Abstract
For differential detection of Taenia solium, Taenia saginata, and Taenia asiatica, loop-mediated isothermal amplification (LAMP) assay targeting the cytochrome c oxidase subunit 1 gene has been recently developed and shown to be sensitive, specific, and effective. However, to achieve differential identification, one specimen requires three reaction mixtures containing a primer set of each Taenia species separately, which is complex and time consuming and increases the risk of cross-contamination. In this study, we developed a simple differential identification of human Taenia species using multiplex LAMP (mLAMP) in combination with dot enzyme-linked immunosorbent assay (dot-ELISA). Forward inner primers of T. solium, T. saginata, and T. asiatica labeled with fluorescein isothiocyanate (FITC), digoxigenin (DIG), and tetramethylrhodamine (TAMRA), respectively, and biotin-labeled backward inner primers were used in mLAMP. The mLAMP assay succeeded in specific amplification of each respective target gene in a single tube. Furthermore, the mLAMP product from each species was easily distinguished by dot-ELISA with an antibody specific for FITC, DIG, or TAMRA. The mLAMP assay in combination with dot-ELISA will make identification of human Taenia species simpler, easier, and more practical.
Introduction
Cysticercosis caused by Taenia solium larvae is a serious emerging threat to public health in many developing countries in Latin America, sub-Saharan Africa, and Asia.1–4 Neurocysticercosis, an infection of the central nervous system, has been increasingly recognized as a lethal helminthic disease in both developing1,5,6 and developed countries.7–9 Among the Taeniae of humans, including T. solium, Taenia saginata, and Taenia asiatica, which cause taeniasis,10–13 only T. solium causes cysticercosis in humans. The lack of diagnostic tools available for use in the field to differentiate T. solium from other human Taenia species hinders the control of cysticercosis in endemic areas. Therefore, it is desirable to develop a simple molecular diagnostic tool that greatly contributes to control strategies.
The loop-mediated isothermal amplification (LAMP) assay, which does not require expensive devices, is a powerful and innovative technology that improves the differential diagnosis of infectious diseases.14,15 In addition, LAMP has high resistance to inhibition of polymerase chain reaction (PCR)–based DNA amplification by substances present in biological specimens, such as fecal samples. The LAMP assay targeting the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene developed for the differentiation of human Taenia species16,17 showed a higher sensitivity for the differential detection of Taenia DNA in fecal samples than the multiplex PCR.18 Furthermore, because the LAMP assay can be performed in a simple incubator, such as a thermos bottle, if the reaction temperature can be maintained during incubation and the results can be inspected with the naked eye, this assay is highly useful in field surveys for rapid identification of tapeworms recovered from taeniasis carriers.19
Although the LAMP assay has provided diagnostic help in identifying infectious agents causing cysticercosis, there are some issues to be resolved for its practical use in the field and clinical site. Specifically, to achieve the differential identification of human Taenia parasites, three separate reaction mixtures containing T. solium, T. saginata, and T. asiatica primer sets for each specimen must be prepared, increasing the risk of contamination and making the assay more time intensive. A simpler LAMP assay to be used as a point-of-care diagnostic in endemic areas is needed.
In this study, we developed a multiplex LAMP (mLAMP) assay that targets the cox1 gene in combination with a dot enzyme-linked immunosorbent assay (dot-ELISA) for easy and rapid differentiation of human Taenia species. Differential identification by the mLAMP assay can be achieved by either agarose gel electrophoresis or dot-ELISA. The mLAMP assay in conjunction with the dot-ELISA will provide new opportunities for practical use as a fast field-based test for the real-time identification of Taenia species.
Materials and Methods
DNA samples.
For testing mLAMP/dot-ELISA, genomic DNA extracted from proglottids or cysticerci was used. Copro-DNA samples prepared from fecal samples were also used. These specimens had been collected from patients in endemic areas and previously identified by multiplex PCR12,13,20 and LAMP16,18,19 assays (Table 1 ).
Table 1.
Genomic DNA samples used in this study
| Taenia spp. | Developmental stage (no. of samples analyzed) | Country |
|---|---|---|
| Taenia solium (102) | Cysticercus (4) | Cameroon |
| Proglottid (75) | China | |
| Cysticercus (1) | Ecuador | |
| Cysticercus (8) | India | |
| Cysticercus (1) | Mozambique | |
| Cysticercus (5) | Nepal | |
| Cysticercus (1) | South Africa | |
| Cysticercus (1) | Tanzania | |
| Proglottid (5) | Thailand | |
| Cysticercus (1) | Vietnam | |
| Taenia saginata (102) | Proglottid (2) | Belgium |
| Proglottid (1) | Brazil | |
| Proglottid (1) | Cameroon | |
| Proglottid (73) | China | |
| Proglottid (1) | Ecuador | |
| Proglottid (15) | Indonesia | |
| Cysticercus (3)* | Indonesia | |
| Proglottid (1) | Korea | |
| Proglottid (5) | Thailand | |
| Taenia asiatica (34) | Proglottid (24) | China |
| Proglottid (1) | Indonesia | |
| Cysticercus (1)* | Indonesia | |
| Proglottid (4) | Philippines | |
| Cysticercus (1) | Taiwan | |
| Proglottid (3) | Thailand |
The cysticerci were prepared from nonobese diabetic/severe combined immunodeficiency mice intraperitoneally injected with in vitro hatched oncospheres.21
LAMP primers.
The LAMP primers for T. saginata and T. asiatica that we previously reported16 were slightly modified, and a new LAMP primer set was designed for T. solium (Table 2 , Figure 1A and B ). Four primers were designed for each species using PrimerExplorer V4 software (Fujitsu, Tokyo, Japan) (http://primerexplorer.jp/). These primers included a forward inner primer (FIP), a backward inner primer (BIP), and two outer primers (F3 and B3). FIP consists of the sense sequence of F2 at the 3′ end and the F1c region at the 5′ end that is complementary to the F1 region. BIP consists of a B2 region at the 3′ end that is complementary to the B2c region and the same sequence as the B1c region at the 5′ end. FIP primers of T. solium, T. saginata, and T. asiatica were labeled at the 5′ end with fluorescein isothiocyanate (FITC), digoxigenin (DIG), and tetramethylrhodamine (TAMRA) (Life Technologies, New York, NY), respectively. BIP primers of each Taenia species were labeled at the 5′ end with biotin (Table 2).
Table 2.
Labeled primer sets targeting the cox1 gene
| Taenia spp. | Primers type | Sequence (5′→3′) |
|---|---|---|
| Taenia solium | F3 | ACATCATATGTTTACGGTTGG |
| B3 | GAACTTTATCTAATACACAAGCAGA | |
| FITC-FIP (F1c + F2) | FITC-TCCCCGTAGGCACTCCAATTAGTTAGATGTTAAGACGGCTGTA | |
| Biotin-BIP (B1c + B2) | Biotin-CGTGTTAATAAGAGTGATCCGGTTCCGGTTACACCACCAAAT | |
| Taenia saginata | F3 | TCGGCAAATATTTAATTCCTTTG |
| B3 | CCAAATGCAACGAAAACATC | |
| DIG-FIP (F1c + F2) | DIG-GCCATCGAAGGAATCAATAACCAATCTGATTTGAATTTACCCCG | |
| Biotin-BIP (B1c + B2) | Biotin-GGTGCTGGTATAGGGTGGACTCATTACTACTTGAAAATAATGATG | |
| Taenia asiatica | F3 | GATTTTCTTTTTTTTGATGCCCA |
| B3 | CGAGAACATCAAAAAATCCAC | |
| TAMRA-FIP (F1c + F2) | TAMRA-AGGCAAGTTTAAATCAGATAACCCATTTTGATAGGTGGTTTTGGTAA | |
| Biotin-BIP (B1c + B2) | Biotin-GGTGCTGGTATAGGGTGAACTCGTTACTACTTGAAAATAATGACG |
BIP = backward inner primer; DIG = digoxigenin; FIP = forward inner primer; FITC = fluorescein isothiocyanate; TAMRA = tetramethylrhodamine.
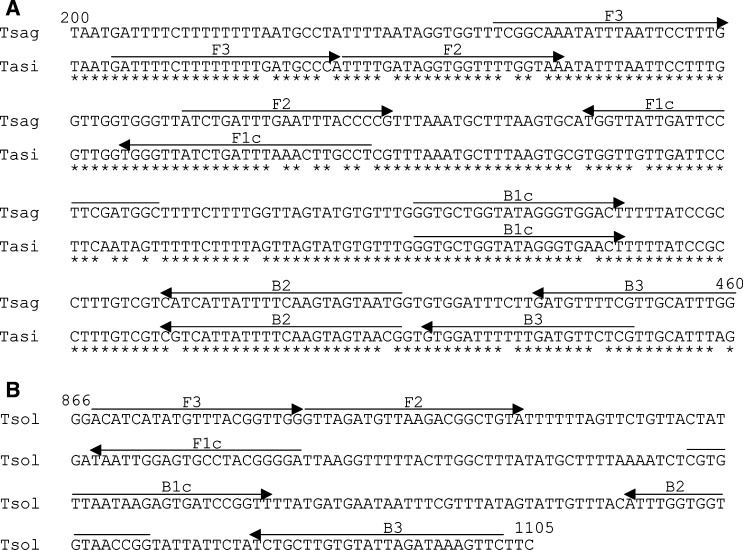
Figure 1.
Nucleotide sequence alignment of the primer region on the cox1 gene for (A) Taenia saginata (AB271695) and Taenia asiatica (AB107234) and (B) Taenia solium (DQ089663). The locations of the primer recognition sites are indicated by arrows. Nucleotide positions of the primer region are shown for T. saginata and T. asiatica (200–460) and T. solium (866–1105) on the cox1 gene.
Preparation of standard plasmid for the cox1 gene.
The cox1 gene of each Taenia parasite was amplified by PCR using forward primer 5′-ATGAATGTCAAATATTTGT-TAAGTT-3′ and reverse primer 5′-CTAAAAGACCATTTCACACGCGAAT-3′ for T. solium, forward primer 5′-ATGAGTGTTAAATTTTTATTAAGTT-3′ and reverse primer 5′-TTAAACTAAAAAACCACGGGCAGGC-3′ for T. saginata, and forward primer 5′-ATGAGTGTTAAATTTTTATTAAGTT-3′ and reverse primer 5′-TTAAACTAAAAAACCACGAGCAAAC-3′ for T. asiatica. The amplification of the cox1 gene was performed with 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 90 seconds, followed by a final extension at 72°C for 5 minutes. The amplified products of the cox1 target of each species were cloned into a pGEM T-vector (Promega, Madison, WI) and the plasmid clone was sequenced on an ABI PRISM 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA) with BigDye terminator 3.1 (Applied Biosystems). The standard plasmid purified from Escherichia coli using a QIAprep Spin Miniprep kit (Qiagen, Hilden, Germany) was used to determine the specificity and sensitivity of the mLAMP/dot-ELISA.
LAMP assay.
The LAMP assay was performed as previously described.16 The reaction was performed in a 25-μL volume of reaction mixture containing primer sets for each Taenia species, including 40 pmol each of FIP and BIP, 5 pmol each of F3 and B3, 8 U of Bst 2.0 WarmStart DNA polymerase (New England Biolabs, Beverly, MA), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 8 mM MgSO4, 10 mM (NH4)2 SO4, 0.1% Tween 20 (Nacalai Tesque, Kyoto, Japan), 0.8 M betaine (Sigma, St. Louis, MO), and 1.4 mM of each dNTP and 50 ng of plasmid DNA or genomic DNA. The reaction mixture was incubated at 60°C for 60 minutes (standard plasmid and genomic DNA from parasite tissue) or for 90 minutes (copro-DNA samples), followed by 80°C for 5 minutes. The LAMP products were electrophoresed on 2.5% agarose gel and detected by staining with ethidium bromide.
Multiplex LAMP assay.
The mLAMP assay was performed in a single tube using a 20-μL reaction mixture containing each of the specific primer sets for each Taenia species (a total of 12 primers: FIPs, BIPs, F3s, and B3s for each of the Taenia species), including 8 pmol each of FITC-FIP, DIG-FIP, and TAMRA-FIP, 8 pmol each of biotin-BIP, 8 pmol each of unlabeled FIP and BIP, 2 pmol each of F3 and B3, 6.4 U of Bst 2.0 WarmStart DNA polymerase (New England Biolabs), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 8 mM MgSO4, 10 mM (NH4)2 SO4, 0.1% Tween 20, 0.8 M betaine (Sigma), and 1.4 mM of each dNTP and 50 ng of plasmid DNA or genomic DNA. The reaction was performed at 60°C for 60 minutes (standard plasmid and genomic DNA from parasite tissue) or for 90 minutes (copro-DNA samples) and then terminated at 80°C for 5 minutes. The mLAMP products were electrophoresed on 2.5% agarose gel and detected by staining with ethidium bromide.
The specificity of the mLAMP assay was assessed with genomic DNA extracted from Hymenolepis nana, Hymenolepis diminuta, Ascaris lumbricoides, and Enterobius vermicularis, with each reaction performed as described above.
Analytical sensitivity.
The sensitivities of the LAMP and mLAMP assays were evaluated using a standard plasmid diluted from 106 copies to one copy/reaction. Each assay was performed as described above.
Dot-ELISA.
Spotting grids with 3 × 3 mm squares for each square were designed on a nitrocellulose membrane (0.45-μm pore size, Bio-Rad Laboratories, Munich, Germany). Anti-FITC rabbit IgG (100 ng), 50 ng of anti-DIG rabbit IgG, 150 ng of anti-TAMRA rabbit IgG (Life Technologies), and 100 ng of mouse IgG diluted with phosphate-buffered saline (PBS) were separately spotted in each grid square on the membrane. The membrane was air-dried for 30 minutes and blocked with blocking solution (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1.0% casein, and 0.1% Tween 20) at room temperature for 20 minutes. After the membrane was washed twice with PBS-Tween 20, it was cut into strips, and each strip was incubated with 5 μL of mLAMP products from each reaction mixture in 150 μL of Tris-buffered saline (50 mM Tris-HCl, 150 mM NaCl; pH 7.5) containing 0.05% Tween 20 (TBST) at room temperature for 15 minutes. The membrane was washed 3 × 1 minute with 1 mL of TBST, followed by incubation with Strep-Tactin alkaline phosphatase conjugate (Bio-Rad) diluted 1:500 in TBST for 10 minutes at room temperature. After the 3 × 1 minute washes with TBST, nitroblue tetrazolium chloride and 5-bromo-4-chloro-3′-indolyl-phosphate p-toluidine (Thermo Scientific, Rockford, IL) substrates were added for color development at room temperature for 5 minutes. The reaction was stopped by washing with distilled water. The development of well-defined purple dots on the membrane was considered a positive result and uncolored dots as a negative result.
Results
LAMP and mLAMP assays.
The BIP and B3 primers previously described16 for T. saginata and T. asiatica were redesigned (Figure 1A) and a new primer set for T. solium was designed from a different region of the cox1 gene (Figure 1B). The LAMP reaction with each specific unlabeled primer set resulted in specific amplification of the target gene from the respective standard plasmid DNA (Figure 2A ). The LAMP method with these primers could detect up to one copy of target DNA, and the LAMP assay with labeled primer sets showed identical results to that with unlabeled primers (Figure 2B).
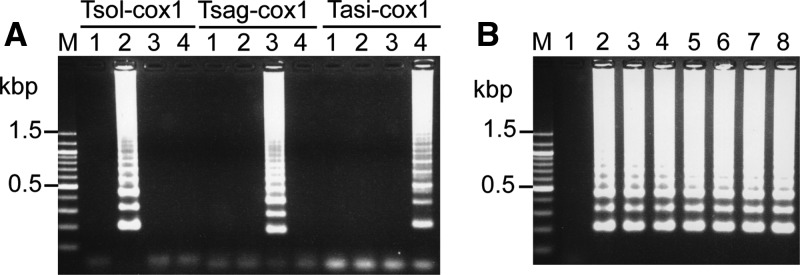
Figure 2.
(A) Loop-mediated isothermal amplification (LAMP) results and (B) sensitivity of LAMP method with Taenia solium cox1 primer set. LAMP products were separated on a 2.5% agarose gel. Lane M, 100 bp DNA ladder (Nacalai Tesque, Kyoto, Japan). (A) Lane 1, negative control without DNA; lane 2, plasmid for T. solium cox1 gene; lane 3, plasmid for Taenia saginata cox1 gene; and lane 4, plasmid for Taenia asiatica cox1 gene. Tsol-cox1, results of LAMP with primer set from T. solium cox1 gene; Tsag-cox1, results of LAMP with primer set from T. saginata cox1 gene; Tasi-cox1, results of LAMP with primer set from T. asiatica cox1 gene. (B) The standard plasmids were serially diluted from 106 copies to one copy per reaction. Lane 1, negative control and lanes 2–8 represent 106, 105, 104, 103, 102, 10, and one copy (ies)/reaction, respectively.
Next, the mLAMP reaction was performed in a single tube with FIP primers of T. solium, T. saginata, and T. asiatica labeled with FITC, DIG, and TAMRA, respectively, and biotin-labeled BIP primers (Table 2). The mLAMP reaction specifically produced the LAMP amplicons of ladder patterns from the respective target gene (Figure 3A ). The mLAMP products showed distinguishable DNA fragment pattern among Taenia species on the agarose gel electrophoresis, with LAMP amplicons from T. saginata species showing the shortest lengths (196 base pairs [bp]) followed by T. solium (225 bp) and then T. asiatica species (247 bp). The mLAMP assay showed analytical sensitivity identical to that of the LAMP assay with each single primer set (data not shown).
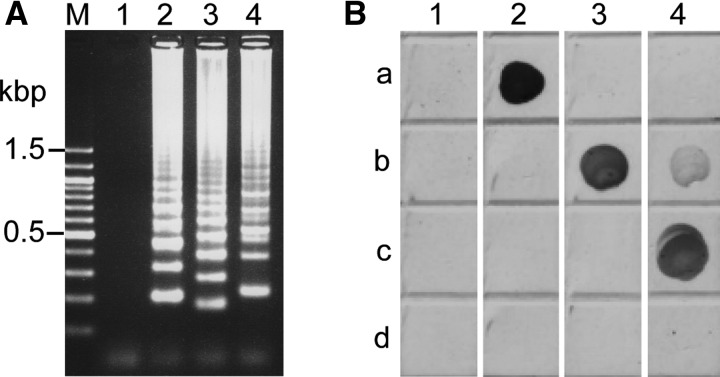
Figure 3.
(A) Results of multiplex loop-mediated isothermal amplification (mLAMP) with standard plasmid carrying cox1 gene of each Taenia species and (B) dot enzyme-linked immunosorbent assay (dot-ELISA). (A) The mLAMP products were separated on a 2.5% agarose gel. Lane M, 100 bp DNA ladder (Nacalai Tesque, Kyoto, Japan); lane 1, negative control without DNA; lane 2, standard plasmid for Taenia solium cox1 gene; lane 3, standard plasmid for Taenia saginata cox1 gene; and lane 4, standard plasmid for Taenia asiatica cox1 gene. (B) The mLAMP product from each Taenia species was incubated with antibodies for fluorescein isothiocyanate (FITC), digoxigenin (DIG), tetramethylrhodamine (TAMRA), and mouse IgG immobilized on the nitrocellulose membrane. Row a, FITC antibody; row b, DIG antibody; row c, TAMRA antibody; and row d, mouse IgG. Lane 1, negative control of mLAMP reaction without DNA; lane 2, mLAMP products from plasmid for the T. solium cox1 gene; lane 3, mLAMP products from plasmid for the T. saginata cox1 gene; and lane 4, mLAMP products from plasmid for the T. asiatica cox1 gene.
Dot-ELISA.
The mLAMP products from each plasmid containing the cox1 genes for T. solium, T. saginata, and T. asiatica were captured by antibodies specific for FITC, DIG, and TAMRA, respectively, on a nitrocellulose membrane and the captured mLAMP products were detected using Strep-Tactin alkaline phosphatase conjugate and chromogenic substrate. As shown in Figure 3B, a positive dot-ELISA result was characterized as a visible colored dot on the membrane. None of the mLAMP products reacted with the mouse IgG, and the negative control without DNA (distilled water) of the mLAMP assay did not show a positive response to any antibody. The mLAMP products from T. asiatica showed a weak signal with the antibody for DIG.
Specificity of mLAMP/dot-ELISA.
The specificity of mLAMP/dot-ELISA was validated with parasite genomic DNA extracted from proglottids and cysticerci (Table 1). As shown in Figure 4A , the mLAMP products from genomic DNAs of T. solium, T. saginata, and T. asiatica could be detected with the antibodies for FIP, DIG, and TAMRA, respectively, indicating that mLAMP/dot-ELISA specifically identifies the three Taenia species. The specificity of the mLAMP assay was further assessed with genomic DNAs extracted from H. nana, H. diminuta, A. lumbricoides, and E. vermicularis, and all DNA samples were negative by the mLAMP assay (data not shown).
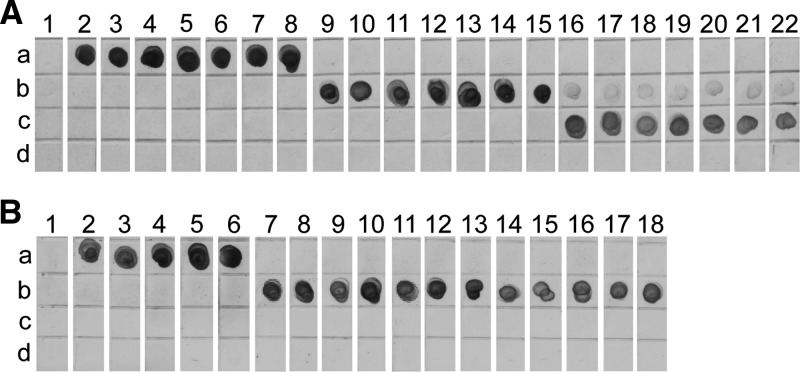
Figure 4.
Assessment of multiplex loop-mediated isothermal amplification (mLAMP)/dot enzyme-linked immunosorbent assay (dot-ELISA) with (A) genomic DNA of each Taenia species and (B) copro-DNA samples. Each antibody was spotted on a separate grid space on the nitrocellulose membrane as described above. (A) A representative subset of results of mLAMP/dot-ELISA with genomic DNA from Table 1. Lane 1, negative control without DNA; lanes 2–8, Taenia solium genomic DNA; lanes 9–15, Taenia saginata genomic DNA; and lanes 16–22, Taenia asiatica genomic DNA. (B) Copro-DNA samples were extracted from taeniid egg-positive feces from China. Lane 1, negative control of mLAMP reaction; lanes 2–6, copro-DNA samples from T. solium taeniasis carriers; and lanes 7–18, copro-DNA samples from T. saginata taeniasis carriers.
Differential identification of Taenia species in copro-DNA.
Copro-DNA extracted from taeniid egg–positive fecal samples collected in endemic areas in China from T. saginata (N = 12) or T. solium (N = 5) taeniasis carriers, each of whom had expelled a single tapeworm, were used to test mLAMP/dot-ELISA. The mLAMP products from T. solium and T. saginata copro-DNA samples had ladder patterns identical to those from their genomic DNA (data not shown) and were detected with the antibodies for FITC and DIG, respectively (Figure 4B).
Discussion
In this study, previously described16 BIP and B3 primers for T. saginata and T. asiatica were redesigned, and a new primer set for T. solium was designed from a different region of the cox1 gene. Because previous LAMP primer sets produced primer dimers in the mLAMP reaction that could be detected by dot-ELISA, a few changes were made to the primer regions to eliminate primer-dimer formation. This study showed that the LAMP assay with these primers had a detection limit similar to that of previous LAMP primer sets.16 The mLAMP reaction performed in a single tube with labeled primers of each Taenia species provided specific amplification of each respective target gene. Differentiation among Taenia species appeared after agarose gel electrophoresis of the mLAMP products based on the size difference of amplified DNA fragments of each Taenia species in agreement with the predicted sizes. The mLAMP assay and subsequent agarose gel electrophoresis showed potential for simple differential detection of Taenia species. However, because it is difficult to perform agarose gel electrophoresis under poor electrical conditions such as in the field, a simple detection method for each mLAMP product by dot-ELISA was developed.
The developed dot-ELISA showed that mLAMP products from genomic DNAs of T. solium, T. saginata, and T. asiatica could be specifically detected with antibodies for FITC, DIG, and TAMRA, respectively. A weak signal with the antibody for DIG was observed with LAMP products from T. asiatica. It may be because the T. saginata DIG-FIP primer annealed to the T. asiatica LAMP products, given that the T. saginata DIG-FIP primer with high identities (F1 region, 86%; F2 region 80%) to T. asiatica DNA sequence was designed inside the amplified region of the T. asiatica gene. In contrast, because the T. asiatica TAMRA-FIP primer with high identities (F1 region, 87%; F2 region, 86%) to T. saginata DNA sequence was designed outside the amplified region of T. saginata gene, the T. asiatica TAMRA-FIP primer did not anneal to T. saginata LAMP products, and the anti-TAMRA antibody did not capture T. saginata LAMP products. Although this interaction between the T. saginata DIG-FIP primer and T. asiatica LAMP products was observed, it was easy to differentiate T. asiatica from T. saginata by dot-ELISA because the signal intensity with anti-TAMRA antibody was stronger than that with anti-DIG antibody. Because dot-ELISA is rapid, is easy to perform and interpret, and does not require expensive equipment or portable field equipment, this technique combined with a molecular approach is a suitable method for species confirmation of infectious agents, offering great advantage for the screening and surveillance of infectious diseases in countries where laboratory facilities and resources are rare.
The evaluation of the specificity of mLAMP/dot-ELISA confirmed that Taenia species from genomic and copro-DNA samples were successfully identified, giving 100% concordance with the results obtained by both multiplex PCR and LAMP methods.12,16,18,20 In addition, there was no cross-reaction with DNA from other intestinal parasites. However, the sample number of other intestinal parasites being small, further evaluation of the specificity of the mLAMP assay should be performed with a larger number of samples. Although we did not perform this assay with copro-DNA from T. asiatica taeniasis carriers because we lacked specimens, we expect a result similar to that for T. asiatica genomic DNA. The LAMP assay was more sensitive than the multiplex PCR technique for the differential detection of Taenia species in stool specimens.18 mLAMP/dot-ELISA will provide efficient identification owing to its high sensitivity and rapidity. However, in the field, this assay may not be appropriate for the investigation of fecal samples to identify taeniasis carriers because it is difficult to prepare copro-DNA under poor electrical situations. To apply this assay to fecal samples under field conditions, we need simple and efficient copro-DNA preparation.
mLAMP/dot-ELISA developed in this study was shown to be sensitive, specific, simple, rapid, practical, and easily reproducible and will accordingly be highly useful for pathogen identification in simplified testing systems that may be appropriate in developing countries where Taenia infections are endemic. Although several other strategies combining mLAMP assay with restriction enzyme digestion analysis, ELISA, or other methods have proved their field practicality for detecting infectious agents, ancillary equipment is still required.22–26 Our dot-ELISA format is simple and can be handily deployed in the field without specific devices or detection system. However, simpler detection of the mLAMP products, such as by lateral flow dipstick,27–29 will be the most appropriate test for easy use in the field. In fact, recent studies have shown that the detection of mLAMP amplicons by label-based lateral flow dipstick is applicable and adequate for use in point-of-care settings, eliminating the use of skilled personnel.30,31
In this study, the mLAMP assay followed by the dot-ELISA method provided accurate, sensitive, and specific results for the differential detection of human Taenia species. In addition, this cost-effective approach yielded a specimen-to-result diagnosis, with a low risk of contamination and within a shorter time for the analysis of several samples than needed by the previous LAMP method.16 The ease of use and production of a visually detectable readout are additional features promoting the feasibility of this molecular approach in resource-constrained settings. Because differentiation of T. solium from T. saginata and T. asiatica is important for the control and prevention of cysticercosis in endemic areas of developing countries, the mLAMP assay in conjunction with dot-ELISA will render the rapid identification of human Taenia species in the field.
ACKNOWLEDGMENTS
All parasite specimens were obtained through more than 20 years of international joint projects in Asia, America, and Africa. We sincerely thank all collaborators.
Footnotes
Financial support: This work was supported by the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Foreign Researchers (no. 26-04103) to Agathe Nkouawa, by the Grant-in-Aid for Scientific Research (B) (15H05273) to Yasuhito Sako, (B) (21406009, 24406011) to Munehiro Okamoto, and (A) (21256003, 24256002) to Akira Ito from JSPS; by the JSPS-Asia/Africa Scientific Platform Fund (2006–2011); and by the Special Coordination Fund for Promoting Science and Technology from the Ministry of Education, Japan (2003–2005 and 2010–2012) to Akira Ito.
Authors' addresses: Agathe Nkouawa, Yasuhito Sako, and Akira Ito, Department of Parasitology, Asahikawa Medical University, Asahikawa, Japan, E-mails: ankouawa@gmail.com, yasusako@asahikawa-med.ac.jp, and akiraito@asahikawa-med.ac.jp. Munehiro Okamoto, Center for Human Evolution Modeling Research, Primate Research Institute, Kyoto University, Inuyama, Japan, E-mail: okamoto.munehiro.6w@kyoto-u.ac.jp.
References
- 1.Garcia HH, Gonzalez AE, Evans CA, Gilman RH. Cysticercosis Working Group in Peru Taenia solium cysticercosis. Lancet. 2003;362:547–556. doi: 10.1016/S0140-6736(03)14117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito A, Nakao M, Wandra T, Suroso T, Okamoto M, Yamasaki H, Sako Y, Nakaya K. Taeniasis and cysticercosis in Asia and the Pacific: present state of knowledge and perspectives. Southeast Asian J Trop Med Public Health. 2005;36((Suppl 4)):S123–S130. [PubMed] [Google Scholar]
- 3.World Health Organization . First WHO Report on Neglected Tropical Diseases: Working to Overcome the Global Impact of Neglected Tropical Diseases. Geneva, Switzerland: World Health Organization; 2010. pp. 97–102. [Google Scholar]
- 4.Winkler AS. Neurocysticercosis in sub-Saharan Africa: a review of prevalence, clinical characteristics, diagnosis, and management. Pathog Glob Health. 2012;106:261–274. doi: 10.1179/2047773212Y.0000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T, Craig PS, Ito A, Chen X, Qiu D, Qiu J, Sato MO, Wandra T, Bradshaw H, Li L, Yang Y, Wang Q. Taeniasis/cysticercosis in a Tibetan population in Sichuan Province, China. Acta Trop. 2006;100:223–231. doi: 10.1016/j.actatropica.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Ito A, Wandra T, Li T, Dekumyoy P, Nkouawa A, Okamoto M, Budke CM. The present situation of human taeniases and cysticercosis in Asia. Recent Pat Antiinfect Drug Discov. 2014;9:173–185. doi: 10.2174/1574891x10666150410125711. [DOI] [PubMed] [Google Scholar]
- 7.Schantz PM, Moore AC, Munoz JL, Hartman BJ, Schaefer JA, Perasaud D, Sarti E, Wilson M, Flisser A, Aron AM. Neurocysticercosis in an Orthodox Jewish community in New York City. N Engl J Med. 1992;327:692–695. doi: 10.1056/NEJM199209033271004. [DOI] [PubMed] [Google Scholar]
- 8.Sorvillo F, Wilkins P, Shafir S, Eberhard M. Public health implications of cysticercosis acquired in the United States. Emerg Infect Dis. 2011;17:1–6. doi: 10.3201/eid1701.101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagida T, Sako Y, Nakao M, Nakaya K, Ito A. Taeniasis and cysticercosis due to Taenia solium in Japan. Parasit Vectors. 2012;5:18. doi: 10.1186/1756-3305-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murrell KD. Epidemiology of taeniosis and cysticercosis. In: Murrell KD, editor. WHO/FAO/OIE Guidelines for the Surveillance, Prevention and Control of Taeniosis/Cysticercosis. Paris, France: OIE, WHO, FAO; 2005. pp. 27–43. [Google Scholar]
- 11.Flisser A, Craig PS, Ito A. Cysticercosis and taeniosis: Taenia solium, Taenia saginata, and Taenia asiatica. In: Palmer SR, Soulsby L, Torgerson PR, Brown DWG, editors. Oxford Textbook of Zoonoses, Biology, Clinical Practice and Public Health Control. Oxford, United Kingdom: Oxford University Press; 2011. pp. 627–644. [Google Scholar]
- 12.Li T, Chen X, Yanagida T, Wang H, Long C, Sako Y, Okamoto M, Wu Y, Giraudoux P, Raoul F, Nkouawa A, Nakao M, Craig PS, Ito A. Detection of human taeniases in Tibetan endemic areas, China. Parasitology. 2013;140:1602–1607. doi: 10.1017/S003118201300111X. [DOI] [PubMed] [Google Scholar]
- 13.Wandra T, Ito A, Swastika K, Dharmawan NS, Sako Y, Okamoto M. Taeniases and cysticercosis in Indonesia: past and present situations. Parasitology. 2013;140:1608–1616. doi: 10.1017/S0031182013000863. [DOI] [PubMed] [Google Scholar]
- 14.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP), a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother. 2013;19:404–411. doi: 10.1007/s10156-013-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nkouawa A, Sako Y, Nakao M, Nakaya K, Ito A. Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J Clin Microbiol. 2009;47:168–174. doi: 10.1128/JCM.01573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sako Y, Nkouawa A, Yanagida T, Ito A. Chapter 9. Loop-mediated isothermal amplification method for a differential identification of human Taenia tapeworms. Nuclear Acid Detection, Methods and Protocols, Methods in Molecular Biology. In: Kolpashchikov DM, Gerasimova YV, editors. Vol. 1039. New York, NY: Humana Press; 2013. pp. 109–120. [DOI] [PubMed] [Google Scholar]
- 18.Nkouawa A, Sako Y, Li T, Chen X, Wandra T, Swastika IK, Nakao M, Yanagida T, Nakaya K, Qiu D, Ito A. Evaluation of a loop-mediated isothermal amplification method using fecal specimens for differential detection of Taenia species from humans. J Clin Microbiol. 2010;48:3350–3352. doi: 10.1128/JCM.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nkouawa A, Sako Y, Li T, Chen X, Nakao M, Yanagida T, Okamoto M, Giraudoux P, Raoul F, Nakaya K, Xiao N, Qiu J, Qiu D, Craig PS, Ito A. Loop-mediated isothermal amplification method for a differential identification of Taenia tapeworms from human: application to a field survey. Parasitol Int. 2012;61:723–725. doi: 10.1016/j.parint.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Yamasaki H, Allan JC, Sato MO, Nakao M, Sako Y, Nakaya Y, Qiu D, Mamuti W, Craig PS, Ito A. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J Clin Microbiol. 2004;42:548–553. doi: 10.1128/JCM.42.2.548-553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakaya K, Mamuti W, Xiao N, Sato MO, Wandra T, Nakao M, Sako Y, Yamasaki H, Ishikawa Y, Craig PS, Schantz PM, Ito A. Usefulness of severe combined immunodeficiency (scid) and inbred mice for studies of cysticercosis and echinococcosis. Parasitol Int. 2006;55((Suppl)):S91–S97. doi: 10.1016/j.parint.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Iseki H, Alhassan A, Ohta N, Thekisoe OM, Yokoyama N, Inoue N, Nambota A, Yasuda J, Igarashi I. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J Microbiol Methods. 2007;71:281–287. doi: 10.1016/j.mimet.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Lee MF, Chen YH, Peng CF. Evaluation of reverse transcription loop-mediated isothermal amplification in conjunction with ELISA-hybridization assay for molecular detection of Mycobacterium tuberculosis. J Microbiol Methods. 2009;76:174–180. doi: 10.1016/j.mimet.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Lee MF, Chen YH, Hsu HJ, Peng CF. One-tube loop-mediated isothermal amplification combined with restriction endonuclease digestion and ELISA for colorimetric detection of resistance to isoniazid, ethambutol and streptomycin in Mycobacterium tuberculosis isolates. J Microbiol Methods. 2010;83:53–58. doi: 10.1016/j.mimet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J Clin Virol. 2013;58:127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki W, Mioulet V, Murray L, Madi M, Haga T, Misawa N, Horii Y, King DP. Development and evaluation of multiplex RT-LAMP assays for rapid and sensitive detection of foot-and-mouth disease virus. J Virol Methods. 2013;192:18–24. doi: 10.1016/j.jviromet.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Njiru ZK. Rapid and sensitive detection of human African trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn Microbiol Infect Dis. 2011;69:205–209. doi: 10.1016/j.diagmicrobio.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 28.Kaewphinit T, Arunrut N, Kiatpathomchai W, Santiwatanakul S, Jaratsing P, Chansiri K. Detection of Mycobacterium tuberculosis by using loop-mediated isothermal amplification combined with a lateral flow dipstick in clinical samples. BioMed Res Int. 2013;2013:926230. doi: 10.1155/2013/926230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yongkiettrakul S, Jaroenram W, Arunrut N, Chareanchim W, Pannengpetch S, Suebsing R, Kiatpathomchai W, Pornthanakasem W, Yuthavong Y, Kongkasuriyachai D. Application of loop-mediated isothermal amplification assay combined with lateral flow dipstick for detection of Plasmodium falciparum and Plasmodium vivax. Parasitol Int. 2014;63:777–784. doi: 10.1016/j.parint.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Jung JH, Oh SJ, Kim YT, Kim SY, Kim WJ, Jung J, Seo TS. Combination of multiplex reverse-transcription loop-mediated isothermal amplification with an immunochromatographic strip for subtyping influenza A virus. Anal Chim Acta. 2015;853:541–547. doi: 10.1016/j.aca.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nurul Najian AB, Engku Nur Syafirah EA, Ismail N, Mohamed M, Yean CY. Development of multiplex loop mediated isothermal amplification (m-LAMP) label-based gold nanoparticles lateral flow dipstick biosensor for detection of pathogenic Leptospira. Anal Chim Acta. 2016;903:142–148. doi: 10.1016/j.aca.2015.11.015. [DOI] [PubMed] [Google Scholar]