Abstract
Despite the pivotal role of MYC in the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL) and many other cancers, the mechanisms underlying MYC-mediated tumorigenesis remain inadequately understood. Here we utilized a well-characterized zebrafish model of Myc-induced T-ALL for genetic studies to identify novel genes contributing to disease onset. We found that heterozygous inactivation of a tricarboxylic acid (TCA) cycle enzyme, dihydrolipoamide S-succinyltransferase (Dlst), significantly delayed tumor onset in zebrafish without detectable effects on fish development. DLST is the E2 transferase of the α-ketoglutarate (α-KG) dehydrogenase complex (KGDHC), which converts α-KG to succinyl-CoA in the TCA cycle. RNAi knockdown of DLST led to decreased cell viability and induction of apoptosis in human T-ALL cell lines. Polar metabolomics profiling revealed that the TCA cycle was disrupted by DLST knockdown in human T-ALL cells, as demonstrated by an accumulation of α-KG and a decrease of succinyl-CoA. Addition of succinate, the downstream TCA cycle intermediate, to human T-ALL cells was sufficient to rescue defects in cell viability caused by DLST inactivation. Together, our studies uncovered an important role for DLST in MYC-mediated leukemogenesis and demonstrated the metabolic dependence of T-lymphoblasts on the TCA cycle, thus providing implications for targeted therapy.
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) is a malignant disease of developing thymocytes affecting individuals of all ages. Despite improvements in treatment regimens, T-ALL remains fatal in 20% of pediatric patients and >50% of adult patients, underscoring the urgent need to identify efficacious and selective therapies.1–3 A better understanding of the molecular mechanisms underlying T-ALL transformation, maintenance and/or progression should facilitate development of effective therapeutics.
The proto-oncogene MYC has been implicated in the pathogenesis of many human cancers, including hematological and solid malignancies.4 In the majority of T-ALL cases, MYC is aberrantly expressed downstream of activated NOTCH1 mutations.5–10 Studies using murine and zebrafish transgenic models firmly established the requirement of MYC for T-ALL initiation, maintenance and progression.11–16 For instance, overexpression of the murine Myc gene under a lymphocyte-specific promoter, rag2, promotes rapid T-ALL development in zebrafish,17 recapitulating the major subtype of human T-ALL with LMO1/2 and SCL overexpression.18
When aberrantly expressed, MYC serves as a transcriptional amplifier to promote expression of a multitude of genes that control cell metabolism, growth, proliferation and differentiation.19–21 To meet the increased energy and nutrient demands during the malignant transformation and tumor progression, MYC reprograms cellular metabolism to promote both glycolysis and glutaminolysis.22–27 The enhanced glutaminolysis leads to elevated levels of tricarboxylic acid (TCA) cycle intermediates,28,29 and cells with aberrant MYC expression rely heavily on mitochondrial oxidative phosphorylation for energy production and macromolecule synthesis.30 In the context of T-ALL, glutaminolysis is critical for leukemic cell growth downstream of NOTCH1.31 Despite these observations, it remains unclear whether the TCA cycle contributes to MYC-mediated tumorigenesis.
Here we combine the genetic capacities of the zebrafish model of Myc-induced T-ALL together with analysis of human T-ALL cells to identify novel genes contributing to MYC-mediated leukemogenesis. We found that heterozygous loss of a gene encoding dihydrolipoamide S-succinyltransferase (Dlst) significantly delayed the onset of Myc-induced T-ALL in zebrafish without affecting fish development. Furthermore, partial inactivation of DLST reduced cell viability and induced apoptosis in human T-ALL cell lines. DLST functions as a transferase in the α-ketoglutarate (α-KG) dehydrogenase complex (KGDHC), which is critical for energy production and macromolecule synthesis in the TCA cycle.32 Taken together, our studies identify DLST as an important mediator of MYC-driven leukemogenesis and provide compelling evidence for the metabolic dependence of T-ALL cells on the TCA cycle. Importantly, these studies provide strong rationale to develop and test therapeutic strategies that target DLST and other TCA cycle enzymes for T-ALL treatment.
METHODS
Fish husbandry
Zebrafish (Danio rerio) husbandry was performed as described33 in zebrafish facilities at the Dana-Farber Cancer Institute (DFCI) and the Boston University School of Medicine, in accordance with IACUC-approved protocols. We included male and female AB fish (from embryos to adult stage) in this research.
Zebrafish genetic studies and fish genotyping
The Myc transgenic fish were bred with 17 different fish lines with heterozygous disruption of known genes (Table 1; from at least two independent experiments).34 For each mutant line, we obtained at least 15 of Myc progeny that gave us the probability of ~ 0.70 to detect significant difference. Compound fish (Myc;A+/* and Myc;A+/+) were monitored at 6 days postfertilization (dpf) for the presence of an enhanced green fluorescent protein-positive (EGFP+) thymus to determine potential alterations in T-cell development and to confirm Mendelian segregation of the Myc transgene. The fish were subsequently scored for the presence of thymic tumors at 60 dpf and the percentage of T-ALL fish was determined. To confirm the tumor-suppressive effect of dlst heterozygous loss, Myc fish were bred to dlst heterozygous fish and their progeny were continuously monitored for tumor development over a course of 3 months. Specifically, fry with unknown genotype were raised and screened blindly once every 5 days starting at 21 dpf to determine the time of tumor onset according to the criteria previously defined.35 Fish were imaged on both brightfield and EGFP channels using a fluorescent dissecting microscope (Nikon SMZ1500, Melville, NY, USA). All tumor-bearing fish were isolated at the time of tumor onset, raised individually and examined 1 week later to confirm tumor development. All transgenic and mutant fish were genotyped by gene-specific PCR using DNA isolated from individual fish.35,36 The primer information is included in Supplementary Table S3.
Table 1.
Zebrafish screens identify dlst as a genetic suppressor for Myc-induced T-ALL
| Zebrafish lines examined | Affected gene (protein encoded) | Percentage of Myc fish with T-ALL at 60 dpf (n) |
|---|---|---|
| AB | None | 100 (n = 1000) |
| hi1055B | tip49/pontin (ruvb-like AAA ATPase 1) | 100 (n = 53) |
| hi550 |
smarca5 (swi/snf-related, matrix associated, actin-dependent regulator of chromatin, subfamily a, member 5) |
100 (n = 51) |
| hi2628 | hdac1 (histone deacetylase 1) | 100 (n = 62) |
| hi2618 | rpa1 (replication protein A1) | 100 (n = 52) |
| hi1386 | utp15 (U3 small nucleolar RNA-associated protein 15) | 100 (n = 55) |
| hi2648 | emi1 (foxb5, F-box protein 5) | 100 (n = 122) |
| hi2293 | chrd (chordin) | 100 (n = 15) |
| hi2639b | suds3 (suppressor of defective silencing 3) | 100 (n = 35) |
| hi3970 | skp1 (S-phase kinase-associated protein 1) | 100 (n = 15) |
| hi3471 | ufd1 (ubiquitin fusion degradation 1-like) | 100 (n = 40) |
| hi1116A | tsr2 (20S rRNA accumulation, homolog) | 100 (n = 40) |
| hi3593 | psmc5 (proteasome 26S subunit ATPase, 6) | 100 (n = 22) |
| L4-6 | smo (smoothened) | 98 (n = 50) |
| hi1727 | ddx18 (myc-regulated DEAD/H box 18 RNA helicase) | 93 (n = 57) |
| hi2109 | tbx16/spadetail (T-box 16) | 91 (n = 53) |
| hi1841B | odc (ornithine decarboxylase 1) | 75 (n = 16) |
| hi1903C | dlst (dihydrolipoamide S-succinyltransferase) | 52 (n = 77) |
Abbreviations: dpf, days postfertilization; T-ALL, T-cell acute lymphoblastic leukemia.
Flow cytometric analysis of zebrafish cells
For cell cycle analysis, zebrafish thymocytes and tumor cells were collected by dissection and incubated with Vybrant DyeCycle Ruby stain (Invitrogen, Carlsbad, CA, USA) at room temperature. DAPI (4,6-diamidino-2-phenylindole) stain was used to distinguish dead vs live cells. Cell cycle analysis was performed on a LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Quantification of DNA content was performed with the ModFit software (Verity Software House, Topsham, ME, USA).
To analyze the hematopoietic precursors, the kidney marrows of zebrafish were harvested and dissociated into single cells in phosphate-buffered saline with 1% fetal bovine serum. Cells were stained with DAPI and then run on a LSRII flow cytometer, where hematopoietic precursors were assessed for characteristics of light scatter properties (forward and side scatter) to determine the percentage of each cell population: erythrocyte, lymphocyte, granulocyte/monocyte, and progenitors, as described previously.37
Cryosectioning and immunostaining
Fish were fixed with 4% paraformaldehyde and embedded in agar/sucrose for cryosectioning. Sections were immunostained by conventional protocols using antibodies against phospho-histone 3 (PH3, Cell Signaling, Danvers, MA, USA).38
Quantitative real-time PCR (q-RT-PCR) and pulse chase
Trizol reagent (Invitrogen) was used to extract total RNA from EGFP+ thymocytes or tumor cells that were sorted by fluorescence-activated cell sorting (FACS). cDNA was synthesized using a Reverse Transcription Kit (Qiagen, Valencia, CA, USA). About 50 ng of cDNA was used for each q-RT-PCR reaction, performed with the SYBR green PCR master mix (Qiagen) and a Step-One PCR instrument (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s manual. All reactions were performed in triplicate, using β-actin as controls. The primer information is included in Supplementary Table S2.
Zebrafish thymocytes and T-ALL cells were collected by dissection, dissociated and treated with cycloheximide (50 µg/ml, Sigma, St Louis, MO, USA) for pulse-chase analysis. Cells were collected at 0, 4, 8 and 10 h after treatment, and proteins were extracted for western blotting analysis of MYC, Dlst and Actin levels.
Patient samples
Human bone marrow specimens were collected with informed consent and with approval of the DFCI Institutional Review Board from pediatric patients with T-ALL who were enrolled at the DFCI for clinical trials. All samples were analyzed with approval of the DFCI Institutional Review Board without linked identifiers. Primary T-ALL cells were also collected from patient-derived murine xenografts housed at the University of Massachusetts. Mononuclear tumor cells were purified by Ficoll-Hypaque density centrifugation.
Protein extraction and western blotting analysis
Zebrafish and human T-ALL cells were lysed in RIPA buffer supplemented with Halt proteinase and phosphatase inhibitor cocktail (Thermo Scientific, Cambridge, MA, USA). The primary antibodies included anti-MYC (Santa Cruz, Dallas, TX, USA; or Cell Signaling), anti-ICN1 (Cell Signaling, Val1744), anti-PTEN (Cell Signaling), anti-DLST (Abnova, Walnut, CA, USA), anti-SHMT2 (Aviva, London, UK) and anti-ACTIN (Sigma). Secondary antibodies included horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Pierce, Cambridge, MA, USA). Autoradiographs were imaged with a G:BOX chemi XT4 (Syngene, Frederick, MD, USA) or a LAS-3000 imaging system (Fuji, Valhalla, NY, USA) and a CCD camera and then subjected to quantification analysis with Syngene GeneTools or Fuji software.
Lentivirus transduction and succinate rescue
The control and DLST shRNA hairpins (shCONTROL/shLUC: 5′-CTTCGAAATGTCCGTTCGGTT-3′, shDLST1: 5′-CCCTAGTGCTGGTATACTATA-3′ and shDLST2: 5′-TGTCTCATAGCCTCGAATATC-3′) were cloned into the pLKO. 1-puro vector, and lentivirus production was conducted as previously described.39 The shMYC/pLKO.1 clone was from the TRC-Hs1.0 (Human) library (TRCN0000010390, Open Biosystems, Lafayette, CO, USA). After lentivirus transduction, cells were maintained in medium containing puromycin (0.5–1.0 µg/ml). The DLST or MYC knockdown was verified by western blotting.
The T-ALL cell lines (DSMZ, authenticated and tested for mycoplasma free) were transduced with shRNA hairpins and shCONTROL, shDLST1 or shDLST2. At 36 h posttransduction, 2 mm of monomethyl succinate (Me-succinate) (Sigma) was supplemented in cell culture media and subsequently added every 2 days thereafter. The growth rates of cells were determined at 4 and 9 days posttransduction.
Apoptosis was assessed after staining cells with Annexin V (BD Biosciences, San Jose, CA, USA) and propidium iodide (PI; 50 µg/ml; Sigma) for 20 min. For cell cycle analysis, cells were fixed in ethanol and stained in PI solution (phosphate-buffered saline, 50 µg/ml PI and 100 µg/ml RNase A) for 20 min at 4 °C. Apoptosis and cell cycle analyses were performed by flow cytometry (FACSCalibur, BD Biosciences) and subsequently analyzed with either the FlowJo or the ModFit software (Verity Software House, Topsham, ME, USA).
Polar metabolomic profiling
Polar metabolites were isolated from human T-ALL cells and analyzed by liquid chromatography–mass spectrometry as previously described.40 The resulting data were subsequently analyzed with the MetaboAnalyst software (https://www.metaboanalyst.ca).
Statistical analysis
Kaplan–Meier analysis and the log-rank test were used to compare times to tumor onset between Myc;dlst+/+ and Myc;dlst+/− fish. Student’s t-test was used to analyze differences in cell cycle, cell size, the number of PH3+ cells, DLST protein levels, Annexin-V-stained cells and cell viability among groups of cells with different genotypes. P-values ≤ 0.05 were considered statistically significant. P-values were not adjusted for multiple comparisons and the variation was similar between the groups that were compared.
RESULTS
Heterozygous loss of dlst partially suppresses Myc-induced T-ALL in zebrafish
To identify genes that contribute to MYC-mediated tumorigenesis, we performed a genetic screen using the zebrafish model of Myc-induced T-ALL that has been previously characterized and demonstrated its relevance to human disease (Figure 1a).17,18,41 Heterozygous inactivation of ornithine decarboxylase (ODC) impairs Myc-induced lymphomagenesis and skin tumorigenesis in mice.42,43 Consistent with the murine studies, we found that heterozygous loss of odc partially suppressed the onset of Myc-induced T-ALL in zebrafish (Table 1). Among the mutants tested, the hi1903C mutant showed the strongest tumor-suppressive effect. In contrast to tumor development in 100% of wild-type Myc progeny and 75% of progeny from the odc*/+ crossed to Myc fish, only 52% (40 of the 77) of the progeny resulting from the hi1903C mutant crossed to Myc fish developed tumors at 60 dpf (Table 1). The hi1903C mutant harbors a retroviral insertion in the first intronic region of a gene encoding dlst.34 When we crossed the dlst heterozygous fish to the Myc transgenic line and monitored their progeny for tumor onset over time, we found that tumor development was significantly suppressed in the dlst heterozygotes compared with their wild-type siblings (63% of Myc;dlst+/− fish vs 100% of Myc;dlst+/+ fish at 63 dpf, P = 0.0003; n = 19 and 26, respectively; Figures 1b–f). Western blotting analysis confirmed that dlst allelic loss led to ~ 50% reduction of Dlst protein in MYC tumor cells (Figures 1g and h).
Figure 1.
Heterozygous loss of dlst partially suppresses Myc-induced T-ALL. (a) Schematic of the rag2:EGFP-mMyc and rag2:GFP constructs that drove the development of EGFP+ T-ALL in the zebrafish model of Myc-induced T-ALL. (b) Tumor induction rates indicate that Myc-induced T-ALL is delayed with heterozygous loss of the dlst allele (n = 19 for Myc; dlst+/− and n = 26 for Myc; dlst+/+; P = 0.0003). (c) EGFP-labeled T-cells in a control rag2:EGFP fish (71 days old) define the normal thymus boundary. (d) Leukemia in a 63-dpf-old Myc;dlst+/+ fish. (e, f) Myc;dlst+/− fish (97 and 100 days old) with no tumor development (e) or early-stage lymphoma (f) based on thymic EGFP expression. (g, h) Western blotting analysis confirmed ~ 50% reduction of Dlst protein levels in T-lymphoblasts from Myc;dlst+/− compared with their Myc;dlst+/+ siblings (mean ± s.d. Dlst-to-Actin ratio; 0.82 ± 0.24 vs 2.42 ± 0.49; P = 0.0069; n = 4 per group). Scale bar for panels (b–d) = 1 mm.
dlst heterozygous loss in Myc-overexpressing tumor cells decreases cell mass and slows cell cycle progression
To examine whether heterozygous loss of dlst affects lymphoblast morphology, we analyzed normal thymocytes and Myc-overexpressing T-ALL cells with or without dlst heterozygous loss: (i) EGFP;dlst+/+; (ii) EGFP;dlst+/−; (iii) Myc;dlst+/+, and (iv) Myc;dlst+/− (Figures 2a–d). Cytospins of FACS-sorted EGFP+ cells were prepared and stained with May-Grunwald/Giemsa. In the absence of Myc overexpression, heterozygous loss of dlst did not alter the size or morphology of the thymocytes (EGFP;dlst+/+ vs EGFP;dlst+/−; Figures 2a and b). However, when Myc was overexpressed, heterozygous loss of dlst resulted in decreased tumor cell size (Myc;dlst+/+ vs Myc;dlst+/−; Figures 2c and d). These findings were confirmed by forward scatter analysis of flow cytometric data from these cells (Figures 2e and f). Next we determined whether heterozygous loss of dlst affected cell cycle in normal thymocytes and Myc-overexpressing T-ALL cells. Heterozygous loss of dlst did not cause any cell cycle changes in non-malignant thymocytes that were collected from young (3-month-old) fish (EGFP;dlst+/+ vs EGFP;dlst+/−; Figure 2g). However, Myc;dlst+/− tumor cells showed a significant shift in their cell cycle profile. Compared with Myc;dlst +/+ tumor cells, Myc;dlst+/− cells had an increased cell fraction in G0/G1 and a decreased fraction in the S phase (Figure 2h). To determine the effect of dlst heterozygous loss on proliferative status of premalignant thymocytes, cryosectioned tissues from tumor-free Myc;dlst+/+ and Myc;dlst+/− fish were stained with anti-PH3 antibody. In the presence of Myc overexpression, heterozygous loss of dlst significantly decreased proliferation of the premalignant thymocytes (Myc;dlst+/+ vs Myc;dlst+/−; Figures 2i–k). Together, these data demonstrate that, in the context of Myc overexpression, heterozygous loss of dlst reduces cell size and decreases cell proliferation.
Figure 2.
Heterozygous loss of dlst results in decreased cell size and a delay in cell cycle progression of tumor cells with Myc overexpression. FACS-sorted EGFP+ cells stained with May-Grunwald–Giemsa: (a) EGFP;dlst+/+; (b) EGFP;dlst+/−; (c) Myc;dlst+/+; and (d) Myc;dlst+/− cells (n = 4 per group). Forward scatter plots of EGFP+ population of EGFP;dlst+/+ vs EGFP;dlst+/− (e) and Myc;dlst+/− vs Myc;dlst+/+ (f). Statistical comparison by Fisher’s exact test demonstrated that Myc;dlst+/− tumor cells were significantly smaller than Myc;dlst+/+ T-ALL cells: P = 0.0022; n = 3 per group. Cell cycle distribution of EGFP;dlst+/+ vs EGFP;dlst+/− thymocytes (g; n = 4 per group) and Myc;dlst+/− vs Myc;dlst+/+ tumor cells (h; n = 3 per group). Percentages of cells in G1/G0, S or G2/M stages of the cell cycle are shown. (i and j) The overlay of anti-PH3 and GFP images of premalignant thymocytes from Myc;dlst+/+ (i) vs Myc;dlst+/− (j) fish. (k) Quantification of PH3+ premalignant thymocytes (GFP+) from Myc;dlst+/+ vs Myc;dlst+/− fish (mean ± s.d. PH3+ cells per area: 6 ± 0.97 vs 0.33 ± 0.21; P = 0.02; n = 3 per group). Fish at ~ 3 months of age were used for analyses in panels (a–h) and fish at ~ 1 month of age were used for analyses in panels (i and j). Myc-overexpressing T-ALL cells in panels (c, d, f and h) were dissected outside the thymic regions of the fish. Scale bar for panels (a–d) = 10 µm and for panels (i and j) = 50 µm.
dlst heterozygous loss does not influence zebrafish development
To determine how extensively dlst heterozygous loss affects fish development, we examined both embryonic and adult zebrafish. By monitoring embryonic development, we found that dlst+/− fish lacked visible morphological abnormalities or hematopoietic defects (compare Supplementary Figures S1b and e with Supplementary Figures S1a and d and Supplementary Figure S1h with Supplementary Figure S1g). However, dlst−/− homozygous mutant embryos failed to increase in body size and exhibited hematopoietic defects by 14 dpf (Supplementary Figures S1c, f, i and j and data not shown), and were no longer viable by 21 dpf (data not shown). To determine whether fish show morphological defects later in development, we fixed and serially sectioned 4-month-old dlst heterozygous and wild-type siblings and stained them with hematoxylin and eosin. Histological analysis of sections from adult dlst+/+ and dlst+/− fish failed to reveal any gross abnormalities in multiple tissues and organs of heterozygous fish, including eye, brain, thymus, kidney, ovary, intestine and muscle, compared with findings in dlst+/+ fish (Figures 3a–j and data not shown). To further determine whether heterozygous inactivation of dlst affects hematopoiesis, we performed flow cytometric analysis of whole kidney marrow (equivalent to mammalian bone marrow) to assess the relative abundance of the major blood cell lineages, including erythrocytes, lymphocytes, monocytes/granulocytes and progenitors. We found that the major blood lineages were unaltered in dlst+/− compared with dlst+/+ fish (Figure 3k) and that neither kidney smears (Figures 3l and m) nor blood smears (data not shown) showed morphological changes. Together, these data indicate that normal cells can tolerate dlst allelic loss.
Figure 3.
dlst heterozygous loss does not affect zebrafish development or normal hematopoiesis. Histological sections of adult (4-month-old) zebrafish: comparison of wild-type (dlst+/+) (a–e) and dlst+/− (f–j) revealed no gross morphological changes in the thymus, kidney, intestine, ovary or muscle (n = 4 per group). Normal hematopoiesis is unaltered in adult zebrafish with dlst heterozygous loss (k–m). When analyzed by flow cytometry, the frequency distribution of blood cell populations (erythrocytes, lymphocytes, monocyte/granulocytes and progenitors) was unaltered by heterozygous dlst loss (k; erythroid: P = 0.4547; lymphoid: P = 0.728; precursor: P = 0.8077; granulocyte/monocyte: P = 0.3914; n = 6 per group) nor was the morphology of these blood cells: dlst+/+ (l) vs dlst+/− (m) from 5-month-old fish stained with May-Grunwald–Giemsa (n = 6 per group). Scale bar for panels (a–f) = 0.2 mm and for panels (l–m) = 10 µm.
DLST protein levels are elevated in both zebrafish and primary patient T-ALL cells
Given that Dlst protein levels correlate with MYC-driven tumor development in zebrafish (Figure 1), we asked whether dlst expression is elevated in MYC-driven T-ALL cells. Western blotting revealed significant upregulation of Dlst protein levels in zebrafish Myc-overexpressing T-ALL cells, compared with those in normal thymocytes (Figures 4a and b). As MYC is a master transcriptional regulator that binds to promoters across the genome,20 we determined whether it transcriptionally regulates DLST. Q-RT-PCR analyses for dlst and β-actin on normal thymocytes (EGFP;dlst+/+) and Myc-overexpressing T-ALL cells (Myc;dlst+/+) failed to show significant differences of dlst transcript levels in these two types of cells (Figure 4c). Nevertheless, in the same T-ALL cells we detected significantly elevated transcript levels of shmt2 (serine hydroxymethyltransferase 2) (Figure 4d), a known MYC transcriptional target.44 These findings indicate that MYC does not transcriptionally regulate DLST. To understand why Dlst protein levels are elevated in Myc-overexpressing T-ALL cells, we next examined the half-life of Dlst in control thymocytes vs T-ALL cells in zebrafish. Pulse-chase analysis revealed that Dlst is more stable in Myc-overexpressing T-ALL cells, compared with that in control thymocytes (Figure 4e).
Figure 4.
Dlst protein levels are elevated in zebrafish T-ALL cells posttranscriptionally. (a) Western blotting showing protein levels of EGFP-MYC, Dlst and Actin in zebrafish T-ALL samples (Myc;dlst+/+ and Myc;dlst+/−), compared with the control thymus samples (EGFP;dlst+/+) of zebrafish. (b) Dlst vs Actin protein ratios demonstrating that Dlst levels are significantly higher in T-ALL cells from both Myc;dlst+/+ and Myc;dlst+/− fish, compared with control thymocytes (mean ± s.d. of Dlst-to-Actin ratio: 6.47 ± 1.9 vs 0.26 ± 0.09; P = 0.017 for EGFP;dlst+/+ vs Myc;dlst+/+; and 1.88 ± 0.5 vs 0.26 ± 0.09; P = 0.019 for EGFP;dlst+/+ vs Myc;dlst+/−; n = 4 per group). (c, d) Q-RT-PCR analysis revealing significantly elevated transcript levels of shmt2 (d; P = 0.0084; n = 3 per group) but not dlst (c; P = 0.225; n = 4 per group) in tumor cells (Myc;dlst+/+), compared with those in normal thymus (EGFP;dlst+/+). (e) Pulse-chase analysis of Dlst half-life showing that Dlst is more stable in tumor cells (Myc;dlst+/+), compared with that in normal thymus (dlst+/+) (>10 vs ≤ 8 h; n = 4 per group). CHX: Cycloheximide. Dlst protein amounts (relative to Actin) are shown in the bottom of panel (e). Because of the degradation of Actin and other proteins, the relative amounts of Dlst in Myc;dlst+/+ T-ALL cells increase over time.
To determine DLST protein levels in primary T-ALL patient samples, we performed western blotting analysis of normal thymus and primary T-ALL patient samples and found that DLST protein levels were also elevated in the T-ALL patient samples analyzed, compared with normal thymus controls (Figures 5a and b). Besides NOTCH1 mutations and aberrant MYC expression, other genetic alterations (for example, phosphatase and tensin homolog (PTEN) loss) can also contribute to human T-ALL pathogenesis.45,46 Hence, we examined the protein levels of cleaved NOTCH1 (ICN1), MYC, PTEN and DLST in human primary T-ALL samples and three T-ALL cell lines (JURKAT, MOLT3 and PEER), and found that DLST protein levels do not correlate with MYC protein levels in human T-ALL cells (Figures 5c and d). To determine whether MYC regulates DLST in human T-ALL cells, we inactivated MYC using an shRNA hairpin that efficiently depletes MYC in human JURKAT cells that overexpress BCL-2 and thus are apoptosis-resistant. Importantly, MYC inactivation led to downregulation of DLST protein levels, as well as SHMT2 (a known MYC target) at 5 days postinfection (Figure 5e). Taken together, these data indicate that T-ALL cells increase DLST protein levels, compared with normal T cells, and MYC may regulate DLST posttranscriptionally.
Figure 5.
DLST protein expression in primary patient T-ALL cells and human T-ALL cell lines. (a) Western blotting showing protein levels of DLST and ACTIN in primary pediatric T-ALL patient samples, compared with normal thymus samples. (b) DLST vs ACTIN protein ratios demonstrating that DLST levels are significantly higher in T-ALL patient samples compared with control thymocytes (P = 0.05; mean ± s.d. of DLST to ACTIN ratio: 0.7 ± 0.2 vs 0.08 ± 0.039; n = 3 for thymus and n = 4 for T-ALL patient samples). (c) Western blotting analysis of protein levels of ICN1 (the intracellular domain of NOTCH1), MYC, PTEN, DLST and ACTIN in primary pediatric T-ALL patient samples. (d) Western blotting analysis of protein levels of ICN1 (the intracellular domain of NOTCH1), MYC, PTEN, DLST and ACTIN in three human T-ALL cell lines (JURKAT, MOLT3 and PEER). (e) Western blotting analysis of MYC, DLST, SHMT2 and ACTIN protein levels in BCL-2-overexpressing JURKAT cells that are transduced with either control shLuciferase or shMYC. Normalized protein amounts for DLST and SHMT2 are shown as numbers below the western blotting panels.
DLST genetic inactivation decreases cell growth and leads to apoptosis in human T-ALL cell lines
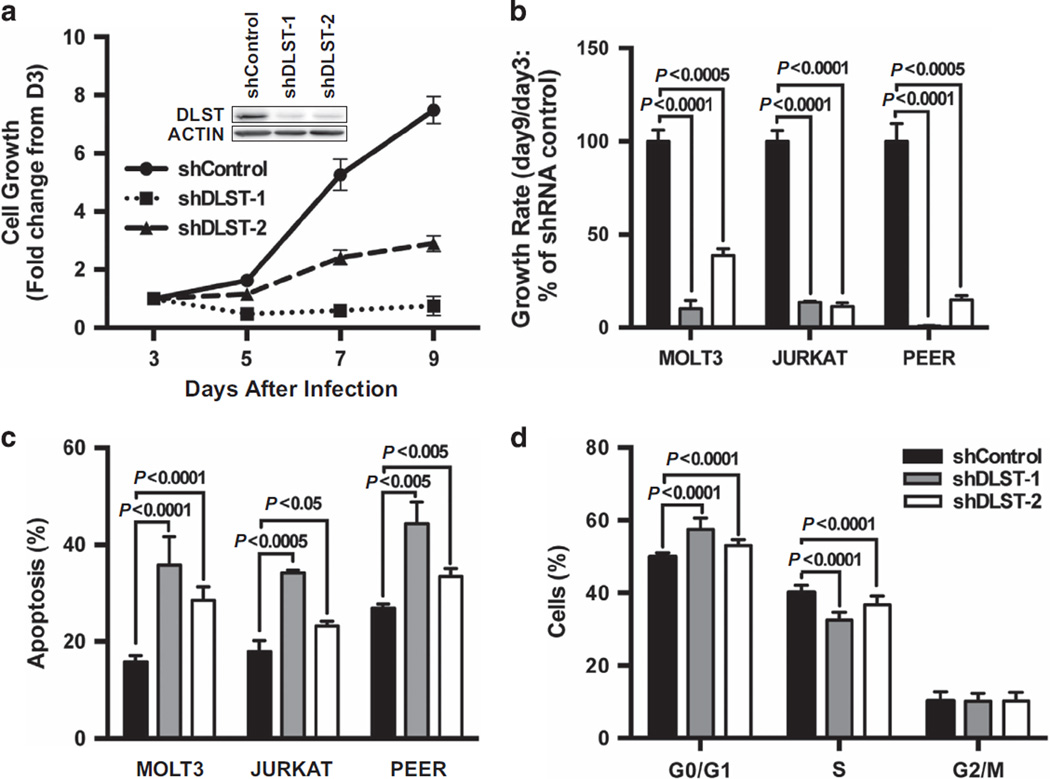
To provide evidence for DLST’s role in the proliferation and survival of human T-ALL cells, we inactivated DLST using two independent shRNA hairpins that efficiently target human DLST (see western blotting insert in Figure 6a and Supplementary Figure S2). Compared with the control hairpin, both shDLST1 and shDLST2 hairpins significantly inhibited the growth of multiple T-ALL cell lines, including: MOLT3, JURKAT, and PEER, all of which express DLST and MYC but with different PTEN status (Figures 5d and 6a and b). The severity of cell growth defect correlated with the degree of DLST knockdown in MOLT3 cells (Supplementary Figure S3). To determine the cellular mechanisms underlying the decrease in cell growth upon DLST knockdown, we stained cells with Annexin-V and found that an increased number of cells were undergoing apoptosis in all three T-ALL cell lines (Figure 6c). At 4 days postinfection, DLST knockdown in MOLT3 cells induced statistically significant changes in the cell cycle profile (Figure 6d). These data indicate that human T-ALL cells depend on DLST for proliferation and survival.
Figure 6.
DLST knockdown by RNAi in human T-ALL cells leads to slowed cell growth and apoptosis. (a, b) The growth kinetics of human T-ALL cells were analyzed after transduction with either a control shRNA or two shRNA hairpins targeting DLST. (a) Relative growth kinetics of the MOLT3 cell line. The insert in panel (a) revealed that two independent shDLST hairpins efficiently target human DLST. (b) Relative growth rate of human MOLT3, JURKAT and PEER T-ALL cells. (c) Apoptosis was induced by shRNA knockdown of DLST in MOLT3, JURKAT and PEER T-ALL cells. Annexin-V staining was performed on cells isolated 8 days after viral infection. (d) Genetic knockdown of DLST induces statistically significant cell cycle changes in MOLT3 cells. At 4 days postinfection, cells were fixed and then stained with PI and analyzed by flow cytometry. Representative data from over four independent experiment repeats are shown.
DLST inactivation blocks α-KG conversion to succinyl-CoA in human T-ALL cells
To investigate mechanisms underlying DLST-mediated cell proliferation and survival, we employed polar metabolomic profiling to assess the metabolic consequences upon DLST inactivation. Given that DLST is part of the KGDHC enzymatic complex that converts α-KG to succinyl-CoA in the TCA cycle (Figure 7a), we examined the cellular levels of these metabolites upon DLST knockdown to determine whether this conversion process was impaired. Utilizing liquid chromatography–mass spectrometry to measure polar metabolites, we found that DLST depletion in human MOLT3 T-ALL cells results in a significant loss of succinyl-CoA and a marked increase of α-KG (Figure 7b), indicating that DLST inhibition efficiently disrupts the activity of KGDHC and the TCA cycle. Additionally, Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of our metabolomics data revealed that multiple pathways involved in lipid and amino-acid metabolism are significantly downregulated by DLST inactivation (Supplementary Table S1), suggesting that the TCA cycle’s role in biosynthesis is disrupted. Similar metabolic changes were also observed in PEER T-ALL cells upon DLST knockdown (Supplementary Table S2). Thus our data show that DLST inactivation disrupts the TCA cycle and macromolecule biosynthesis in human T-ALL cells.
Figure 7.
DLST knockdown by RNAi results in disruption of the TCA cycle that can be rescued by addition of succinate. (a) Schematic of the TCA cycle. (b) DLST inhibition leads to a disruption of the TCA cycle in MOLT3 cells, resulting in an accumulation of α-KG and loss of succinyl-CoA. (c) Succinate rescues MOLT3 cells from slowed cellular growth induced by DLST knockdown. At 36 h postinfection, MOLT3 cells transduced with shCONTROL or shDLST hairpin were supplemented with 2 mm of Me-succinate in their culture media.
Succinate rescues DLST genetic inactivation in human T-ALL cells
To determine whether disruption of the TCA cycle contributes to the growth defect of T-ALL cells, we next examined the effect of restoring the TCA cycle through addition of cycle intermediates on the cell growth phenotype associated with DLST inactivation. Given that DLST knockdown disrupts the conversion of α-KG to succinyl-CoA (Figures 7a and b), we selected succinate as the immediate downstream intermediate in the TCA cycle. Me-succinate has been previously demonstrated capable of entering the TCA cycle to functionally substitute succinate.47 Thus we supplemented this membrane-permeable form of succinate, Me-succinate (2 mm), into the culture media in MOLT3, JURKAT and PEER cells transduced with shCONTROL or shDLST hairpin. The addition of Me-succinate was sufficient to rescue the defect in cell growth resulting from DLST knockdown in MOLT3 cells (Figure 7c), as well as in JURKAT and PEER cells (Supplementary Figure S4). These results demonstrate that α-KG conversion to succinyl-CoA mediated by DLST/KGDHC in the TCA cycle is an important metabolic step critical for the survival and proliferation of human T-ALL cells.
DISCUSSION
Metabolic reprogramming is a hallmark of cancer, allowing tumor cells to meet the increased nutrient and energy demands imposed by rapid and uncontrolled proliferation.48,49 The TCA cycle has a pivotal role in cellular metabolism, where it fulfills the bioenergetic, biosynthetic and redox balance requirements of cells. DLST is a TCA cycle transferase functioning in the KGDHC that catalyzes the conversion of α-KG to succinyl-CoA for energy production and macromolecule synthesis.32 Our studies implicate DLST as an important mediator of MYC-mediated leukemogenesis. Interestingly, we observed elevated DLST protein levels in zebrafish Myc-overexpressing T-ALL cells as well as in primary T-ALL patient samples, compared with their respective thymus controls. This suggests that there is a selective pressure imposed on T-lymphoblasts to reprogram their cellular metabolism through overexpression of DLST. Although MYC may regulate DLST posttranscriptionally, we observed elevated DLST protein levels in human T-ALL cells that lack MYC overexpression, suggesting that genes other than MYC can also regulate DLST. Importantly, merely 50% reduction of Dlst is sufficient to significantly delay the onset of Myc-induced T-ALL in zebrafish. This delay in tumor onset is accompanied with decreased proliferation of Myc;dlst+/− T-lymphoblasts, compared with Myc;dlst+/+ tumor cells. Additionally, partial DLST inhibition mediated by shRNA decreased cell viability and promoted apoptosis in human T-ALL cell lines. These data demonstrate the metabolic dependence of T-lymphoblasts on the TCA cycle for proliferation and survival. Interestingly, despite the significant delay in tumor onset, most Myc;dlst+/− zebrafish eventually developed T-ALL, whereas acute DLST inactivation in human T-ALL cells induced apoptosis. These data suggest that alternative pathways may have been activated in zebrafish T-lymphoblasts over time to compensate for dlst loss.
Aberrant MYC activity enhances glutamine anaplerosis, resulting in increased levels of glutamine-derived TCA cycle intermediates in both in vitro and in vivo models.28,29 Importantly, the conversion of α-KG to succinyl-CoA governed by DLST/KGDHC is not only important for oxidative phosphorylation but also critical for the entry of glutamine into the TCA cycle in the form of α-KG.50 Indeed, T-ALL cells utilize glutaminolysis for cell growth mediated by NOTCH1.31 Hence, the disrupted TCA cycle due to DLST loss could inhibit glutamine anaplerosis, which is predicted to reduce the availability of TCA cycle intermediates required to produce the lipids, amino acids and nucleotides needed for cell growth and proliferation. Our in vitro studies in human T-ALL cell lines demonstrate that DLST inhibition disrupts the TCA cycle, as evidenced by the accumulation of α-KG and reduction of succinyl-CoA. This TCA cycle disruption subsequently led to defects in macromolecule anabolism. Consistent with the biochemical changes detected in these human T-ALL cells, upon DLST genetic inactivation, we observed smaller cell size and growth defects of lymphoblasts that were associated with reduced cell cycle progression in zebrafish and increased apoptosis in human T-ALL cells. Other than glutamine, the TCA cycle also utilizes glucose and fatty acids; hence, it is likely that DLST loss may also affect the catabolism of these nutrients. Notably, when the TCA cycle function is restored by the addition of a downstream cycle intermediate, succinate, DLST inhibition no longer has effects on cell viability in MOLT3 and JURKAT T-ALL cells and has less severe effects on cell viability in PEER cells. These data indicate that the biochemical conversion of α-KG to succinyl-CoA governed by DLST/KGDHC is critical for T-ALL proliferation and survival and demonstrate that the TCA cycle is important for MYC-mediated tumorigenesis.
In our zebrafish T-ALL model, heterozygous inactivation of Dlst delays Myc-induced T-ALL onset. This delayed tumor onset is associated with slower growth and proliferation of Myc;dlst+/− tumor cells. The allelic loss of Dlst is predicted to reduce the function of DLST/KGDHC in both normal and T-ALL cells. Interestingly, a smaller cell size and delayed cell cycle progression is only observed in Myc;dlst+/− cells, compared with Myc;dlst+/+ tumor cells, but not in control thymocytes with or without dlst allelic loss. Additionally, approximately 50% reduction of DLST protein levels is sufficient to significantly decrease viability and induce apoptosis in both human MOLT3 and PEER T-ALL cells regardless of their PTEN status. These results implicate that MYC-dependent leukemic cells are more sensitive to the TCA cycle disruption than cells lacking enforced MYC overexpression (for example, normal thymocytes and hematopoietic precursors). Thus these results provide rationale to test the strategy of targeting DLST/KGDHC to treat T-ALL.
Taken together, our in vitro and in vivo data demonstrate that MYC-driven tumor cells rely on DLST/KGDHC and the TCA cycle for cell growth and survival. As multiple types of tumors, including B-cell lymphoma and liver cancer cells, also utilize the TCA cycle,28,29 it will be important to determine whether DLST inhibition also impairs the proliferation and survival of cancer cells other than T-ALL cells. Unfortunately, efficacious and selective DLST inhibitors are currently unavailable. Further studies are needed to develop and test anti-DLST/KGDHC agents to determine whether targeting DLST/KGDHC represents a productive approach to treat T-ALL and perhaps other tumors as well.
Supplementary Material
Acknowledgments
We thank John Gilbert and Joseph Hirsch for editorial assistance; Nicholas Nagykery and Lu Zhang for technical assistance; Bethany M. Moore, Trevor Grant, Dr Nicolay Brandon, Dr Adam Lerner, Dr Enxuan Jing, Dr Arthur JL Cooper, Dr Gromoslaw Smolen and Dr Keith Tornheim for helpful discussions and critical review of the manuscript; and Julia Etchin for reagents. We also thank Gregory Molind, John Lyons and Derek Walsh for zebrafish husbandry. HF was supported by a K99CA134743/R00CA134743 award (National Institute of Health), a Karin Grunebaum faculty fellowship from the Karin Grunebaum Cancer Foundation, a Ralph Edwards Career Development Professorship from Boston University, a St Baldrick Scholar Award from the St Baldrick's Foundation, an Institutional grant (IRG –72-001-36-IRG) from the American Cancer Society and a Young Investigator grant from the Leukemia Research Foundation. ATL receives funding support from a 1R01 CA176746 grant (National Institute of Health). IH and DL acknowledge support through a NHLB1 T32 HL007501 training grant. HLP receives support from a NSFC 81200368 grant. The content of this research is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
NMA and HF designed and performed experiments and wrote the manuscript; DL, FJL, HLP, MA, EG, TC, DJH, IH, YS and ML performed experiments; MRS, TS, AA, TTD and AS provided intellectual input to the project and experiments; JER and MAK provided patient samples from murine xenografts. DSN checked statistical analysis and provided intellectual input; ATL and HF supervised the project.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 2.Ko RH, Ji L, Barnette P, Bostrom B, Hutchinson R, Raetz E, et al. Outcome of patients treated for relapsed or refractory acute lymphoblastic leukemia: a Therapeutic Advances in Childhood Leukemia Consortium study. J Clin Oncol. 2010;28:648–654. doi: 10.1200/JCO.2009.22.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marks DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993) Blood. 2009;114:5136–5145. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 5.Koch U, Radtke F. Notch in T-ALL: new players in a complex disease. Trends Immunol. 2011;32:434–442. doi: 10.1016/j.it.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Palomero T, Ferrando A. Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas. Clin Cancer Res. 2008;14:5314–5317. doi: 10.1158/1078-0432.CCR-07-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzoneva G, Ferrando AA. Recent advances on NOTCH signaling in T-ALL. Curr Top Microbiol Immunol. 2012;360:163–182. doi: 10.1007/82_2012_232. [DOI] [PubMed] [Google Scholar]
- 9.Weng AP, Ferrando AA, Lee W, JPt Morris, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 10.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart M, Cameron E, Campbell M, McFarlane R, Toth S, Lang K, et al. Conditional expression and oncogenicity of c-myc linked to a CD2 gene dominant control region. Int J Cancer. 1993;53:1023–1030. doi: 10.1002/ijc.2910530628. [DOI] [PubMed] [Google Scholar]
- 12.Leder A, Pattengale PK, Kuo A, Stewart TA, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 13.Morse HC, 3rd, Hartley JW, Fredrickson TN, Yetter RA, Majumdar C, Cleveland JL, et al. Recombinant murine retroviruses containing avian v-myc induce a wide spectrum of neoplasms in newborn mice. Proc Natl Acad Sci USA. 1986;83:6868–6872. doi: 10.1073/pnas.83.18.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez A, Grebliunaite R, Feng H, Kozakewich E, Zhu S, Guo F, et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med. 2011;208:1595–1603. doi: 10.1084/jem.20101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood. 2006;108:653–661. doi: 10.1182/blood-2006-01-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roderick JE, Tesell J, Shultz LD, Brehm MA, Greiner DL, Harris MH, et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood. 2014;123:1040–1050. doi: 10.1182/blood-2013-08-522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 18.Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoepfler PS. Myc goes global: new tricks for an old oncogene. Cancer Res. 2007;67:5061–5063. doi: 10.1158/0008-5472.CAN-07-0426. [DOI] [PubMed] [Google Scholar]
- 20.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 24.Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O'Donnell KA, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy TA, Dang CV, Young JD. Isotopically nonstationary 13C flux analysis of Myc-induced metabolic reprogramming in B-cells. Metab Eng. 2013;15:206–217. doi: 10.1016/j.ymben.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herranz D, Ambesi-Impiombato A, Sudderth J, Sanchez-Martin M, Belver L, Tosello V, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21:1182–1189. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheu KF, Blass JP. The alpha-ketoglutarate dehydrogenase complex. Ann NY Acad Sci. 1999;893:61–78. doi: 10.1111/j.1749-6632.1999.tb07818.x. [DOI] [PubMed] [Google Scholar]
- 33.Westerfield M. The Zebra fish Book: A Guide for the Laboratory Use of Zebra fish (Brachydanio rerio) Eugene, OR, USA: University of Oregon Press; 1994. [Google Scholar]
- 34.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, et al. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br J Haematol. 2007;138:169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- 36.Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 37.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 38.Macdonald R. Zebrafish immunohistochemistry. Methods Mol Biol. 1999;127:77–88. doi: 10.1385/1-59259-678-9:77. [DOI] [PubMed] [Google Scholar]
- 39.Sanda T, Tyner JW, Gutierrez A, Ngo VN, Glover J, Chang BH, et al. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013;3:564–577. doi: 10.1158/2159-8290.CD-12-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackburn JS, Liu S, Raiser DM, Martinez SA, Feng H, Meeker ND, et al. Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia. 2012;26:2069–2078. doi: 10.1038/leu.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Cleveland JL, O'Brien TG. Haploinsufficiency for odc modifies mouse skin tumor susceptibility. Cancer Res. 2005;65:1146–1149. doi: 10.1158/0008-5472.CAN-04-3244. [DOI] [PubMed] [Google Scholar]
- 44.Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heart E, Yaney GC, Corkey RF, Schultz V, Luc E, Liu L, et al. Ca2+, NAD(P)H and membrane potential changes in pancreatic beta-cells by methyl succinate: comparison with glucose. Biochem J. 2007;403:197–205. doi: 10.1042/BJ20061209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward PS, Thompson CB. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol. 2012;4:a006783. doi: 10.1101/cshperspect.a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.