Abstract
Introduction
MicroRNAs (miRNAs) are non-coding RNAs that regulate multiple cell processes during cancer progression. Renal cell carcinoma (RCC) is a malignancy with a poor prognosis. In this study, we aimed to investigate the roles of miR-630 in RCC progression.
Material and methods
Expression of miR-630 was evaluated by quantitative real-time polymerase chain reaction (qRT-PCR) in four renal cancer cell lines (786-O, ACHN, Caki-1, and Caki-2) and one normal human proximal tubule epithelial cell line (HK-2). Next, miR-630 inhibitor was used to inhibit miR-630 expression in 786-O cells. Finally, its effects on cell proliferation, apoptosis, migration, and invasion were evaluated.
Results
The expression level of miR-630 was higher in renal cancer cell lines 786-O, ACHN, Caki-1, and Caki-2 than that in the normal renal cell line HK-2 (p < 0.05). Furthermore, a proliferation assay, apoptosis assay, migration assay and invasion assay were performed, and the results showed that down-regulation of miR-630 expression by miR-630 inhibitor significantly inhibited cell proliferation, migration, and invasion, which meanwhile induced cell apoptosis of the renal cancer cell line 786-O.
Conclusions
This is the first time that miR-630 expression has been shown to be associated with renal cancer progression, and down-regulation of miR-630 can inhibit tumor progression, which provides a potential therapeutic target for renal cancer treatment.
Keywords: renal cell carcinoma, miR-630, proliferation, migration, invasion
Introduction
Renal cell carcinoma (RCC) is the most common solid cancer of adult kidney, accounting for approximately 90% of kidney neoplasms and 3% of all adult malignancies [1]. Among the subtypes of RCC, clear cell renal cell carcinoma (ccRCC) is the most abundant (∼75%) and was over-represented (∼90%) in a series of metastatic RCC [2]. Renal cell carcinoma responds poorly to chemotherapy and radiotherapy, and surgery remains the only curative treatment [3]. Early detection is of great importance for patient outcome. The 5-year survival for patients diagnosed with organ-confined disease is approximately 90%, whereas the prognosis of patients with distant metastasis remains poor, with a 5-year survival rate of less than 10% [4, 5]. At present, the treatment of RCC, especially metastatic RCC, remains a big challenge. Therefore, identification of specific and effective therapeutic targets for RCC would be of great value.
MicroRNA (miRNA) is a short non-coding RNA of 18–25 nucleotides that regulates gene expression post-transcriptionally [6]. It has been predicted that as much as 30% of the human genome is regulated by miRNAs, and each miRNA can regulate the translation of hundreds of target mRNAs [7, 8]. Many miRNAs are abnormally expressed in different types of clinical cancer and play important regulatory roles in a variety of biological processes, such as cell proliferation, apoptosis, differentiation, initiation, and metastasis [9, 10]. Some highly expressed miRNAs act as oncogenes, whereas low level miRNAs could act as tumor suppressors by negatively regulating oncogenes [11]. Among them, miR-630 is one of the most frequently studied miRNAs in cancer biology. Up-regulation of miR-630 has been reported extensively in human cancers, suggesting that miR-630 may function as an oncogene in a variety of tumors such as gastric cancer [12]. Our previous study successfully identified that miR-630 was significantly up-regulated in RCC, and miR-630 was associated with RCC histologic grade, lymph node metastasis, and distant metastasis [13]. However, the biological roles and underlying mechanism of miR-630 in RCC remain unclear. In this study, we investigated the roles of miR-630 in the proliferation, apoptosis, migration and invasion of renal cancer cells 786-O.
Material and methods
Cell culture and treatment with reagents
Human renal cancer cell lines 786-O, ACHN, Caki-1, and Caki-2 were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (CCCAS, China). Immortalized normal human proximal tubule epithelial cell line HK-2 was purchased from the American Type Culture Collection (ATCC, USA). HK-2 cells were cultured in KSFM medium (Gibco), and other cells were cultured in RPMI-1640 medium (HyClone) with 10% fetal bovine serum (FBS, Gibco), 50 U/ml of penicillin and 50 µg/ml of streptomycin. All cells were cultured in a sterile incubator maintained at 37°C in 5% CO2.
786-O cells were transfected with miR-630 inhibitor or miRNA inhibitor negative control (Invitrogen) according to the reagent protocol. The final concentration was 30 nM and cells were transfected using Lipofectamine 2000 (Invitrogen) as described before [13].
Cell proliferation assay
Renal cancer cells 786-O were transfected with miR-630 inhibitor or miRNA inhibitor negative control for 48 h, and then were reseeded into 96-well plates. Cell density was adjusted to 5 × 103/well, and the final volume was 150 µl/well. MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) solution (20 µl) was added to the plates 24, 48, 72, and 96 h later. The cells were cultured for 4 h at 37°C. Then, the medium was discarded and 150 µl of DMSO was added and oscillated for 15 min. Optical density (OD) was detected at a wavelength of 490 nm using an enzyme-labeled analyzer. Three independent experiments (triplicate in each) were performed.
Cell apoptosis assay
Renal cancer cells 786-O were cultured in 6-well plates (1 × 106 cells/well) in antibiotic-free medium, after being transfected with miR-630 inhibitor or miRNA inhibitor negative control for 48 h. Cells for apoptosis analysis were collected, washed twice, and stained with FITC-Annexin V and PI, using the FITC-Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's manual. Apoptotic cells were analyzed using a FACSCalibur flow cytometer (BD Bioscience). Three independent experiments were performed.
Wound healing assay
To determine cell migration, 786-O cells transfected with miR-630 inhibitor or miRNA inhibitor negative control were seeded in 12-well plates, incubated in their respective complete culture medium, and grown to confluence overnight. Wounds were made with a sterile white pipette tip, and the debris was washed three times with PBS and then cells were cultured. The speed of wound closure was monitored and photographed at 0 and 48 h. Three independent experiments were performed.
Transwell invasion assay
The cell invasion assays were performed in a 24-well transwell chamber, which was precoated with 100 µg of Matrigel. 786-O cells transfected with miR-630 inhibitor or the miRNA inhibitor negative control in each group were collected and re-suspended in serum-free medium at a concentration of 1 × 105 cells/ml. Then, 200 µl cell suspensions were added into the upper chamber, and the bottom chamber was filled with 500 µl of culture medium containing 10% FBS. After incubation for 48 h at 37°C, 5% CO2, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed and stained using 0.1% crystal violet. The numbers of invaded cells were counted in five randomly selected fields under a microscope (Olympus).
RNA isolation and qRT-PCR
Total RNA was isolated from cells using TRIZOL reagent according to the manufacturer's protocol (Invitrogen). RNA was reverse transcribed using SuperScript First Strand cDNA System (Invitrogen) according to the manufacturer's instructions. The PCR amplification was performed for 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, on an Applied Biosystems 7900HT (Applied Biosystems) with 1.0 µl of cDNA and SYBR Green real-time PCR Master Mix (Takara). Data were collected and analyzed by SDS2.3 software (Applied Biosystems). The expression level of miR-630 was internally normalized against that of RNU44. The relative value was expressed by the 2–ΔΔCt method. Each experiment was performed in triplicate and repeated three times.
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 software (IBM, Chicago). One-way analysis of variance (ANOVA) and Student's t test were used as appropriate [14], and values of p < 0.05 were considered statistically significant.
Results
miR-630 shows high expression in renal cancer cell lines
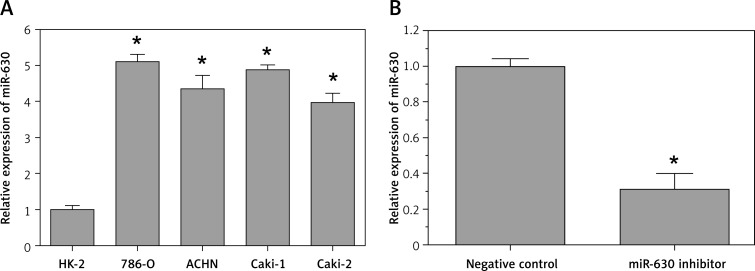
The qRT-PCR results showed that the expression level of miR-630 in four renal cancer cell lines (786-O, ACHN, Caki-1, and Caki-2) was at least 4-fold higher than that in a normal human proximal tubule epithelial cell line (HK-2) (p < 0.05, Figure 1 A).
Figure 1.
Expression of miR-630 in renal cancer cell lines. The relative miR-630 expression levels were determined using qRT-PCR. A – Expression of miR-630 was significantly higher in renal cancer cell lines (786-O, ACHN, Caki-1 and Caki-2) than that in the non-malignant renal cell line HK-2. B – miR-630 inhibitor significantly decreased miR-630 expression
Results are expressed as mean ± SD for three replicate determinations; *p < 0.05.
In addition, when 786-O cells were transfected with miR-630 inhibitor, the expression of miR-630 was significantly decreased compared with the miRNA inhibitor negative control group (p < 0.05, Figure 1 B). The results indicated that the expression of miR-630 in renal cancer cells 786-O was successfully knocked down.
Effect of miR-630 on cell proliferation
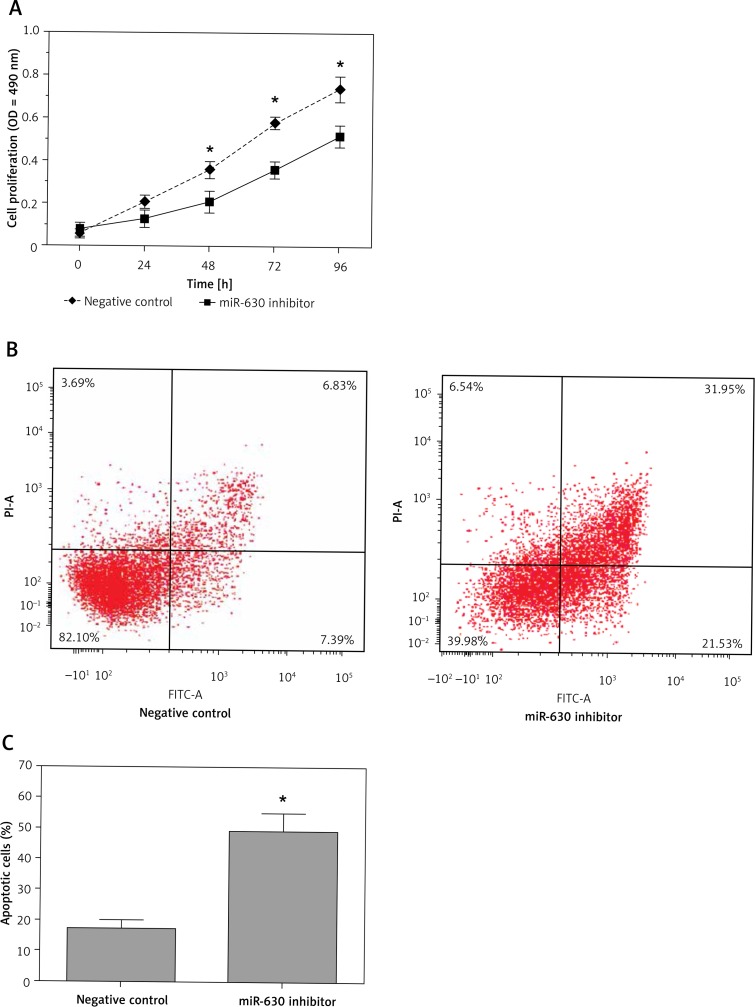
The MTT assay showed that down-regulation of miR-630 significantly inhibited the proliferation of 786-O cells compared with the miRNA inhibitor negative control group (p < 0.05, Figure 2 A), which indicated that knockdown of expression of miR-630 had an inhibitory effect on the proliferation of renal cancer cells. Next we explored the potential mechanisms of cell proliferation inhibition by miR-630 inhibitor. The apoptosis assay showed that 786-O cells transfected with miR-630 inhibitor had an increased apoptosis rate, which increased from 17.3% (inhibitor negative control group) to 49.2% (miR-630 inhibitor group) (p < 0.05, Figures 2 B, C). Our results demonstrated that down-regulation of miR-630 had an inhibitory effect on the growth of renal cancer cells by increasing cell apoptosis.
Figure 2.
Down-regulation of miR-630 inhibited cell proliferation of 786-O cells. A – MTT assay showed that 786-O cells transfected with miR-630 inhibitor displayed significantly lower proliferation ability compared with the inhibitor negative control group. B, C – Cell apoptosis assay showed that 786-O cells transfected with miR-630 inhibitor displayed a significantly higher apoptotic rate compared with the inhibitor negative control group
Results are expressed as mean ± SD for three replicate determinations; *p < 0.05.
Effect of miR-630 on cell migration and invasion
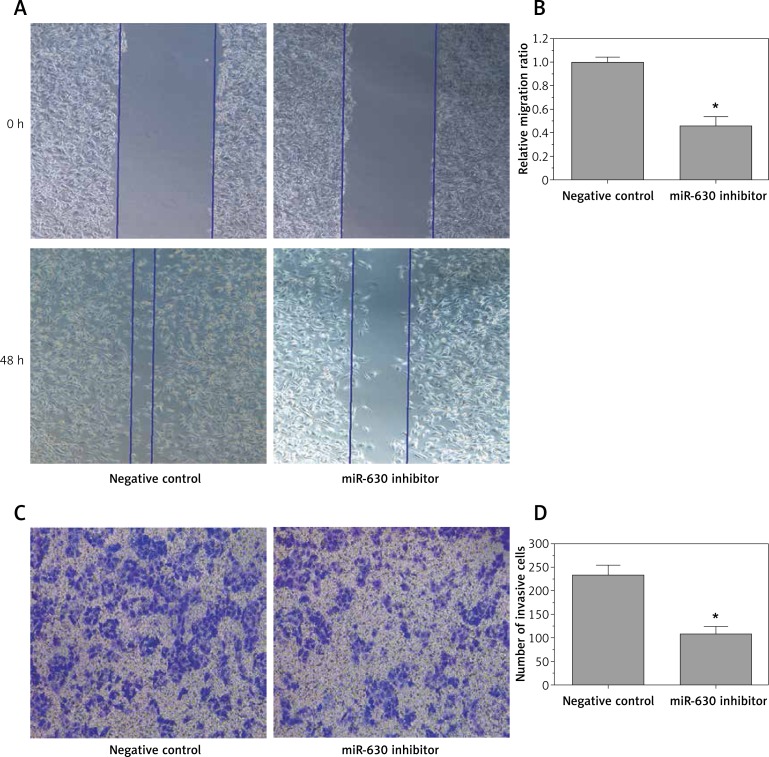
The wound healing assay showed that the migratory rate of 786-O cells transfected with miR-630 inhibitor was significantly decreased compared with the miRNA inhibitor negative control group (0.46 ±0.08 vs. 1.00 ±0.04) (p < 0.05, Figures 3 A, B). The transwell invasion assay indicated that the invasion of 786-O cells transfected with miR-630 inhibitor was significantly decreased compared with the miRNA inhibitor negative control group (108 ±16 vs. 233 ±21) (p < 0.05, Figures 3 C, D). Our study indicated that down-regulation of miR-630 could resist the migration and invasion ability of 786-O cells in vitro and the up-regulation of miR-630 may be a cause of renal cancer with high migratory and invasiveness.
Figure 3.
Down-regulation of miR-630 inhibited cell migration and invasion of 786-O cells. A, B – Wound healing assay showed that 786-O cells transfected with miR-630 inhibitor displayed significantly lower migration capacity compared with the inhibitor negative control group. C, D – Transwell invasion assay showed that 786-O cells transfected with miR-630 inhibitor displayed significantly lower invasion capacity compared with the inhibitor negative control group
Results are expressed as means ± SD for three replicate determinations; *p < 0.05.
Discussion
Renal cell carcinoma (RCC) is the most lethal genitourinary tumor in the world. Currently, some progress in clinical and basic research of RCC has been made, but the molecular genetic mechanism involved in RCC is still unclear [15]. Moreover, objective and reliable diagnostic biomarkers and effective targeted therapeutic agents are also lacking.
Previous studies have shown that more than 1,900 human miRNAs have been identified, which may regulate over 60% of genes in mammals [16]. Due to their great importance in the regulation of gene expression, it has been widely accepted that miRNAs are involved in maintaining normal cellular physiology. The dysregulation of miRNA expression has been associated with cancer initiation and progression by regulating expression of tumor suppressors and oncogenes [17].
Limited studies to date have reported on the role of miR-630 in cancer. Kuo et al. found that miR-630 was decreased in lung cancer and inversely correlated with advanced stage, higher lymph node metastasis, higher grade invasion, poor overall survival, and poor disease-free survival in lung cancer [18]. However, Chu et al. showed that miR-630 expression level was significantly elevated in gastric cancer and associated with gastric cancer invasion, lymph node metastasis, distant metastasis, and TNM stage [12]. In our previous study, we also found that miR-630 was significantly higher in renal cancer and associated with renal cancer histologic grade, lymph node metastasis, distant metastasis and TNM stage, which indicated that miR-630 may play an important role in development and progression of renal cancer [13]. To our knowledge, the function of miR-630 in renal cancer carcinogenesis is still not clear.
Huang et al. found that induced miR-630 expression reduced levels of anti-apoptotic genes, whereas inhibition of miR-630 in head and neck squamous cell carcinoma cells up-regulated these genes and was associated with increased cell survival [19]. Galluzzi et al. observed that up-regulation of miR-630 expression could arrest A549 cells in the G0-G1 phase, and reduced cell proliferation [20]. Farhana et al. reported that up-regulation of miR-630 in pancreatic cancer cells could induce apoptosis by targeting IGF-1R [21]. Corcoran et al. found that inhibition of miR-630 could induce cellular motility by increase of migration and invasion ability in breast cancer cells, Conversely, up-regulation of miR-630 expression in breast cancer cells could decrease the cellular motility [22]. However, since individual miRNAs in different cancers may have a large number of different gene targets, they have different functions in various malignancies. Thus, the functional discovery of individual miRNAs may allow a deeper insight into regulation of gene expression and complexity of cancer progression. Chu's study indicated that miR-630 expression was increased in gastric cancer cells and has an oncogenic role in gastric cancer [12]. In the present study, we investigated miR-630 mRNA expression by qRT-PCR assay in four renal cancer cell lines (786-O, ACHN, Caki-1 and Caki-2) and one normal human proximal tubule epithelial cell line (HK-2). The results showed that miR-630 expression was increased in renal cancer cell lines compared with that in the HK-2 cell line. To further understand the function of miR-630 in renal cancer, we knocked down miR-630 expression by miR-630 inhibitor on 786-O cells, and found that decrease of miR-630 expression in 786-O cells could significantly inhibit renal cancer cell proliferation and induce cell apoptosis. Next, we explored the possible effect of migration and invasion in 786-O cells after inhibition of miR-630. We found that down-regulation of miR-630 could significantly inhibit the migration and invasion capacity of renal cancer 786-O cells. These results suggest that miR-630 acts as an oncogene in renal cancer.
In conclusion, this study is the first to examine the function of miR-630 in renal cancer tumorigenesis and progression. Our work suggests that blocking miR-630 activity in renal cancer is a potential novel therapeutic approach. miR-630 is involved in many areas of tumor progression, including cell proliferation, apoptosis, migration and invasion. Intervention of miR-630 function by agents may have potential therapeutic value in the prevention of renal cancer. Thus, our findings may not only provide a molecular basis for the role of miR-630 in renal cancer but also suggest a novel therapeutic target for the treatment of renal cancer.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ljungberg B, Campbell SC, Cho HY, et al. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615–21. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B, Cowan NC, Hanbury DC, et al. EAU guidelines on renal cell carcinoma: the 2010 update. Eur Urol. 2010;58:398–406. doi: 10.1016/j.eururo.2010.06.032. [DOI] [PubMed] [Google Scholar]
- 3.Chow WH, Devesa SS, Warren JL, et al. Rising incidence of renal cell cancer in the United States. JAMA. 1999;281:1628–31. doi: 10.1001/jama.281.17.1628. [DOI] [PubMed] [Google Scholar]
- 4.Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med. 1996;335:865–75. doi: 10.1056/NEJM199609193351207. [DOI] [PubMed] [Google Scholar]
- 5.Novara G, Ficarra V, Antonelli A, et al. Validation of the 2009 TNM version in a large multi-institutional cohort of patients treated for renal cell carcinoma: are further improvements needed? Eur Urol. 2010;58:588–95. doi: 10.1016/j.eururo.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–97. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Baek D, Villén J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol. 2011;223:102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catto JWF, Alcaraz A, Bjartell AS, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–81. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Liu M, Xu YF, et al. Combination of quercetin and hyperoside has anticancer effects on renal cancer cells through inhibition of oncogenic microRNA-27a. Oncol Rep. 2014;31:117–24. doi: 10.3892/or.2013.2811. [DOI] [PubMed] [Google Scholar]
- 12.Chu D, Zhao Z, Li Y, et al. Increased MicroRNA-630 expression in gastric cancer is associated with poor overall survival. PloS One. 2014;9:e90526. doi: 10.1371/journal.pone.0090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao JJ, Chen PJ, Duan RQ, et al. Up-regulation of miR-630 in clear cell renal cell carcinoma is associated with lower overall survival. Int J Clin Exp Pathol. 2014;7:3318–23. [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Liu Y, Gui Y, et al. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–9. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 15.Teh BT, Yang XJ, Teh BS, et al. Renal cell carcinoma[M]//Encyclopedia of Molecular Mechanisms of Disease. Berlin Heidelberg: Springer; 2009. pp. 1822–3. [Google Scholar]
- 16.Grange C, Collino F, Tapparo M, et al. Oncogenic micro-RNAs and renal cell carcinoma. Front Oncol. 2014;4:49. doi: 10.3389/fonc.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–84. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 18.Kuo TC, Tan CT, Chang YW, et al. Angiopoietin-like protein 1 suppresses SLUG to inhibit cancer cell motility. J Clin Investig. 2013;123:1082–95. doi: 10.1172/JCI64044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Chuang A, Hao H, et al. Phospho-deltaNp63alpha is a key regulator of the cisplatin-induced microRNAome in cancer cells. Cell Death Differ. 2011;18:1220–30. doi: 10.1038/cdd.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galluzzi L, Morselli E, Vitale I, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 21.Farhana L, Dawson MI, Murshed F, et al. Upregulation of miR-150* and miR-630 induces apoptosis in pancreatic cancer cells by targeting IGF-1R. PloS One. 2013;8:e61015. doi: 10.1371/journal.pone.0061015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corcoran C, Rani S, Breslin S, et al. miR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol Cancer. 2014;13:71. doi: 10.1186/1476-4598-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]