Abstract
The fungal pathogen Sclerotinia sclerotiorum causes stem rot of oilseed rape (Brassica napus) worldwide. In preparation for genome-wide association mapping (GWAM) of sclerotinia resistance in B. napus, 152 accessions from diverse geographical regions were screened with a single Canadian isolate, #321. Plants were inoculated by attaching mycelium plugs to the main stem at full flower. Lesion lengths measured 7, 14 and 21 days after inoculation were used to calculate the area under the disease progress curve (AUDPC). Depth of penetration was noted and used to calculate percent soft and collapsed lesions (% s + c). The two disease traits were highly correlated (r = 0.93). Partially resistant accessions (AUDPC <7 and % s + c <2) were identified primarily from South Korea and Japan with a few from Pakistan, China and Europe. Genotyping of accessions with 84 simple sequence repeat markers provided 690 polymorphic loci for GWAM. The general linear model in TASSEL best fitted the data when adjusted for population structure (STRUCTURE), GLM + Q. After correction for positive false discovery rate, 34 loci were significantly associated with both disease traits of which 21 alleles contributed to resistance, while the remaining enhanced susceptibility. The phenotypic variation explained by the loci ranged from 6 to 25 %. Five loci mapped to published quantitative trait loci conferring sclerotinia resistance in Chinese lines.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-016-0496-5) contains supplementary material, which is available to authorized users.
Keywords: Genome-wide association mapping, Brassica napus, Oilseed rape, Canola, Simple sequence repeats, SSR, Stem rot, Sclerotinia sclerotiorum
Introduction
Brassica napus L. and other Brassica species are grown worldwide. In temperate regions, B. napus varieties low in glucosinolates and erucic acid (canola) provide vegetable oil for human consumption, while the protein-rich meal is added to animal feed rations. In some parts of the world, B. napus is traditionally grown as a leafy vegetable, whereas biofuel production is a relative new use for this crop in other regions (Abbadi and Leckband 2011).
The plant pathogenic fungus Sclerotinia sclerotiorum (Lib.) de Bary has a wide host range including numerous dicot crop species, such as oilseed rape where the disease causes symptoms known as stem rot or white mold. The pathogen forms sclerotia (resting bodies of mycelium) that can survive in the soil for several years. The sclerotia germinate and produce apothecia from which ascospores are dispersed within the crop. Spores infect plants by first colonizing senescent petals adhering to stems and leaves followed by direct penetration of the plant’s epidermis. Yield loss results predominantly from colonization of the main stem which blocks transport of water and nutrients to the developing seeds. It has been estimated that for each 1 % infected plants there is a 0.5–0.7 % yield reduction (Morral et al. 1984; Del Río et al. 2007; Kirkegaard et al. 2006; Koch et al. 2007). Fungicides have to be applied prophylactically which makes it inherently difficult to obtain consistent economical returns. Consequently, genetic resistance for controlling S. sclerotiorum is desirable both economically and environmentally.
Plant breeders in China were the first to develop partially resistant varieties, such as Zhongyou 821 and Zhongshuang 9 (Li et al. 1999; Wang et al. 2004). Quantitative resistance loci (QTL) were subsequently mapped in several Chinese cultivars and breeding lines (Zhao and Meng 2003; Zhao et al. 2006; Yin et al. 2010; Wu et al. 2013), one European cultivar (Wei et al. 2014) and a wild Brassica species B. incana (Mei et al. 2013) all demonstrating that sclerotinia resistance is a complex trait. However, the level of resistance was relatively low and prompted researchers to screen more B. napus varieties, breeding lines and land races (Zhao et al. 2004; Bradley et al. 2006), other brassicas (Li et al. 2007; Mei et al. 2011) and crucifer species (Uloth et al. 2013). Introgression of resistance from distantly related species into oilseed rape has been undertaken (Garg et al. 2010; Navabi et al. 2010; Ding et al. 2013), but is hampered by linkage drag of undesirable traits, varying levels of sterility and occasionally insurmountable crossing barriers.
To date, mapping of sclerotinia resistance loci has been carried out in populations derived from a cross between susceptible and resistant parents. However, marker–trait associations can also be identified in a collection of unrelated lines using the genome-wide association mapping (GWAM) approach. This method exploits the non-random association between phenotypic traits and molecular markers in genetically diverse germplasm in which random recombination events have accumulated over millennia (Flint-Garcia et al. 2003; Rafalski 2010). In B. napus, GWAM has so far been used to map glucosinolate content (Hasan et al. 2008), oil content (Zou et al. 2010), phenolic compounds (Rezaeizad et al. 2011) and several seed traits (Honsdorf et al. 2010). Jestin et al. (2011) used association mapping to identify loci conferring quantitative resistance to the fungal pathogen Leptosphaeria maculans causing blackleg in oilseed rape. Recently, GWAM was employed to map resistance to sclerotinia in B. napus, sunflower and soybean (Wei et al. 2015; Fusari et al. 2012; Bastien et al. 2014). Wei et al. (2015) were able to identify three loci associated with resistance in primarily Chinese B. napus lines using a disease screening method that involved wounding of the plant tissue. We set out to screen a more diverse collection of B. napus germplasm from various parts of the world using an inoculation method that resembles natural infection. The objectives were to identify new sources of sclerotinia resistance and to utilize the GWAM approach at various stringencies to demarcate a larger set of molecular markers contributing to resistance.
Materials and methods
Plant material
Initially, more than 400 B. napus accessions from diverse geographical regions of the world were screened for resistance to S. sclerotiorum. A subset of 152 accessions was selected consisting of lines with a consistent sclerotinia phenotype among single plants including the most resistant and susceptible lines. These lines represented three important productions areas of the world, Canada, China and Europe, as well as South Korea and Japan which are rarely examined in other studies. A large number of lines had been received by scientists at the Saskatoon Research Station (SRS prefix) over the years (Table S1). Seeds of other lines were obtained from three gene banks, the Plant Gene Resources of Canada (CN), the Nordic Gene Bank (NGB) and the Gene Bank in Prague, Czechoslovakia (15O0). The countries with number of accessions in brackets were as follows: Argentina (1), Australia (1), Canada (11), China (14), eleven countries in Europe (31), Japan (19), South Korea (58), Pakistan (15) and two of unknown origin (Table S1).
Sclerotinia phenotyping
A single S. sclerotiorum isolate #321 was used for all inoculations. This isolate was collected in 1992 and represents a common clone identified in oilseed rape fields in the province of Alberta, Canada (Kohli et al. 1995). The aggressiveness of the isolate was maintained by inoculating the susceptible Canadian variety, Westar, every 5–6 months. Re-isolated cultures were stored either as mycelium at −80 °C in cryo-freezer solution (5 % skim milk in 20 % glycerol) or as sclerotia at 4 °C until needed for inoculation.
Seed of the 152 B.napus accessions was sown in a soilless mixture supplemented with slow-release fertilizer with one seed per 12.5-cm pot and five pots per line. The pots were placed randomly on a greenhouse bench in a 16-/8-h day/night cycle with light provided by high-pressure sodium lamps. The temperature was maintained between 21 and 24 °C during the day and 17–18 °C during the night. Pots were drip-irrigated and after 3 weeks fertilized weekly with NPK 20–20–20 (3 g/L). When plants were at full flower, the main stem was inoculated with S. sclerotinia as originally described by Buchwaldt et al. (2005) as follows. Isolate #321 from a stock culture was plated in 9-cm Petri dishes on glucose media (glucose 20 g, malic acid 3 g, NH4NO3 2 g, KH2PO4 1 g, NaOH 1 g, MgSO47H2O 0.1 g, agar 20 g, distilled water 1000 mL) modified from Cruickshank (1983). Plates were incubated at 22/18 °C in a 16-/8-h day/night cycle with light provided by white fluorescent bulbs. After 4–6 days, and before hyphae reached the edge of the Petri dish, mycelium plugs (5–7 mm diameter) were cut from the actively growing margin. Each plug was placed on a 3 × 6 cm piece of Parafilm and attached to the main stem with mycelium facing the epidermis. Each stem was inoculated at two internodes with two un-inoculated internodes inbetween to avoid the developing lesions growing together. The length of each stem lesion was measured 7, 14, and 21 days after inoculation (dai). On these dates, the depth of penetration into the stem was assessed by slightly pressing each lesion and recording it as either firm, soft (stem not girdled) or collapsed (stem completely girdled). Percent soft plus collapsed lesions (% s + c) was calculated for each line. The area under the disease progress curve (AUDPC) was calculated using the lesion lengths from the three dates as follows where Li is lesion length on day i, t the number of days between observations and n the number of observations. The 152 B. napus accessions were screened in batches of 30 lines with the Canadian variety Westar as a susceptible control included in every test. Five plants of each accession were screened in one or two tests for a total of 10–20 stem lesions per line.
Genotyping with SSR markers
Seeds of 152 B. napus lines were grown as described above, and leaf tissue was collected from single 4–5-week-old plants and immediately frozen in liquid nitrogen. The tissues were freeze-dried in a Freezone 6 dryer (Labconco Corp, KS, USA) for 48 h and then ground in a Mixture Mill 300 (Retsch Inc, PA, USA). DNA was extracted using a CTAB method, quantified using a NanoVue Plus spectrophotometer (GE Healthcare, NJ, USA) as per the manufacturer’s instruction, diluted to 10 ng/μL and kept at −20 °C until needed for genotyping.
Around 700 primer pairs flanking simple repeat DNA sequences in B. napus were developed at Agriculture and Agri-Food Canada beforehand, and the intervening DNA sequence of each locus was known. Eighty-four of these simple sequence repeat (SSR) markers were selected based on their polymorphism in selected B. napus lines, even distribution across the genome and high reproducibility in previous tests. A list of forward and reverse primers for the 84 SSRs were published previously (Gyawali et al. 2013). The forward primer (5′ end) of each SSR marker was labeled with either a 6-FAM (6-carboxyfluorescein), HEX (hexachloro-6-carboxyfluorescein) or TET (tetrachloro-fluorescein) fluorescent tag. DNA was amplified by PCR in 384-well plates using a C1000 thermal cycler (BioRad Life Sciences, CA, USA). Reactions were carried out in 12 µL volume containing 0.25 mM dNTPs, 1× PCR buffer (containing 15 mM MgCl2), 0.5 μM forward and reverse primer, 0.25 units Taq DNA polymerase and 20 ng genomic DNA. PCR was initiated with a hot start at 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s and 68 °C for 1 min, with a final extension step of 68 °C for 5 min. Amplification products with different fluorescent tags were multiplexed and analyzed by capillary electrophoresis on a MegaBACE 1000 platform (GE Healthcare, NJ, USA). Data were analyzed using Fragment Profiler version 1.2 (GE Healthcare, NJ, USA). The SSR alleles were scored with GeneMapper v 3.7 (Applied Biosystems) according to their size using a 550 bp size standard included in all samples. Each peak was visually confirmed at least twice. SSR alleles were scored as dominant markers with 1 = present and 0 = absent, since B. napus is an allotetraploid species with two sub-genomes, AACC (2n = 38), and because homozygous and heterozygous loci could not be differentiated. A total of 1359 microsatellite alleles were identified, which was reduced to 690 after filtering for low allele frequencies of <5 %.
Analysis of population structure Q and ΔK
In preparation for GWAM, both population structure (Q) and kinship (K) in the B. napus collection were analyzed to reduce the possibility of false positive trait–marker associations. Population structure was analyzed using the STRUCTURE version 2.3 software program (Evanno et al. 2005). It uses a Bayesian clustering method to obtain a Q-matrix that describes the percentage of parentage for each genotype by placing individuals randomly into k-number of subpopulations. The set of 690 SSR scoring data was analyzed from k = 1 to k = 10 with five independent iterations for each value. The Markov Chain Monte Carlo model (MCMC) was used with admixture ancestry and independent allele frequency parameters as suggested by Pritchard and Wen (2004). To reduce the effect of the initial configuration, a burn-in period of 105 iterations was selected followed by another 105 iterations. STRUCTURE HARVESTER version 6.7 (Earl and vonHoldt 2012) was used to visualize the likelihood of each number of subpopulations, ΔK, which is a modification of the kinship value K, according to the method suggested by Evanno et al. (2005). The most likely number of subpopulations was selected, and the average of the five corresponding iterations resulted in a final Q value for each B. napus accession which was used to assign the lines to the different subpopulations (Table S1). The population structure was also examined by principal coordinate analysis (PCoA) in NtSYS version 2.2 using the DECENTER and EIGEN functions with the first two coordinates plotted in a graph. Each B. napus accession was labelled with a letter for country of origin and consecutive numbers (Table S1).
GWAM
The marker–trait relationship was analyzed with a general linear model (GLM) and the mixed linear model (MLM) in TASSEL version 3.0 (Bradbury et al. 2007). The mixed model approach was used as described in detail by Yu et al. (2006). Thus, two models were tested in order to obtain the best association between SSR alleles and the two disease traits, namely GLM + Q and MLM + Q + K, both followed by a correction for positive false discovery rate, pFDR. The matrices were combined as follows: Sclerotinia resistance = STRUCTURE [Q] + SSR marker + error, sclerotinia resistance = STRUCTURE [Q] + relative kinship [K] + SSR marker + error. The pFDR (q < 0.05) was applied in order to test the statistical significance of all detected markers (Storey 2002). The output data included percent phenotypic variation explained (R2) by each significant SSR allele and the additive effect, where negative values indicate the presence of the alleles that enhance resistance, and positive values indicate the presence of the alleles that enhance susceptibility.
Co-location of SSR in GWAM and published QTL from linkage mapping
Possible co-location of SSR loci in the present study and published quantitative trait loci (QTLs) conferring sclerotinia resistance was assessed using the physical chromosome location of markers. The full DNA sequence of all SSR markers in the present study was known. These sequences were queried against the B. napus genome sequence in the Genoscope database (http://www.genoscope.cns.fr/brassicanapus) (Chalhoub et al. 2014) using the default BLAT (BLAST-like alignment tool) algorithm thereby obtaining the chromosome location measured in nucleotides (Kent 2002). DNA sequence information was obtained for 220 markers underlying 65 QTLs in five publications, Zhao and Meng (2003), Zhao et al. (2006), Wu et al. (2013), Wei et al. (2014) and Mei et al. (2013). The markers consisted of either sequenced RFLP probes or forward and reverse primers of SSR and AFLP markers, ranging from 15 to 28 nucleotides in length. The RFLP sequences were queried directly against the B. napus genome using the BLAT algorithm as described above. Since the BLAT algorithm is unreliable with sequences shorter than 33 nucleotides, the SSR and AFLP primer sequences were queried against the B. napus genome sequence in the Brassica database (BRAD) (http://brassicadb.org/brad/index.php) using the BLASTn program. If no hits were recorded, the B. rapa and B. oleracea genomes were queried. The resulting intervening DNA sequences were subsequently used to query the B. napus genome in Genoscope as described above. In most cases, markers mapped to the expected chromosome based on the published information, and the most likely nucleotide location was selected based on the highest BLAT score (>100) and identity (>90 %), while hits to homologous chromosomes were discounted. Only in a few cases was the hit entirely to an alternate chromosome. SSR loci contributing to sclerotinia resistance in the present study were considered close to previously published markers if located within 500,000 nucleotides of the QTL peak, or in cases when the peak marker did not map to the expected chromosome, between the closest pair of flanking markers. The limit of 500 kb flanking the QTL peak was a conservative value selected based on the median of previously published estimates of linkage disequilibrium (LD) decay in B. napus (Qian et al. 2014).
Results
Partially resistant accessions
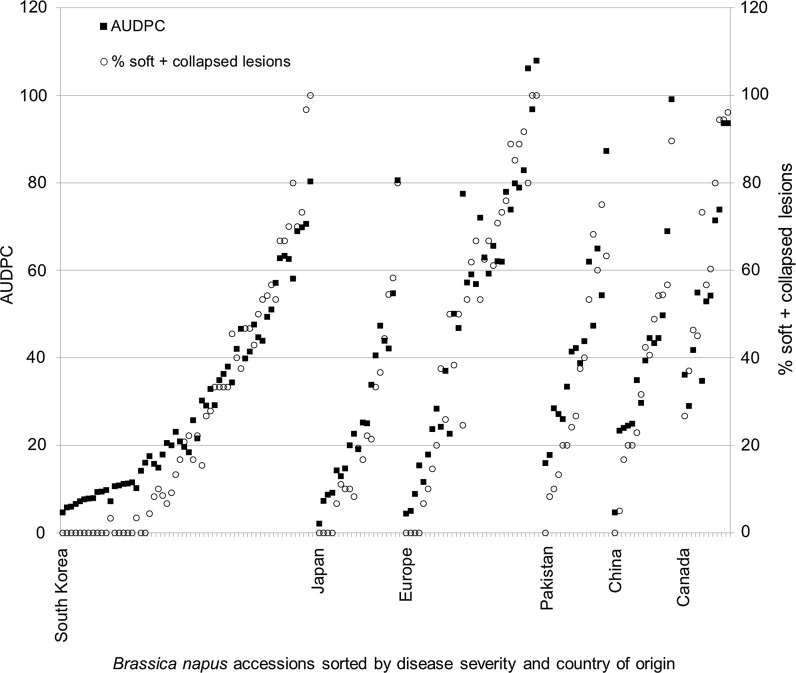
Disease symptoms caused by S. sclerotiorum on the 152 B. napus accessions ranged from 2.1 to 107.9 AUDPC and from 0 to 100 % soft + collapsed lesions (Fig. 1; Table S1). A high positive correlation was found between the two disease traits by regression analysis, r = 0.93 (P < 0.001). The following accessions had the highest level of partial resistance to S. sclerotiorum isolate #321: Buk Wuk 3, 12, 13 and 20, Kuju 18, 19, 22, 27, 35 and 40, Dong Hae 3 and 6 from South Korea; Norin 16, Tokiwa natane, Kinki 22 and 30 from Japan; Tzuyechein 32 from China; SRS1632 from Poland and Tanto from France (Table S1). Based on a cutoff of AUDPC <10 and % s + c <10, the frequency of partially resistant lines was South Korea 19 %, Japan 21 %, Europe 10 %, China 7 % and Pakistan 6 %, while none of the Canadian lines were partially resistant. The data set used for GWAM was therefore somewhat biased for resistant lines originating from the neighboring countries, South Korea and Japan (Fig. 1).
Fig. 1.
Results from screening a world collection of 152 B. napus accessions for resistance to the fungal pathogen S. sclerotiorum
Subpopulations and kinship
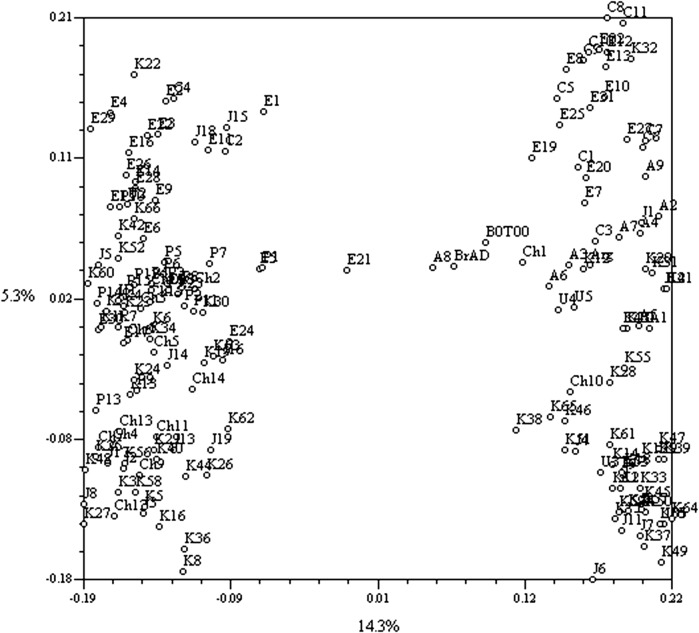
Analysis of population structure (Q) based on the clustering method in STRUCTURE, the ad hoc estimation of ΔK and PCoA all established the presence of two genetically distinct subpopulations. The result from PCoA is shown in Fig. 2. Based on a threshold of Q < 0.85 membership probability, 55 % (84) accessions belonged to subpopulation one (SP1), 38 % (58) to subpopulation two (SP2), while only 7 % (10) were placed in an admixed group (AD) (Table S1). Semi-winter-type accessions from China and Pakistan were unique to SP1, while all Canadian accessions were unique to SP2. Accessions from Europe, Japan and South Korea consisted of both semi-winter and spring types.
Fig. 2.
Principal coordinate plot of the first three variables based on polymorphism in simple sequence repeat (SSR) in a world collection of 152 B. napus accessions. The group on the left-hand side consists primarily of lines with semi-winter-type growth habit and the group on the right-hand side is mostly spring types
SSR markers associated with sclerotinia resistance
Rare SSR alleles, which were identified in <5 % of the 152 B. napus lines, were eliminated to reduce detection of false marker–trait associations in subsequent data analysis. The allele frequency of the remaining 690 SSRs ranged from 5.3 to 94.5 % (Table 1). The GLM + Q model resulted in identification of 43 SSR loci significantly associated with resistance to S. sclerotiorum of which 34 were shared by both disease traits, AUDPC and % s + c. The P values of the remaining nine markers were low for one or the other trait (Table 1). The phenotypic effect (R2) explained by each marker ranged from 6 to 25 % for both traits, and the additive effect ranged from 18 to 41 % (Table 1). Of the 34 common loci, the additive effect was negative for 21 loci and positive for 13 loci indicating that their presence enhanced resistance and susceptibility, respectively.
Table 1.
Simple sequence repeat (SSR) markers associated with resistance to S. sclerotiorum in B. napus identified by genome-wide association mapping in the GLM + Q model followed by a test of positive false discovery rate (q)
| SSR marker allele | Linkage group (cM)a | Physical positionb | Alleles | Area under the disease progress curvec | % Soft + collapsed lesions | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Absent | Present | P value | R 2 | Additive effectd | P valuee | R 2 | Additive effect | |||
| SR12355_213 | A1: 79.6 | A1: 22,709,214 | 49 | 82 | 2.8E−04*,b | 9.8 | −16.0 | |||
| SN3761_179 | A2: 33.3 | A2: 191,624 | 91 | 39 | 0.0016 | 4.9E−04* | 9.2 | 20.2 | ||
| SR0612_466 | A2: 69.1 | A2: 2,208,155 | 70 | 57 | 6.6E−05** | 12.1 | −17.1 | 4.0E−04* | 9.6 | −18.4 |
| SR12015_304 | na | A3: 9,607,501 | 41 | 90 | 5.4E−06** | 15.0 | 20.2 | 1.2E−04** | 10.9 | 20.7 |
| SN0569_188 | A3: 63.8 | A3: 13,046,378 | 102 | 28 | 7.1E−04* | 8.7 | 26.0 | |||
| SR0536_237 | A3: 93.5 | A3: 23,044,269 | 109 | 19 | 9.2E−04* | 8.4 | −20.1 | 1.2E−05* | 14.2 | −31.5 |
| SN13034_133 | A4: 20.7 | A4: 7,856,881 | 65 | 66 | 9.0E−06** | 14.3 | −18.4 | 1.8E−05** | 13.4 | −21.4 |
| SN4276_480 | A5: 45.2 | A5: 4,793,850 | 91 | 39 | 9.1E−05** | 11.4 | −18.0 | 8.3E−05** | 11.5 | −21.9 |
| SR12348I_180 | A5: 18.8 | A5: 8,755,826 | 108 | 22 | 2.1E−05** | 13.3 | −24.1 | 2.8E−04* | 9.9 | −25.0 |
| SN1914_326 | A6: 47.3 | na | 67 | 63 | 1.9E−08** | 22.1 | −22.8 | 3.5E−09** | 24.1 | −28.7 |
| SN11795R_86 | A6: 10.8 | A6: 1,810,358 C6: 8,058,522 |
83 | 47 | 9.3E−08** | 20.2 | 22.7 | 6.3E−08** | 20.6 | 27.8 |
| SN1914J_250 | A6: 47.3 | A6: 6,240,231 | 46 | 84 | 7.9E−05** | 11.6 | 17.3 | 2.4E−04* | 10.1 | 19.4 |
| SS1949_205 | A6: 67.6 | A6: 19,329,293 | 91 | 40 | 4.8E−04* | 9.1 | 15.9 | 8.5E−04* | 8.4 | 18.3 |
| SN12508_220 | A6: 69.9 | A6: 20,236,736 | 93 | 36 | 3.5E−05** | 12.7 | 19.5 | 1.3E−05** | 14.0 | 24.5 |
| SS2141_137 | A7: 63.3 | A7: 1,881,404 | 89 | 40 | 1.7E−06** | 16.7 | −23.2 | 7.3E−07** | 17.7 | −28.7 |
| SR7223_481 | A7: 32.1 | A7: 14,440,743 | 100 | 30 | 5.1E−04* | 9.1 | −17.3 | 3.5E−04* | 9.6 | −21.4 |
| SR12173_203 | A7: 54.9 | A7: 16,297,589 | 68 | 56 | 8.4E−04* | 8.8 | 14.6 | 2.5E−04* | 10.5 | 19.2 |
| SS2288_235 | C8: 64.2 | A9: 11,306,600 | 122 | 7 | 7.9E−04* | 8.6 | −31.5 | 2.2E−04* | 10.3 | −41.4 |
| SN3560_346 | na | A10: 11,967,893 | 90 | 39 | 2.9E−05** | 13.0 | −19.9 | 1.3E−05** | 14.0 | −24.9 |
| SORH62_283 | na | A10: 15,400,691 | 98 | 33 | 1.9E−04* | 10.3 | −18.5 | 1.6E−05** | 13.5 | −25.5 |
| SR11329I_302 | na | C1: rand_659,665 | 92 | 39 | 4.9E−05** | 12.1 | 18.4 | 8.2E−06** | 14.4 | 24.2 |
| SN12788_290 | C1: 54.8 | C1: 29,218,479 | 63 | 66 | 4.3E−06** | 15.5 | −19.3 | 8.9E−05** | 11.5 | −20.0 |
| SR12095I_442 | A2: 0.0 | C2: 1,738,778 | 92 | 36 | 2.4E−04* | 10.3 | −18.2 | 7.7E−05** | 11.8 | −23.5 |
| SR12040_178 | na | C2: 31,573,592 | 72 | 56 | 4.9E−04* | 9.3 | −18.1 | |||
| SN0218_405 | C2: 51.7 | C2: 39,285,406 | 93 | 33 | 1.5E−04* | 11.1 | 18.2 | 1.4E−04* | 11.1 | 22.1 |
| SN3821_495 | C3: 40.9 | C3: 8,220,706 | 95 | 35 | 2.3E−04* | 10.2 | 18.1 | 1.4E−04* | 10.8 | 22.5 |
| SORD79_127 | C3: 47.9 | C3: 13,142,903 | 66 | 64 | 1.9E−05** | 13.4 | 17.8 | 2.0E−04* | 10.3 | 18.8 |
| SNRA56_319 | C3: 46.6 | C3: 15,610,433 | 100 | 31 | 1.9E−04* | 10.3 | −22.2 | |||
| SN0569_371 | A3: 63.8 | C3: 19,294,001 | 112 | 18 | 9.0E−04* | 8.3 | 20.3 | |||
| SN2179F_226 | C3: 66.9 | C3: 23,259,732 | 10 | 121 | 1.4E−06** | 16.7 | −37.2 | 4.2E−04* | 9.3 | −33.4 |
| SR12324I_336 | C3: 79.3 | C3: 34,968,604 | 93 | 37 | 1.3E−05** | 14.0 | −20.3 | 3.8E−06** | 15.5 | −25.9 |
| SR1157_487 | C4: 19.4 | C4: 2,598,847 | 81 | 48 | 2.3E−06** | 16.3 | 20.3 | 9.2E−05** | 11.5 | 20.5 |
| SN12745_278 | C4: 29.0 | C4: 6,035,809 | 25 | 106 | 6.4E−04* | 8.7 | −18.2 | 6.9E−04* | 8.6 | −21.7 |
| SN0857_287 | na | C4: 6,035,809 | 89 | 42 | 0.0046 | 6.1 | −12.8 | 4.4E−04* | 9.2 | −19.0 |
| SN1944_308 | C5: 29.9 | C5: 10,903,693 | 74 | 52 | 3.7E−04* | 9.8 | 15.2 | |||
| SN0866_278 | C8: 83.4 | C5: 22,635 | 112 | 18 | 0.0011 | 1.9E−04* | 10.4 | −27.1 | ||
| SORH06_416 | C6: 41.5 C6: 43.2 | C5: 27,616,764 | 73 | 56 | 1.9E−07** | 19.4 | −22.0 | 1.5E−05* | 13.8 | −22.3 |
| SR6083_413 | C6: 32.8 | C6: 1,792,933 | 118 | 9 | 9.7E−05** | 11.5 | −32.8 | 1.4E−04* | 11.1 | −38.7 |
| SN11542_199 | C8: 18.2 | C8: 8,182,817 | 100 | 24 | 9.3E−04* | 8.7 | −18.2 | 5.5E−04* | 9.4 | −22.9 |
| SR12348I_114 | na | C8: 24,799,307 | 70 | 60 | 5.9E−05** | 12.0 | 16.8 | 3.6E−05** | 12.6 | 20.8 |
| SN1944_280 | C8: 83.8 | C8: 38,176,584 | 94 | 32 | 2.7E−04* | 10.2 | −21.2 | |||
| SR12386_286 | A5: 52.5 | C8: 31,566,379 | 116 | 14 | 3.6E−05** | 12.6 | 27.8 | 1.9E−04* | 10.4 | 30.4 |
| SN2016_270 | na | C9: 13,616,587 | 54 | 72 | 2.6E−09** | 25.1 | −24.4 | 1.0E−07** | 20.6 | −26.7 |
aCentimorgan (cM) distances of SSR markers according to the integrated map by Wang et al. (2011); the locus with the strongest sequence homology is shown; na = information not available
bLocation in nucleotides identified by DNA sequence homology in Genoscope (Chalhoub et al. 2014) presented as the mid-point of the sequence regardless of overall SSR size
cAUDPC calculated from the stem lesion length measured 7, 14 and 21 days after inoculation
dNegative values specify markers contributing to resistance while positive values specify susceptibility
e* q < 0.05; ** q < 0.001
The density of loci varied between chromosomes A1–C9 as shown in the graph of negative log of probability in Fig. 3. Highest densities were found on chromosome A1, A3, A5, C3 and C6 and lowest on A4 and A9. SSR associated with sclerotinia resistance was equally distributed between the A and C sub-genomes (Table 1; Fig. 3). Only 44 and 20 % of the SSR markers associated with lesion length and % s + c, respectively, were significant in the MLM + Q + K model. Moreover, the positive false discovery rate (q) further declared all loci spurious except two.
Fig. 3.
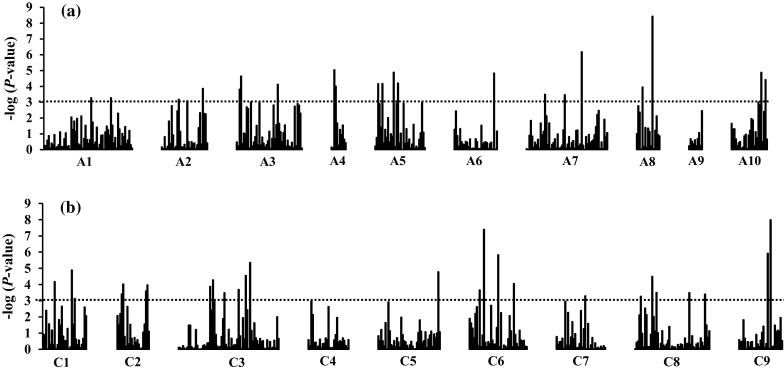
Graphic representation of negative log P values of SSR loci in B. napus associated with partial resistance to S. sclerotiorum by chromosome in the A and C genomes. The dotted line is the significance threshold level of P < 0.001 used to declare SSR loci significant in association mapping using the GLM + Q model. a Genome AA, b genome CC.
Co-location of SSR in GWAM and published QTL from linkage mapping
Of the 220 markers representing 65 previously published QTLs conferring sclerotinia resistance, 198 could be assigned to chromosome and nucleotide position in the B. napus genome. The remaining 22 markers were AFLP and SSRs without any sequence or primer information. Of 34 SSR loci obtained in the present study, 26 were associated with both AUDPC and % soft + collapsed sessions (Table 1). Of these 26 SSR, five were physically mapped within 500 kb of a published QTL peak marker or between the closest pair of flanking markers as follows: SR12095I on chromosome A2 in the present study mapped close to marker BGO153 and within the QTL region, BGO150-CB10022, identified by stem inoculation of B. napus line J7005 (Wu et al. 2013); SN12788 on C1 mapped close to SWUC150 identified by cut stem inoculation of B. incana line C01 (Mei et al. 2013), and within the QTL region, SWUC130-FITO083, identified by cut stem inoculation of B. napus cultivar Express from Germany (Wei et al. 2014); SR12040 on C2 mapped close to pW251 identified by petiole inoculation of B. napus line Huadbl2 (Zhao et al. 2006) and within QTL region, CB10316-Ol09A06, identified by cut stem inoculation of B. napus cultivar Express from Germany (Wei et al. 2014); SR12324 on C3 mapped close to FITO110 and SWUC455 identified by cut stem inoculation of B. incana line C01 (Mei et al. 2013); and SN12475 on C4 mapped within QTL region, pX105gE-pX116bH, identified by petiole inoculation of B. napus line Huadbl2 (Zhao et al. 2006).
Discussion
The stem inoculation method used to phenotype the B. napus collection for resistance to S. sclerotiorum was first described by Buchwaldt et al. (2005). It was purposely designed to mimic the infection that causes the most serious yield loss in commercial fields. Moreover, inoculation using mycelium simulated the so-called infection cushions that are formed by fungal hyphae on senescent petals which adhere to the plants under wet weather conditions. The gradual spread of infection allowed rating of symptom development over several weeks. The relative difference among accessions became more obvious over time and was maintained up to maturity. The disease data collected 3 weeks after inoculation resulted in more significant marker-resistance associations than after 1 and 2 weeks (data not shown). Confidence in the association with the molecular markers identified in the present study was ensured by selecting 152 lines with consistent sclerotinia phenotype from a larger collection of 400 lines previously screened for resistance. Good separation in lesion length (measured as AUDPC) and penetration into the vascular tissue (measured as % soft and collapsed lesions) between resistant and susceptible lines also added to the confidence. The most resistant lines (AUDPC <20, % s + c <10) comprised 28 % of the mapping population, while the most susceptible lines (AUDPC >60, % s + c >70) comprised 20 %.
The advantage of disease rating 3 weeks after inoculation was also demonstrated by Li et al. (2007). The stem inoculation method has been adopted by others for screening of various brassica germplasm and crucifer species (Li et al. 2007; Garg et al. 2010; Navabi et al. 2010; Wu et al. 2013; Uloth et al. 2013). Other stem inoculation techniques have used insertion of toothpicks with mycelium into the stem (Zhao and Meng 2003; Yin et al. 2010), or placing cut stems in trays before inoculation (Ding et al. 2013; Mei et al. 2013; Wei et al. 2014). However, in the present study wounding of the plant was deliberately avoided, so as not to interfere with the expression of defense genes and molecular pathways in the epidermis, mesophyll and vascular bundles. Simpler and faster screening techniques have been employed, such as inoculation of cotyledons (Garg et al. 2008), detached leaves (Zhao and Meng 2003; Bradley et al. 2006; Wu et al. 2013) or placing of mycelium on the cut end of petioles of young plants (Zhao et al. 2004, 2006; Bradley et al. 2006). However, none of the methods resemble field infection, and the rapid deterioration of infected tissues in all cases necessitated assessment of symptoms after only 2–8 days.
In preparation for GWAM, more than 400 B. napus accessions were screened for resistance against S. sclerotiorum. Subsequently, the most resistant and susceptible lines within certain geographical regions were selected for a total of 152 lines. Since partial resistance was known to exist in germplasm from China, a large portion of accessions were selected from countries in Asia. However, it was not possible to create a balanced number of resistant and susceptible lines in all regions. Examination of genetic diversity and population structure of an almost identical collection of 173 B. napus accessions was published earlier, but without analysis of association to sclerotinia resistance (Gyawali et al. 2013). Both the previous and the present study showed the presence of two subpopulations primarily based on spring or winter growth habits as was discussed in detail previously. Subpopulations in B. napus are primarily based on growth habit, plant morphology related to crop utilization such as oilseed rape, swedes for fodder, and vegetables (Hasan et al. 2008; Chen et al. 2008; Bus et al. 2011; Rezaeizad et al. 2011).
As is apparent from GWAM studies in B. napus and other plant species, the best fitting model for association analysis is identified on a case by case basis, and the trade-off between dismissing false positives and accepting false negatives is somewhat arbitrary. In the present study, the GLM + Q model best fitted the data and identified 21 SSR alleles contributing to the reduction in lesion growth along the stem and depth of penetration into the stem explaining 6–25 % of the phenotypic variation, while the presence of 13 other SSR alleles contributed to susceptibility. In contrast, only two markers were significant in the MLM + Q + K model. Thus, this model increased the number of false negatives at the cost of reducing false positives, which should be avoided in an exploratory analysis like association mapping, as it is more important to identify significant alleles than avoid false positives (Honsdorf et al. 2010). False positives can be effectively controlled by applying a double statistics method that uses population structure matrix as a covariate in the regression model, followed by a correction for positive false discovery rate, pFDR (Hasan et al. 2008; Jestin et al. 2011; Rezaeizad et al. 2011). Jestin et al. (2011) identified 49 and 60 markers associated with blackleg resistance in B. napus using the GLM + Q and MLM + K models, respectively, while none of the markers remained significant when the MLM + Q + K model was applied. Yu et al. (2006) showed the MLM + Q + K model better described their data compared to GLM, GLM + Q and MLM + K, in accounting for false positives. Association mapping of sclerotinia resistance in sunflower was undertaken using DNA polymorphisms in selected defense genes. There, the MLM model was suitable and was not greatly affected by either Q or K matrices (Fusari et al. 2012). The MLM + Q + K model gave the best fit for mapping of sclerotinia resistance in soybean (Bastien et al. 2014).
An investigation was undertaken to determine whether the SSR loci identified by GWAM in the current study coincided with published sclerotinia resistance QTLs identified by linkage analysis using biparental (resistant and susceptible), doubled haploid mapping populations. Zhao and Meng (2003) mapped sclerotinia resistance QTL in the Chinese line Ning RS-1 by inoculating detached leaves rated 2 dai and by inserting toothpicks with mycelium into stems rated 5 dai. Three QTLs for each method were identified explaining 13–23 % of the phenotype, but none were shared by the methods. Zhao et al. (2006) mapped QTLs in both Huadbl2 (China) and Major (France) by cut petiole inoculation rated 4–8 dai, which resulted in eight and one QTLs, respectively, explaining 6–22 % of the phenotype. Yin et al. (2010) mapped QTLs in the Chinese variety Zhongyou 821 using stem inoculation with three types of inoculum (mycelium grown on either PDA, toothpicks or petals) with symptoms rated 3–7 dai. Although up to ten QTLs were identified for each method explaining 10–31 %, none of the loci were shared. Wu et al. (2013) mapped QTLs in the Chinese line J7005 grown at three field locations. The stems were inoculated with mycelium plugs and lesion length rated 7 dai. Ten QTLs were identified explaining between 7 and 32 % of resistance, of which markers on chromosome A9 and C6 were consistently significant over test locations. Also, detached leaves were inoculated with mycelium plugs and lesions rated 2 dai, but the three resultant QTLs were different from the stem QTLs. Wei et al. (2015) mapped QTL in the European winter-type variety Express both under natural sclerotinia infection and by mycelium plug inoculation of stems placed in trays rated 3 dai. Six and five QTL were identified using the two methods explaining 5–20 %, and one QTL on chromosome C2 was shared. Li et al. (2015) produced an integrated map using 540 SSR markers from these five publications and determined the location of 35 sclerotinia QTLs using the B. napus genome sequence in Genoscope (Chalhoub et al. 2014). Subsequently, they concluded that two sclerotinia QTL were conserved across studies, one on A9 (Wu et al. 2013; Wei et al. 2014) and C6 (Wu et al. 2013; Zhao et al. 2006), respectively. Examination of possible co-location of molecular markers underlying 65 QTLs in the same five publications with SSR loci in the present study showed that five loci (7.6 %) on A2, C1, C2, C3 and C4 were likely captured by GWAM. We conclude that despite differences in origin of B. napus accessions and phenotyping methods between studies, the overlapping map locations indicate that some of the defense mechanisms against S. sclerotiorum could be shared by different resistant B. napus lines. In our study, the GWA identified 21 loci conferring resistance to S. sclerotiorum and 13 loci conferring susceptibility mapped to 12 of the 19 B. napus chromosomes with almost equally distribution between the A and C genomes. Furthermore, none of the SSR markers were linked when the conservative 500-kb cutoff was applied. The GWA study conducted by Wei et al. (2015) identified three loci on A8, C3 and C6. Although the map location of individual resistance loci was determined using the same B. napus genome sequence in Genoscope (Chalhoub et al. 2014), none of the resistance loci were shared between the two GWA studies. The discrepancy can be explained by different inoculation and disease rating methodologies; the first study used inoculation of intact plant stems without wounding and measurement of stem lesion length over 21 days, whereas the second study used wounding and measurement of lesion length after only 3 days. Wounding of the stem surface may circumvent molecular or mechanical defense mechanisms during initial infection as also discussed by Wei et al. (2015). We conclude that differences in phenotyping method, B. napus germplasm, marker technology and association analysis all contribute to variation between the two studies.
Predictably, most markers associated with sclerotinia resistance in the present study were new, primarily because many resistance accessions were evaluated for the first time. Also, whereas the published QTL were detected in Chinese lines, the partially resistance accessions in the GWAM study originated mainly from South Korea (Buk Wuk 3, 12, 13 and 20; Kuju 18, 19, 22, 27, 35 and 40; Dong Hae 3 and 6) and Japan (Norin 16, Tokiwa natane, Kinki 22 and 30), and single accessions were identified from China (Tzuyechein 32), Poland (SRS1632) and France (Tanto). The new sources of resistant B. napus accessions identified in the present study are suitable starting points for development of improved oilseed rape varieties. However, screening with isolates from the area of planned utilization is prudent, since evidence of isolates with different levels of aggressiveness and host preference is accumulating (Ge et al. 2012; Taylor et al. 2015).
Since a relatively low coverage of the B. napus genome was achieved with 690 SSR loci in the present study, further research is underway to more accurately map loci conferring sclerotinia resistance including GWAM utilizing markers based on single nucleotide polymorphisms and QTL mapping in biparental, doubled haploid populations derived from some of the partially resistant lines from Pakistan, China, South Korea and Japan. In the future, this additional marker information will aid transfer of stem rot resistance to adapted oilseed rape varieties.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful for access to the Brassica napus seed of lines labeled SRS (Saskatoon Research Station) which were mainly donated by Drs. Gerhard Rakow and Keith Downey and maintained by Richard Gugel. We appreciate funding received from the Saskatchewan Ministry for Agriculture (ADF program), Saskatchewan Canola Development Commission and Agriculture and Agri-Food Canada (AIP program).
Author’s contributions
LB, IP and DH secured funding, designed and supervised the experiments. LB and MH phenotyped the accessions. SG, DB and KH genotyped the accessions. SG and JD analyzed the data. SG and LB wrote the manuscript. All authors have reviewed the manuscript.
References
- Abbadi A, Leckband G. Rapeseed breeding for oil content, quality, and sustainability. Eur J Lipid Sci Technol. 2011;113:1198–1206. doi: 10.1002/ejlt.201100063. [DOI] [Google Scholar]
- Bastien M, Sonah H, Belzile F. Genome wide association mapping of Sclerotinia sclerotiorum resistance in soybean with genotyping-by-sequencing approach. Plant Genome. 2014 doi: 10.1186/s12870-020-02401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Bradley CA, Henson RA, Porter PM, LeGare DG, Del Río LE, Khot SD. Response of canola cultivars to Sclerotinia sclerotiorum in controlled and field environments. Plant Dis. 2006;90:215–219. doi: 10.1094/PD-90-0215. [DOI] [PubMed] [Google Scholar]
- Buchwaldt L, Li R, Hegedus DD, Rimmer SR (2005) Pathogenesis of Sclerotinia sclerotiorum in relation to screening for resistance. In: 13th international sclerotinia workshop, Monterey, CA, USA, p 22
- Bus A, Körber N, Snowdon RJ, Stich B. Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor Appl Genet. 2011;123:1413–1423. doi: 10.1007/s00122-011-1676-7. [DOI] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;22:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- Chen S, Nelson MN, Ghamkhar K, Fu T, Cowling WA. Divergent patterns of allelic diversity from similar origins: the case of oilseed rape (Brassica napus L.) in China and Australia. Genome. 2008;51:1–10. doi: 10.1139/G07-095. [DOI] [PubMed] [Google Scholar]
- Cruickshank RH. Distinction between Sclerotinia species by their pectic zymograms. Trans Br Mycol Soc. 1983;80:117–119. doi: 10.1016/S0007-1536(83)80171-5. [DOI] [Google Scholar]
- Del Río LE, Bradley CA, Henson RA, Endres GJ, Hanson BK, McKay K, Halvorson M, Porter PM, Le Gare DG, Lamey HA. Impact of sclerotinia stem rot on yield of canola. Plant Dis. 2007;91:191–194. doi: 10.1094/PDIS-91-2-0191. [DOI] [PubMed] [Google Scholar]
- Ding Y, Mei J, Li Q, Liu Y, Wan H, Wang L, Becker HC, Qian W. Improvement of Sclerotinia sclerotiorum resistance in Brassica napus by using B. oleracea. Genet Resour Crop Evol. 2013 [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- Fusari CM, Di Rienzo JA, Troglia C, Nishinakamasu V, Moreno MV, Maringolo C, Quiroz F, Alvarez D, Escande A, Hopp E, Heinz R, Lia VV, Paniego NB. Association mapping in sunflower for sclerotinia head rot. BMC Plant Biol. 2012;12:93–105. doi: 10.1186/1471-2229-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg H, Sivasithamparam K, Banga SS, Barbetti MJ. Cotyledon assay as rapid and reliable method of screening for resistance against Sclerotinia sclerotiorum in Brassica napus genotypes. Australas Plant Pathol. 2008;37:106–111. doi: 10.1071/AP08002. [DOI] [Google Scholar]
- Garg H, Atri C, Sandhu PS, Kaur B, Renton B, Banga SK, Singh H, Singh C, Barbetti MJ, Banga S. High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crops Res. 2010;117:51–58. doi: 10.1016/j.fcr.2010.01.013. [DOI] [Google Scholar]
- Ge, et al. Delineation of Sclerotinia sclerotiorum pathotypes using differential resistance responses on Brassica napus and B. juncea genotypes enables identification of resistance to prevailing pathotypes. Field Crops Res. 2012;127:248–258. doi: 10.1016/j.fcr.2011.11.022. [DOI] [Google Scholar]
- Gyawali S, Hegedus DD, Parkin IAP, Poon J, Higgins E, Horner K, Bekkaoui D, Coutu C, Buchwaldt L. Genetic diversity and population structure in a world collection of Brassica napus accessions with emphasis on South Korea, Japan, and Pakistan. Crop Sci. 2013;53:1537–1545. doi: 10.2135/cropsci2012.10.0614. [DOI] [Google Scholar]
- Hasan M, Friedt W, Pons-Kühnemann J, Freitag NM, Link K, Snowdon RJ. Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus) Theor Appl Genet. 2008;116:1035–1049. doi: 10.1007/s00122-008-0733-3. [DOI] [PubMed] [Google Scholar]
- Honsdorf N, Becker HC, Ecke W. Association mapping for phenological, morphological, and quality traits in canola quality winter rapeseed (Brassica napus L.) Genome. 2010;53:899–907. doi: 10.1139/G10-049. [DOI] [PubMed] [Google Scholar]
- Jestin C, Lodé M, Vallée P, Domin C, Falentin C, Horvais R, Coedel S, Manzanares-Dauleux MJ, Delourme R. Association mapping of quantitative resistance for Leptosphaeria maculans in oilseed rape (Brassica napus L.) Mol Breed. 2011;27:271–287. doi: 10.1007/s11032-010-9429-x. [DOI] [Google Scholar]
- Kent JW. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202.ArticlepublishedonlinebeforeMarch2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard JA, Robertson MJ, Hamblin P, Sprague SJ. Effect of blackleg and sclerotinia stem rot on canola yield in the high rainfall zone of southern New South Wales, Australia. Aust J Agric Res. 2006;57:201–212. doi: 10.1071/AR05179. [DOI] [Google Scholar]
- Koch S, Dunker S, Kleinhenz B, Röhrig M, Von Tiedemann A. A crop loss-related forecasting model for sclerotinia stem rot in winter oilseed rape. Phytopathology. 2007;97:1186–1194. doi: 10.1094/PHYTO-97-9-1186. [DOI] [PubMed] [Google Scholar]
- Kohli Y, Brunner LJ, Yoell H, Milgroom MG, Anderson JB, Morrall RA, Kohn LM. Clonal dispersal and spatial mixing in populations of the plant pathogenic fungus, Sclerotinia sclerotiorum. Mol Ecol. 1995;4:69–77. doi: 10.1111/j.1365-294X.1995.tb00193.x. [DOI] [Google Scholar]
- Li Y, Chen J, Bennett R, Kiddle G, Wallsgrove R, Huang Y, He Y (1999) Breeding, inheritance, and biochemical studies on Brassica napus cv. Zhongyou 821: tolerance to Sclerotinia sclerotiorum (stem rot). In: Wratten N, Salisbury PA (eds) 10th international Rapeseed Congress: new horizons for an old crop. TRI Publ Canberra, Australia
- Li CX, Siddique AB, Sivasithamparam K, Salisbury P, Banga SS, Banga S, Chattopadhyay C, Kumar A, Singh R, Singh D, Agnihotri A, Liu SY, Li YC, Tu J, Wang YF, Barbetti MJ. The importance of the type and time of inoculation and assessment in the determination of resistance in Brassica napus and B. juncea–Sclerotinia sclerotiorum. Aust J Agric Res. 2007;58:1198–1203. doi: 10.1071/AR07094. [DOI] [Google Scholar]
- Li J, Zhao Z, Hayward A, Cheng H, Fu D. Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica. 2015;205:483–489. doi: 10.1007/s10681-015-1417-0. [DOI] [Google Scholar]
- Mei J, Qian L, Disi JO, Yang X, Li Q, Li J, Frauen M, Cai D, Qian W. Identification of resistant sources against Sclerotinia sclerotiorum in Brassica species with emphasis on B. oleracea. Euphytica. 2011;177:393–399. doi: 10.1007/s10681-010-0274-0. [DOI] [Google Scholar]
- Mei J, Ding Y, Lu K, Wei D, Liu Y, Disi JO, Li J, Liu L, Liu S, McKay J, Qian W. Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor Appl Genet. 2013;126:549–556. doi: 10.1007/s00122-012-2000-x. [DOI] [PubMed] [Google Scholar]
- Morral RRA, Dueck J, Verma P. Yield losses due to Sclerotinia stem rot in western Canadian rapeseed. Can J Plant Pathol. 1984;6:265. [Google Scholar]
- Navabi ZK, Strelkov SE, Good AG, Thiagarajah MR, Rahman MH. Brassica B-genome resistance to stem rot (Sclerotinia sclerotiorum) in a doubled haploid population of Brassica napus × Brassica carinata. Can J Plant Pathol. 2010;32:237–246. doi: 10.1080/07060661.2010.484229. [DOI] [Google Scholar]
- Pritchard JK, Wen W. Documentation for STRUCTURE software. Chicago: The University of Chicago Press; 2004. [Google Scholar]
- Qian L, Qian W, Snowdon RJ. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genomics. 2014;15:1170. doi: 10.1186/1471-2164-15-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA. Association genetics in crop improvement. Curr Opin Plant Biol. 2010;13:174–180. doi: 10.1016/j.pbi.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Rezaeizad A, Wittkop B, Snowdon R, Hasan M, Mohammadi V, Zali A, Friedt W. Identification of QTLs for phenolic compounds in oilseed rape (Brassica napus L.) by association mapping using SSR markers. Euphtica. 2011;177:335–342. doi: 10.1007/s10681-010-0231-y. [DOI] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Taylor A, Coventry E, Jones JE, Clarkson JP. Resistance to a highly aggressive isolate of Sclerotinia sclerotiorum in Brassica napus diversity set. Plant Pathol. 2015 [Google Scholar]
- Uloth MB, You MP, Finnegan PM, Banga SS, Banga SK, Sandhu PS, Yi H, Salisbury PA, Barbetti MJ. New sources of resistance to Sclerotinia sclerotiorum for crucifer crops. Field Crops Res. 2013;154:40–52. doi: 10.1016/j.fcr.2013.07.013. [DOI] [Google Scholar]
- Wang HZ, Liu GH, Zheng YB, Wang XF, Yang Q. Breeding of the Brassica napus cultivar Zhongshuang 9 with high-resistance to Sclerotinia sclerotiorum and dynamics of its important defense enzyme activity. Sci Agric Sin. 2004;37:23–28. [Google Scholar]
- Wang J, Lydiate DJ, Parkin IAP, Falentin C, Delourme R, Carion PWC, King GJ. Integration of linkage maps for the amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics. 2011;12:101. doi: 10.1186/1471-2164-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Mei J, Fu Y, Disi JO, Li J, Qian W. Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol Breed. 2014;34:1797–1804. doi: 10.1007/s11032-014-0139-7. [DOI] [Google Scholar]
- Wei L, Jian H, Lu K, Filardo F, Yin N, Liu L, Qu C, Li W, Du H, Li J. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J. 2015 doi: 10.1111/pbi.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cai G, Tu J, Li L, Liu S, Luo X, Zhou L, Fan C, Zhou Y. Identification of QTLs for resistance to Sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS One. 2013;8(7):e67740. doi: 10.1371/journal.pone.0067740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Yi B, Chen W, Zhang W, Tu J, Fernando WGD, Fu T. Mapping of QTLs detected in a Brassica napus DH population for resistance to Sclerotinia sclerotiorum in multiple environments. Euphytica. 2010;173:25–35. doi: 10.1007/s10681-009-0095-1. [DOI] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhao J, Meng J. Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.) Theor Appl Gene. 2003;106:759–764. doi: 10.1007/s00122-002-1171-2. [DOI] [PubMed] [Google Scholar]
- Zhao J, Peltier AJ, Meng J, Osborn TC, Grau CR. Evaluation of sclerotinia stem rot resistance in oilseed Brassica napus using a petiole inoculation technique under greenhouse conditions. Plant Dis. 2004;88:1033–1039. doi: 10.1094/PDIS.2004.88.9.1033. [DOI] [PubMed] [Google Scholar]
- Zhao J, Udal JA, Quijada PA, Grau CR, Meng J, Osborn TC. Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor Appl Genet. 2006;112:509–516. doi: 10.1007/s00122-005-0154-5. [DOI] [PubMed] [Google Scholar]
- Zou J, Jiang C, Cao Z, Li R, Long Y, Chen S, Meng J. Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome. 2010;53:908–916. doi: 10.1139/G10-075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.