II prior to analysis by Flow Cytometry. The experiment was performed in two independent experiments from different mature b12 mAb transgenic mice(Ota et al., 2013).
Animal Inoculations
NewZealand white female rabbits were inoculated subcutaneously with 25 μg of protein as either soluble or conjugated to 4% DGS-NTA(Ni) liposomes and formulated in 20% of Adjuplex™ (Advanced BioAdjuvants) in a total volume of 150 μl. For the control group, blank 4% DGS-NTA(Ni) liposomes with MPLA and R848 were formulated in 20% of Adjuplex™. The liposomes contained 30 μg MPLA and 6.5 μg R848 per each injection. Test bleeds were collected two weeks after each inoculation. To determine in vivo GC formation, 3 groups of 6 weeks old C57BL/6 mice (5 mice per group) were subcutaneously inoculated in the hind legs by hock injection(Kamala, 2007) with either PBS, 10 μg of soluble JRFL SOSIP trimeric protein formulated in 1 unit of ISCOMATRIX, 10 μg of JRFL SOSIP trimeric protein conjugated to 4% DGS-NTA(Ni) liposomes formulated in 1 unit of ISCOMATRIX, or 4% DGS-NTA(Ni) liposomes formulated in 1 unit of ISCOMATRIX in a total volume of 100 μl. Fourteen days following inoculation, the draining popliteal lymph nodes were isolated and prepared as single cell suspensions and subjected to staining with mAbs as described below.
ELISA
ELISAs were performed in 96-well MaxiSorp plates (Nalgene Nunc International). Plates were coated for 4 hrs at RT with anti-His tag mAb (2 μg/ml; R&D Systems). After blocking the plates with non-fat milk and fetal bovine serum (FBS) for overnight at 4°C, the plates were incubated with JRFL SOSIP trimeric protein at 2 μg/ml for 2 hr at room temperature. Next, the plates were incubated with five-fold serial dilutions of the immune sera starting at 1:200, and after 1 hr were washed with buffer, followed by incubation with HRP-conjugated anti-rabbit IgG (1:5000) or HRP-conjugated anti-rabbit IgM (1:5000) for detection. The plates were developed by a chromogenic substrate for HRP, 3,3′, 5, 5′-tetramethylbenzidine (Life Technologies). Reactions were stopped by the addition of sulfuric acid and absorbance was measured at 450 nm. For ex-vivo studies, culture supernatants were collected and cytokine ELISAs were performed using DuoSet ELISA Development Kits (R&D Systems) according to manufacturer’s instructions.
Neutralization assay
The pseudoviruses were prepared and neutralization assays were performed as described previously(Li et al., 2005). Briefly, rabbit sera were diluted and pre-incubated with virus (200,000 RLU) for 30 min at 37C before adding to 10,000 TZM-bl reporter cells per well. These cells contain an integrated luciferase gene under the control of the Tat-sensitive HIV LTR. Virus was incubated for cells the cells for 48 hrs to allow infection and potential luciferase induction, after which the cells were lysed, and relative luciferase units were measured by a Victor luminometer (PerkinElmer).
Flow cytometry of mouse LN-derived B cells
Murine lymph nodes were gently disrupted through a 70 μm cell sieve, followed by extensive washing. All cells were labeled with live/dead cell viability reagent (Invitrogen) followed by blocking with anti-mouse CD16/CD32 (BD Pharmingen). Next, the cells were incubated with APC anti-mouse CD19 and FITC anti-mouse GL7 (BioLegend) and post-fixed with paraformaldehyde before acquiring cells on an LSRII (Becton Dickinson) to determine fluorescent mAb binding. Data were analysed with FlowJo software (TreeStar).
Statistical Data Analysis
For statistical analysis of cell-surface activation and cytokine secretion from the trimer-activated B cells (Figure 5), values from each group were statistically compared by a two-tailed paired t-test with 95% confidence interval. Individual activation markers or cytokine values were compared between the soluble trimers versus the trimer-conjugated liposomes at the same concentration of stimulant. Values for the B cell GC marker, GL7 (Figure 6), induced by either the soluble trimers or the trimer-conjugated liposomes were statistically analyzed using the two-tailed unpaired t-test with 95% confidence interval. For the rabbit immunogenicity studies analyzing IgG binding and serum neutralization (Figure 7B and 7D), the soluble trimer and trimer-conjugated liposome values were statistically compared by two-tailed Mann Whitney test. All statistical analysis were performed using Prism (GraphPad Software, Inc) and significance was defined as P<0.05.
Supplementary Material
Highlights.
Well-ordered HIV envelope glycoprotein trimers are arrayed on synthetic liposomes.
The trimers maintain structural integrity as determined by biophysical means and EM.
The trimer-conjugated liposomes better activate B cells compared to soluble trimers.
Trimer conjugation enhances the generation of germinal center B cells in vivo.
Acknowledgments
This work was funded by the HIVRAD grant number P01 AI104722, CHAVI-ID grant number AI100663 (JI and RW), R01AI073148 (DN) and AI098602 (AS and MZ) and by the International AIDS Initiative (IAVI) and its generous donors (RW). IAVI’s work is made possible by generous support from many donors including: the Bill & Melinda Gates Foundation; the Ministry of Foreign Affairs of Denmark; Irish Aid; the Ministry of Finance of Japan; the Ministry of Foreign Affairs of the Netherlands; the Norwegian Agency for Development Cooperation (NORAD); the United Kingdom Department for International Development (DFID), and the United States Agency for International Development (USAID). The full list of IAVI donors is available at www.iavi.org. This study is made possible by the generous support of the Bill & Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery and the American people through USAID. The contents are the responsibility of the International AIDS Vaccine Initiative and do not necessarily reflect the views of USAID or the United States Government. We would like to thank Oleksandr Kalyuzhniy and Richard Wilson for technical assistance and William Schief, James Paulson, Matthew McCauley and Darrell Irvine for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
J.I., R.W. designed the research; J.I. performed research; A.S. designed ex-vivo experiments; D.N. provided b12 mice; S.K.S provided JRFL NFL construct; J.G. provided JRFL SOSIP construct; J.I., R.W., D.N. and M.Z. wrote the manuscript.
References
- Arsov Z, Quaroni L. Direct interaction between cholesterol and phosphatidylcholines in hydrated membranes revealed by ATR-FTIR spectroscopy. Chem Phys Lipids. 2007;150:35–48. doi: 10.1016/j.chemphyslip.2007.06.215. [DOI] [PubMed] [Google Scholar]
- Beddows S, Franti M, Dey AK, Kirschner M, Iyer SP, Fisch DC, Ketas T, Yuste E, Desrosiers RC, Klasse PJ, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360:329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nature Immunology. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Caldeira Jdo C, Medford A, Kines RC, Lino CA, Schiller JT, Chackerian B, Peabody DS. Immunogenic display of diverse peptides, including a broadly cross-type neutralizing human papillomavirus L2 epitope, on virus-like particles of the RNA bacteriophage PP7. Vaccine. 2010;28:4384–4393. doi: 10.1016/j.vaccine.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield S, Lee Y, Schroder A, Rothman P. Cutting edge: IL-4 induces suppressor of cytokine signaling-3 expression in B cells by a mechanism dependent on activation of p38 MAPK. J Immunol. 2005;174:2494–2498. doi: 10.4049/jimmunol.174.5.2494. [DOI] [PubMed] [Google Scholar]
- Deml L, Kratochwil G, Osterrieder N, Knuchel R, Wolf H, Wagner R. Increased incorporation of chimeric human immunodeficiency virus type 1 gp120 proteins into Pr55gag virus-like particles by an Epstein-Barr virus gp220/350-derived transmembrane domain. Virology. 1997;235:10–25. doi: 10.1006/viro.1997.8669. [DOI] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner C, Mirzabekov T, Sodroski J, Wyatt R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. Journal of virology. 2002;76:3511–3521. doi: 10.1128/JVI.76.7.3511-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, et al. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. Journal of virology. 2015 doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamala T. Hock immunization: a humane alternative to mouse footpad injections. J Immunol Methods. 2007;328:204–214. doi: 10.1016/j.jim.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JM, Noeldeke E, Ha HJ, Peng H, Rits-Volloch S, Harrison SC, Chen B. Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18542–18547. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of virology. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, O’Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, et al. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. Journal of virology. 2012;86:11231–11241. doi: 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Doyle-Cooper C, Cooper AB, Doores KJ, Aoki-Ota M, Le K, Schief WR, Wyatt RT, Burton DR, Nemazee D. B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J Immunol. 2013;191:3179–3185. doi: 10.4049/jimmunol.1301283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejawar-Gaddy S, Kovacs JM, Barouch DH, Chen B, Irvine DJ. Design of lipid nanocapsule delivery vehicles for multivalent display of recombinant Env trimers in HIV vaccination. Bioconjug Chem. 2014;25:1470–1478. doi: 10.1021/bc5002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plzakova L, Kubelkova K, Krocova Z, Zarybnicka L, Sinkorova Z, Macela A. B cell subsets are activated and produce cytokines during early phases of Francisella tularensis LVS infection. Microb Pathog. 2014;75:49–58. doi: 10.1016/j.micpath.2014.08.009. [DOI] [PubMed] [Google Scholar]
- Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquir Immune Defic Syndr. 1993;6:7–14. [PubMed] [Google Scholar]
- Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, et al. Influences on the Design and Purification of Soluble, Recombinant Native-Like HIV-1 Envelope Glycoprotein Trimers. Journal of virology. 2015;89:12189–12210. doi: 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaeian M, Porras C, Pan Y, Kreimer A, Schiller JT, Gonzalez P, Lowy DR, Wacholder S, Schiffman M, Rodriguez AC, et al. Durable antibody responses following one dose of the bivalent human papillomavirus L1 virus-like particle vaccine in the Costa Rica Vaccine Trial. Cancer Prev Res (Phila) 2013;6:1242–1250. doi: 10.1158/1940-6207.CAPR-13-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J, Chackerian B. Why HIV virions have low numbers of envelope spikes: implications for vaccine development. PLoS Pathog. 2014;10:e1004254. doi: 10.1371/journal.ppat.1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller JT, Lowy DR. Raising expectations for subunit vaccine. J Infect Dis. 2015;211:1373–1375. doi: 10.1093/infdis/jiu648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, et al. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Journal of virology. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


