Abstract
Because of genetic heterogeneity present in idiopathic scoliosis, we previously defined clinical subsets (a priori) from a sample of families with idiopathic scoliosis to find genes involved with spinal curvature. Previous genome-wide linkage analysis of seven families with at least two individuals with kyphoscoliosis found linkage (P-value = 0.002) in a 3.5-Mb region on 5p13.3 containing only three known genes, IRX1, IRX2, and IRX4. In this study, the exons of IRX1, IRX2, and IRX4, the conserved noncoding elements in the region, and the exons of a nonprotein coding RNA, LOC285577, were sequenced. No functional sequence variants were identified. An intrafamilial test of association found several associated noncoding single nucleotide variants. The strongest association was with rs12517904 (P = 0.00004), located 6.5 kb downstream from IRX1. In one family, the genotypes of nine variants differed from the reference allele in all individuals with kyphoscoliosis, and two of three individuals with scoliosis, but did not differ from the reference allele in all other genotyped individuals. One of these variants, rs117273909, was located in a conserved noncoding region that functions as an enhancer in mice. To test whether the variant allele at rs117273909 had an effect on enhancer activity, zebrafish transgenesis was performed with overlapping fragments of 198 and 687 bp containing either the wild type or the variant allele. Our data suggests that this region acts as a regulatory element; however, its size and target gene(s) need to be identified to determine its role in idiopathic scoliosis.
Keywords: idiopathic scoliosis, IRX genes, zebrafish transgenesis, conserved noncoding regions, kyphoscoliosis
Idiopathic scoliosis is defined as a lateral curvature of the spine greater than ten degrees (°) documented by radiographic analysis, and present in the late juvenile or adolescent period in otherwise normal individuals. The prevalence of idiopathic scoliosis in the general population is estimated to be 2–3% (Bunnell 1986; Lonstein and Carlson 1984). Both sporadic [idiopathic scoliosis (IS)] and familial forms [familial idiopathic scoliosis (FIS)] exist. Several genetic analyses have reported linkage of FIS to various candidate regions, including chromosomes 6p (LOD = 1.42), 10q (LOD = 1.60), and 18q (ATA82B02, LOD = 8.26) (Wise et al. 2000); 17p11.2 (D17S799, LOD = 3.2) (Salehi et al. 2002); 19p13.3 (D19S922, LOD = 4.087), and 2q (LOD = 1.72) (Chan et al. 2002); Xq23-26 (GATA172D05, LOD = 2.23) (Justice et al. 2003); 6p25-22 (D6S1031, P-value = 0.0032), 6q14-16 (D6S1031, P-value = 0.0092), 9q32-34 (D9S915, P-value = 0.0005), 16q11-q12 (D16S2623, P-value = 0.0005) and 17p11-q11 (D16S2623, P-value = 0.0005) (Miller et al. 2005); 8q12 (D8S1136, LOD = 2.77) (Gao et al. 2007); 9q31.2-q34.2 (D9S2157, LOD = 3.64) and 17q25.3-qtel (AAT095, LOD = 4.08) (Ocaka et al. 2008); 12p (GATA49D12, LOD = 3.5) (Raggio et al. 2009) and 3q12.1 (D3S2462, LOD = 3.01), and 5q13.3 (D5S203, LOD = 3.0) (Edery et al. 2011). Population based genome-wide associations of IS have been reported to rs11190870 (P-value = 1.24 × 10−19) on 10q24.31 near LBX1 (Takahashi et al. 2011), rs6570507 (P-value = 1.27 × 10−14) on 6q24.1 in GPR126 (Kou et al. 2013), and rs12946942 (P-value = 4 × 10−8) on 17q24.3 near SOX9 and KCNJ2 (Miyake et al. 2013). These findings, some of which have been independently replicated, suggest that IS and/or FIS is a complex genetic disorder with substantial clinical and genetic heterogeneity.
In an effort to reduce the genetic heterogeneity present in FIS, we previously created subsets from our collection of FIS families based on either the most-likely mode of inheritance or on different clinical criteria (Justice et al. 2003; Miller et al. 2005, 2006). One of our clinically defined subgroups, the kyphoscoliosis (KS) group, consisted of non-Hispanic white families (seven families, 53 individuals) with two or more individuals having a scoliotic curve ≥ 10° in combination with a thoracic curve ≥ 40° (Miller et al. 2006). The prevalence of KS in the United States is estimated to be one in 1000 people for lateral and posterior spinal curvatures > 35°, and one in 10,000 for more severe KS, defined by lateral and posterior curvatures > 70° (Bergofsky 1979). Previous linkage analysis of the KS subgroup identified candidate regions on chromosomes 5p15.3, 13q13.3, and 13q32, and analyses of single nucleotide variants (SNVs) narrowed the linkage region on 5p15.3 to about 3.5 Mb (P-value = 0.002) (Miller et al. 2006). The only genes present in this region are IRX1, IRX2, IRX4, and C5orf38, a protein precursor in the promoter region of IRX2, which has coordinated expression with IRX2 and lacks homology to any known protein in the public databases (Wu et al. 2006).
The Iroquois (IRX) genes are members of a highly conserved gene family, the TALE (three-amino acid-loop extension) homeobox genes that encode homeoproteins (Bürglin 1997). Vertebrate IRX genes have been found to play a role in neural tube, heart, and ectoderm patterning (Cavodeassi et al. 2001; Gómez-Skarmeta and Modolell 2002). In humans, the IRX genes are clustered into two groups of three genes believed to be the result of a segmental duplication, because IRX1, IRX2, and IRX4 on 5p are paralogs of IRX3, IRX5, and IRX6 respectively, on chromosome 16q (Bosse et al. 2000; Gómez-Skarmeta and Modolell 2002; Ogura et al. 2001; Peters et al. 2000). This region on 16q was previously found to be linked to FIS (P-value < 0.0005) in a sample of 202 FIS families when the threshold for scoliosis was ≥ 30° (Miller et al. 2005).
Our previous linkage results suggest the involvement of the IRX gene family and/or transcriptional enhancers within the highly conserved noncoding elements (CNE) surrounding the IRX genes on 5p in the expression of FIS (Miller et al. 2006). The IRX genes are surrounded by a large amount of CNEs, which share sequence similarity between regions on the same chromosome, as well as with regions on 16q (Bejerano et al. 2004; de la Calle-Mustienes et al. 2005; McEwen et al. 2006; Sandelin et al. 2004; Woolfe et al. 2005). Several vertebrate CNE regions have been tested in vivo in zebrafish and mouse, and many of these have been shown to function as tissue-specific enhancers (de la Calle-Mustienes et al. 2005; Goode et al. 2005; McEwen et al. 2006; Nobrega et al. 2003; Woolfe et al. 2005).
In this study, we examined whether the IRX genes or the surrounding regulatory elements are involved in the development of FIS by sequencing the exons and the CNEs located 500 kb upstream and downstream from IRX1, IRX2, and IRX4. Intrafamilial tests of association were used to identify significantly associated SNVs. In addition, a SNV in a highly conserved noncoding region associated with FIS in one family was selected to test for functional significance. Zebrafish transgenesis was carried out in order to determine if the CNE surrounding this SNV, rs117273909, acts as an enhancer in vivo, and if the alternate allele affects the enhancer function.
Materials and Methods
Subjects
All probands and their relatives were clinically characterized by a single orthopedic surgeon. Written informed consent was obtained for all study participants, in accordance with the Institutional Review Board of the participating institutions.
Primer design for genes and conserved noncoding elements
Primers covering the IRX exons genes, the CNEs, and a long intergenic nonprotein RNA in the linkage region, LOC285577, were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). CNEs were selected for sequencing if they had a LOD score > 100 based on the PhastCons Placental Mammal Conserved Elements, 28-way Multiz Alignment (http://www.genome.ucsc.edu), and 500 kb ± from IRX1, IRX2, and IRX4 (Supplemental Material, Figure S1 and Table S1). For rs117273909, genotyping of the controls was performed by sequencing. Sequences of primers are available on request.
PCR and sequencing
For mutational analysis of the genes, PCR was performed on DNA from 53 individuals from seven FIS families from the KS clinical subset using the HotStarTaq amplification protocol (Qiagen). For the sequencing of CNEs and LOC285577, PCR from 46 of these individuals was carried out using the KAPA 2G Fast HS ReadyMix PCR Kit (KAPA Biosystems, Wilmington, MA). The reactions were analyzed on 3730 DNA Sequencers (Applied Biosystems, Grand Island, NY).
DNA isolated from blood samples of 100 controls consisting of individuals who married into FIS families, and who did not have FIS, were amplified with rs117273909 primers using GeneAmp High Fidelity PCR System (Applied Biosystems, Grand Island, NY). The products were sequenced on an Applied Biosystems / Hitachi 3730 Genetic Analyzer. Sequencing analysis was performed using Sequencing Analysis version 5.2, and Sequence Scanner version 1.0 (both from Applied Biosystems, Grand Island, NY). Alignments of DNA sequences were done with SeqScape (Applied Biosystems, Grand Island, NY), Sequencher (Gene Codes Corporation, Ann Arbor, MI), and CodonCode Alignment software (v 3.7.1.1).
Statistical methods
Data cleaning was carried out on 344 CNE SNVs and 70 insertions/deletions from 46 individuals. Individuals with a genotype missing rate > 10%, and variants (SNVs, insertions and deletions) with either a Polyphred (v6.11) value < 99, a missing rate > 10%, and/or two or more Mendelian inconsistencies were removed. The Polyphed program identifies heterozygous single nucleotide substitutions, and assigns scores ranging from 99 to 0 to each heterozygous site, where a score of 99 indicates a very good fit and stands for a true positive rate of > 97%. Mendelian inconsistencies were tested using PEDCHECK (O’Connell and Weeks 1998). SNVs that became monomorphic after these steps were removed. Data cleaning reduced the number of individuals for analysis to 38, the number of SNVs to 197, and the number of insertions/deletions to 40. Of the remaining individuals, 22 had scoliosis (of which 14 had KS), 12 were unaffected and four had no curvature information. Tests of Hardy-Weinberg equilibrium (Wigginton et al. 2005) identified one SNV (hg19, chr5:3188028) with a P-value < 0.0001, which was retained for analysis because our sample was ascertained for familial idiopathic scoliosis and removal of SNVs in this situation can remove causative SNVs.
All SNVs were tested for association to the quantitative trait using ASSOC [S.A.G.E., v.6.0.1], a likelihood-based test of association that compares the likelihood of the data in models with and without a marker, and uses the phenotype and genotype information of the entire family. The degree of lateral curvature was analyzed as a quantitative phenotype, and genotypic (a/a, a/A, or A/A) and allelic (presence of minor allele) tests of association were performed.
Cloning of putative regulatory elements into zebrafish enhancer detection vector
Site-directed mutagenesis was used to change the wild type rs117273909 C allele to the variant T allele (Bioinnovatise, Rockville, MD). Four constructs (198bp C allele, 198bp T allele, 687bp C allele, and 687bp T allele) were cloned into the zebrafish enhancer detection (ZED) vector (Bessa et al. 2009), which has insulators that prevent false positive expression due to position effects. Tol2 mRNA was synthesized with the mMessage mMachine SP6 kit (Ambion, Grand Island, NY) using NotI linearized pCS2FA-transposase vector as template DNA (Kwan et al. 2007).
Zebrafish transgenesis
Microinjections of all four constructs were performed into one- to two-cell stage zebrafish embryos using 25–45 pg of plasmid DNA mixed with 50 pg of Tol2 mRNA. Embryos were incubated at 28° with 0.003% PTU (1-phenyl 2-thiourea) to suppress pigmentation. Embryos with transgene integration were identified by red fluorescent protein (RFP) expression in the skeletal muscle at 48 hr postfertilization (hpf), and green fluorescent protein (GFP) expression was observed in RFP positive embryos from 48 hpf to 5 d post fertilization (dpf). Embryos positive for both RFP and GFP were grown to adulthood. Several germline transmitting founders were identified for each construct, and their progeny were evaluated for patterns of GFP expression from 48 hpf to 5 dpf. Embryos from F2 generation of germline transmitting founders were also evaluated for GFP expression by crossing F1 adult fish with wildtype fish. The only difference in sequence between the C and T allele constructs was at the rs117273909 locus. Screening and imaging of embryos were performed using the Zeiss SteREO Lumar.V12 stereomicroscope with an AxioCam HRC color camera or Zeiss Axio Observer Z1 inverted microscope with an AxioCam MRm black and white camera.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Variant analysis of IRX genes exons and CNEs
Sequencing of the IRX1, IRX2, and IRX4 exons in 53 individuals did not identify any functional sequence variants. Sequencing of the CNEs surrounding the IRX genes in 46 individuals identified 197 SNVs suitable for analysis, of which 16 were ± 500 kb of IRX4, 70 were ± 500 kb of IRX2, and 160 were ± 500 kb of IRX1, with eight SNVs overlapping the conserved regions of IRX4 and IRX2 and 41 SNVs overlapping the conserved regions of IRX1 and IRX2. Of the 197 SNVs, 23 were novel.
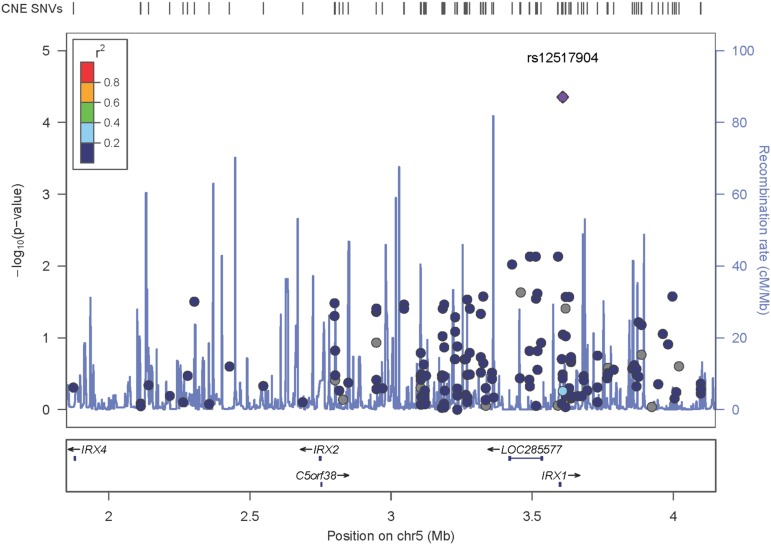
Intrafamilial tests of association between the genotypes of the 197 SNVs and the quantitative trait (scoliotic curvature) resulted in 30 SNVs (Table 1) with nominal significance (P-values < 0.05). Only rs12517904 (P-value = 0.00004, Figure 1) was significant after adjusting for multiple testing (Bonferroni correction P-value = 0.00013). No association was identified with the 40 insertions/deletions.
Table 1. Association analysis results (P-values < 0.05).
| SNP | Positiona | MAFb | Allelic Association | Genotypic Association |
|---|---|---|---|---|
| rs67250895 | 2303160 | 0.22 | 0.03124 | 0.14792 |
| rs139215365 | 2427684 | 0.07 | 0.25035 | 0.03999 |
| rs16870466 | 2799648 | 0.11 | 0.04946 | 0.05745 |
| rs1497457 | 2799759 | 0.31 | 0.03285 | 0.19374 |
| rs2934527 | 2947498 | 0.48 | 0.04338 | 0.09179 |
| rs2934528 | 2947877 | 0.32 | 0.03903 | 0.07172 |
| rs183225473 | 3045672 | 0.04 | 0.03913 | 0.03913 |
| rs146531224 | 3046004 | 0.20 | 0.03438 | 0.03438 |
| rs117273909 | 3182971 | 0.44 | 0.03913 | 0.03913 |
| rs111916055 | 3187995 | 0.03 | 0.03438 | 0.03438 |
| rs16871553 | 3269602 | 0.39 | 0.02924 | 0.02924 |
| rs73733752 | 3278780 | 0.54 | 0.03913 | 0.03913 |
| rs61712864 | 3317668 | 0.18 | 0.04626 | 0.25960 |
| rs73733769 | 3326276 | 0.03 | 0.02653 | 0.02653 |
| rs73032754 | 3428448 | 0.04 | 0.00948 | 0.00965 |
| novel | 3459187 | 0.13 | 0.03913 | 0.03913 |
| rs117494736 | 3459291 | 0.10 | 0.02326 | 0.02326 |
| rs537539844 | 3491460 | 0.03 | 0.03893 | 0.03893 |
| rs35450818 | 3491486 | 0.03 | 0.00737 | 0.03819 |
| rs34560950 | 3512343 | 0.50 | 0.00737 | 0.00737 |
| rs62336074 | 3513171 | 0.47 | 0.02838 | 0.02838 |
| rs35155570 | 3518273 | 0.03 | 0.02409 | 0.02409 |
| rs828332 | 3591325 | 0.03 | 0.00737 | 0.00737 |
| rs12517904 | 3608064 | 0.03 | 0.00004 | 0.16992 |
| rs76205392 | 3617948 | 0.03 | 0.03893 | 0.03893 |
| rs71577554 | 3618245 | 0.03 | 0.02686 | 0.02686 |
| rs10475220 | 3618387 | 0.22 | 0.02686 | 0.02686 |
| rs10475221 | 3618393 | 0.03 | 0.02686 | 0.02686 |
| rs78040936 | 3630480 | 0.20 | 0.02686 | 0.02686 |
| novel | 4003967 | 0.04 | 0.02645 | 0.02645 |
Map positions obtained from NCBI (GRCh37/hg19).
Maximum likelihood estimate of minor allele frequency from founders (FREQ [S.A.G.E., v6.0.1]).
Figure 1.
Regional allelic association plots of SNVs in CNEs surrounding IRX genes. The position of each SNV is indicated by the circles; diamond indicates the SNV with the most significant association (rs12517904, P-value = 0.00004). The color of the circles is the linkage disequilibrium (r2) between each SNV and rs12517904, indicated by the color scale on the left. Gray circles indicate a lack of linkage disequilibrium between the SNV and rs12517904. Genes in the region are indicated in the lower box. Recombination rates (cm/Mb) are depicted by the purple vertical lines. The plot was created using Locus Zoom (https://statgen.sph.umich.edu/locuszoom).
We searched for variants cosegregating with KS. In one family, the genotypes of nine SNVs differed from the reference allele in all individuals affected with KS, and two out of three individuals with FIS (Table 2); unaffected family members, and all other individuals genotyped for this study, did not differ from the reference allele for any of these nine SNVs. The one family member with FIS that did not differ from the reference allele had a scoliotic curve of 36°, and no hyperkyphosis. Three of these SNVs were located in highly conserved regions, based on whole-genome alignment of vertebrates. Of these three, only rs117273909 (P-value = 0.039 for association analysis using all families genotyped in this study) was found to be conserved in all 96 vertebrate species in which this SNV and the corresponding CNE were present (www.genome.ucsc.edu). Rs117273909 is located in the ∼841 kb gene desert between IRX1 (413 kb downstream) and IRX2 (431 kb downstream), and thus could function as an enhancer for either IRX1 or IRX2, or for both.
Table 2. SNV genotypes for family 1.
| SNV | bp | Conserved Regiona | P-Valueb | Allelesc | MAFd | Unaffected | Scoliosis | Kyphoscoliosis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs114080324 | 2947499 | Yes | 0.116 | G/A | 0.006 | GG | GG | GG | — | GG | AG | AG | AG | AG | AG |
| rs183225473 | 3045672 | No | 0.039 | G/C | 0.006 | GG | GG | GG | CG | GG | CG | CG | CG | CG | CG |
| rs10068728 | 3045983 | Yes | 0.034 | T/G | 0.27 | TT | TT | TT | TG | TT | — | TG | TG | TG | TG |
| rs146531224 | 3046004 | No | 0.039 | G/A | 0.006 | GG | GG | GG | AG | GG | AG | AG | AG | AG | AG |
| rs117273909 | 3182971 | Yes | 0.039 | C/T | 0.005 | CC | CC | CC | CT | CC | CT | CT | CT | CT | CT |
| rs111916055 | 3187995 | No | 0.034 | A/G | 0.028 | AA | AA | AA | AG | AA | — | AG | AG | AG | AG |
| Novel | 3188045 | No | 0.083 | T/G | NA | TT | TT | TT | AT | TT | — | AT | AT | — | AT |
| rs16871553 | 3269602 | No | 0.029 | C/T | 0.109 | TT | TT | TT | CT | TT | CT | CT | CT | CT | — |
| rs73733752 | 3278780 | No | 0.039 | A/G | 0.03 | AA | AA | AA | AG | AA | AG | AG | AG | AG | AG |
Based on GERP score > 2 (www.genome.uscs.edu).
P-values the same for genotypic and allelic association tests, using all families in study.
Reference allele/alternate allele.
Minor allele frequency, from dbSNP website (www.ncbi.nlm.nih.gov/SNP).
We determined the frequency of the variant T allele of rs117273909 in a Caucasian population by sequencing 100 controls consisting of individuals who married into our FIS families, and who did not have a history of FIS. Of these 100 controls, 90 matched the reference sequence (C), one was heterozygous (C/T), and nine failed to amplify, resulting in a T allele frequency of 0.00515. In the 1000 Genomes Project (www.1000genomes.org), rs117273909 was found to be C/T in 27 of 5008 genotypes (T allele frequency of 0.00539), and heterozygous A/T in three out of 5008 genotypes.
Based on the association with KS (P-value = 0.039), the presence of a nonreference allele cosegregation with KS in a single family, the fact that rs117273909 is located in a noncoding fragment (element_603, hg19 chr5:3182218-3183271,VISTA Enhancer Browser, http://enhancer.lbl.gov), which drives expression in the hindbrain in five out of 10 transgenic mice embryos (Visel et al. 2007), and the highly conserved nature of this SNV, we gave this SNV priority in our effort to determine if this SNV was functional. We then used zebrafish transgenesis to evaluate the effect of a C to a T allele substitution (rs117273909).
Regulatory activity of CNE containing rs117273909
We performed zebrafish transgenesis to determine if changing the allele at rs117273909 from a C (wild type allele) to a T would result in a change in its regulatory activity. Tena et al. (2011) tested a 1172bp fragment encompassing rs117273909, which did not show any regulatory activity in Xenopus, possibly due to the presence of repressors: therefore, we chose to test smaller fragments for regulatory activity. For the zebrafish transgenesis assay, the wild type (C allele) and variant (T allele) versions of a 198 bp fragment (hg19, chr5:3182938-3183135), and an overlapping larger 687 bp fragment (hg19, chr5:3182466-3183152), both selected based on strong conservation across vertebrates, were tested for regulatory activity (Figure S2). Despite strong RFP expression, no consistent GFP expression patterns were observed in the embryos injected with any of the four constructs (Figure S3).
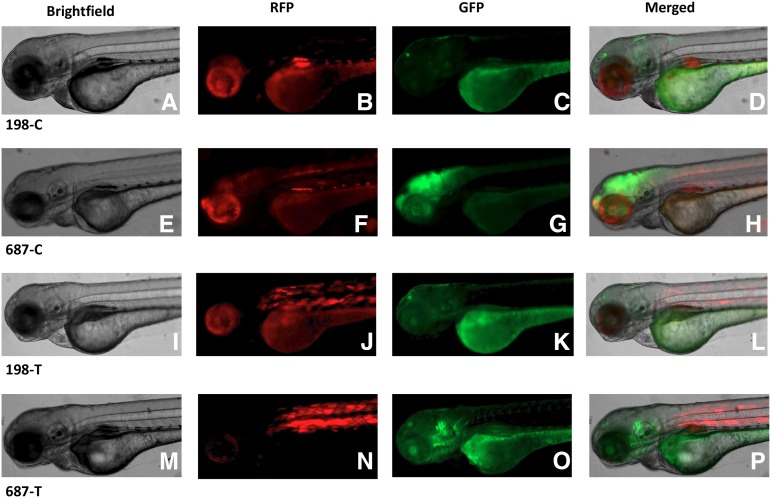
Due to the mosaic nature of the transgene expression during transient transgenesis, we generated multiple stable transgenic lines for all four constructs by screening founders for germline transmission. Variable GFP expression patterns, ranging from weak and diffused expression to strong and specific expression, were observed in the F1 and F2 progeny of multiple germline transmitting founders for each of the four constructs (Figure 2, A–P). These data may indicate positional effects of the transgene integration in independent founders for each of the four constructs. The specific GFP expression patterns were observed in pineal gland, pharyngeal arches, and brain. However, there was no association of these expression patterns with any particular construct.
Figure 2.
Patterns of GFP expression in F1 embryos at 72 hpf. Images of lateral views of representative embryos for each of the four constructs are shown in Brightfield (A, E, I, and M), RFP expression (B, F, J, and N), GFP expression (C, G, K, and O) and merged image of all three images (D, H, L, and P). In all images, embryos are oriented with their anterior to the left.
Discussion
Our results suggest that noncoding fragments as small as 198 bp can act as regulatory elements in zebrafish, which is in contrast to a report that a 1172 bp fragment (TA3235, 131:818307–820479 bp, xenTrot2) overlapping our 198 bp and 697 bp fragments did not drive expression in Xenopus (Tena et al. 2011). At present, there are no guidelines regarding the identification of regulatory regions, the size of these regions, or the type of activity present (repressors vs. enhancers). We were not able to determine in this study whether this regulatory region plays a role in the expression of FIS. Although we observed a strong expression trend with one of the two alleles at rs117273909, we did not observe a common pattern of expression among multiple lines, and thus could not rule out position effects. GFP expression occurring in different regions (the pharyngeal arches, pineal gland and brain) could be due to positional effects, something the ZED vector should minimize, or could be the result of the involvement of this regulatory region with a homeobox gene. The expression of our conserved fragment in the pineal gland is of interest, since pinealectomised chickens develop scoliosis (Machida et al. 1993).
There appeared to be differences in the level of expression and the timing of GFP expression between the wild type and alternate allele at rs117273909. de la Calle-Mustienes et al. (2005) noticed that DNA sequence differences in zebrafish and mouse CNEs did not result in differences in where expression occurred, but affected the level and/or timing of the transcription. The change in timing and regulation may be due to the sequence degeneration, which interferes with loop formation. Tena et al. (2011) identified a three-dimensional architecture that forms through CCCTC-binding, present in the Irx clusters of mice, zebrafish, and Xenopus, which brings the Irx1/3 and Irx2/5 promoters together, and demonstrated that cis-regulatory elements in the Irx clusters interact with more than one Irx promoter, up to distances of 1.6 Mb. This looping mechanism may help facilitate the delivery of RNA polymerase, transactivators, and transcription factors to the promoter to the right tissue at the correct time.
In summary, tests of association identified several significant SNVs that may lie in regulatory regions that influence gene expression. These SNVs may play a role in the phenotype, but the relevance of each individual SNV can be determined only by in vitro and in vivo assays. It is also possible that unidentified SNVs in highly conserved regulatory regions further upstream from the IRX family may disrupt developmental patterning and be responsible for the variation of the degree of lateral and thoracic curvature in individuals in these families. Future work will focus on completely sequencing the region with the most significant association, as well as performing additional in vivo assays of regulatory regions surrounding the other associated SNVs identified in this study, including rs1251709, and the SNVs in LOC285577, of which little is known to date. Once we have a better understanding of what tissues are involved in the scoliosis phenotype, we will be able to define the regulatory regions using both in vitro and in vivo assays.
Supplementary Material
Acknowledgments
The ZED vector was kindly provided by Jose Bessa (Centro Andaluz de Biología del Desarrollo, Universidad Pablo de Olavide, Carretera de Utrera Km1, 41013 Sevilla, Spain). We greatly appreciate the helpful suggestions provided by Valer Gotea, Laura Elnitski, and Tyra Wolfsberg. We also would like to thank the families who participated in this study. This research was supported, in part, by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (NIH). Some of the results were obtained with the program S.A.G.E., which was supported by grant 1 P41 RR03655 from the National Center for Research Resources. Grant funding was provided by the Scoliosis Research Society, the National Scoliosis Foundation, Scoliosis Association, Inc., the Institute de France Foundation Yves Cotrel, the Orthopedic Pediatric Society of North America, and the Orthopedic Research and Education Foundation, and NIH grant 1-R01-AR048862-01A1. The control samples were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core (http://DNASequencingCore.ucdenver.edu), which is supported by a NIH/National Cancer Institute Cancer Center Core Support Grant (P30 CA046934).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.029975/-/DC1
Communicating editor: V. Vieland
Literature Cited
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W. J., et al. , 2004. Ultraconserved elements in the human genome. Science 304(5675): 1321–1325. [DOI] [PubMed] [Google Scholar]
- Bergofsky E. H., 1979. Respiratory failure in disorders of the thoracic cage. Am. Rev. Respir. Dis. 119(4): 643–669. [DOI] [PubMed] [Google Scholar]
- Bessa J., Tena J. J., de la Calle-Mustienes E., Fernandez-Minan A., Naranjo S., et al. , 2009. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 238(9): 2409–2417. [DOI] [PubMed] [Google Scholar]
- Bosse A., Stoykova A., Nieselt-Struwe K., Chowdhury K., Copeland N. G., et al. , 2000. Identification of a novel mouse Iroquois homeobox gene, Irx5, and chromosomal localisation of all members of the mouse Iroquois gene family. Dev. Dyn. 218(1): 160–174. [DOI] [PubMed] [Google Scholar]
- Bunnell W. P., 1986. The natural history of idiopathic scoliosis before skeletal maturity. Spine 11(8): 773–776. [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25(21): 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F., Modolell J., Gomez-Skarmeta J. L., 2001. The Iroquois family of genes: from body building to neural patterning. Development 128(15): 2847–2855. [DOI] [PubMed] [Google Scholar]
- Chan V., Fong G. C., Luk K. D., Yip B., Lee M. K., et al. , 2002. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am. J. Hum. Genet. 71(2): 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E., Feijóo C. G., Manzanares M., Tena J. J., Rodríguez-Seguel E., et al. , 2005. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 15(8): 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P., Margaritte-Jeannin P., Biot B., Labalme A., Bernard J. C., et al. , 2011. New disease gene location and high genetic heterogeneity in idiopathic scoliosis. Eur. J. Hum. Genet. 19(8): 865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Gordon D., Zhang D., Browne R., Helms C., et al. , 2007. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am. J. Hum. Genet. 80(5): 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta J. L., Modolell J., 2002. Iroquois genes: genomic organization and function in vertebrate neural development. Curr. Opin. Genet. Dev. 12(4): 403–408. [DOI] [PubMed] [Google Scholar]
- Goode D. K., Snell P., Smith S. F., Cooke J. E., Elgar G., 2005. Highly conserved regulatory elements around the SHH gene may contribute to the maintenance of conserved synteny across human chromosome 7q36.3. Genomics 86(2): 172–181. [DOI] [PubMed] [Google Scholar]
- Justice C. M., Miller N. H., Marosy B., Zhang J., Wilson A. F., 2003. Familial idiopathic scoliosis: evidence of an X-linked susceptibility locus. Spine 28(6): 589–594. [DOI] [PubMed] [Google Scholar]
- Kou I., Takahashi Y., Johnson T. A., Takahashi A., Guo L., et al. , 2013. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat. Genet. 45(6): 676–679. [DOI] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., et al. , 2007. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236(11): 3088–3099. [DOI] [PubMed] [Google Scholar]
- Lonstein J. E., Carlson J. M., 1984. The prediction of curve progression in untreated idiopathic scoliosis during growth. J. Bone Joint Surg. Am. 66(7): 1061–1071. [PubMed] [Google Scholar]
- Machida M., Dubousset J., Imamura Y., Iwaya T., Yamada T., et al. , 1993. An experimental study in chickens for the pathogenesis of idiopathic scoliosis. Spine 18(12): 1609–1615. [DOI] [PubMed] [Google Scholar]
- McEwen G. K., Woolfe A., Goode D., Vavouri T., Callaway H., et al. , 2006. Ancient duplicated conserved noncoding elements in vertebrates: a genomic and functional analysis. Genome Res. 16(4): 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N. H., Justice C. M., Marosy B., Doheny K. F., Pugh E., et al. , 2005. Identification of candidate regions for familial idiopathic scoliosis. Spine 30(10): 1181–1187. [DOI] [PubMed] [Google Scholar]
- Miller N. H., Marosy B., Justice C. M., Novak S. M., Tang E. Y., et al. , 2006. Linkage analysis of genetic loci for kyphoscoliosis on chromosomes 5p13, 13q13.3, and 13q32. Am. J. Med. Genet. A. 140(10): 1059–1068. [DOI] [PubMed] [Google Scholar]
- Miyake A., Kou I., Takahashi Y., Johnson T. A., Ogura Y., et al. , 2013. Identification of a susceptibility locus for severe adolescent idiopathic scoliosis on chromosome 17q24.3. PLoS One 8(9): e72802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M. A., Ovcharenko I., Afzal V., Rubin E. M., 2003. Scanning human gene deserts for long-range enhancers. Science 302(5644): 413. [DOI] [PubMed] [Google Scholar]
- Ocaka L., Zhao C., Reed J. A., Ebenezer N. D., Brice G., et al. , 2008. Assignment of two loci for autosomal dominant adolescent idiopathic scoliosis to chromosomes 9q31.2-q34.2 and 17q25.3-qtel. J. Med. Genet. 45(2): 87–92. [DOI] [PubMed] [Google Scholar]
- O’Connell J. R., Weeks D. E., 1998. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63(1): 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K., Matsumoto K., Kuroiwa A., Isobe T., Otoguro T., et al. , 2001. Cloning and chromosome mapping of human and chicken Iroquois (IRX) genes. Cytogenet. Cell Genet. 92(3–4): 320–325. [DOI] [PubMed] [Google Scholar]
- Peters T., Dildrop R., Ausmeier K., Rüther U., 2000. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 10(10): 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggio C. L., Giampietro P. F., Dobrin S., Zhao C., Dorshorst D., et al. , 2009. A novel locus for adolescent idiopathic scoliosis on chromosome 12p. J. Orthop. Res. 27(10): 1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi L. B., Mangino M., De Serio S., De Cicco D., Capon F., et al. , 2002. Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Hum. Genet. 111(4–5): 401–404. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Bailey P., Bruce S., Engström P. G., Klos J. M., et al. , 2004. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5(1): 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Kou I., Takahashi A., Johnson T. A., Kono K., et al. , 2011. A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat. Genet. 43(12): 1237–1240. [DOI] [PubMed] [Google Scholar]
- Tena J. J., Alonso M. E., de la Calle-Mustienes E., Splinter E., de Laat W., et al. , 2011. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat. Commun. 2: 310. [DOI] [PubMed] [Google Scholar]
- Visel A., Minovitsky S., Dubchak I., Pennacchio L. A., 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35(Database issue): D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton J. E., Cutler D. J., Abecasis G. R., 2005. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 76(5): 887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise C. A., Barnes R., Gillum J., Herring J. A., Bowcock A. M., et al. , 2000. Localization of susceptibility to familial idiopathic scoliosis. Spine 25(18): 2372–2380. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., et al. , 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3(1): e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Tommerup N., Ming Wang S., Hansen L., 2006. A novel primate specific gene, CEI, is located in the homeobox gene IRXA2 promoter in Homo sapiens. Gene 371(2): 167–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.