Abstract
Despite the recommendation for the use of merozoite surface protein 1 (msp1), merozoite surface protein 2 (msp2), and glutamate-rich protein (glurp) genes as markers in drug efficacy studies by World Health Organization and their limited use in Bangladesh, the circulating Plasmodium falciparum population genetic structure has not yet been assessed in Bangladesh. This study presents a comprehensive report on the circulating P. falciparum population structure based on msp1, msp2, and glurp polymorphic gene markers in Bangladesh. Among the 130 pretreatment (day 0) P. falciparum samples from seven malaria-endemic districts, 14 distinct genotypes were observed for msp1, 20 for msp2, and 13 for glurp. Polyclonal infection was reported in 94.6% (N = 123) of the samples. Multiplicity of infection (MOI) for msp1 was the highest (1.5) among the MOIs of the markers. The heterozygosity for msp1, msp2, and glurp was 0.89, 0.93, and 0.83, respectively. Data according to different malaria-endemic areas are also presented and discussed. Bangladesh is considered as a malaria-hypoendemic country. However, the prevalence of polyclonal infection and the genetic diversity of P. falciparum do not represent hypoendemicity.
Introduction
Globally 198 million people, 90% of whom are from Africa, were afflicted by malaria in 2013 resulting in 584,000 deaths.1 Among the 10 malaria-endemic countries of the World Health Organization (WHO) southeast Asian region, Bangladesh is considered as hypoendemic for malaria,2 and 90% of malaria is caused by Plasmodium falciparum. Of the 64 administrative districts of Bangladesh, 13 districts bordering India and Myanmar are malaria endemic, and of these 13, almost 80% of the malaria cases are reported from Bandarban, Khagrachari, and Rangamati (collectively called Chittagong hill tracts [CHT]).3 However, from 2009 to 2013, a > 68.2% decrease in malaria incidences has been reported in Bangladesh as it declined from 84,690 confirmed malaria cases in 2008 to 26,891 confirmed malaria cases in 2013.4
It is noted that virulence of a parasite attenuates with evolution; however, this is not applicable for P. falciparum.5 Genetic diversity of the parasite arises during adjustment with different environmental and immunological factors such as drugs, host immune systems, and transmission intensity, and it contributes to the parasite's virulence.6,7
Several P. falciparum genes have been found to show extensive genetic polymorphism. This phenomenon has been exploited for assessing the genetic diversity and population dynamics. High polymorphism in merozoite surface protein 1 (msp1), merozoite surface protein 2 (msp2), and glutamate-rich protein (glurp) genes have been shown in genetic diversity studies in different malaria-endemic countries from Asia to Africa.7–17 Large allelic polymorphism has been reported in the block 2 of the msp1, the central repetitive domain of the msp2, and region II of the glurp. In msp1, three distinct allelic families have been described: K1, MAD20, and RO33 while in msp2, two allelic families, FC27 and 3D7/IC, have been described.8,13,18 These markers have been used to distinguish recrudescence from new infections in various drug trial studies7,11,12 including studies from CHT areas of Bangladesh.19,20 Though, msp1, msp2, and glurp have been used in a limited approach in drug resistance studies, the parasite population genetic profile has not been assessed systematically in Bangladesh yet.
Understanding of the genetic diversity within the circulating population of the parasite can aid in designing successful malaria control and prevention strategy because of its effect on pathological and immunological mechanisms, drug efficacy, and disease transmission.21 Therefore, the purpose of the study was to reveal the genetic diversity in the circulating P. falciparum population based on the polymorphism of msp1, msp2, and glurp genes in Bangladesh.
Methods
Study samples.
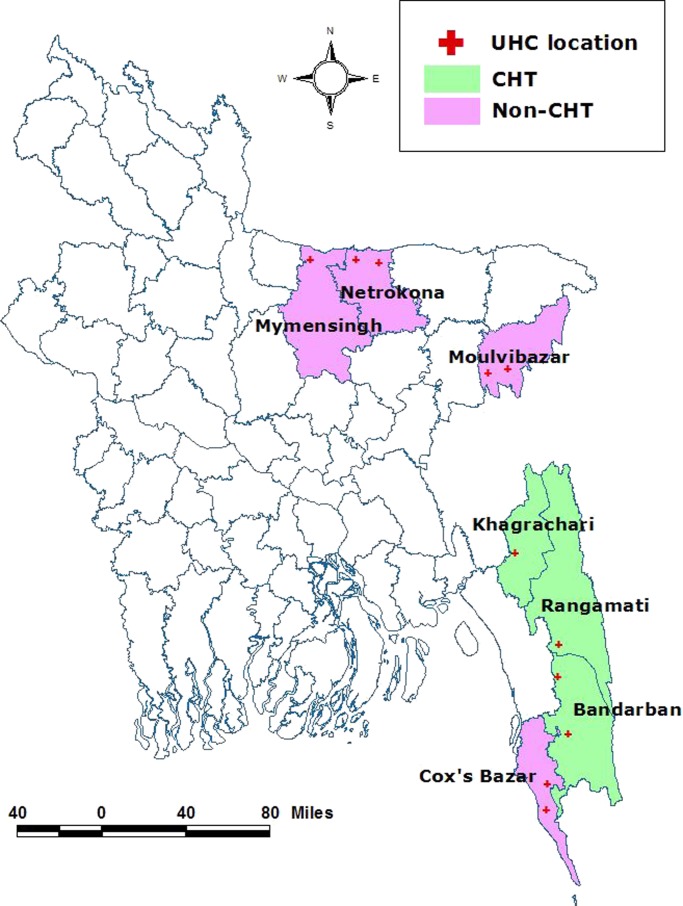
Whole blood samples obtained from febrile patients with clinical symptoms were collected from corresponding upazila (subdistrict) health complexes of seven malaria-endemic districts in Bangladesh: Bandarban, Rangamati, Khagrachari, Netrokona, Mymensingh, Moulvibazar, and Cox's Bazar (Figure 1 ). Of these seven districts, three were from CHT region (Bandarban, Rangamati, and Khagrachari) referred to as CHT areas, and four were from other endemic areas (Netrokona, Mymensingh, Moulvibazar, and Cox's Bazar), referred to as non-CHT areas. A total of 130 (69 from CHT areas and 61 from non-CHT areas) pretreatment (day 0) P. falciparum monoinfected samples confirmed by microscopy and nested polymerase chain reaction (PCR) and/or real-time PCR were selected for this study. All samples were taken from a study that has been described elsewhere.22 Nested-PCR and real-time PCR were done as described previously.23,24 The study was approved by the Institutional Ethics Review Committee of the International Center for Diarrhoeal Disease Research, Bangladesh. All participants or legal guardians signed informed consent before participant enrollment and sample collection. Complete anonymity was maintained at each stage of the study.
Figure 1.
Map of the study location. UHC = upazila (subdistrict) health complexes.
Genotyping PCR.
Extracted DNA from whole blood was used as the template for the analysis of the block 2 of msp1 (K1, MAD20, and RO33 allelic families), central polymorphic region of msp2 (FC27 and 3D7/IC allelic families), and region II of glurp. Nested-PCR-based amplification of the DNA and genotype analysis were performed following the protocol as previously described13 with modifications as described below. The positive controls for these three genes were obtained from the Swiss Tropical and Public Health Institute (Basel, Switzerland). The positive control samples were provided in FTA card from in vitro cultures, and DNA was isolated in the laboratory. DNA from whole blood and FTA (Whatman Inc., Cliffin, NJ) card was extracted using Qiagen Blood Mini Kit (Qiagen, Valencia, CA). All PCR amplifications were performed on a MyCycler thermal cycler (BioRad Laboratories, Inc., Hercules, CA). All the amplification reactions were carried out in 25 μL volume.
For the initial amplification reaction for msp1 and msp2, the reaction mixture contained 300 nM (for msp1) or 120 nM (for msp2) of each primer, 200 μM each of four deoxyribonucleotide triphosphates (Invitrogen™, Life Technologies, Carlsbad, CA), 3.5 mM MgCl2, 1× bovine serum albumin (BSA), and 1.25 U of Taq DNA Polymerase (New England Biolabs, Inc., Ipswich, MA). In the amplification reaction, 2 μL of DNA was used as template. The amplified product from the initial reaction was diluted six times with nuclease-free water, and 2 μL of the diluted product was used as template for the nested round of reaction for both msp1 and msp2. The nested round reaction mixture contained 300 nM (msp1) or 120 nM (msp2) of each allelic family specific primer, 200 μM each of four dNTPs, 3.5 mM MgCl2, 1× BSA, and 0.75 U (msp1) or 0.8 U (msp2) of Taq DNA Polymerase. The cycling conditions for both initial and nested-PCR were an initial denaturation at 95°C for 3 minutes followed by 35 cycles of denaturation at 95°C for 1 minute, annealing at 54°C (initial amplification) or 61°C (for nested) for 1 minute, extension at 68°C for 1 minute with a final extension at 68°C for 5 minutes.
For glurp, in the initial amplification of the semi-nested PCR, 2 μL of purified DNA was amplified with 100 nM of each primer, 150 μM each of four dNTPs, 3.5 μM MgCl2, 1× BSA, and 0.8 U of Taq DNA Polymerase. The product from the first step of the PCR was diluted five times with deionized water, and 1 μL of the diluted product was used for the second step. The second step of the PCR was done with 100 nM of each primer, 200 μM each of four dNTPs, 3.5 μM MgCl2, 1× BSA, and 0.8 U of Taq DNA Polymerase. The cycle conditions for the initial and semi-nested PCR amplification were an initial denaturation of 3 minutes at 95°C followed by 35 cycles of denaturation at 95°C for 1 minute, annealing at 60°C for 1 minute, extension at 68°C for 2 minutes with final extension at 68°C for 5 minutes. All the nested or semi-nested round reactions were done in duplicate for each sample, for each marker.
Allele detection and estimation of molecular weight.
The amplified nested-PCR products for the three genes were either stored at +4°C or analyzed immediately after PCR by electrophoresis on a 1.5% ethidium bromide–stained agarose gel and visualized under ultraviolet transillumination (GelDoc®; BioRad Laboratories, Inc.). Band sizing and molecular weights were estimated against appropriate DNA ladder marker for unique bands by visual inspection. Then the fragments were grouped in to 25 bp (for msp1 and msp2) and 50 bp (for glurp) “bins.” All fragments falling within the range of the bin were considered as same genotype.
Mean multiplicity of infection and expected heterozygosity.
The mean multiplicity of infection (MOI), a quantitative variable, was calculated as the quotient of the total number of genotypes for each marker and the number of PCR-positive samples. MOI for CHT and non-CHT regions, as well as the MOI for the country, was calculated. The expected heterozygosity (HE), a measure for genetic diversity, was calculated by using the formula: HE = [n/(n − 1)] [(1 − ΣPi2)], where n is the number of isolates analyzed and Pi the frequency of the ith allele in the population.12 The HE values estimate the ratio of all parasites that would be heterozygous for any of the three marker genes.
Statistical analysis.
The proportion of alleles observed at each genetic locus within each group (CHT and non-CHT) was compared using two-sample independent proportion test. The data analysis was conducted in STATA version 13 (StataCorp LP, College Station, TX). A P value of < 0.05 was considered significant.
Results
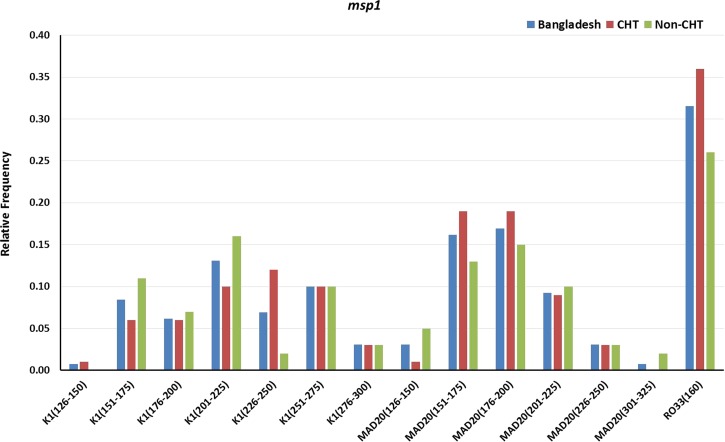
Of 130 samples, 111 (85.4%) were successfully amplified for msp1. Fourteen msp1 genotypes were observed in Bangladesh, representing K1 (seven genotypes), MAD20 (six genotypes), and RO33 (one genotype) allelic families. A total of 168 clones were observed of sizes ranging from 126 to 375 bp where, 93 (55.4%) were from CHT areas and the remaining 75 (44.6%) were from non-CHT areas. Figure 2 shows the relative frequency of the msp1 genotypes by areas. For K1 allelic family, clones of 201–225 bp genotype had the highest (0.13) relative frequency in Bangladesh, and for MAD20, the highest (0.17) relative frequency was observed in 176–200 bp genotype. The RO33 allelic family being monomorphic showed only one genotype of size 160 bp.
Figure 2.
Genetic diversity and frequency of genotypes of merozoite surface protein 1 (msp1) gene in general and in Chittagong hill tracts (CHT) and non-CHT areas in Bangladesh.
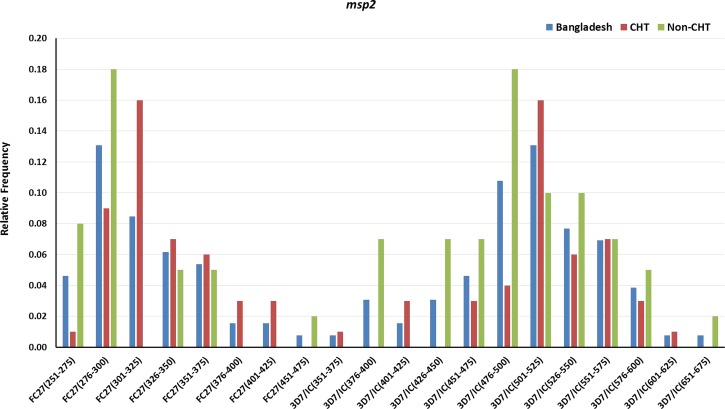
For msp2, 102 (78.5%) of 130 samples were successfully amplified. Of the 69 samples from the CHT area 51 (73.9%) and 51 (83.6%) of the 61 non-CHT samples contained msp2 clones. A total of 128 clones of 20 msp2 genotypes (eight of FC27 allelic family and 12 of 3D7/IC allelic family) were observed from Bangladesh with molecular size ranging from 251 to 675 bp. Of these 128 clones, 62 (48.4%) were from CHT areas and the remaining 66 (51.6%) were from non-CHT areas. Figure 3 shows the relative frequency of the msp2 genotypes by areas. For the FC27 allelic family, clones of 276–300 bp genotype had the highest relative frequency (0.13), whereas the highest (0.13) relative frequency of clones for 3D7/IC were observed for the 501–525 bp genotype in Bangladesh.
Figure 3.
Genetic diversity and frequency of genotypes of merozoite surface protein 2 (msp2) gene in general and in Chittagong hill tracts (CHT) and non-CHT areas in Bangladesh.
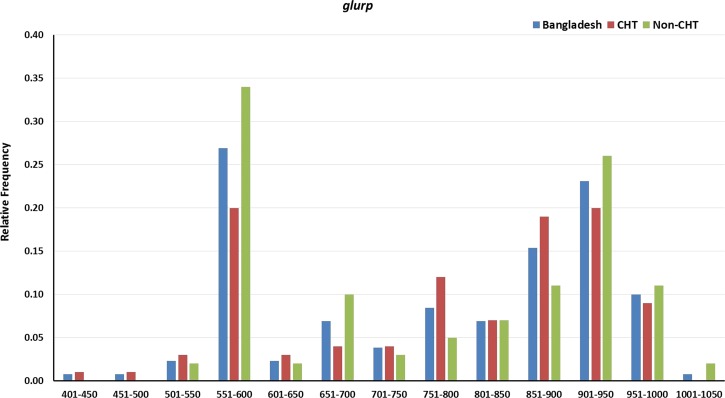
All the samples were amplified for glurp. A total of 141 clones belonging to 13 genotypes with sizes ranging from 401 to 1,050 bp were observed in Bangladesh. Of the 141 clones, 72 (51.1%) were from CHT areas and remaining 69 (48.9%) were from non-CHT areas. Relative frequencies of glurp genotypes are in Figure 4 . The highest (0.27) relative frequency of clones for glurp was observed for 551–600 bp genotype in Bangladesh.
Figure 4.
Genetic diversity and frequency of genotypes of glutamate-rich protein (glurp) gene in general and in Chittagong hill tracts (CHT) and non-CHT areas in Bangladesh.
Among the samples, 94.6% (N = 123; 97.1% in CHT and 91.8% in non-CHT) were polyclonal (i.e., harbored two or more clones). Of the 111 msp1-positive samples, 40.5% (N = 45; 42.4% in CHT and 38.5% in non-CHT) samples were polyclonal, whereas of the 102 msp2-positive samples, 21.6% (N = 22; 21.6% in CHT and 21.2% in non-CHT) samples harbored two or more clones. Among the glurp-positive samples, 8.5% (N = 11; 4.4% in CHT and 13.1% in non-CHT) samples contained two or more clones.
Two-sample independent proportion test to compare CHT and non-CHT showed that there was no significant difference (P > 0.05) in the distribution for clones of K1, MAD20, and RO33 allelic families of msp1, FC27 allelic family of msp2, and glurp. However, significant difference (P < 0.05) in the distribution for clones of 3D7/IC allelic family of msp2 between CHT and non-CHT has been observed.
The calculated mean MOI using the data from each of the three markers are presented in Table 1. The table also shows generally very high HE values. MOI for msp1 was the highest (1.51) among the MOIs of the markers, while msp2 showed the highest (0.93) HE value in Bangladesh.
Table 1.
Mean MOI and HE
| Area | msp1 | msp2 | glurp | |||
|---|---|---|---|---|---|---|
| MOI | HE | MOI | HE | MOI | HE | |
| CHT | 1.58 | 0.76 | 1.22 | 0.93 | 1.04 | 0.86 |
| Non-CHT | 1.44 | 0.84 | 1.29 | 0.90 | 1.11 | 0.78 |
| Bangladesh | 1.51 | 0.80 | 1.25 | 0.93 | 1.08 | 0.83 |
CHI = Chittagong hill tracts; HE = expected heterozygosity; glurp = glutamate-rich protein; MOI = multiplicity of infection; msp1 = protein merozoite surface protein 1; msp2 = merozoite surface protein 2.
Discussion
For P. falciparum, the polymorphism of msp1, msp2, and glurp have been in use for parasite genotyping in antimalarial drug trials and efficacy studies as a recommended marker by the WHO, and it has been in use for population diversity studies in different malaria-endemic regions.7–17 Since treatment with drugs of malaria patients is an integral part of malaria control and prevention strategy and the selected markers can be used as drug efficacy markers, knowing the baseline population structure of circulating parasites in an endemic country will aid in monitoring change in transmission pattern as well as efficacy of drugs that are in use. In this study, pretreatment patients were enrolled and genotyped to know parasite population structure and provide a baseline data for future reference.
This study showed that in Bangladesh msp2 has more genotypes (20) than msp1 and glurp. In CHT areas, higher genotype variation was reported for msp2 and glurp compared with the non-CHT areas, while the number of genotypes was the same for msp1 in both areas. K1 was the predominant allelic family of msp1 and 3D7/IC for msp2. These findings are similar to the previous reports from Pakistan, India, Yemen, and sub-Saharan African countries.10,11,16,25 A topography-based variation in the distribution of msp1, msp2, and glurp allelic families within the same area was described in previous studies,8,10,16 which corroborates the finding of variations in allelic families in CHT and non-CHT areas in this study. Though the findings are corroborated by previous studies, in this study no significant differences were observed in the distribution of clones between the CHT and the non-CHT sample groups except for clones of 3D7/IC allelic family of msp2. This may reflect the low number of samples that were assessed in the study. The K1 allelic family was observed to be predominant in a previous drug trial study in the CHT areas of Bangladesh where msp1 and glurp were used as markers.19
Since polyclonal infection can enhance the crossing between gametes of different clones to generate novel genotypes by meiotic recombination during sexual reproduction in mosquitoes,5,6,18,26 prevalence of polyclonal infection was evaluated in this study, which was 94.6%. Usually, the prevalence of polyclonal infections ranges from 20% to 30% for areas with hypoendemicity, 50% in hypo/mesoendemic areas to almost 100% in some holoendemic areas.25
A positive association between MOI and endemicity of P. falciparum has been reported in several studies,8,9,11,12,17,23 although some exceptions where high MOI was observed in hypoendemic areas are known.14,19 In the present study, MOI was 1.51, 1.25, and 1.08 for msp1, msp2, and glurp, respectively. In southeast Asia, MOI of P. falciparum for hypoendemic regions ranges from 1.00 to 2.70.9,16,17
A high level of heterozygosity (0.80–0.93) for all the three markers in Bangladesh was recorded in this study. This finding is higher than the heterozygosity range (0.51–0.65) reported earlier in the southeast Asia/Pacific region but quite comparable to some malaria-hyperendemic African countries (0.76–0.98).8,11,12,27 It is considered that in areas with decreasing malaria endemicity, the heterozygosity of P. falciparum genotypes will also decrease,8,27,28 but this is not necessarily true for a malaria-hypoendemic country like Bangladesh with a declining trend in malaria incidence,1 rather heterozygosity represents diversity of a hyperendemic circulating parasite population in Bangladesh. Diversity in the parasite population might be an effect of several factors. Administration of antimalarial drugs may force the local parasite population toward a new resistant phenotype by mutation in certain genes,6 such mutation has been reported from two of the samples of this study.22 Since the study areas are populated by people of different ethnic groups,2 this might present different host immune systems for the parasite to withstand, which could generate high genetic diversity.6
Despite WHO recommendations for the use of msp1, msp2, and glurp as markers in drug efficacy studies29 and their limited use in the drug efficacy studies in CHT areas of Bangladesh,19,20 the circulating P. falciparum population genetic structure is still unknown. The present study, to the best of our knowledge, is the first comprehensive report of the circulating P. falciparum population genetic diversity in Bangladesh. Malaria caused by P. falciparum in Bangladesh is mostly due to polyclonal infection with high genetic diversity, which do not represent hypoendemicity.
In the current national malaria strategic plan of Bangladesh, there is no policy to address asymptomatic malaria cases. Therefore, the inferences that are made about population-wide genetic heterogeneity only based on the symptomatic cases may not represent the actual P. falciparum population structure in Bangladesh.
ACKNOWLEDGMENTS
We appreciate the valuable contribution of Gulam Musawwir Khan, Shariar Mustafa, Khaja Md. Mohiuddin, Khalid Eakbal Anik, Md. Imtiaz Khalil, and Sultan Mahmud in this study. We also acknowledge the Malaria Research and Reference Reagent Resource Center (MR4) for providing the primers that were used in the study. We thank Astrid K. Dier for her comments on earlier versions of this manuscript. icddr,b is thankful to the Government of Australia, Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support. icddr,b also acknowledges with gratitude the commitment of Swiss Academy of Medical Science (SAMS) and Velux Foundation to its research efforts. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Footnotes
Financial support: This study was funded by the SAMS and the Velux Foundation.
Authors' addresses: Mohammad Shafiul Alam, Hasan Mohammad Al-Amin, Mohammad Golam Kibria, Wasif A. Khan, and Rashidul Haque, International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b), Dhaka, Bangladesh, E-mails: shafiul@icddrb.org, alamin@icddrb.org, golam.kibria@icddrb.org, wakhan@icddrb.org, and rhaque@icddrb.org. Rubayet Elahi, Department of Biochemistry, Virginia Tech, Blacksburg, VA, E-mail: rubayet@vt.edu. Abu Naser Mohon, Department of Microbiology and Infectious Disease, Cumming School of Medicine, University of Calgary, Alberta, Canada, E-mail: manmohon@ucalgary.ca. Hamida Khanum, Parasitology Branch, Department of Zoology, University of Dhaka, Dhaka, Bangladesh, E-mail: hamida_khanum@yahoo.com.
References
- 1.World Health Organization . World Malaria Report 2014. Geneva, Switzerland: WHO Press; 2014. [Google Scholar]
- 2.Khan WA, Sack DA, Ahmed S, Prue CS, Alam MS, Haque R, Khyang J, Ram M, Akter J, Nyunt MM, Norris D, Glass G, Shields T, Haq MZ, Cravioto A, Sullivan DJ., Jr Mapping hypoendemic, seasonal malaria in rural Bandarban, Bangladesh: a prospective surveillance. Malar J. 2011;10:124. doi: 10.1186/1475-2875-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haque U, Overgaard HJ, Clements ACA, Norris DE, Islam N, Karim J, Roy S, Haque W, Kabir M, Smith DL, Glass GE. Malaria burden and control in Bangladesh and prospects for elimination: an epidemiological and economic assessment. Lancet Glob Health. 2014;2:e98–e105. doi: 10.1016/S2214-109X(13)70176-1. [DOI] [PubMed] [Google Scholar]
- 4.National Malaria Control Program . Malaria National Strategic Plan 2015–2020. Dhaka; Bangladesh: 2015. Government of Bangladesh. [Google Scholar]
- 5.Hartl DL. The origin of malaria: mixed messages from genetic diversity. Nat Rev Microbiol. 2004;2:15–22. doi: 10.1038/nrmicro795. [DOI] [PubMed] [Google Scholar]
- 6.McConkey GA, Waters AP, McCutchan TF. The generation of genetic diversity in malaria parasites. Annu Rev Microbiol. 1990;44:479–498. doi: 10.1146/annurev.mi.44.100190.002403. [DOI] [PubMed] [Google Scholar]
- 7.Kiwuwa MS, Ribacke U, Moll K, Byarugaba J, Lundblom K, Farnert A, Fred K, Wahlgren M. Genetic diversity of Plasmodium falciparum infections in mild and severe malaria of children from Kampala, Uganda. Parasitol Res. 2013;112:1691–1700. doi: 10.1007/s00436-013-3325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atroosh WM, Al-Mekhlafi HM, Mahdy MA, Saif-Ali R, Al-Mekhlafi AM, Surin J. Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on MSP-1 and MSP-2 genes. Parasit Vectors. 2011;4:233. doi: 10.1186/1756-3305-4-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta P, Singh R, Khan H, Raza A, Yadavendu V, Bhatt RM, Singh V. Genetic profiling of the Plasmodium falciparum population using antigenic molecular markers. Scientific World Journal. 2014;2014:140867. doi: 10.1155/2014/140867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain MM, Sohail M, Kumar R, Branch OH, Adak T, Raziuddin M. Genetic diversity in merozoite surface protein-1 and 2 among Plasmodium falciparum isolates from malarious districts of tribal dominant state of Jharkhand, India. Ann Trop Med Parasitol. 2011;105:579–592. doi: 10.1179/2047773211Y.0000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck HP, Snounou G, Felger I, Olliaro P, Mugittu K. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malar J. 2011;10:79. doi: 10.1186/1475-2875-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoepflin S, Valsangiacomo F, Lin E, Kiniboro B, Mueller I, Felger I. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar J. 2009;8:250. doi: 10.1186/1475-2875-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 14.Baruah S, Lourembam SD, Sawian CE, Baruah I, Goswami D. Temporal and spatial variation in MSP1 clonal composition of Plasmodium falciparum in districts of Assam, northeast India. Infect Genet Evol. 2009;9:853–859. doi: 10.1016/j.meegid.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Congpuong K, Sukaram R, Prompan Y, Dornae A. Genetic diversity of the msp-1, msp-2, and glurp genes of Plasmodium falciparum isolates along the Thai-Myanmar borders. Asian Pac J Trop Biomed. 2014;4:598–602. doi: 10.12980/APJTB.4.2014APJTB-2014-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghanchi NK, Martensson A, Ursing J, Jafri S, Bereczky S, Hussain R, Beg MA. Genetic diversity among Plasmodium falciparum field isolates in Pakistan measured with PCR genotyping of the merozoite surface protein 1 and 2. Malar J. 2010;9:1. doi: 10.1186/1475-2875-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang JM, Moon SU, Kim JY, Cho SH, Lin K, Sohn WM, Kim TS, Na BK. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131. doi: 10.1186/1475-2875-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pratt-Riccio LR, Perce-da-Silva Dde S, Lima-Junior JC, Theisen M, Santos F, Daniel-Ribeiro CT, de Oliveira-Ferreira J, Banic DM. Genetic polymorphisms in the glutamate-rich protein of Plasmodium falciparum field isolates from a malaria-endemic area of Brazil. Mem Inst Oswaldo Cruz. 2003;108:523–528. doi: 10.1590/0074-0276108042013022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akter J, Thriemer K, Khan WA, Sullivan DJ, Jr, Noedl H, Haque R. Genotyping of Plasmodium falciparum using antigenic polymorphic markers and to study anti-malarial drug resistance markers in malaria endemic areas of Bangladesh. Malar J. 2012;11:386. doi: 10.1186/1475-2875-11-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Broek IVF, van der Wardt S, Talukder L, Chakma S, Brockman A, Nair S, Anderson TC. Drug resistance in Plasmodium falciparum from the Chittagong Hill Tracts, Bangladesh. Trop Med Int Health. 2004;9:680–687. doi: 10.1111/j.1365-3156.2004.01249.x. [DOI] [PubMed] [Google Scholar]
- 21.Kiwanuka GN. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997–2007. J Vector Borne Dis. 2009;46:1–12. [PubMed] [Google Scholar]
- 22.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, Pillai DR. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013) Malar J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203. doi: 10.1385/1-59259-271-6:189. [DOI] [PubMed] [Google Scholar]
- 24.Alam MS, Mohon AN, Mustafa S, Khan WA, Islam N, Karim MJ, Khanum H, Sullivan DJ, Jr, Haque R. Real-time PCR assay and rapid diagnostic tests for the diagnosis of clinically suspected malaria patients in Bangladesh. Malar J. 2011;10:175. doi: 10.1186/1475-2875-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-abd NM, Mahdy MA, Al-Mekhlafi AM, Snounou G, Abdul-Majid NB, Al-Mekhlafi HM, Fong MY. The suitability of P. falciparum merozoite surface proteins 1 and 2 as genetic markers for in vivo drug trials in Yemen. PLoS One. 2013;8:e67853. doi: 10.1371/journal.pone.0067853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J. 2008;7:17. doi: 10.1186/1475-2875-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 28.Anthony TG, Conway DJ, Singh JC, Matusop A, Ratnam S, Shamsul S, Singh B. Fragmented population structure of Plasmodium falciparum in a region of declining endemicity. J Infect Dis. 2005;191:1558–1564. doi: 10.1086/429338. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organization . Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy: Genotyping to Identify Parasite Populations. Geneva, Switzerland: WHO Press; 2008. [Google Scholar]