Abstract
Molecular techniques based on real-time polymerase chain reaction (qPCR) allow the detection and quantification of DNA but are unable to distinguish between signals from dead or live cells. Because of the lack of simple techniques to differentiate between viable and nonviable cells, the aim of this study was to optimize and evaluate a straightforward test based on propidium monoazide (PMA) dye action combined with a qPCR assay (PMA-qPCR) for the selective quantification of viable/nonviable epimastigotes of Trypanosoma cruzi. PMA has the ability to penetrate the plasma membrane of dead cells and covalently cross-link to the DNA during exposure to bright visible light, thereby inhibiting PCR amplification. Different concentrations of PMA (50–200 μM) and epimastigotes of the Maracay strain of T. cruzi (1 × 105–10 parasites/mL) were assayed; viable and nonviable parasites were tested and quantified by qPCR with a TaqMan probe specific for T. cruzi. In the PMA-qPCR assay optimized at 100 μM PMA, a significant qPCR signal reduction was observed in the nonviable versus viable epimastigotes treated with PMA, with a mean signal reduction of 2.5 logarithm units and a percentage of signal reduction > 98%, in all concentrations of parasites assayed. This signal reduction was also observed when PMA-qPCR was applied to a mixture of live/dead parasites, which allowed the detection of live cells, except when the concentration of live parasites was low (10 parasites/mL). The PMA-qPCR developed allows differentiation between viable and nonviable epimastigotes of T. cruzi and could thus be a potential method of parasite viability assessment and quantification.
Introduction
Trypanosoma cruzi is a parasitic protozoan that causes Chagas disease in human beings and other mammals, and is transmitted by triatomine vectors in endemic zones, as well as by nonvector routes, including vertically from mother to newborn, organ transplantation, blood transfusion, and other less common ways such as laboratory accidents. This systemic chronic illness represents the third highest parasitic disease burden after malaria and schistosomiasis, and is a serious public health issue in 21 endemic Latin American countries, with an estimated 8 million people already infected and about 50,000 new cases per year.1–3 With increasing globalization, Chagas disease is also becoming a health threat worldwide. Cases arising from blood transfusion, organ transplantation, and vertical transmission are being increasingly detected in non-endemic areas, including the United States, Canada, several European countries, and Oceania, as a consequence of large-scale migration of infected individuals from Latin America.4–8 Moreover, oral infection of Chagas disease is currently considered as an important transmission pathway in endemic areas, even though where vectorial transmission has been successfully interrupted,9 with high mortality rates.4 In the Brazilian Amazon region, several outbreaks of Chagas disease have been described due to oral transmission, involving more than 1,500 patients.10 Chagas disease could therefore be classified as a foodborne infection, principally associated with the consumption of infected food such as wild animal meat, homemade juices, and artisan beverages contaminated with the parasite-infected vector.9,11,12
In an effort to achieve a more sensitive detection of T. cruzi than provided by conventional parasitological techniques, in the last 20 years, PCR technology has been applied to identify T. cruzi DNA in blood samples or biopsies from chagasic patients,13,14 which has opened new possibilities in diagnosis and follow-up assessment of chemotherapy.15–17 PCR has also proved useful for T. cruzi detection in vector and reservoir studies.18,19 However, a drawback of the PCR technique is that it cannot distinguish between DNA signals from live or dead parasites and consequently a positive result does not imply pathogen viability. Methods to evaluate T. cruzi viability based on RNA detection are not routinely used due to their high handling complexity. Likewise, axenic culture presents low sensitivity and takes a long time to provide conclusive results.20,21 Saavedra and others have developed a hybrid PCR and xenodiagnosis (XD) methodology to evaluate parasite viability in chronic chagasic patients and improve the sensitivity of diagnosis by XD, but as stated before, the classic techniques are time consuming, and require trained personal and special laboratory conditions, among other drawbacks.22
Recently developed photochemical dyes can be applied in combination with PCR to detect viable cells, based on cell membrane integrity. Ethidium monoazide (EMA) and propidium monoazide (PMA) have the capacity to bind to free DNA/RNA, but not protected nucleic acid, as they are cell membrane impermeable. In the case of damaged or dead cells, the dyes can pass through the membrane and covalently cross-link to organic molecules, including DNA, under exposure to bright visible light.23,24 This covalent binding prevents subsequent amplification of DNA by PCR, thereby indicating cell nonviability. The efficiency of these techniques depends on a variety of factors, including the type and concentration of the dye, the light source, incubation conditions, and the microorganism, all of which need to be considered during optimization.25
EMA-PCR and PMA-PCR assays have been applied to a wide variety of microorganisms, including bacterial vegetative cells, bacterial spores, fungi, viruses, and yeast, principally in the fields of environment and food.26–31 Lately, they have also been applied in bacterial studies on clinical samples, indicating that this method constitutes a potential alternative to diagnosis by microscopy and culture, as well as in monitoring early treatment response32,33 or in drug experimental assays.34,35 However, to date, this methodology has had only scant application in parasites, for example, oocysts of Cryptosporidium, cysts of Giardia duodenalis and trophozoites and cysts of Acanthamoeba castellani in clinical and environmental samples.29,31,36,37 The aim of this study was to evaluate if PMA-qPCR assay can differentiate between live and dead epimastigotes of T. cruzi and thus have potential application in parasite viability assessment and quantification.
Materials and Methods
Epimastigotes of T. cruzi and inactivation treatment.
Epimastigotes of T. cruzi (Maracay strain) were grown in liver infusion tryptose medium (LIT) at 28°C until the logarithmic growth phase. Twenty milliliters of the culture was centrifuged at 1,800 × g for 10 minutes; the pellet was suspended in the same medium and viable parasites were counted in a Rosenthal hemocytometer chamber with trypan blue dye. The stock was serially diluted 10-fold from 1 × 105 to 10 parasites/mL with LIT. Each dilution was equally divided to make two sets of parasite suspension. One set of parasites was used for live parasite studies and the other set was subjected to inactivation by exposure to isopropanol (final concentration of 70%) for 10 minutes. Isopropanol was removed by harvesting epimastigotes using centrifugation at 10,000 × g for 5 minutes before resuspension in LIT. These assays were performed in duplicate. As a control, an aliquot of each set of parasites was cultured to ensure the viability or nonviability of the parasites in LIT for 2 weeks.
PMA treatment.
PMA (GenIUL, Barcelona, Spain) was dissolved in water of molecular biology grade (Sigma-Aldrich, Saint Louis, MO) to obtain a stock solution of 2,000 μM, which was stored at 4°C in darkness for no longer than 2 months. All subsequent steps using PMA were performed under minimal light conditions. PMA stock solution was added at a final concentration of 50, 100, and 200 μM to a total volume of 300 μL of 1 × 105, 1 × 104, and 1 × 103 parasites/mL, both live and dead, to determine the optimal final PMA concentration. The resulting suspension was incubated for 30 minutes at room temperature in darkness, mixing every 10 minutes. Live and dead parasites not treated with PMA were used as a control. All the samples (treated and not treated with PMA) were then photoactivated for 15 minutes in constant mode using a light-emitting diode (LED) source that emits light in the blue range of the visible spectra (464–467 nm, 60 W; Phast Blue PhotoActivation System; GenIUL, Barcelona, Spain). The assay was performed in duplicate.
DNA extraction.
Immediately after the photoactivation, the samples were pelleted by centrifugation at 13,000 × g for 5 minutes, and the remaining supernatant was discarded to achieve a final volume of 200 μL. DNA extraction was done with the High Pure PCR Template Preparation kit (Roche, Mannheim, Germany) and eluted in 200 μL of elution buffer (10 mM Tris-HCl, pH 8.5) according to the manufacturer's instructions. The concentration of eluted DNA was measured in a NanoDrop (ND-1000, ThermoScientific, Wilmington, DE) and stored at −20°C for qPCR analysis. For the extraction negative control (ENC), LIT was used without a template.
Real time PCR (qPCR) assay.
Five microliters of extracted DNA was amplified by qPCR in a thermocycler (LightCycler 480, Roche) in duplicate. The primers, probes, and conditions of the technique were as described by Piron and others17 with some modifications. Briefly, the following were used: Cruzi 1 and Cruzi 2 primers, and a Cruzi 3 probe, which was labeled with 6-carboxyfluorescein and a minor groove binder. The final concentrations in the PCR mixture were as follows: 1 × LightCycler 480 Probes Master (Roche), 750 nM of each T. cruzi primer and 250 nM of the T. cruzi probe in 20 μL reaction volume. The amplification was run in 45 cycles and the annealing temperature was 58°C.
A standard curve was constructed with 1/10 serial dilutions, in elution buffer (10 mM Tris-HCl, pH 8.5), of total DNA extracted from the Maracay strain from 1 × 105 to 1 parasites/mL. Molecular biology-grade water (Sigma-Aldrich) and ENC were used as negative controls.
The parasitic load of every sample was calculated using LightCycler 480 software by the second derivative maximum method. The limit of detection (LOD) of the technique was calculated in 2 parasite equivalents/mL.17
Statistical analysis.
The data were analyzed with IBM SPSS Statistics. Comparisons were carried out with a one-way analysis of variance and post hoc Tukey's honestly significant difference. Differences with P values < 0.05 were considered statistically significant.
Results
qPCR on T. cruzi epimastigotes.
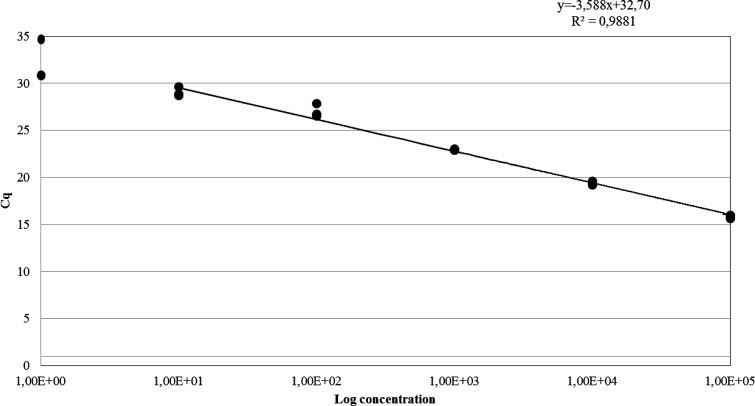
The qPCR standard curve showed a linearity of 0.9881 between the log concentrations of epimastigotes and the Cq value, with a dynamic range from 1 × 105 to 10 parasites/mL and an efficiency of 90% (Figure 1 ). Before the PMA treatment, the serial 10-fold diluted stock from 1 × 105 to 10 parasites/mL of live and dead T. cruzi epimastigotes were measured by qPCR in Cq values, and expressed in parasite equivalents/mL (Figure 2 ). Values for live and dead parasites were quite similar at all concentrations, indicating that minimal quantities of DNA were lost in the washing steps of the procedure. All the control cultures of the epimastigotes were positive, except 10 parasites/mL and those treated with isopropanol.
Figure 1.
Standard curve constructed by plotting the mean Cq values, studied in triplicate, with respect to the logarithm10 of the Trypanosoma cruzi DNA concentrations (10-fold serial dilutions 1 × 105–1 parasite/mL).
Figure 2.
Results by real time polymerase chain reaction of different concentrations of live and dead parasites before propidium monoazide treatment (A = 1 × 105; B = 1 × 104; C = 1 × 103; D = 1 × 102; E = 10 parasites/mL). (A) Results expressed in Cq values. (B) Results quantified in parasite equivalents/mL.
PMA-qPCR optimization on T. cruzi epimastigotes.
The optimum concentration of PMA that provided the greatest difference in qPCR values between treated viable and nonviable parasites was expressed in ΔCq values (Cq PMA-treated dead parasites − Cq PMA-treated live parasites). Three concentrations of parasites/mL (1 × 103–1 × 105) and three concentrations of PMA (50, 100, and 200 μM) were assayed. The best differentiation between live and dead cells was achieved by 100 μM in all the parasite concentrations assayed, and increasing the PMA concentration did not improve the results (Figure 3 ).
Figure 3.
Propidium monoazide (PMA) real-time polymerase chain reaction optimization results, expressed in ΔCq obtained at different PMA concentrations (50, 100, and 200 μM) when different parasite concentrations were tested (1 × 103–1 × 105 parasites/mL).
Treated and nontreated with 100 μM PMA concentrations of live T. cruzi epimastigotes (1 × 105, 1 × 104, 1 × 103, 1 × 102, 10 parasites/mL) were studied by qPCR and no significant signal reduction in PMA-treated versus nontreated living cells (P > 0.05) was observed, indicating no significant effect of PMA on live T. cruzi epimastigotes.
To assess the PMA impact on the reduction of the qPCR signal of dead versus live parasites, the same concentrations of dead and live T. cruzi epimastigotes were tested (Table 1). A higher shift was observed for 1 × 105–1 × 103 parasites/mL with a ΔCq between 9 and 7.5, which is equivalent to a fall of 2.5–3.2 log units between the viable and nonviable parasites; for 1 × 102 and 10 parasites/mL, a ΔCq of around 2.7–3.7 was detected, which is equivalent to a fall of 1.8 log units. A significant (P < 0.002) PMA-qPCR signal reduction was observed for all parasite concentrations studied, except for 10 parasites/mL. The percentage of signal reduction was between 98.4% and 99.9% for all concentrations studied. Despite the high level of reduction, a number of parasites were still detected. The concentration of dead parasites dropped below 1 parasite equivalent/mL in concentrations ≤ 103 parasites/mL, but at higher parasite concentrations, the reduction exceeded the LOD of the technique (Figure 4 ).
Table 1.
qPCR results of different concentrations of viable and nonviable epimastigotes, treated with 100 μM PMA. Comparison of Cq values, parasite concentration, and percentage of PMA-qPCR signal reduction
| Parasites/mL* | Mean Cq values | Mean parasite concentration† | ||||||
|---|---|---|---|---|---|---|---|---|
| Dead parasites | Live parasites | ΔCq | Dead parasites | Live parasites | Δ Concentration§ | Signal reduction‡ | P value | |
| 1.00E + 05 | 24.79 | 15.82 | 9.0 | 3.023E + 02 | 1.021E + 05 | 1.018E + 05 | 99.7 | 0.001 |
| 1.00E + 04 | 28.12 | 19.46 | 8.7 | 1.165E + 01 | 9.815E + 03 | 9.803E + 03 | 99.8 | 0.000 |
| 1.00E + 03 | 30.55 | 23.09 | 7.5 | 5.600E − 01 | 9.575E + 02 | 9.569E + 02 | 99.9 | 0.000 |
| 1.00E + 02 | 30.21 | 26.48 | 3.7 | 8.500E − 01 | 5.927E + 01 | 5.842E + 01 | 98.5 | 0.002 |
| 1.00E + 01 | 31.76 | 29.04 | 2.7 | 9.000E − 02 | 5.890E + 00 | 5.800E + 00 | 98.4 | 0.241 |
PMA = propidium monoazide; qPCR = real-time polymerase chain reaction.
Concentration of viable and nonviable epimastigotes of Trypanosoma cruzi.
qPCR results expressed in parasite equivalents/mL.
Expressed in percentage.
Δ Concentration calculated as live parasites − dead parasites.
Figure 4.
Propidium monoazide (PMA) real-time polymerase chain reaction results expressed in parasite equivalents/mL of different concentrations of live and dead parasites, both treated with PMA. Epimastigotes were studied at A = 1 × 105; B = 1 × 104; C = 1 × 103; D = 1 × 102; E = 10 parasites/mL. Limit of detection (LOD) = 2 parasite equivalents/mL.
Quantification of live T. cruzi from live/dead parasite mixtures.
To observe the effect of a mixture of live and dead epimastigotes on the qPCR-PMA, a set of 10-fold dilutions of live parasites ranging from 1 × 105 to 10 parasites/mL was mixed with a concentration of 1 × 105 dead parasites. This assay was performed in duplicate, treated and nontreated with PMA. The qPCR results indicated that the concentrations expressed in parasites equivalent/mL were similar for all parasite concentrations when untreated (Figure 5 ). These results reflect the maximum concentration of dead parasites in a sample (1 × 105 parasites/mL), and fail to reflect the real number of live parasites in the mixture. In contrast, the qPCR values of the PMA-treated mixture indicate a linear relationship between Cq and the number of viable cells, which was only affected when the concentration of live cells was low, demonstrating that the concentration results basically reflect the amount of live parasite DNA in the mixture.
Figure 5.
Quantification in parasites equivalent/mL of Trypanosoma cruzi from mixture of live (1 × 105–10 parasites/mL) and dead (1 × 105 parasites/mL) parasites, treated and nontreated with 100 μM propidium monoazide.
Discussion
Although molecular methods such as PCR can help to detect and quantify parasites with high sensitivity and specificity, PCR by itself is unable to differentiate between live and dead parasites, which can undermine the value of the results. Recently, qPCR has been tested in combination with PMA in a variety of microorganisms, principally those affecting the food industry and more recently in clinical pathogens, to assess treatment effectiveness. In this study, we assessed the performance of PMA treatment in minimizing detection signals by qPCR from nonviable epimastigotes of T. cruzi and propose it as a potential tool for viability quantification.
The photochemical dye PMA at 100 μM significantly reduced the qPCR signal from nonviable epimastigotes, effectively separating them from viable parasites. The PMA treatment appeared to have no toxic effect on the epimastigotes, since no significant differences in concentration were observed between live PMA-treated and nontreated parasites. This suggests that PMA does not penetrate the membrane of living T. cruzi epimastigotes at the tested levels. Similar PMA concentrations have been used without significant cytotoxic effects on protozoa such as Cryptosporidium spp. oocysts36 and Mycobacterium tuberculosis bacteria.38
A considerable signal reduction in dead T. cruzi epimastigotes was achieved by PMA-qPCR, with a decrease in detection of > 98% at all parasite concentrations studied. Nevertheless, at high parasite concentrations (1 × 105–1 × 104 parasites/mL), despite the very high signal reduction (a maximum of 99%), a remaining qPCR signal in the dead PMA-treated parasites generated a false positive. These results are consistent with other studies, where PMA was unable to completely eliminate the qPCR signal of dead Salmonella serovar Enteritidis, Mycobacterium avium, and Listeria innocua.39–41 In contrast, in Acanthamoeba spp. (1 × 106 cysts and trophozoites killed by autoclave), the PMA-qPCR signal was successfully reduced to zero by enhancing the PMA concentration to 200 μM, after 100 μM proved ineffective in differentiating between viable and nonviable parasites.29 In our study, increasing the PMA concentration did not further reduce the qPCR signal of dead epimastigotes of T. cruzi. Similarly, Barbau-Piednoir and others (2014) found that, Cq values for all tested dilutions of dead bacteria did not differ between 75 and 150 μM of PMA.
To reduce or avoid both false-negative and false-positive qPCR signals,25 a variety of factors should be taken into account when optimizing the technique, such as the type and concentration of dye, the light source, the type of microorganism, or the amplicon length. Of the two types of dye used in this field, PMA is described as more effective in differentiating between live and dead cells, whereas EMA is slightly more efficient in signal suppression, although with the disadvantage that it can penetrate the living cells of some microorganisms.24,42 The light source, as mentioned above, is another factor in the generation of false positives. In particular, studies using halogen lamps without an emission wavelength specific for PMA show fluctuating efficiency, due to variable light activation and the intense heat emitted. To minimize this variability, we used a commercial LED-based system designed especially for the exposure of cell suspensions to light, with the advantage that LEDs emit light in the blue range of visible spectra, allowing for optimal dye activation without heat generation. Therefore, this factor was ruled out as the cause of the persistent qPCR signal.
Some authors suggest the technique can be further improved by studying the effect of PCR amplicon length, an important experimental parameter when analyzing samples treated with viability dyes.43 Alonso and others (2014) reported a more effective exclusion of dead cysts of Giardia duodenalis in a qPCR assay with longer amplicons. Likewise, Li and Chen (2013) found a good correlation between amplicon length and the signal inhibitory effect of PMA treatment on dead cells of Salmonella spp, concluding that the best qPCR signal reduction was obtained with the larger amplicon, albeit with a slight loss of technique efficiency. As suggested by Soejima and others (2011)44 and Contreras and other (2011),45 the beneficial effect of targeting longer DNA sequences is likely due to the increased probability of dye binding in the targeted region, resulting in a stronger inhibition of the amplification. The sensitivity of the qPCR assay is lower when using larger amplicons, which could lead to false-negative results if the signal falls below the LOD.31,46 Therefore, optimizing a technique involves attaining a balance between the reduction of the qPCR signal in the dead cells and sensitivity. In our study, the qPCR technique, previously validated for diagnosis,17 used a set of primers that amplify a region of satellite DNA 166 bp long. Therefore, a larger target gene would probably help to completely eliminate the remaining signal of the dead parasites observed, although at the risk of reducing the sensitivity of the technique. Also, some studies suggest that the sequence of the target gene may influence the noncomplete amplification signal suppression from dead cells.47,48 Further research on enhancing the removal of the remaining signal of the dead parasites is necessary.
Some authors indicate that the ratio between live and dead cells can affect the performance of the method, considering that dead cells cannot exceed live cells by a factor of 1 × 103 without impacting on the PMA-qPCR.25 Other authors, such as Pan and Breidt, have reported that the linear relationship between Cq and the number of viable cells of Listeria monocytogenes was affected when the ratio of dead cells exceeded 1 × 104 and the concentration of live cells was less than 1 × 103 CFU/mL.49 Our results showed that the technique was capable of differentiating live T. cruzi from a live/dead parasite mixture, despite the high number of dead cells present in all samples. The technique failed only when a low concentration of live parasites was combined with a high concentration of dead parasites, which could be explained by a saturation of PMA by this high number of dead cells.25
The PMA-qPCR strategy optimized in this study effectively differentiated between viable and nonviable epimastigotes of T. cruzi, with a significant reduction in the qPCR signal. This method has potential application in viability assessment and quantification due to its various advantages: it requires only a few hours to carry out, in comparison with an axenic culture that takes at least 20–30 days; its handling is simple and straightforward compared with RNA detection techniques; the use of the highly sensitive and specific TaqMan probe renders it suitable for application in, for example, raw food matrices, as it would avoid interference from bacteria or fungus, a problem in culture techniques. Nevertheless, this method has its limitations, as the reagents are expensive, highly trained personnel are required for its application, and it is not always available in laboratories. It would be desirable if the method was tested by other laboratories and on other trypanosomatids.
The described method could therefore be especially useful in differentiating DNA from viable parasites in fields such as food security to prevent oral infections or study outbreaks, diagnostics to evaluate chemotherapeutic efficacy, research on vectors and reservoirs, or antitrypanosomal drug activity assays.
ACKNOWLEDGMENTS
We thank Francesc Codony of the Laboratori de Microbiologia Sanitària i Mediambiental, Universitat Politècnica de Catalunya for providing his expertise in the design of the experiments and Lucy Brzoska for her invaluable advice on the preparation of the manuscript.
Footnotes
Financial support: This work is part of a research study supported by the National R&D+i Plan 2008–2011 and ISC III -Subdirección General de Evaluación y Fomento de la Investigación (PI 10/00533), was in part funded by CONICYT Becas Chile (72130155) and is part of the project 2014 SGR 1241 de la Generalitat de Catalunya.
Authors' addresses: Beatriz Cancino-Faure, Roser Fisa, M. Magdalena Alcover, and Cristina Riera, Laboratori de Parasitologia, Departament de Microbiologia i Parasitologia Sanitàries, Facultat de Farmacia, Universitat de Barcelona, Barcelona, Spain, E-mails: mbcancino@gmail.com, rfisa@ub.edu, mmagdalenaalcoveramengual@ub.edu, and mcriera@ub.edu. Teresa Jimenez-Marco, Fundació Banc de Sang i Teixits de les Illes Balears, Palma de Mallorca, Balearic Islands, Spain, and IUNICS Institut Universitari d'Investigació en Ciències de la Salut, Universitat de les Illes Balears, Cra. de Valldemossa, Balearic Islands, Spain, E-mail: tjimenez@fbstib.org.
Reprint requests: Roser Fisa, Laboratori de Parasitologia, Departament de Microbiologia i Parasitologia Sanitàries, Facultat de Farmàcia, Universitat de Barcelona, Avinguda Joan XXIII s/n. E-08028, Barcelona, Spain, E-mail: rfisa@ub.edu; Tel: +34 934024502; Fax: +34 934024504.
References
- 1.Steverding D. The history of Chagas disease. Parasit Vectors. 2014;7:1–8. doi: 10.1186/1756-3305-7-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Research priorities for Chagas disease, human African trypanosomiasis and leishmaniasis. World Health Organ Tech Rep Ser. 2012;975:1–100. [PubMed] [Google Scholar]
- 3.Bern C, Montgomery SP. An estimate of the burden of Chagas disease in the United States. Clin Infect Dis. 2009;49:e52–e54. doi: 10.1086/605091. [DOI] [PubMed] [Google Scholar]
- 4.Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375:1388–1402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- 5.Jackson Y, Gétaz L, Wolff H, Holst M, Mauris A, Tardin A, Sztajzel J, Besse V, Loutan L, Gaspoz JM, Jannin J, Albajar P, Luquetti A, Chappuis F. Prevalence, clinical staging and risk for blood-borne transmission of Chagas disease among Latin American migrants in Geneva, Switzerland. PLoS Negl Trop Dis. 2010;4:e592. doi: 10.1371/journal.pntd.0000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bern C, Montgomery SP, Katz L, Caglioti S, Stramer SL. Chagas disease and the US blood supply. Curr Opin Infect Dis. 2008;21:476–482. doi: 10.1097/QCO.0b013e32830ef5b6. [DOI] [PubMed] [Google Scholar]
- 7.Cancino-Faure B, Fisa R, Riera C, Bula I, Girona-Llobera E, Jimenez-Marco T. Evidence of meaningful levels of Trypanosoma cruzi in platelet concentrates from seropositive blood donors. Transfusion. 2015;55:1249–1255. doi: 10.1111/trf.12989. [DOI] [PubMed] [Google Scholar]
- 8.Roca Saumell C, Soriano-Arandes A, Solsona Díaz L, Gascón Brustenga J. Documento de consenso sobre el abordaje de la enfermedad de Chagas en atención primaria de salud de áreas no endémicas. Aten Primaria. 2015;47:308–317. doi: 10.1016/j.aprim.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nóbrega AA, Garcia MH, Tatto E, Obara MT, Costa E, Sobel J, Araujo WN. Oral transmission of Chagas disease by consumption of Açaí palm fruit, Brazil. Emerg Infect Dis. 2009;15:653–655. doi: 10.3201/eid1504.081450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coura JR, Junqueira ACV. Ecological diversity of Trypanosoma cruzi transmission in the Amazon basin. The main scenaries in the Brazilian Amazon. Acta Trop. 2015;151:51–57. doi: 10.1016/j.actatropica.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Toso MA. Oral transmission of Chagas disease. Rev Med Chil. 2011;139:258–266. [PubMed] [Google Scholar]
- 12.Barreto-de-Albuquerque J, Silva-dos-Santos D, Pérez AR, Berbert LR, Santana-van-Vliet ED, Farias-de-Oliveira DA, Moreira OC, Roggero E, Carvalho-Pinto CE, Jurberg J, Cotta-de-Almeida V, Bottasso O, Savino W, Meis JD. Trypanosoma cruzi infection through the oral route promotes a severe infection in mice: new disease form from an old infection? PLoS Negl Trop Dis. 2015;9:e0003849. doi: 10.1371/journal.pntd.0003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Britto C, Cardoso MA, Monteiro Vanni CM, Hasslocher-Moreno A, Xavier SS, Oelemann W, Santoro A, Pirmez C, Morel CM, Wincker P. Polymerase chain reaction detection of Trypanosoma cruzi in human blood samples as a tool for diagnosis and treatment evaluation. Parasitology. 1995;110:241–247. doi: 10.1017/s0031182000080823. [DOI] [PubMed] [Google Scholar]
- 14.Virreira M, Torrico F, Truyens C, Alonso-Vega C, Solano M, Carlier Y, Svoboda M. Comparison of polymerase chain reaction methods for reliable and easy detection of congenital Trypanosoma cruzi infection. Am J Trop Med Hyg. 2003;68:574–582. doi: 10.4269/ajtmh.2003.68.574. [DOI] [PubMed] [Google Scholar]
- 15.Schijman AG, Altcheh J, Burgos JM, Biancardi M, Bisio M, Levin MJ, Freilij H. Aetiological treatment of congenital Chagas' disease diagnosed and monitored by the polymerase chain reaction. J Antimicrob Chemother. 2003;52:441–449. doi: 10.1093/jac/dkg338. [DOI] [PubMed] [Google Scholar]
- 16.Sánchez G, Coronado X, Zulantay I, Apt W, Gajardo M, Solari S, Venegas J. Monitoring the efficacy of specific treatment in chronic Chagas disease by polymerase chain reaction and flow cytometry analysis. Parasite. 2005;12:353–357. doi: 10.1051/parasite/2005124353. [DOI] [PubMed] [Google Scholar]
- 17.Piron M, Fisa R, Casamitjana N, López-Chejade P, Puig L, Vergés M, Gascón J, Gómez i Prat Jordi, Portús M, Sauleda S. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 2007;103:195–200. doi: 10.1016/j.actatropica.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Cominetti MC, Csordas BG, Cunha RC, Andreotti R. Geographical distribution of Trypanosoma cruzi in triatomine vectors in the State of Mato Grosso do Sul, Brazil. Rev Soc Bras Med Trop. 2014;47:747–755. doi: 10.1590/0037-8682-0234-2014. [DOI] [PubMed] [Google Scholar]
- 19.Herrera CP, Licon MH, Nation CS, Jameson SB, Wesson DM. Genotype diversity of Trypanosoma cruzi in small rodents and Triatoma sanguisuga from a rural area in New Orleans, Louisiana. Parasit Vectors. 2015;8:1–9. doi: 10.1186/s13071-015-0730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiari E, Dias JC, Lana M, Chiari CA. Hemocultures for the parasitological diagnosis of human chronic Chagas' disease. Rev Soc Bras Med Trop. 1989;22:19–23. doi: 10.1590/s0037-86821989000100004. [DOI] [PubMed] [Google Scholar]
- 21.Luz ZMP. Changes in the hemoculture methodology improve the test positivity. Mem Inst Oswaldo Cruz. 1999;94((Suppl 1)):295–298. doi: 10.1590/s0074-02761999000700053. [DOI] [PubMed] [Google Scholar]
- 22.Saavedra M, Zulantay I, Apt W, Martínez G, Rojas A, Rodríguez J. Chronic Chagas disease: PCR-xenodiagnosis without previous microscopic observation is a useful tool to detect viable Trypanosoma cruzi. Biol Res. 2013;46:295–298. doi: 10.4067/S0716-97602013000300011. [DOI] [PubMed] [Google Scholar]
- 23.Nogva HK, Drømtorp SM, Nissen H, Rudi K. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques. 2003;34:804–808. doi: 10.2144/03344rr02. 810, 812–813. [DOI] [PubMed] [Google Scholar]
- 24.Nocker A, Cheung C-Y, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods. 2006;67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Fittipaldi M, Nocker A, Codony F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J Microbiol Methods. 2012;91:276–289. doi: 10.1016/j.mimet.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Agustí G, Codony F, Fittipaldi M, Adrados B, Morató J. Viability determination of Helicobacter pylori using propidium monoazide quantitative PCR. Helicobacter. 2010;15:473–6. doi: 10.1111/j.1523-5378.2010.00794.x. [DOI] [PubMed] [Google Scholar]
- 27.Josefsen MH, Löfström C, Hansen TB, Christensen LS, Olsen JE, Hoorfar J. Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl Environ Microbiol. 2010;76:5097–5104. doi: 10.1128/AEM.00411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andorrà I, Esteve-Zarzoso B, Guillamón JM, Mas A. Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int J Food Microbiol. 2010;144:257–262. doi: 10.1016/j.ijfoodmicro.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Fittipaldi M, Pino Rodriguez NJ, Adrados B, Agustí G, Peñuela G, Morató J, Codony F. Discrimination of viable Acanthamoeba castellani trophozoites and cysts by propidium monoazide real-time polymerase chain reaction. J Eukaryot Microbiol. 2011;58:359–364. doi: 10.1111/j.1550-7408.2011.00557.x. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Chen J-Q. Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food. BMC Microbiol. 2013;13:273. doi: 10.1186/1471-2180-13-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso JL, Amorós I, Guy RA. Quantification of viable Giardia cysts and Cryptosporidium oocysts in wastewater using propidium monoazide quantitative real-time PCR. Parasitol Res. 2014;113:2671–2678. doi: 10.1007/s00436-014-3922-9. [DOI] [PubMed] [Google Scholar]
- 32.Rogers GB, Stressmann FA, Koller G, Daniels T, Carroll MP, Bruce KD. Assessing the diagnostic importance of nonviable bacterial cells in respiratory infections. Diagn Microbiol Infect Dis. 2008;62:133–141. doi: 10.1016/j.diagmicrobio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Miotto P, Bigoni S, Migliori GB, Matteelli A, Cirillo DM. Early tuberculosis treatment monitoring by Xpert(R) MTB/RIF. Eur Respir J. 2012;39:1269–1271. doi: 10.1183/09031936.00124711. [DOI] [PubMed] [Google Scholar]
- 34.De Oliveira-Silva JCV, Machado-de-Assis GF, Oliveira MT, Noguieira Paiva NC, Silva Araujo MS, Martins Carneiro C, Martins OA, Rodrigues Martins H, de Lana M. Experimental benznidazole treatment of Trypanosoma cruzi II strains isolated from children of the Jequitinhonha Valley, Minas Gerais, Brazil, with Chagas disease. Mem Inst Oswaldo Cruz. 2015;110:86–94. doi: 10.1590/0074-02760140260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caldas S, Caldas IS, Cecílio AB, Diniz LF, Talvani A, Ribeiro I, Bahia MT. Therapeutic responses to different anti-Trypanosoma cruzi drugs in experimental infection by benznidazole-resistant parasite stock. Parasitology. 2014;141:1–10. doi: 10.1017/S0031182014000882. [DOI] [PubMed] [Google Scholar]
- 36.Brescia CC, Griffin SM, Ware MW, Varughese EA, Egorov AI, Villegas EN. Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl Environ Microbiol. 2009;75:6856–6863. doi: 10.1128/AEM.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CW, Lu LW, Kuo CL, Hung NT. Density of environmental Acanthamoeba and their responses to superheating disinfection. Parasitol Res. 2013;112:3687–3696. doi: 10.1007/s00436-013-3556-3. [DOI] [PubMed] [Google Scholar]
- 38.De Assunção TM, Batista EL, Deves C, Villela AD, Pagnussatti VE, De Oliveira Dias AC, Kritski A, Rodrigues-Junior V, Basso LA, Santos DS. Real time PCR quantification of viable Mycobacterium tuberculosis from sputum samples treated with propidium monoazide. Tuberculosis (Edinb) 2014;94:421–427. doi: 10.1016/j.tube.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Kralik P, Nocker a, Pavlik I. Mycobacterium avium subsp. paratuberculosis viability determination using F57 quantitative PCR in combination with propidium monoazide treatment. Int J Food Microbiol. 2010;141:S80–S86. doi: 10.1016/j.ijfoodmicro.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Løvdal T, Hovda MB, Björkblom B, Møller SG. Propidium monoazide combined with real-time quantitative PCR underestimates heat-killed Listeria innocua. J Microbiol Methods. 2011;85:164–169. doi: 10.1016/j.mimet.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 41.Barbau-Piednoir E, Mahillon J, Pillyser J, Coucke W, Roosens NH, Botteldoorn N. Evaluation of viability-qPCR detection system on viable and dead Salmonella serovar Enteritidis. J Microbiol Methods. 2014;103:131–137. doi: 10.1016/j.mimet.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Cawthorn DM, Witthuhn RC. Selective PCR detection of viable Enterobacter sakazakii cells utilizing propidium monoazide or ethidium bromide monoazide. J Appl Microbiol. 2008;105:1178–1185. doi: 10.1111/j.1365-2672.2008.03851.x. [DOI] [PubMed] [Google Scholar]
- 43.Ditommaso S, Giacomuzzi M, Ricciardi E, Zotti CM. Viability-qPCR for detecting Legionella: comparison of two assays based on different amplicon lengths. Mol Cell Probes. 2015;29:1–7. doi: 10.1016/j.mcp.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Soejima T, Schlitt-Dittrich F, Yoshida S. Polymerase chain reaction amplification length-dependent ethidium monoazide suppression power for heat-killed cells of Enterobacteriaceae. Anal Biochem. 2011;418:37–43. doi: 10.1016/j.ab.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Contreras PJ, Urrutia H, Sossa K, Nocker A. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J Microbiol Methods. 2011;87:89–95. doi: 10.1016/j.mimet.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 46.Opel KL, Chung D, McCord BR. A study of PCR inhibition mechanisms using real time PCR. J Forensic Sci. 2010;55:25–33. doi: 10.1111/j.1556-4029.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- 47.Chang B, Taguri T, Sugiyama K, Amemura-Maekawa J, Kura F, Watanabe H. Comparison of ethidium monoazide and propidium monoazide for the selective detection of viable Legionella cells. Jpn J Infect Dis. 2010;63:119–123. [PubMed] [Google Scholar]
- 48.Dannelley JM, Boyce L, Gaubatz JW. Efficiency of photoaffinity labeling DNA homopolymers and copolymers with ethidium monoazide. Photochem Photobiol. 1986;43:7–11. doi: 10.1111/j.1751-1097.1986.tb05584.x. [DOI] [PubMed] [Google Scholar]
- 49.Pan Y, Breidt F. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl Environ Microbiol. 2007;73:8028–8031. doi: 10.1128/AEM.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]