To the Editor:
Nuclear factor of activated T cells (NFAT) is a family of transcription factors activated by dephosphorylation mediated by Ca++-activated calcineurin. NFAT coordinates different aspects of T-cell development and activation of T, B, natural killer, and mast cells and is the target of the immunosuppressive drug cyclosporin A.1 We reported recently that targeted deletion of NFATc1 in T cells resulted in inhibition of TH2 and TH17 differentiation.2
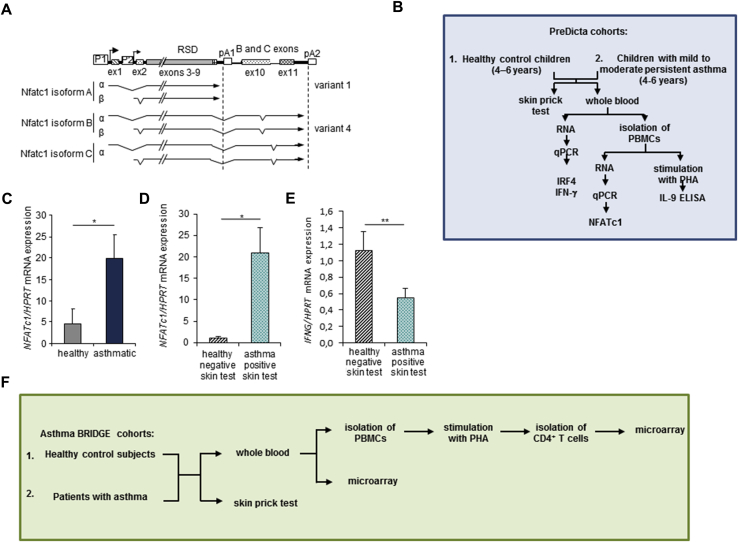
Here we first investigated NFATc1 mRNA expression in PBMCs isolated from healthy and asthmatic children from the PreDicta cohort. Children with allergic asthma expressed significantly more NFATc1 mRNA than healthy control subjects (Fig 1, A-C, and Tables I and II). Furthermore, we found that asthmatic children with a positive skin test result had significantly increased expression of NFATc1 mRNA compared with healthy control subjects (Fig 1, D, and Tables I and II). These results suggested that NFATc1 might have a role in allergic asthma. Consistent with a TH2-inducing function of NFATc1 and a TH2 cytokine inhibitory property of the TH1 cytokine IFN-γ (IFNG), levels of this TH1 cytokine were found to be downregulated in children with asthma and a positive skin test result (Fig 1, E).
Fig 1.
Increased expression of NFATc1 in atopic children with asthma. A,NFATc1 gene transcript isoforms or variants. B, Schematic representation of the PreDicta analysis. C,NFATc1 mRNA expression was analyzed by using quantitative real-time PCR in PBMCs (n = 12-17 children per group). D, Children shown in Fig 1, C, were further subdivided into healthy children with negative skin test results (n = 5) and asthmatic children with positive skin test results (n = 13). E,IFNG mRNA expression analyzed by using quantitative real-time PCR in RNA isolated from whole blood (n = 6 and 15 children per group). F, Schema of the subjects and their obtained gene expression profiles from Asthma BRIDGE Cohort. Statistical significance in this figure was evaluated with the Student t test. *P ≤ .05 and **P ≤ .01. Data are presented as means ± SEMs.
Table I.
Clinical outcome of children with asthma participating in the WP1-UKER cohort in the European PreDicta study
| Group/patient | Age (y) | Sex | PBMCs (106 cells/mL of blood) | Skin prick test | FEV1 actual value (%) | Asthma control | Asthma severity | Cigarette exposure/d | No. of symptomatic episodes before B0 |
|---|---|---|---|---|---|---|---|---|---|
| Asthma | |||||||||
| 1 | 6 | M | 1.75 | Positive | 126 | C | MOPA | 0 | 0 |
| 2 | 6 | M | 2.27 | Positive | 80 | PC | MOPA | 0 | 3 |
| 3 | 5 | F | 2.05 | Positive | 108 | PC | MIPA | 25 | — |
| 4 | 6 | M | 1.98 | Positive | 128 | C | MIPA | 0 | 1 |
| 5 | 5 | M | 2.58 | Positive | 102 | PC | I | 0 | 4 |
| 6 | 5 | F | 2.62 | Positive | 129 | C | I | 0 | 1 |
| 7 | 5 | M | 2.32 | Positive | 143 | PC | I | 0 | 4 |
| 8 | 6 | F | 1.57 | Positive | 94 | PC | I | 0 | 3 |
| 9 | 4 | M | 1.96 | Positive | 115 | PC | MOPA | 0 | 3 |
| 10 | 5 | F | 1.93 | Positive | 92 | U | MOPA | 0 | 15 |
| 11 | 6 | F | 2.05 | Positive | 111 | C | I | 0 | 2 |
| 12 | 5 | M | 1.14 | Positive | 99 | C | I | 0 | 0 |
| 13 | 4 | F | 2.89 | Negative | 135 | C | I | 0 | 4 |
| 14 | 4 | M | 1.91 | ND | 96 | C | I | 5 | 4 |
| 15 | 5 | M | 1.29 | Positive | 80 | C | I | 0 | 2 |
| 16 | 5 | M | 1.83 | Positive | 86 | C | I | 0 | 1 |
| 17 | 5 | M | 2.50 | Positive | 107 | C | I | 0 | 2 |
| 18 | 4 | M | 1.54 | Positive | 71 | PC | I | 0 | 3 |
| 19 | 4 | M | 2.20 | Negative | 86 | C | I | 0 | 3 |
| 20 | 5 | F | 2.27 | ND | 98 | C | I | 0 | 1 |
| 21 | 5 | F | 1.77 | Negative | — | U | MIPA | 0 | 20 |
| 22 | 5 | M | 1.39 | Positive | 81 | C | MIPA | 0 | 3-5 |
| Mean | 5.00 | 14 M | 1.99 | 103.19 | 1.36 | ||||
| SEM | 0.14 | 8 F | 0.09 | 4.20 | 1.15 | ||||
B0, Baseline visit; C, controlled; F, female; I, intermittent; M, male; MIPA, mild persistent asthma; MOPA, moderate persistent asthma; ND, not done; PC, partially controlled; U, uncontrolled; UKER, Uniklinikum Erlangen; WP1, Work Program 1.
Table II.
Clinical outcome of control children participating in the WP1-UKER cohort in the European PreDicta study
| Group/patient | Age (y) | Sex | PBMCs (106 cells/mL of blood) | Skin prick test | FEV1 actual value (%) | Cigarette exposure/d | No. of symptomatic episodes before B0 |
|---|---|---|---|---|---|---|---|
| Healthy subjects | |||||||
| 1 | 6 | M | 1.14 | ND | — | 0 | — |
| 2 | 6 | F | 2.95 | ND | 121 | 0 | — |
| 3 | 5 | F | 1.79 | Negative | 81 | 0 | — |
| 4 | 4 | M | 2.01 | ND | — | 0 | — |
| 5 | 6 | M | 1.41 | ND | 105 | 0 | — |
| 6 | 4 | F | 2.14 | ND | 109 | 0 | — |
| 7 | 6 | M | 2.86 | ND | 87 | 0 | — |
| 8 | 4 | M | 1.73 | Negative | 100 | 0 | — |
| 9 | 5 | F | 1.67 | ND | 112 | 0 | — |
| 10 | 5 | F | 2.01 | Positive | 119 | 0 | — |
| 11 | 4 | M | 2.43 | Positive | 116 | 0 | — |
| 12 | 5 | M | 2.32 | ND | 111 | 0 | — |
| 13 | 4 | M | 2.40 | Negative | 109 | 0 | — |
| 14 | 4 | F | 1.60 | Negative | 109 | 0 | — |
| 15 | 5 | M | 2.22 | Negative | 92 | 0 | — |
| 16 | 4 | M | 2.88 | Negative | — | 0 | — |
| 17 | 4 | M | 1.38 | Negative | 109 | 0 | — |
| 18 | 5 | M | 1.13 | ND | 110 | 0 | — |
| 19 | 4 | M | 1.77 | ND | 118 | 0 | — |
| 20 | 5 | F | 1.50 | ND | — | 0 | — |
| Mean | 4.75 | 13 M | 1.97 | 106.75 | 0 | ||
| SEM | 0.17 | 7 F | 0.12 | 2.48 | 0 | ||
B0, Baseline visit; F, female; M, male; ND, not done; UKER, Uniklinikum Erlangen; WP1, Work Program 1.
As in PBMCs from the PreDicta cohort, NFATc1 showed altered expression levels in peripheral blood CD4+ T cells from 300 children and adults from the Asthma BRIDGE cohort (P < .05; Fig 1, F, and Table III). This cohort was described previously by Raby et al,3 and some of the microarray data regarding genes other than NFATc1 and IRF4 were described before.4 Both the PreDicta study and the Asthma BRIDGE study used the same phenotype definitions. The shorter isoform A (variant 1) lacks the C-terminal extension of approximately 245 amino acids that is present in all other NFAT proteins. This isoform of NFATc1 was found to be significantly overexpressed in peripheral blood CD4+ T cells from asthmatic patients with a positive skin test result compared with both asthmatic patients with a negative skin test result and nonasthmatic subjects with a negative skin test result (Table III). Moreover, the induced isoform D (variant 4) of NFATc1 was also found to be differentially expressed between asthmatic patients with a positive skin test result and nonasthmatic subjects with a negative skin test result (Table III). The log fold change (FC) for NFATc1 expression was 0.21 (1.2-fold increase), with an average expression of 7.44 (P = .013). The log FC for NFATc1 expression was 0.1 (1.1-fold increase), with an average expression of 7.83 (P = .013). Adding the 84 white asthmatic patients with positive skin test results to the model and adjusting for age, race, and sex (146 black plus 84 white vs 39 black subjects; female/male ratio, 127:106) did not change the NFATc1 result (log FC = 0.1, P = .018). Induction of the short isoform A of NFATc1 takes place after activation of T cells and is controlled by promoter 1 (Fig 1, A), a strong inducible promoter. This effect is autoregulated by the NFAT transcription factors.5 NFATc1/A is thought to be needed for exerting effector functions in activated T cells. In contrast to NFATc1/C, NFATc2, and NFATc3 proteins, the short isoform A of NFATc1 is not able to promote cell apoptosis.5 We presume that this function of NFATc1 leads to a prolonged survival of effector T cells in asthmatic patients with a positive skin test result. Moreover, it has been observed that NFATc1/αA (Fig 1, A) is the most prominent NFATc1 protein on receptor stimulation of peripheral B and T cells. In these cells the first activation, through the respective receptor, of the full induction of NFATc1/αA requires 24 hours. By contrast, this isoform is completely induced within a few hours after secondary stimulation.5 This is in line with our findings that NFATc1/A is upregulated in CD4+ T cells of asthmatic patients with a positive skin test result because these patients were already sensitized to an allergen. Moreover, we observed that NFATc1 isoform D (variant 4) was upregulated in asthmatic patients with a positive skin test result. To our knowledge, the function of NFATc1/D has not been described yet.
Table III.
Comparison between different groups of children and adults with or without asthma and atopy (Asthma BRIDGE)
| Group comparisons | Asthma +, skin test + (n = 251) |
|---|---|
| Asthma −, skin test + (n = 44) | P = .1 |
| Asthma +, skin test − (n = 47) | P = .006 (NFATc1 isoform A) |
| Asthma −, skin test − (n = 75) |
P = .002 (NFATc1 isoform A); P = .05 (NFATc1 isoform D) |
For analysis using a microarray, there were 47,009 tag probes to target individual exons, including 4 exon sequences linked to specific NFATc1 isoforms. The particular isoforms of NFATc1 mRNA were as follows: NFATc1 isoform A = variant 1; NM 172390.2; NP 765978.1; NFATc1 isoform D = variant 4; NM 172388.2; NP 765976.1; Gene ID: 4772. P values for association of probes with phenotypes were adjusted by using the Benjamini-Hochberg method for multiple comparisons between groups.
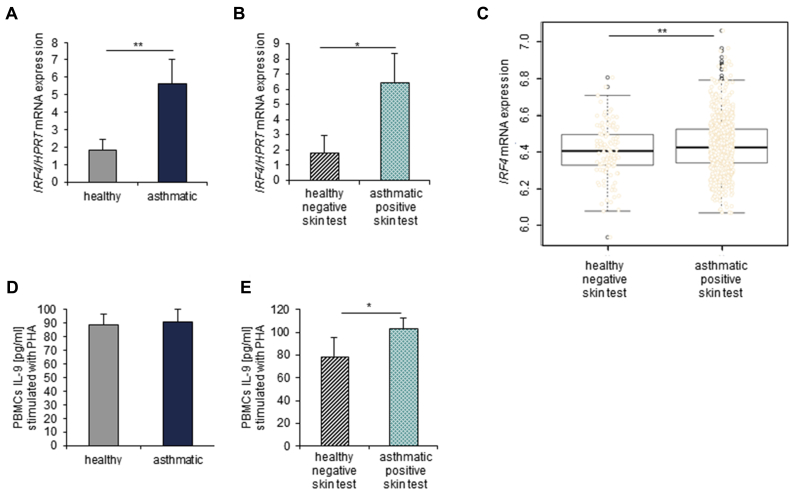
In addition to NFATc1, the transcription factor interferon regulatory factor 4 (IRF4), which is encoded by the IRF4 gene, has been shown to play a role in the differentiation of various T-cell subsets known to have an effect on asthma pathology.6 Therefore we investigated IRF4 mRNA expression in whole blood from healthy and asthmatic children from PreDicta (Fig 2, A). We found that the asthmatic children had a significantly increased expression of IRF4 compared with that seen in healthy control subjects (Fig 2, A), especially when they also had a positive skin test result (Fig 2, B). This observation was further confirmed by analyzing total blood cells in Asthma BRIDGE asthmatic patients with positive skin test results (P = .02; Fig 2, C). IRF4 is known to be an important factor for innate and adaptive immune responses, cooperating with various other transcription factors, including NFAT family members, to act as both a transcriptional repressor and activator.7 Therefore it is likely that IRF4 interacts with NFATc1 to prolong the survival of effector T cells in patients with allergic asthma. Namely, IRF4 would directly promote the production of IL-4, one of the TH2-associated cytokines that contributes to asthma pathogenesis, by binding to the IL-4 promoter in cooperation with NFATc1. In support of this notion, we found reduced IFNG mRNA expression in the group of children with positive skin test results who had higher levels of NFATc1 and IRF4. Consistently, we previously reported that asthmatic mice deficient in NFATc1 in T cells (NFATc1fl/flxCD4Cre) have increased numbers of IFN-γ+CD4+ T cells in their lungs and that these cells expressed less Batf, a transcription factor essential for immunoglobulin class-switching that cooperates with IRF4 at the promoter of different genes relevant for asthma8 and that NFATc1fl/flxCD4Cre mice have reduced serum levels of ovalbumin (OVA)–specific IgE.2 Moreover, these data are also supported by our findings in the basic leucine zipper transcription factor, ATF-like (BATF)–deficient mice, which do not induce IgE, and we demonstrated T cells that produce increased IFN-γ levels9 in a model of allergic asthma.
Fig 2.
Increased mRNA expression and IL-9 production in asthmatic patients with a positive skin test result. A,IRF4 mRNA expression analyzed by using quantitative real-time PCR in RNA isolated from whole blood (n = 13-14 children per group). B and C, Further subdivision of children shown in Fig 2, A, into healthy subjects with negative skin test results (n = 6, PreDicta, Fig 2, B; n = 104, Asthma BRIDGE, Fig 2, C) and asthmatic patients with positive skin test results (n = 8, PreDicta, Fig 2, B; n = 144, Asthma BRIDGE, Fig 2, C). D and E, PBMCs from the PreDicta children were cultured with PHA for 48 hours, and afterward, ELISA was performed for IL-9 production on cell supernatants (Fig 2, D: n = 19 healthy children and n = 20 asthmatic children; Fig 2, E: n = 8 healthy children with negative skin test results and n = 15 asthmatic children with positive skin test results). The Student t test was used to evaluate statistical significance. *P ≤ .05 and **P ≤ .01. Data are presented as means ± SEs.
Because IRF4 is also known to be crucial for IL-9 production,6 we also investigated IL-9 in cultured PBMCs of preschool children stimulated with PHA. We could not find any differences in IL-9 production when we compared asthmatic and healthy control children (Fig 2, D), but we observed a significant increase in IL-9 levels in the supernatants of PBMCs isolated from asthmatic children with an additional positive skin test result compared with those isolated from healthy children with a negative skin test result (Fig 2, E).
Both NFATc1 and IRF4 also positively influence IL-9 production of TH9 cells.6 Consistent with the increased expression of both IRF4 and NFATc1 seen in asthmatic patients with positive skin test results, we also found increased IL-9 levels in their PBMCs.
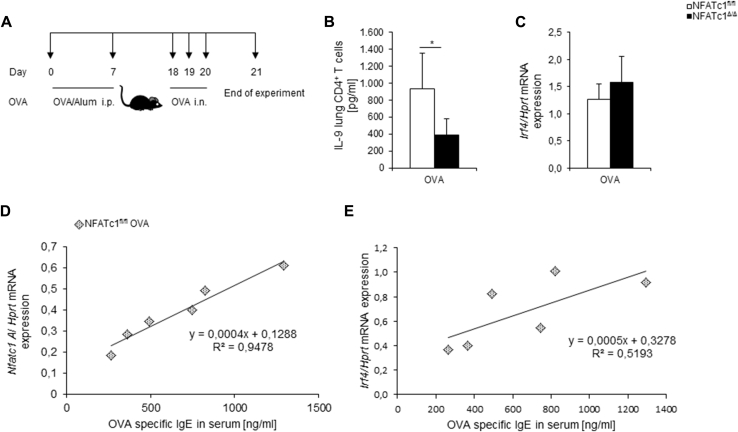
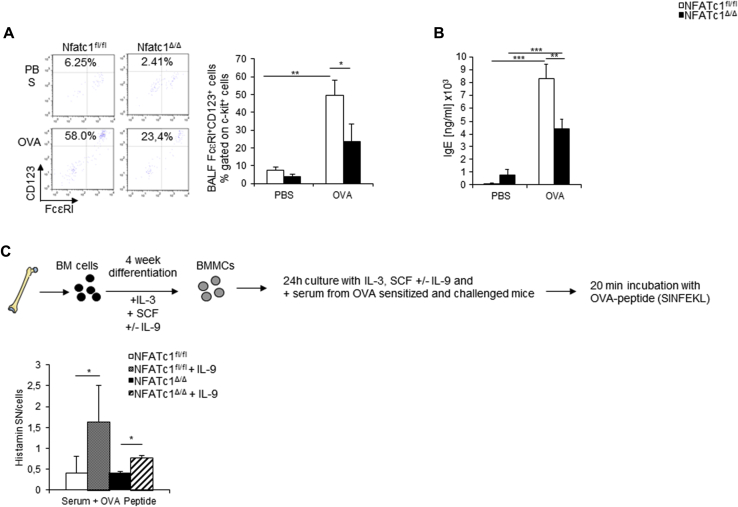
Overall, we found that in PBMCs and CD4+ T cells of asthmatic patients with a positive skin test result, expression of NFATc1 isoforms and IRF4 was increased, suggesting a fundamental role for both transcription factors in the immunologic switch in allergic asthma. In this survey we analyzed subjects from 2 different studies. Although the PreDicta study includes only 42 children, we obtained important indications regarding NFATc1, IRF4, and IL-9 expression in patients with allergic asthma. These results could be verified and confirmed in the bigger cohorts of the Asthma BRIDGE study, in which 300 asthmatic patients and 122 control subjects were included. The functional relevance of NFATc1 in asthmatic patients was supported by our observations in targeted conditional deletion of NFATc1 in T lymphocytes, where Nfatc1/A mRNA expression perfectly correlated with OVA-specific IgE levels (R2 = 0.94) in patients with experimental asthma (see Fig E1, D, and the Methods, Results, and Discussion sections in this article's Online Repository at www.jacionline.org) and resulted in downregulation of IL-9 (see Fig E1, B) and mast cell function (see Figs E2 and E3 and the Methods, Results, and Discussion sections in this article's Online Repository at www.jacionline.org). Therefore targeting NFATc1 in T lymphocytes might ameliorate the allergic phenotype seen in asthmatic patients.
Fig E1.
Decreased IL-9 production in NFATc1fl/flxCD4Cre mice after allergen sensitization and challenge. A, Experimental design of the OVA-induced asthma model. B, IL-9 protein concentrations were measured in supernatants of purified lung CD4+ T cells from allergen-treated mice and cultured for 24 hours with α-CD3 and α-CD28 antibodies (n = 7-11 mice per group). C,Irf4 mRNA expression measured by using quantitative real-time PCR in isolated lung CD4+ T cells (n = 8-13 mice per group). D, Correlation between Nfatc1/A mRNA expression in lung CD4+ T cells and OVA-specific IgE levels in serum of Nfatc1fl/fl OVA-treated mice (n = 6). E, Correlation between Irf4 mRNA expression in lung CD4+ T cells and OVA-specific IgE in serum of Nfatc1fl/fl OVA mice (n = 6). The Student t test was used to evaluate statistical significance. *P ≤ .05. Data are presented as means ± SEMs.
Fig E2.
Decreased mast cell numbers and activation in NFATc1fl/flxCD4Cre mice. A, Mast cell numbers (c-kit+FcεRI+CD123+ cells) were analyzed by using flow cytometry in the lungs of NFATc1fl/fl control mice and NFATc1fl/flx CD4Cre mice (n = 4-11 mice per group). B, IgE serum levels were measured with ELISA (n = 12-19 mice per group). C, Experimental design for bone marrow–derived mast cell (BMMC) differentiation (upper panel) and histamine release (lower panel). Histamine release was measured by means of ELISA (lower panel; n = 4 mice per group). Mast cells differentiated from Nfatc1fl/flxCD4Cre mice received serum from OVA-sensitized and challenged Nfatc1fl/flxCD4Cre mice, whereas mast cells from Nfatc1fl/fl mice received serum from OVA-sensitized and challenged Nfatc1fl/fl mice. Statistical significances in this figure were evaluated with the Student t test. *P ≤ .05, **P ≤ .01, and ***P ≤ .001. Data are presented as means ± SEMs.
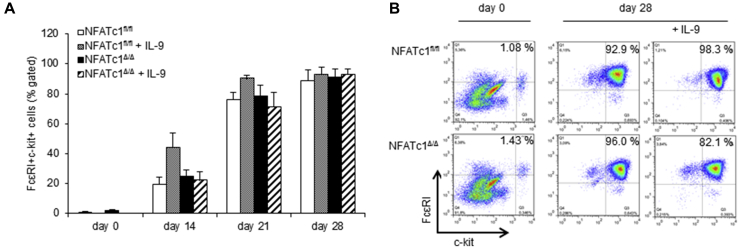
Fig E3.
Mast cell differentiation. A, Flow cytometric analysis of bone marrow–derived mast cell differentiation, as described in the Methods section in this article's Online Repository. FACS analysis for FcεRI+c-kit+ cells was performed every week to monitor the differentiation status of the cultured cells. B, Corresponding dot plots from the first and fourth weeks of mast cell differentiation are shown.
Acknowledgments
We thank Rebekka Springel and Sonja Trump at the Molecular Pneumology Department in Erlangen and Lena Schramm, Ines Yava, and Evelin Muschiol at the Children's Hospital in Erlangen for their technical assistance.
Footnotes
Supported by the European grant PreDicta (no. 4435). Work on Asthma BRIDGE was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH/NHLBI; grant no. RC2 HL101543).
Disclosure of potential conflict of interest: A. Graser has received research and travel support from PreDicta (SFB643) and is employed by University Hospital Erlangen. B. A. Raby is an unpaid board member for CureSpark and receives royalties from UpToDate. S. Finotto has received research and travel support from PreDicta (SFB643), is employed by University Hospital Erlangen, and has received research support from CCC-Erlangen. The rest of the authors declare that they have no relevant conflicts of interest.
Methods
Participants (PreDicta study)
Two cohorts of preschool-aged children with and without asthma at the age of 4 to 6 years were recruited at the Children's Hospital of Erlangen, Department of Pediatrics and Adolescent Medicine. This research was conducted through the European study “Post-infectious immune reprogramming and its association with persistence and chronicity of respiratory allergic diseases” (PreDicta), a multicenter prospective cohort study carried out in 5 different centers in Europe. The study was approved by the ethics committee of the Friedrich-Alexander University Erlangen-Nürnberg, Germany (reference no. 4435), and is also registered in the German Clinical Trial Register (registration no. DRKS00004914). Informed consent was obtained from the parents of all children of the PreDicta study. Twenty-two asthmatic patients and 20 healthy subjects were analyzed.
Inclusion criteria for cases were age of 4 to 6 years, gestational age of 36 weeks or greater, and diagnosis of asthma within the last 2 years confirmed by a doctor of the Children's Hospital in Erlangen of mild-to-moderate persistent severity according to Global Initiative for Asthma (GINA) guidelines (2005). In this study we have assessed the degree of asthma as follows.
Severity level I is defined as intermittent asthma. The children are free of asthma symptoms for at least 2 months with an FEV1 of greater than 80% and a mean expiratory flow (MEF) of greater than 60%.
Severity level II is defined as mild persistent asthma. The children are symptom free for a period of less than 2 months. At the moment, they do not have asthmatic symptoms, with an FEV1 of greater than 80% and an MEF of greater than 60%.
Severity level III is defined as moderate persistent asthma. The children have asthma symptoms several days a week with an FEV1 of less than 80% and an MEF of less than 65%. At the B0 visit, parents were asked to answer questions about asthma control according to 2009 GINA guidelines. Possible answers were as follows: (1) severity I, which means controlled asthma; (2) severity II, which means partly controlled asthma; and (3) severity III, which means uncontrolled asthma.
The GINA guidelines take into account FEV1, limitation of physical activity, frequency of exacerbations, and use of asthma “reliever” medication. Exclusion criteria were severe or brittle asthma, children receiving immunotherapy or more than 6 courses of oral steroids during the preceding 12 months, and children with other chronic respiratory diseases (cystic fibrosis, bronchopulmonary dysplasia, and immunodeficiencies), except allergic rhinitis. Furthermore, children with other chronic diseases or chronic medication use, except atopic eczema, were excluded. The children performed a peak expiratory flow maneuver. Atopy was proved by at least 1 positive skin prick test response (wheal size, ≥3 mm; HAL Allergie GmbH, Düsseldorf, Germany). For skin prick tests, we analyzed allergic reactions to the following allergens: cat, house dust mite, Alternaria species, birch, grass, Ambrosia species, and mold. Asthma was confirmed by a physician's diagnosis of mucus production, airway hyperresponsiveness, and dyspnea. At visit B0, the doctor asked the mother several questions to evaluate the child's asthma symptoms during the last 12 months. The control subjects had no history of asthma, wheezing, or atopic illness.
Isolation and culture of human PBMCs (PreDicta study)
Peripheral blood from control or asthmatic children was collected into venous blood collection tubes with lithium heparin (BD Vacutainer Tubes; BD Biosciences, San Jose, Calif); the heparinized blood was transferred to a sterile 15-mL tube and diluted with an equal volume of warm saline and mixed well. The diluted blood was carefully stratified on top of the density gradient medium Biocoll separating solution (Biochrom AG, Berlin, Germany) and centrifuged. Afterward, the layer of PBMCs, which can be found between plasma and Biocoll, was transferred to a new sterile 15-mL tube. After washing the cells twice with RPMI 1640 medium, cells were used for RNA isolation, as described below. Some of the cells were cultured in complete culture medium for 48 hours with PHA (10 μg/mL). The complete culture medium consisted of RPMI-1640 supplemented with 25 mmol/L HEPES and l-Glutamine (Gibco, Invitrogen, Thermo Fisher Scientific, Waltham, Mass), 100 IU/mL penicillin (Sigma-Aldrich, St Louis, Mo), 100 μg/mL streptomycin (Sigma-Aldrich), 50 μmol/L β-mercaptoethanol (Sigma-Aldrich), 200 mmol/L 1% l-glutamine (Sigma-Aldrich), 1% MEM Vitamin (Sigma-Aldrich), 1% nonessential amino acids (Sigma-Aldrich), 1% sodium pyruvate (Sigma-Aldrich), and 10% H-FBS (Sigma-Aldrich).
RNA isolation from whole blood (PreDicta study)
Additionally, whole blood from each child from the PreDicta cohort was collected into Tempus Blood RNA Tubes (Life Technologies, Carlsbad, Calif) and stored at −80°C. RNA was isolated and purified by using RNA binding beads with a MagMAX for Stabilized Blood Tube RNA Isolation Kit (Ambion, Life Technologies), according to the manufacturer's instructions.
Isolation of human CD4+ T cells (Asthma BRIGDE study)
Asthma BRIDGE is an open-access biorepository for subjects participating in genetic studies of asthma in the EVE Consortium.E1 Sample collection and processing were carried out at each institution according to standardized and validated protocols. These samples were centralized at the Data Coordinating Center at the Channing Division of Network Medicine at Brigham and Women's Hospital at the Harvard Medical School (Boston, Mass). Contributing centers isolating peripheral blood CD4+ T lymphocytes used a modified version of the protocol previously optimized for collections in the Childhood Asthma Management Program study by using anti-CD4+ microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) and column separation. The modification includes isolating PBMCs and then stimulating them with PHA before CD4+ lymphocyte isolation.
NFATc1 and IRF4 microarray analysis of human peripheral blood CD4+ T cells or whole blood cells (Asthma BRIDGE study)
NFATc1 alone or in cooperation with IRF4 is known to be important for the differentiation of different subsets of lymphocytes that play a crucial role in asthma development.E2, E3, E4, E5, E6 To analyze these factors, microarray analysis was used within the Asthma BRIDGE study. Therefore samples for RNA extraction were collected with BD Vacutainer CPT tubes (BD Diagnostics, Franklin Lakes, NJ), placed on ice, and centrifuged within 1 hour (20 minutes at 1700 relative centrifugal force). Total RNA was extracted by using the RNeasy Mini Protocol (Qiagen, Valencia, Calif) and stored at −80°C. Analysis with the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, Calif) confirmed average total RNA yields of 2 μg per collection, with minimal evidence of RNA degradation and 28S/18S ratios approaching 2.0. Some of the microarray data regarding genes other than NFATc1 and IRF4 were described before.E7, E8
For our differential expression analyses, we studied 300 asthmatic Asthma BRIDGE subjects with log2-transformed and quantile-normalized gene expression (n = 47,009 probes) from unstimulated blood-derived CD4+ T cells measured with Illumina Human HT-12 v4 BeadChips (Illumina, San Diego, Calif). Asthma was defined based on having a doctor's diagnosis of the disease with evidence of reversible airflow obstruction with a bronchodilator. The R Bioconductor limma package (version 3.20.1)E9 was used to perform differential expression analysis. A linear model was fitted along with implementation of empiric Bayes statistics to assess whether expression levels of the candidate gene NFATc1 were significantly altered among asthmatic patients after stratifying by race and adjusting for age and sex. Two differential expression analyses were performed: (1) comparing asthmatic patients with positive skin test results (defined as a wheal ≥3 mm in size) with atopic dermatitis (eczema) with those without eczema and (2) comparing asthmatic patients with positive skin test results with those with negative skin test results.
Adult asthmatic patients with positive skin test results (Asthma BIRDGE study)
Eighty-seven white asthmatic patients with positive skin test results were identified within the Asthma BRIDGE study. After removing those asthmatic patients with missing information on unknown conditions of atopic eczema or skin tests, 82 subjects (female/male subjects, 34/48), consisting of 49 asthmatic patients with eczema and positive skin test results and 33 asthmatic patients without eczema and positive skin test results, remained for analysis.
One hundred forty-seven black asthmatic patients with positive and negative skin test results were identified within the Asthma BRIDGE study. After removing 1 subject with missing information, 146 subjects (female/male subjects, 91/55), consisting of 107 asthmatic patients with positive skin test results and 39 asthmatic patients with negative skin test results, remained for analysis.
Mice
NFATc1fl/fl control mice and NFATc1fl/flxCD4Cre mice were on a C57BL/6 background and kept in house under specific pathogen-free conditions. Mice were 6 to 8 weeks old in all experiments. The experiments were performed with approved licenses (23-177-07/G09-1-008 from the ethical review board Rheinland-Pfalz and 54-2532.1-2/10 and 54-2532.1-55/12 from the government of Mittelfranken, Bavaria).
Allergen sensitization and challenge
For the experimental asthma model, mice were sensitized with OVA (100 μg; Calbiochem, San Diego, Calif) complexed with alum (10%; Sigma-Aldrich, Steinheim, Germany) on days 0 and 7 administered intraperitoneally. Allergen challenge was performed with an intranasal treatment of OVA (2 mg/mL) on days 18, 19, and 20, as described previously.E10 Mice were killed on day 21, and lung cells were isolated.
Isolation of murine lung CD4+ T cells
CD4+ T cells were purified from isolated total lung cell suspensions, as previously described.E11 Shortly, total lung cells were incubated with anti-mouse CD4 L3T4 microbeads and positively sorted in a magnetic cell sorter system (MACS; Miltenyi Biotech, Germany), according to the manufacturer's protocol. Afterward, CD4+ T cells were cultured in RPMI with plate-bound α-CD3 (2 μg/mL) and soluble α-CD28 (2 μg/mL) antibodies for 24 hours in a density of 106 cells/mL. Supernatants were analyzed by means of ELISA for protein production.
Isolation and differentiation of murine bone marrow–derived mast cells
Hind legs of the mice were prepared, and bone marrow cells were flushed from the tibias and femurs with sterile PBS. Afterward, cells were centrifuged for 5 minutes at 4°C and 1500 rpm. Cells were then resuspended in RPMI 1640 culture medium supplemented with 10% FBS, 1% penicillin/streptomycin, 1% L-glutamine, 1% HEPES, 50 μmol of β-mercaptoethanol, and 10 ng/mL IL-3 and stem cell factor (PeproTech, Hamburg, Germany) and cultured for 4 weeks in 10 mL of the medium in small culture flasks, as previously described.E12 Some of the cells were additionally cultured with 10 ng/mL recombinant IL-9 (PeproTech). Every week, medium with cytokines was replaced, and fluorescence-activated cell sorting (FACS) analysis was performed to control mast cell differentiation.
Histamine release by murine bone marrow–derived mast cells
After 4 weeks of cell culture, mast cells were resuspended in warm Tyrode buffer (1 × 107 cells/mL of buffer) and incubated with 10 μg/mL purified mouse IgE, κ isotype control antibody (BioLegend, Fell, Germany) for 30 minutes at 37°C. Afterward, cells were washed twice with warm Tyrode buffer and centrifuged for 5 minutes at 4°C and 1500 rpm. The cell pellet was then resuspended again in warm Tyrode buffer (5 × 106 cells/mL of buffer) and incubated with TNP-conjugated BSA (Santa Cruz Biotechnology, Heidelberg, Germany) for 10 minutes at 37°C. Thereafter, 1 mL of cold Tyrode buffer was added, and cells were centrifuged for 5 minutes at 4°C and 1500 rpm. Supernatants were collected, and the cell pellet was resuspended in Tyrode buffer. Then, cells and supernatants were boiled for 10 minutes at 99°C and frozen at −80°C until the ELISA for histamine was performed.
Some of the mast cells were plated in a 96-well plate overnight with medium and cytokines and serum (1:1 media and serum) from OVA-sensitized and challenged NFATc1fl/fl control mice and NFATc1fl/flxCD4Cre mice. The next day, cells were resuspended and incubated in warm Tyrode buffer with OVA peptide (500 μg/mL; SINFEKL) for 20 minutes at 37°C. Afterward, 1 mL of cold Tyrode buffer was added, and cells were centrifuged for 5 minutes at 4°C and 1500 rpm. Supernatants were collected, and cell pellets were resuspended in Tyrode buffer. Thereafter, cells and supernatants were boiled for 10 minutes at 99°C and then frozen at −80°C until ELISA for histamine was performed.
Flow cytometric analysis
Brochoalveolar lavage (BAL) of the right lung was performed with 0.8 mL of saline 2 times. Total BAL fluid was collected and centrifuged for 5 minutes at 4°C. Cell pellets were resuspended in 1 mL of PBS and used for FACS analysis. BAL fluid cells and bone marrow–derived mast cells were stained with FACS antibodies for 30 minutes at 4°C, washed in PBS, and then analyzed with a FACSCalibur (BD PharMingen, Heidelberg, Germany). The following antibodies were used: anti-mouse FcεRI, CD123, and c-kit antibodies (eBioscience, Frankfurt, Germany). Afterward, measurements were analyzed with FlowJo software (TreeStar, Ashland, Ore).
ELISA
Human and mouse IL-9 were detected by using a Ready-SET-Go! ELISA kit from eBioscience (sensitivity, 1 pg/mL). Murine serum IgE was detected with the OptEIA sandwich ELISA kit from BD Biosciences (1562-100 ng/mL), and OVA-specific IgE was detected with the LEGEND MAX ELISA kit from BioLegend (sensitivity, 20.7 pg/mL). Histamine was measured with Histamine ELISA Fast Track (sensitivity, 0.2 ng/mL; Labor Diagnostika Nord GmbH, Nordhorn, Germany).
RNA isolation and quantitative real time-PCR
PBMCs were directly lysed, and total RNA was extracted by using PeqGold RNA Pure, according to the manufacturer's protocol (PeqLab, Erlangen, Germany). The RNA concentration was determined by using a spectrophotometer (Nanodrop; Peqlab, Erlangen, Germany). RNA was then reverse transcribed with the first-strand cDNA synthesis kit (MBI Fermentas, St Leon-Rot, Germany). The resulting cDNA was amplified by means of quantitative real-time PCR with SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Munich, Germany). Quantitative real-time PCR was performed with a cycle of 2 minutes at 98°C; 50 cycles of 5 seconds at 95°C, 10 seconds at 60°C, and 5 seconds at 65°C; and 5 seconds at 95°C in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories).
The following primers and sequences were used:
-
•
human hypoxanthine phosphoribosyltransferase 1 (HPRT): 5′-TGACACTGGCAAAACAATGCA-3′ and 5′-GGTCCTTTTCACCAGCAAGCT-3′;
-
•
human IFNG: 5′-TGA CCA GAG CAT CCA AAA GA-3′ and 5′- TGA CCA GAG CAT CCA AAA GA-3′;
-
•
human IRF4: 5′-AGACTGTGCCAGAGCAGGAT-3′ and 5′-GGGTCTGGAAACTCCTCTCC-3′;
-
•
human NFATc1: 5′- GCATCACAGGGAAGACCGTGTC-3′ and 5′-GAAGTTCAATGTCGGAGTTTCTGAG-3′;
-
•
murine Hprt: 5′-GCCCCAAAATGGTTAAGGTT-3′ and 5′-TTGCGCTCATCTTAGGCTTT;
-
•
murine Irf4: 5′-ACGCTGCCCTCTTCAAGGCTT-3′ and 5′-TGGCTCCTCTCGACCAATTCC-3′; and
-
•
murine Nfatc1/A: 5′-ACCTGTGCAAGCCAAATTCC-3′ and 5′-AGAGTTACCATTGGCAGGAAG-3′.
The mRNA expression was normalized to HPRT, and FC differences were calculated by using the ΔΔ cycle threshold method.
Statistical analysis
Differences were evaluated for significance by using the Student 2-tailed t test for independent events (PreDicta study, murine model). The coefficient of correlations was calculated by using statistical analysis in Excel software. Data are presented as means ± SEMs. For the Asthma BRIDGE Study, the R Bioconductor limma package (version 3.20.1) was used to perform differential expression analysis in gene microarray analyses.
Results and Discussion
To further explore our human data on NFATc1, we further analyzed the recently described mice that lack NFAC1 in T cells (NFATc1fl/flxCD4Cre) in a murine model of asthma. IL-9 is a critical cytokine in the pathogenesis of allergic asthma, levels of which were found to be upregulated in asthmatic patients with a positive skin test result. Thus we next examined IL-9 production in a murine model of asthma in the absence of NFATc1 in T cells (Fig E1, A and B). In isolated lung CD4+ T cells we found that IL-9 levels were decreased in allergen-sensitized and challenged NFATc1fl/flxCD4Cre mice compared with those in NFATc1fl/fl control mice (Fig E1, B). By contrast, Irf4 was found not to be regulated by NFATc1 (Fig E1, C). Finally, we found an almost perfect direct correlation between OVA-specific IgE levels in the serum and Nfatc1/A in lung CD4+ T cells (Fig E1, D) in this model of allergic asthma. Irf4 also positively correlated with OVA-specific IgE levels in serum but not as much as Nfatc1/A (Fig E1, E).
Although our data suggest that NFATc1 does not influence the expression of IRF4, it is already known that IRF4 cooperates and interacts with NFAT.E13 It might be that this interaction is reduced in the conditional NFATc1 knockout mice, and this could explain the reduced production of IL-9 in the absence of NFATc1.
IRF4 is known to be essential for IL-9 production, and now we found that NFATc1 also has an effect on IL-9 production either through a direct effect on the promoter of IL-9 (alone or along with IRF4) or through its influence on IL-2E14 or IL-4, which are required for IL-9 production. In fact, TGF-β and IL-4 are required to induce TH9 differentiation.E15
IL-9 has been shown to be a growth factor for mast cells.E16, E17 Thus we investigated the number of mast cells in the BAL fluid of NFATc1fl/fl control and NFATc1fl/flxCD4Cre mice (Fig E2, A). We observed that mast cell numbers are significantly increased after allergen sensitization and challenge in NFATc1fl/fl control mice compared with those in NFATc1fl/flxCD4Cre mice (Fig E2, A). Because the number of mast cells is decreased in the absence of NFATc1 in T cells and because IgE crosslinking on the surfaces of mast cells leads to their activation and degranulation, we also investigated serum levels of IgE. We could observe that sensitized and challenged NFATc1fl/flxCD4Cre mice had lower total levels of IgE in their serum than NFATc1fl/fl control mice (Fig E2, B).
OVA-specific IgE levels are also significantly reduced in our asthma model in Nfatc1fl/flxCD4Cre mice compared with those in control mice.E2 Moreover, we reported here that OVA-specific IgE levels directly correlated with NFATc1 in a model of allergic asthma. In addition, we previously reported that reduced expression of CD40 ligand on CD4 T cells of Nfatc1fl/flxCD4Cre mice indicates a reduced interaction between the T and B cells in these mice, which results in lower IgE production.E2
NFATc1 is known to be expressed in mast cells, where it influences the production of IL-13 and TNF-α after FcεRI signal induction, without influencing the differentiation and activation of these cells.E17 IgE levels in serum were also found to be downregulated in the absence of NFATc1 in asthmatic mice. IL-9 is known to be produced by mast cells in an autocrine manner after IgE crosslinking. It then promotes the release of numerous preformed mediators, such as histamine and IL-1β, which further induce IL-9 production and several cytokines, including IL-5 and IL-13.E18
We then went on to further analyze the effect of IL-9 on mast cell differentiation, expansion, and histamine release. To this aim, we differentiated mast cells from the bone marrow of NFATc1fl/fl control mice and NFATc1fl/flxCD4Cre mice for 4 weeks (Fig E2, C). Mast cells from NFATc1fl/fl control mice were then cultured overnight with serum from OVA-sensitized and challenged NFATc1fl/fl control mice, and mast cells from NFATc1fl/flxCD4Cre mice were cultured with serum from OVA sensitized and challenged NFATc1fl/flxCD4Cre mice. The next day, differentiated mast cells were incubated with OVA peptide (SINFEKL) and analyzed for histamine release (Fig E2, C). We observed that mast cells from both mouse strains incubated with OVA peptide and IL-9 had induced histamine release from mast cells. Additionally, the mast cells from NFATc1fl/flxCD4Cre mice cultured with IL-9 showed reduced histamine release compared with those isolated from NFATc1fl/fl control mice (Fig E2, C). Together, these data demonstrate that mast cell activation with the specific peptide used to sensitize mice leads to increased histamine release in sensitized mast cells in the presence of NFATc1 and IL-9, suggesting that the NFATc1 deficiency in T cells results in defective IgE production, which affects IgE-orchestrated mast cell activation mediated through IL-9.
References
- 1.Muller M.R., Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 2.Koch S., Reppert S., Finotto S. NFATc1 deletion in T lymphocytes inhibits the allergic trait in a murine model of asthma. Clin Exp Allergy. 2015;45:1356–1366. doi: 10.1111/cea.12493. [DOI] [PubMed] [Google Scholar]
- 3.Raby B., Barnes K., Beaty T.H., Bosco A., Carey V.J., Castro M. Asthma Bridge: the Asthma Biorepository for Integrative Genomic Exploration. Am J Respir Crit Care Med. 2011;183:A6189. [Google Scholar]
- 4.Sharma S., Zhou X.B., Thibault D.M., Himes B.E., Liu A., Szefler S.J. A genome-wide survey of CD4(+) lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014;134:1153–1162. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuvpilo S., Jankevics E., Tyrsin D., Akimzhanov A., Moroz D., Jha M.K. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity. 2002;16:881–895. doi: 10.1016/s1074-7613(02)00329-1. [DOI] [PubMed] [Google Scholar]
- 6.Staudt V., Bothur E., Klein M., Lingnau K., Reuter S., Grebe N. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 7.De Silva N.S., Simonetti G., Heise N., Klein U. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev. 2012;247:73–92. doi: 10.1111/j.1600-065X.2012.01113.x. [DOI] [PubMed] [Google Scholar]
- 8.Glasmacher E., Agrawal S., Chang A.B., Murphy T.L., Zeng W.W., Vander Lugt B. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubel C., Sopel N., Graser A., Hildner K., Reinhardt C., Zimmermann T. The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte- and mast cell-driven immune responses in the setting of allergic asthma. J Allergy Clin Immunol. 2014;133:198–206. doi: 10.1016/j.jaci.2013.09.049. e1-9. [DOI] [PubMed] [Google Scholar]
References
- Torgerson D.G., Ampleford E.J., Chiu G.Y., Gauderman W.J., Gignoux C.R., Graves P.E. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S., Reppert S., Finotto S. NFATc1 deletion in T lymphocytes inhibits the allergic trait in a murine model of asthma. Clin Exp Allergy. 2015;45:1356–1366. doi: 10.1111/cea.12493. [DOI] [PubMed] [Google Scholar]
- Muller M.R., Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- Robinson D.S., Hamid Q., Ying S., Tsicopoulos A., Barkans J., Bentley A.M. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med. 1992;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- Hu C.M., Jang S.Y., Fanzo J.C., Pernis A.B. Modulation of T cell cytokine production by interferon regulatory factor-4. J Biol Chem. 2002;277:49238–49246. doi: 10.1074/jbc.M205895200. [DOI] [PubMed] [Google Scholar]
- Rengarajan J., Tang B., Glimcher L.H. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells. Nat Immunol. 2002;3:48–54. doi: 10.1038/ni744. [DOI] [PubMed] [Google Scholar]
- Sharma S., Zhou X.B., Thibault D.M., Himes B.E., Liu A., Szefler S.J. A genome-wide survey of CD4(+) lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014;134:1153–1162. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.T., Chu J.H., Gomez J., Koenigs M., Holm C., He X.X. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am J Respir Crit Care Med. 2015;191:1116–1125. doi: 10.1164/rccm.201408-1440OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Finotto S., Buerke M., Lingnau K., Schmitt E., Galle P.R., Neurath M.F. Local administration of antisense phosphorothioate oligonucleotides to the c-kit ligand, stem cell factor, suppresses airway inflammation and IL-4 production in a murine model of asthma. J Allergy Clin Immunol. 2001;107:279–286. doi: 10.1067/mai.2001.113049. [DOI] [PubMed] [Google Scholar]
- Sauer K.A., Scholtes P., Karwot R., Finotto S. Isolation of CD4+ T cells from murine lungs: a method to analyze ongoing immune responses in the lung. Nat Protoc. 2006;1:2870–2875. doi: 10.1038/nprot.2006.435. [DOI] [PubMed] [Google Scholar]
- Ubel C., Sopel N., Graser A., Hildner K., Reinhardt C., Zimmermann T. The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte- and mast cell-driven immune responses in the setting of allergic asthma. J Allergy Clin Immunol. 2014;133:198–206. doi: 10.1016/j.jaci.2013.09.049. e1-9. [DOI] [PubMed] [Google Scholar]
- De Silva N.S., Simonetti G., Heise N., Klein U. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev. 2012;247:73–92. doi: 10.1111/j.1600-065X.2012.01113.x. [DOI] [PubMed] [Google Scholar]
- Kajiyama Y., Umezu-Goto M., Kobayashi N., Takahashi K., Fukuchi Y., Mori A. IL-2-induced IL-9 production by allergen-specific human helper T cell clones. Int Arch Allergy Immunol. 2007;143:71–75. doi: 10.1159/000101409. [DOI] [PubMed] [Google Scholar]
- Staudt V., Bothur E., Klein M., Lingnau K., Reuter S., Grebe N. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 Cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Wiener Z., Falus A., Toth S. IL-9 increases the expression of several cytokines in activated mast cells, while the IL-9-induced IL-9 production is inhibited in mast cells of histamine-free transgenic mice. Cytokine. 2004;26:122–130. doi: 10.1016/j.cyto.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Klein M., Klein-Hessling S., Palmetshofer A., Serfling E., Tertilt C., Bopp T. Specific and redundant roles for NFAT transcription factors in the expression of mast cell-derived cytokines. J Immunol. 2006;177:6667–6674. doi: 10.4049/jimmunol.177.10.6667. [DOI] [PubMed] [Google Scholar]
- Tete S., Nicoletti M., Saggini A., Maccauro G., Rosati M., Conti F. Interleukin-9 and Mast Cells. J Biol Regul Homeost Agents. 2012;26:319–326. [PubMed] [Google Scholar]