Abstract
Telomeres are nucleoprotein complexes at the end of eukaryotic chromosomes. Many telomere-binding proteins bind to telomeric repeat sequences and further generate T-loops in animals. However, it is not clear if they regulate telomere organization using epigenetic mechanisms and how the epigenetic molecules are involved in regulating the telomeres. Here, we show direct interactions between the telomere-binding protein, AtTRB2 and histone deacetylases, HDT4 and HDA6, in vitro and in vivo. AtTRB2 mediates the associations of HDT4 and HDA6 with telomeric repeats. Telomere elongation is found in AtTRB2, HDT4 and HDA6 mutants over generations, but also in met1 and cmt3 DNA methyltransferases mutants. We also characterized HDT4 as an Arabidopsis H3K27 histone deacetylase. HDT4 binds to acetylated peptides at residue K27 of histone H3 in vitro, and deacetylates this residue in vivo. Our results suggest that AtTRB2 also has a role in the regulation of telomeric chromatin as a possible scaffold protein for recruiting the epigenetic regulators in Arabidopsis, in addition to its telomere binding and length regulation activity. Our data provide evidences that epigenetic molecules associate with telomeres by direct physical interaction with telomere-binding proteins and further regulate homeostasis of telomeres in Arabidopsis thaliana.
INTRODUCTION
Chromatin structure and remodeling are important components of genetic and epigenetic regulations of gene expression (1). In higher eukaryotes, post-translational modification on the N-terminal tails of histones and DNA methylation play crucial roles in determining whether chromatin is in the active euchromatin or a silent heterochromatin; and these chromatin states affect the expression of genes involved in numerous biological processes such as growth and development, regeneration and oncogenesis (2).
Histone acetylation levels are determined by the action of histone acetyltransferases and histone deacetylases (HDACs) (3). Plant HDACs can be grouped into three major families; RPD3/HDA1, SIR2 and HD2 (4). HDA6 is a well-studied RPD3-type HDAC (5–16). In contrast to RPD3- and SIR2-type, HD2-type histone deacetylases are plant specific (17,18). A few studies have showed interactions between HD2- and RPD3-type deacetylases (5,7); however, whether this is functionally relevant is unclear.
DNA methylation is conserved in eukaryotic organisms. 5-methyl cytosine DNA methylation is found at symmetrical CpG sites, and CpNpG sites in plants, as well as at asymmetric CpHpH sites (19). In Arabidopsis, several methyltransferase proteins that affect chromatin structures have been identified. Among those, MET1, homologous to mammalian Dnmt1, is a CpG methyltransferase (20,21). The chromomethylase CMT3 is unique in plants and important for CpNpG and asymmetric methylation (22,23). DRMs, Domain Rearranged Methyltransferases which is homologous to mammalian Dnmt3, also function in asymmetric methylation and locus-specific CpNpG methylation, and are required for de novo DNA methylation (24–27).
Telomeres are nucleoprotein complexes at the physical ends of linear eukaryotic chromosomes (28). Telomeres protect the chromosome ends from fusion and degradation to avoid the loss of genetic information (29). Telomeres consist of repetitive G-rich DNA tightly regulated by specialized proteins such as telomerase and telomere-binding proteins.
Telomerase is a specialized reverse transcriptase complex that can add telomeric repeats to the absolute ends of chromosomes using its own internal RNA template, thereby effectively stabilizing the telomere length (30,31). Telomere-binding proteins have been identified in many species, in yeasts, animals and plants (28,32,33). It is known that telomere-binding proteins facilitate the formation of specialized telomeric structure (T-loop), and that these proteins interact with other proteins involved in DNA recombination and repair (32). In Arabidopsis, several telomere-binding proteins are identified and divided into TRFL (TRF-like) family and SMH (Single-Myb-Histone)-like family (33). Among them, SMH proteins are relatively small and share a unique tripartite structure consisting of an N-terminal Myb domain, a central histone H1-like globular domain and a C-terminal coiled-coil region. AtTRB proteins belong to the SMH-like protein family and are characterized as telomere-binding proteins in Arabidopsis (34–36).
Recent studies in Arabidopsis have revealed that subtelomeric regions and interstitial telomeric sequences (ITSs) are heterochromatic, whereas telomeres exhibit euchromatic features. Furthermore, histone methyltransferases and chromatin remodeling protein control subtelomeric heterochromatin formation (37). Moreover, it was reported that telomeric chromatin structure is regulated by non-coding telomeric RNAs and RNA-dependent DNA methylation (RdDM) pathway in Arabidopsis (38).
Previously, several studies indicated that there are involvements of several histone modifiers in telomeres, for example SIN3-LIKE1 (SNL1) in Arabidopsis (39), SIR complex and Rpd3 HDAC proteins in budding yeast (40,41) and SIRT6 in human (42,43). However, the relationships between telomere-binding proteins and these epigenetic regulators are not understood yet.
In this study, we report direct interactions between the Arabidopsis telomere-binding protein, AtTRB2 and histone deacetylases, HDT4 and HDA6. We also characterize the function of HDT4; HDT4 is a putative H3 lysine 27 deacetylase in Arabidopsis. Furthermore, we observe that enzymes involved in histone modifications and DNA methylations associate with telomeres through the interaction with AtTRB2 and possibly regulate lengthening of telomeres in Arabidopsis. Our data provide evidences to support possible epigenetic events in chromosome termini. We propose that telomere-binding proteins function as capping proteins for protecting the telomeres, but also as scaffold proteins for recruiting epigenetic regulators, and that these epigenetic regulators are easily accessible to telomeres through interaction with telomere-binding proteins, implying that these proteins contribute to maintaining the homeostasis of telomeres in Arabidopsis thaliana.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana plants (ecotype Columbia and Wassilewskija) were cultivated at 22°C in a growth chamber under a 16/8 h light/dark photoperiod. A. thaliana seeds were surface sterilized in 30% bleach solution and plated on solid 1× Murashige and Skoog (MS) medium or transferred to soil.
Mutant identification
We screened the Institut National de la Recherche Agronomique Versailles Arabidopsis T-DNA insertion collection (ecotype Wassilewskija), using the flanking insertion site FLAG sequence database (44), and identified putative mutant plants with a T-DNA insertion; FST 242F11 for AtTRB2 gene and FST 374G03 for HDT4 gene. The progeny of plants heterozygous for a T-DNA insertion into the AtTRB2 and HDT4 genes were genotyped by polymerase chain reaction (PCR), using the primers L1 (T-DNA left border) (5′-CTACAAATTGCCTTTTCTTATCGAC-3′). Plants homozygous for the AtTRB2 and HDT4 disruption (referred to as G1) were self-pollinated to obtain subsequent generations.
Complementation analysis
For the complementation test of attrb2–1−/− plants, AtTRB2 gene was amplified and cloned the DNA fragment into a binary vector pFP101-HA (45), and then introduced into the attrb2–1−/− G5 mutants via in planta transformation (46). The complementation vector was introduced into the Agrobacterium tumefaciens strain, GV3101. The transformants were selected using a seed-expressed fluorescent marker (45) and then genotyped and analyzed for AtTRB2 mRNA expression and telomere length.
Nucleic acid isolation
Total RNA was isolated from 10-day-old seedlings and tissues from mature plants using the easy-BLUE Total RNA Extraction Kit (iNtRON Bio Technology Co., Ltd). A. thaliana genomic DNA was prepared from 10-day-old seedlings using nucleic acid extraction buffer (0.2 M Tris–HCl, pH 8.0, 0.4 M LiCl, 1% sodium dodecyl sulphate (SDS) and 25 mM ethylenediaminetetraacetic acid (EDTA)) and isolated using a standard method (47).
Reverse transcription PCR
We obtained the cDNAs of AtTRB1, AtTRB2, AtTRB3, HDT1, HDT2, HDT3, HDT4, HDA6 and HDA19 genes from total RNA prepared from 10-day-old seedlings by RT-PCR. PCR conditions were as follows: 10 min at 94°C, 28 cycles of 30 s at 94°C, 45 s at 58°C and 90 s at 72°C, 10 min at 72°C. Three independent cDNA clones were identified and used in this study. The primers used in this experiment are listed in the supplementary document.
Yeast two-hybrid assay
Yeast two-hybrid screening of AtTRB2 bait was performed by Panbionet Corp. (www.panbionet.com) with Arabidopsis cDNA activation domain (AD) library using yeast strain PBN204 (MATa ura3–52 his3–200 ade2–101 trp-901 leu2–3,112 gal4D gal80D ura3::kanMX6-pGAL1-URA3 pGAL1-lacZ ade2::pGAL2-ADE2) containing three reporters (URA3, lacZ and ADE2). The Arabidopsis cDNA inserts were cloned into EcoRI/XhoI-digested pGADT7 vector in three different frames. AtTRB2 was cloned into EcoRI/BamHI sites of pGBKT vector, which vector contains DNA binding domain of GAL4 (GAL4DB). Yeast transformants of the AtTRB2 bait and an Arabidopsis cDNA AD library were spread on selection media [SD-leucine, tryptophan, uracil (SD-LWU)] that support growth of yeasts with bait and prey plasmids yielding proteins interacting each other.
Expression and purification of proteins
Coding regions of HDAC and AtTRB genes were cloned into pGEX-5X-1 (GE Healthcare) and pET22b (Novagen), respectively. Glutathione S-transferase (GST)-fused full and truncated HDAC proteins were expressed in Escherichia coli BL21 (DE3)/RIL cells (Stratagene) using the pGEX-5X-1 expression vector, between the EcoRI and/or XhoI sites (Fermentas, Thermo Scientific) and purified on a GSTrap™ column, as directed by the manufacturer (GE Healthcare). Hexa-histidine-AtTRB proteins were cloned into pET22b (Novagen) and expressed in E. coli BL21 (DE3)/RIL cells (Stratagene) by the same method. Purified proteins were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentrations were determined by the Bradford assay and samples were stored in aliquots at −80°C.
In vitro pull-down assay
AtTRB-6xHis proteins were incubated with Ni+-charged beads (GE Healthcare) at 4°C for 3 h and washed three times with IP buffer (50 mM Tris–HCl, pH7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40 plus protease inhibitors) and further incubated with purified GST-HDAC proteins in IP buffer overnight at 4°C with rotation. After the incubation, the protein-bound beads were washed with the same buffer at least six times and subjected to western blot analysis.
In vitro histone binding assay
A total of 1–5 μg of GST and GST-tagged HDAC proteins were incubated with 1–5 μg of calf thymus total histones (Worthington) in binding buffer (50 mM Tris–HCl, pH 7.5, 1M NaCl, 1% NP-40, 0.5 mM EDTA, 1 mM phenylmethyl sulphonyl fluoride (PMSF) with protease inhibitors (Roche)) at 4°C for 3 h, followed by an additional incubation for 1 h with glutathione beads (GE Healthcare), as described previously (48). The protein-bound beads were washed extensively with binding buffer. Bound proteins were analyzed by SDS-PAGE and western blot analysis.
In vitro histone peptide binding assay
Histone peptides about 20 amino acids long conjugated with biotin were synthesized (Abclone). Histone peptide binding assay was performed similar to as described (48). Five micrograms of the biotinylated histone peptides were incubated with 5 μg of the purified GST-HDAC proteins in buffer (50 mM Tris–HCl, pH7.5, 300 mM NaCl, 0.1% NP-40, 1 mM PMSF with protease inhibitors) overnight at 4°C with rotation, followed by an additional incubation for 1 h with streptavidin beads (GE Healthcare) and six washes with the same buffer. Bound proteins were analyzed by SDS-PAGE and western blot analysis.
Total histone protein blot analysis
Ten-day-old seedlings were ground to a fine powder in liquid nitrogen, resuspended in 3× SDS sample buffer, sonicated six times for 10 s, and then boiled for 10 min as described previously (49). After centrifugation for 10 min at 15 000 g, supernatants were used for protein blot using antibodies.
Transient expression of the AtTRB2 and HDAC genes in tobacco epidermal cells
To verify the interaction between AtTRB2 and HDT4 proteins, we used the pSPYNE-35S and pSPYCE-35S vectors for the bimolecular fluorescence complementation (BiFC) assay (50). The XbaI and SpeI sites of the two vectors were used for cloning. To perform the co-immunoprecipitation (co-IP) assay, we used the modified pCAMBIA1390 binary vectors for ectopic expression of the AtTRB2, HDT4 and HDA6 genes in plants. The modified binary vectors contain the additional DNA sequences for FLAG or HA tag into the original pCAMBIA1390 binary vector. These modified vectors were donated from Dr H.-S. Pai. The coding regions of AtTRB2, HDT4 and HDA6 genes were inserted into the EcoRI and NcoI sites of the modified pCAMBIA1390 vectors. For infiltration of Nicotiana benthamiana, the A.tumefaciens strain C58C1 was infiltrated into the abaxial air space of 3- to 4-week-old plants. The p19 protein of tomato bushy stunt virus was used to suppress gene silencing. Co-infiltration of Agrobacterium strains containing the constructs and the p19 silencing plasmid was carried out at OD600 of 0.7:0.7:1.0. Epidermal cell layers of tobacco leaves were assayed for fluorescence 1–3 days after infiltration using a fluorescence microscope (BX51 Research Microscope, Olympus).
Co-immunoprecipitation assay
FLAG- and HA-fused modified pCAMBIA1390 binary vectors were used for ectopic expression of AtTRB2 and HDAC genes. We harvested and ground the infiltrated tobacco leaves in liquid nitrogen and lysed the tissues in IP buffer (50 mM Tris–HCl, pH7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40 and protease inhibitors). Cell extracts were immunoprecipitated with anti-Flag M2 or anti-HA monoclonal antibody-conjugated agarose beads (Sigma Aldrich) overnight at 4°C with rotation. Protein-bound beads were washed extensively with IP buffer and subjected to SDS-PAGE and western blot analysis.
Preparation of transgenic plants for ectopic expression of HDT4
Transformation of A.thaliana (Col-0) with Agrobacterium (GV3101) was carried out using the same constructs as were used in the co-IP assay. Transgenic plants were screened in hygromycin-containing 1× MS solid media.
Telomere length analysis
A terminal restriction fragment (TRF) assay was performed as described previously (51). Genomic DNA was extracted from 10-day-old seedlings. Ten micrograms of genomic DNA was digested with the Tru9I (Promega) restriction enzyme, and then separated by electrophoresis on a 0.8% agarose gel and blotted onto a nylon membrane. Radio-labeled [TTTAGGG]70 oligonucleotides were used as probes. Hybridization was done in 0.5 M NaHPO4 (pH 7.2), 7% SDS, 1 mM EDTA, 1% bovine serum albumin at 60°C. Membranes were washed under the following conditions: low stringency = 2× SSC, 1% SDS, at 60°C; high stringency = 1× SSC, 0.1% SDS, at 60°C.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed in accordance with the protocol (52). We used the tobacco leaves expressed proteins by infiltration. One milligram of the each samples were harvested, crosslinked with 1% formaldehyde and DNA was precipitated with antibodies. Eluted DNA was dot-blotted onto a nylon membranes and hybridized with 32P-labeled [TTTAGGG]70 oligonucleotides probe. Hybridization and the washing of the membranes were carried out according to the method described in TRF assay.
DNA methylation analysis
We extracted the genomic DNA from 10-day-old seedlings. A total of 10 μg of genomic DNA was digested with the methylation sensitive restriction enzymes, 20 U of MspI, HpaII, BstNI and EcoRII (Fermentas, Thermo Scientific) and then separated by electrophoresis in a 0.8% agarose gel. We blotted the digested and separated genomic DNA onto a nylon membrane and analyzed by Southern blot analysis using 32P-labeled [TTTAGGG]70 oligonucleotides as a probe. Hybridization and the washing of the membranes were carried out according to the method described in TRF assay.
Antibodies
The following antibodies were used: anti-FLAG, anti-FLAG-HRP, anti-HA, anti-HA-HRP (Sigma Aldrich), anti-FLAG, anti-HA, anti-GST (abm), anti-GST, anti-His (Santa cruz biotechnology), anti-H1, H2A, H2B (Abcam) and anti-H3, H4, H3K9Ac, H3K27Ac, H3K4me3, H3K9me2, H3K27me2 (Upstate).
Accession numbers
Arabidopsis Genome Initiative locus identifiers are as follows: AtTRB1 (AT1G49950), AtTRB2 (AT5G67580), AtTRB3 (AT3G49850), HDT1 (AT3G44750), HDT2 (AT5G22650), HDT3 (AT5G03740), HDT4 (At2G27840), HDA6 (AT5G63110), HDA19 (AT4G38130), MET1 (AT5G49160), and CMT3 (AT1G69770).
RESULTS
Arabidopsis telomere-binding protein AtTRB2 interacts with HD2-type histone deacetylase HDT4
Our recent biochemical and structural studies revealed that AtTRB2 has unique telomere binding activity with high affinity to both Arabidopsis- and human-type telomeric repeats (35,36). In order to understand the additional role of AtTRB2 protein beyond telomeric repeat binding, we screened the A.thaliana cDNA library (1.4 × 107) using the yeast two-hybrid system with AtTRB2 as bait to identify AtTRB2-interacting proteins. Among the real positives, we identified HDT4 (NM_128344) as an AtTRB2-interacting protein and subsequently analyzed it by sequencing.
HDT4 was previously identified as a putative HDAC belonged to the HD2 class in Arabidopsis (53,54). We confirmed the interaction between AtTRB2 and HDT4 using a yeast two-hybrid system (Figure 1A). co-IP experiments revealed that AtTRB2 interacts with HDT4 in vivo (Figure 1B). Direct interaction of AtTRB2 with HDT4 was further confirmed by a bimolecular fluorescence complementation (BiFC) assay (Figure 1C). Tobacco was used as a heterologous transient expression system because this plant species has cells with a larger nucleus and much longer telomeres (40–160 kb) than those of A. thaliana. Strong fluorescence signals were observed in the nucleus of tobacco epidermal cells when YFPN-AtTRB2 and YFPC-HDT4 were co-expressed (Figure 1C). To confirm direct interaction between these two proteins, we performed an in vitro pull-down assay. As illustrated in Figure 1D, AtTRB2 interacted specifically with HDT4 among four different HD2-type HDACs (HDT1–4). Taken together, these results indicate that AtTRB2 interacts specifically with HDT4 in vivo and in vitro.
Figure 1.

AtTRB2 interacts physically with the HD2-type histone deacetylase, HDT4, in vivo and in vitro. (A) Yeast two-hybrid assay showing direct interaction between AtTRB2 and HDT4 (left panel). Right panel indicates the positive- and negative control in this assay. Growth on the SD-LWAU plate and the blue signal on filter assay indicate activation of the reporter genes, Ade2/Ura3 and LacZ, respectively. L, leucine (-); W, tryptophan (-); A, adenine (-); U, uracil (-). The upper panel indicates the combination of constructs used in this assay. (B) Analysis of AtTRB2 binding to HDT4 in plant cells determined by immunoblot. Empty vector control and IP with IgG were used to test the specificity of interactions between proteins and antibody. (C) Bimolecular fluorescence complementation (BiFC) analysis detected fluorescence signals generated by the interaction between AtTRB2 and HDT4 in tobacco epidermal cells (Nicotiana benthamiana); YFPN + YFPC, the negative control using empty vectors (a, b, c); YFPN-AtTRB2 + YFPC-AtTRB2, the positive control (d, e, f); YFPN-AtTRB2 + YFPC-HDT4 (g, h, i). DAPI-stained image showing DNA (b, e, h) and bright field image (c, f, i). Bar = 10 μm. (D) In vitro pull-down assay showing the exclusive interaction of AtTRB2 with HDT4 among HD2 class histone deacetylases in Arabidopsis. Asterisk marks represent the non-specific signals detected by a-GST antibody in this assay.
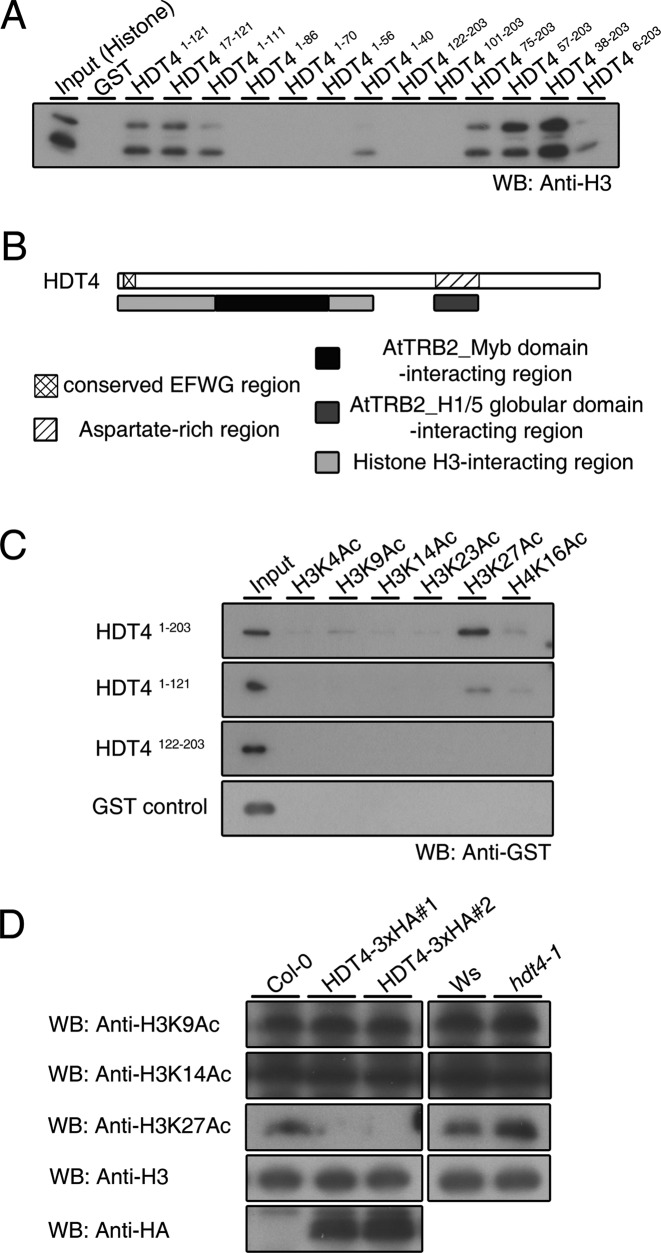
To map the regions in HDT4 that interact with AtTRB2, we performed a pull-down assay using several truncated forms of AtTRB2 (Figure 2A) and HDT4 (Figure 2B). Western blot analysis revealed that Myb domain-containing AtTRB21–97 was able to bind to 10 truncated HDT4 protein variants; the exceptions were HDT41–40, HDT4101–203 and HDT4122–203 (Figure 2C and D). In contrast, H1/5-like globular domain-containing AtTRB297–201 did not interact with HDT41–40 or an additional truncated HDT4159–203 protein (Figure 2E). These results indicate that the Myb domain of AtTRB2 interacts with the N-terminal region of HDT4 (HDT440–86 approximately) and that the histone H1/5-like globular domain of AtTRB2 interacts with the C-terminal region of HDT4 (HDT4133–154), which is annotated as an aspartate-rich region. The domains in HDT4 responsible for interaction with the domains of AtTRB2 are schematically diagrammed in Figure 2F.
Figure 2.
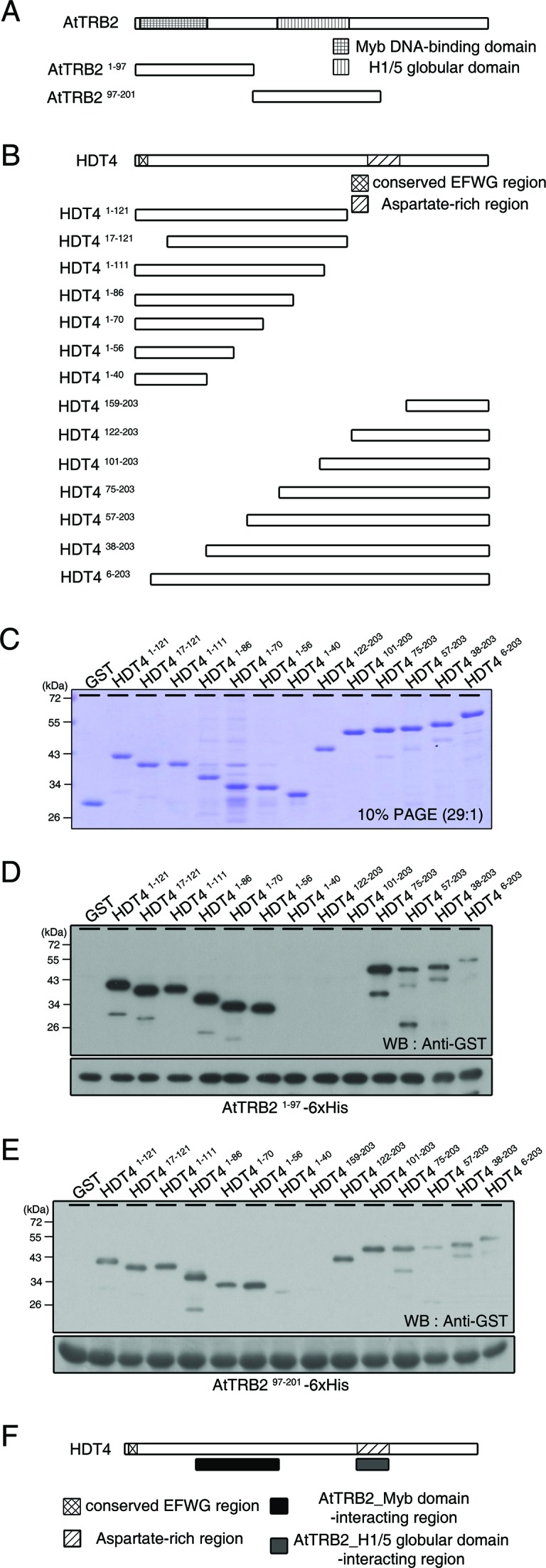
Mapping the interaction domains on AtTRB2 and HDT4. (A and B) Schematic representation of several truncated AtTRB2 (A) and HDT4 protein constructs (B). AtTRB2 has a Myb DNA-binding domain in the N-terminal and a H1/5 globular domain in the central region. HDT4 has a conserved EFWG motif and an aspartate-rich region in the N-terminal and C-terminal region, respectively. (C) PAGE shows the purified truncated HDT4 recombinant proteins. (D and E) Analysis of HDT4 recombinant proteins binding to the domains of AtTRB2 monitored by immunoblot. (F) Scheme of the regions of HDT4 that interact with AtTRB2.
HDT4 is a putative histone H3 lysine 27 deacetylase
We ascertained whether HDT4 can interact with histone proteins in vitro using calf thymus total histone proteins. Compared with GST alone, GST-HDT4 showed binding activity to histone H3 (Supplementary Figure S1A) and we also demonstrated that the N-terminal region of HDT4 (HDT41–121) is involved in this interaction (Supplementary Figure S1B). When several truncated HDT4 proteins were used (Figure 2C), we found that the N-terminal and central regions of HDT4 (HDT41–40 and HDT487–100) interacted with histone H3 (Figure 3A), implying that these histone H3-interacting regions and AtTRB2-interacting regions are located side-by-side with each other, but do not overlap (Figure 3B).
Figure 3.
HDT4 is a putative histone H3 lysine 27 deacetylase in Arabidopsis. (A) Analysis to map the H3-interacting region on HDT4 in vitro. (B) Scheme of the region of HDT4 involved in the interaction with histone H3. (C) In vitro peptide binding assay showing binding of HDT4 to acetylated lysine residues of histone H3. (D) Acetylated lysine status of histone H3 in two independent HDT4-HA-expressing transgenic plants and hdt4–1 mutant plants determined by various antibodies in vivo. Immunoblotting with H3 antibody was performed to ensure equal sample loading.
Next, we identified which acetylated amino acid residue of histone H3 protein is recognized by HDT4. As illustrated in Figure 3C, full-length and N-terminal truncated GST-HDT4 proteins bound to acetylated H3 lysine 27 (H3K27Ac) exclusively. To further evaluate the deacetylase activity of HDT4 in vivo, H3 lysine acetylation status in two independent HDT4–3xHA tagging lines and hdt4–1 mutant plants was determined by immunoblot analysis with antibodies. Ectopic expression of HDT4–3xHA resulted in a decrease in H3K27Ac levels, but not H3K9Ac or H3K14Ac levels (Figure 3D, left panel), while a loss-of-function mutation of HDT4 resulted in increased accumulation of H3K27Ac compared to wild-type (Figure 3D, right panel). Based on these results, we conclude that HDT4 is a putative histone H3K27 deacetylase in A. thaliana.
HDT4 is associated with Arabidopsis telomeres
To examine whether HDT4 can associate with telomeres, we analyzed telomeric ChIP (T-ChIP) fractions by dot-blot hybridization with a telomeric repeat probe (55). We used tobacco leaves transiently co-expressing HDT4 and AtTRB2 used in the previous co-IP assay (Figure 1B). AtTRB2 associated strongly with telomeric repeat sequences (Figure 4A), while a HDT4-associated telomeric DNA signal was only detected in HDT4/AtTRB2 co-expressing plants but not in plants expressing HDT4 alone (Figure 4B and C). Thus, our T-ChIP data indicate that HDT4 occupies plant telomeric repeat regions only in the presence of AtTRB2, suggesting that HDT4, as a complex with AtTRB2, has a potential role in regulating the chromatin state and organization of telomeres in A. thaliana.
Figure 4.
HDT4 associates with telomeric repeat DNA in an AtTRB2-dependent manner. (A) Telomeric ChIP (T-ChIP) analysis showing the association between AtTRB2 and telomeric chromatin in tobacco. (B) Telomeric ChIP analysis to assess the recruitment of HDT4 in the presence of AtTRB2. [TTTAGGG]70 repeats were used as probes. Empty vectors used for the co-immunoprecipitation assay were used as negative controls. Mock refers beads only without antibody to determine if there was non-specific binding of chromatin to beads. Input DNA loaded represents 5% of total DNA. (C) Quantification of multiple independent T-ChIP experiments as shown in (B). Values represent the T-ChIP signal obtained by subtracting the mock signal from the raw value for background and normalized by the input signal. Error bars indicate the SEM; n = 3.
RPD3-type histone deacetylase HDA6 is a component of the AtTRB2 complex in Arabidopsis
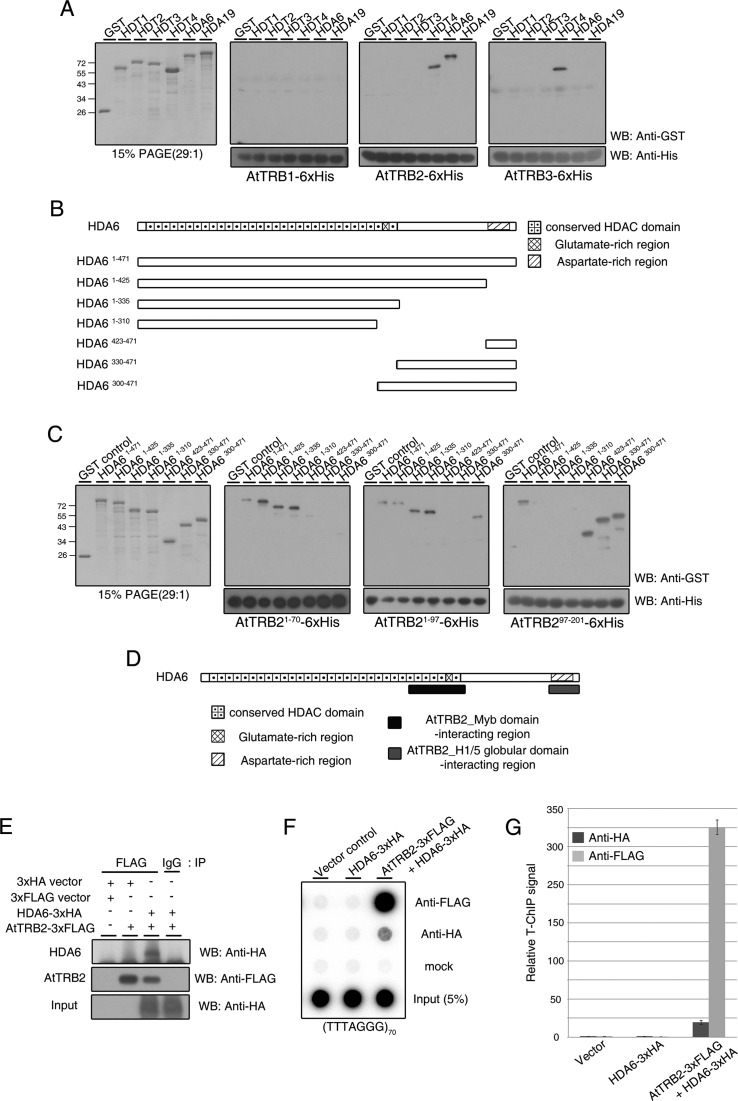
A recent study reported that HDT4 can interact with the RPD3-type HDAC proteins, HDA6 and HDA19, based on BiFC assays, suggesting that these two different types of HDAC proteins may cooperate to modulate chromatin state (7). To ascertain the association of these RPD3-type HDACs with AtTRB proteins, we performed in vitro binding assays with HDA6 and HDA19 proteins (Figure 5A, first panel). Although AtTRB proteins have high amino acid sequence similarity to one another, AtTRB2 was able to bind to both HDT4 and HDA6 (Figure 5A, third panel), while AtTRB3 was able to bind only to HDT4, but not HDA6 (Figure 5A, fourth panel). AtTRB1 did not bind to any HDAC proteins tested in this assay (Figure 5A, second panel). These results indicate that there are selective interactions between AtTRBs and HDACs.
Figure 5.
AtTRB2 also interacts with HDA6, a RPD3-type histone deacetylase. (A) Western blot analysis showing the different patterns of interaction between AtTRB proteins and various HDAC proteins. PAGE shows the purified HDAC proteins used in this assay (first panel). (B) Scheme of various HDA6 truncated proteins fused to GST. HDA6 protein comprises three predicted regions: a conserved HDAC domain in the N-terminal region, a glutamate-rich region in the center of the protein and an aspartate-rich region in the C-terminal region. (C) Pull-down assay to map the regions of HDA6 that interact with AtTRB2. PAGE shows the purified HDA6-truncated proteins shown in (B). (D) Schematic map indicating the region of HDA6 involved in binding to AtTRB2. (E) Western blots demonstrating the interaction between AtTRB2 and HDA6 in vivo. Empty control and IP with IgG were used as negative controls. (F) HDA6-associated telomeric repeat signal was detected in an AtTRB2-dependent manner. (G) Quantification of multiple independent T-ChIP experiments as shown in (F). T-ChiP analysis was performed and values were calculated as described in the legend to Figure 4.
We defined regions in HDA6 involved in the interaction with AtTRB2 using several truncated HDA6 proteins (Figure 5B and C). AtTRB21–97 showed binding activity to five truncated HDA6 proteins, but not HDA6423–471 or HDA6330–471 (Figure 5C, third panel). AtTRB21–70, which is shorter than AtTRB21–97, was able to bind to HDA6 protein variants with C-terminal deletions (Figure 5C, second panel). In contrast, AtTRB297–201 bound to all truncated HDA6 proteins containing the aspartate-rich region (Figure 5C, fourth panel). These results indicate that the Myb domain of AtTRB2 interacts with the central region of HDA6, including the glutamate-rich region and that the histone H1/5-like globular domain of AtTRB2 interacts with the aspartate-rich region of HDA6 (Figure 5D).
In addition, we examined the region of HDA6 that interacts with histone proteins. As shown in Supplementary Figure S2, the HDAC domain and C-terminal region between the glutamate and aspartate-rich regions of HDA6 protein bind to histone H3.
Next, we performed a co-IP assay using tobacco leaves transiently expressing HDA6 and AtTRB2. As shown in the western blot in Figure 5E, HDA6-HA was pulled down by AtTRB2-FLAG, indicating an interaction between AtTRB2 and HDA6 in vivo.
We then performed T-ChIP assay to ascertain whether HDA6 associates with telomeres. Similar to HDT4 in Figure 4, a HDA6-associated telomeric DNA signal was detected in HDA6/AtTRB2 co-expressing plants only (Figure 5F and G). All T-ChIP data indicate that HDT4 and HDA6 associate with telomeric chromatin in an AtTRB2-dependent manner.
Possible epigenetic regulation in telomere homeostasis in A. thaliana
The interaction between AtTRB2, HDT4 and HDA6 raises the questions of whether and how epigenetic modifications affect the organization of telomeres in Arabidopsis. To address these questions, we analyzed telomere lengthening in relevant A. thaliana mutants by TRF analysis. For the TRF assay, we obtained independent individual homozygote lines by self-pollination in order to measure the telomere length over generations. By doing this, we were able to exclude the possibility of misleading results in terms of changes in telomere length that would have been encountered had we used a single plant or a certain generation only. If the change in telomere length after one generation was not large, a bigger change could be observed over several generations.
We obtained transferred-DNA (T-DNA) insertional mutant lines null for AtTRB2 and HDT4 from the INRA Arabidopsis knock-out facility (ecotype Wassilewskija). These lines were designated attrb2–1 and hdt4–1 in this study (Supplementary Figures S3 and S4). No PCR products were generated in reactions with cDNA from mutant plants in RT-PCR assays, confirming that expression of full-length AtTRB2 and HDT4 mRNAs were abolished in attrb2–1 and hdt4–1 mutants, respectively (Figure 6B and D).
Figure 6.
Telomere-binding protein and epigenetic molecules regulate telomere dynamics in Arabidopsis. (A) Terminal Restriction Fragmentation (TRF) analysis demonstrating the regulation of telomere maintenance by AtTRB2 in Arabidopsis. Three individual homozygous lines of attrb2–1 mutants (ecotype Wassilewskija) were analyzed to determine the alteration of telomere length, Generation 5 (G5) mutants of attrb2–1 were used for complementation assay to make sure the recovery of the telomere length by the expression of the additional AtTRB2 gene. (B) Level of AtTRB2 mRNA in mutants and complemented plants was analyzed by RT-PCR. (C) Telomere lengthening in hdt4–1−/− mutants. Four individual homozygous mutant lines were isolated. Two groups of wild-type plants (ecotype Wassilewskija) having different telomere lengths were isolated; shorter and longer telomere. (D) Level of HDT4 mRNA in wild-type and hdt4–1 mutants. The 60S RP gene was used to determine the amount of template to facilitate quantitative comparisons in (B) and (D). (E and F) TRF assay to analyze the telomere alterations in HDA6, CMT3 and MET1 mutants (ecotype Columbia, axe1–5, cmt3 and met1, respectively). Southern blots demonstrating a general tendency toward increased length over seven generations (G7) in axe1–5 mutant (E) and the telomere elongation over five generations (G5) both in cmt3 and met1 mutants (F). Arrowheads in axe1–5 mutants represent the additionally detected signals at G6 and G7 by probe. For TRF analysis, genomic DNAs isolated from a pool of seedlings of wild-type or mutant at each generation were digested with Tru9I. Telomere length is determined by southern hybridization with [TTTAGGG]70 probe.
We randomly chose three independent homozygous AtTRB2-deficient lines from T3 seeds of the T-DNA insertion attrb2–1 mutant. Wild-type siblings generated from the heterozygous line were used for experimental controls. Individual lines were propagated through successive generations by self-pollination. TRF analysis revealed that telomeres in attrb2–1 homozygous mutants increased steadily to 10 kb after seven generations by 0.5–1.0 kb per generation, compared with wild-type (Figure 6A and Supplementary Figure S3). In complementation lines, telomere length recovered to wild-type levels (Figure 6A). These results confirm that AtTRB2 is required for telomere length maintenance in vivo.
TRF assay with hdt4–1 mutants revealed telomere lengthening for up to fifth generations (G5) by 0.5–1.0 kb per generation (Figure 6C). Telomeres of wild-type siblings generated from the heterozygous lines displayed a bimodal size distribution; one group possessed shorter telomeres (3.5–5.5 kb) while the other had longer telomeres (5.5–7 kb) (Figure 6C, data not shown) (56). Ectopically expressed 3xHA-tagged HDT4 transgenic lines showed decreased telomere length (Supplementary Figure S4).
Telomere lengthening was also evaluated in an HDA6 gene mutant (axe1–5). TRF assay revealed that the length of telomeres in axe1–5 increased up to seventh generations (G7), although the extent of increase was not as dramatic as that of telomeres of attrb2–1 and hdt4–1 mutants (Figure 6E). Especially, new bands for the high molecular size were detected in these G6 and G7 axe1–5 mutants. These results indicate that HDT4 and HDA6 are plant HDACs to act as negative regulators of telomere elongation.
HDA6 interacts directly with MET1 in locus-directed heterochromatin silencing (6,8). Chromomethylase CMT3 is involved in non-CpG methylation by recognizing both H3K9me and H3K27me (57), which are possible modifications derived from the action of HDT4 (in this study) and HDA6 (11,13). Based on these notions, we further examined whether DNA methyltransferases are involved in telomere lengthening using mutant lines (met1 for MET1, cmt3 for CMT3). In cmt3 and met1 mutants, the length of telomeres increased over generations (G5) (Figure 6F).
Possible involvement of DNA methylation in chromosome termini in Arabidopsis
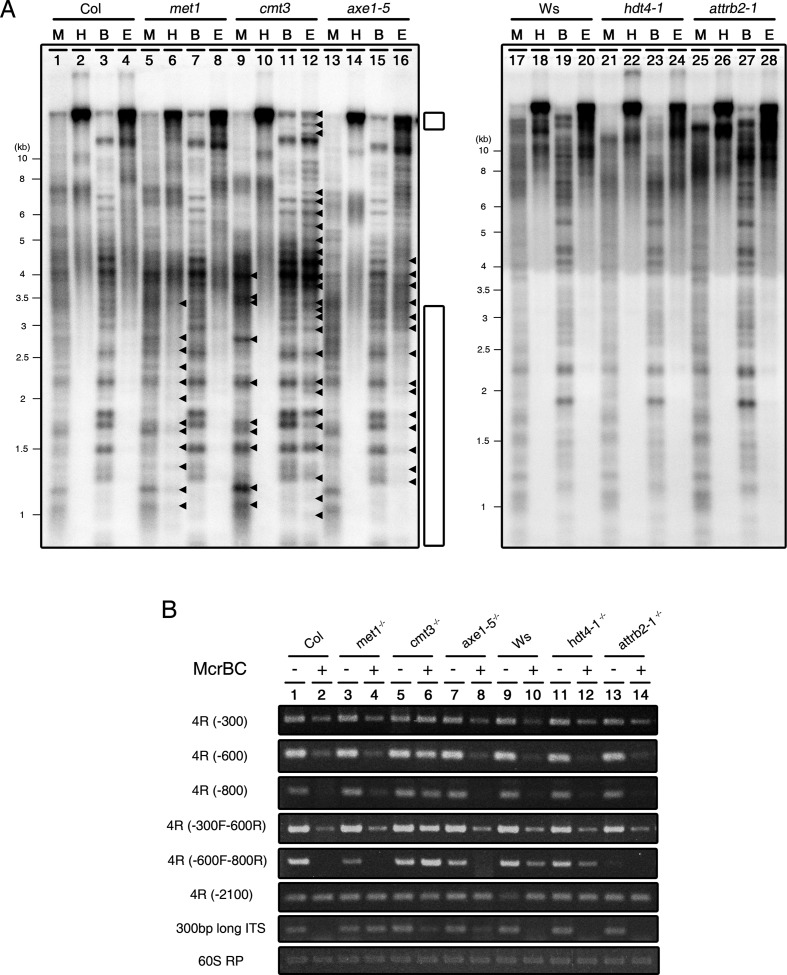
We investigated the pattern of DNA methylation at chromosome termini in these mutants using two different sets of methylation-sensitive restriction enzymes (37). One set of restriction enzymes, MspI and HpaII, recognizes CCGG sequences. MspI is inhibited only by CpNpG methylation, while HpaII is inhibited by both CpG- and CpNpG methylation. Another set of enzymes, BstNI and EcoRII, recognizes CCWGG sequences. Whereas BstNI is insensitive to all DNA methylation, EcoRII is sensitive to CpNpG and CpHpH methylation. Thus, each set of enzymes is able to indicate the presence of CpG- and non-CpG DNA methylation. Chromosome termini consist of telomeric and subtelomeric regions, and a CCGG sequence is frequently found in the subtelomeric region.
The analysis using methylation-sensitive restriction enzymes revealed several features (Figure 7). First, the pattern of the digested genomic DNA detected by the telomeric repeat sequence probes differed between Col and Ws, suggesting that there is an ecotype-specific composition of DNA sequences at the chromosome termini in wild-type Arabidopsis, although ITSs dispersed in Arabidopsis genome can also be detected by telomeric repeat sequence probes. In addition, the patterns of DNA methylation at gDNA fragments detected by telomeric repeat sequence probes in met1 and cmt3 mutants were different from those in the wild-type (Figure 7A, left panel, lanes 1–4 and 17–20). In the met1 mutant, a similar pattern of gDNA fragmentation was observed in both MspI and HpaII digests, but the telomeric repeat signal detected in the HpaII-digested gDNA bands was very weak in the lower molecular weight region (Figure 7A, lane 6). On the other hand, higher molecular weight bands from the cmt3 mutant were degraded to smaller gDNA fragments (Figure 7A, lane 12). Specifically, there were several different molecular weight bands of MspI-digested gDNA in this mutant, which were not found in the wild-type (lane 9). Moreover, BstNI- and EcoRII-treated lanes in cmt3 showed a similar digestion pattern (lanes 11–12). These results indicate that MET1 and CMT3 affect CpG- and non-CpG DNA methylation of telomeric repeat sequence detected gDNA fragments, respectively, although the methylation effect of MET1 is much weaker than that of CMT3. In particular, the hda6 mutant (axe1–5) showed that EcoRII-treated, but not HpaII-treated, gDNA is degraded into gDNA fragments with weak signal intensity (Figure 7A, lane 16). This digestion pattern is similar to that of the cmt3 mutant, suggesting HDA6 contributes to the function of CMT3 in non-CpG DNA methylation in gDNA fragments detected by telomeric repeat sequence probes. On the contrary, there were no alterations in the pattern of DNA methylation in hdt4–1 and attrb2–1 mutants compared to the wild-type (Figure 7A, right panel, lanes 21–28).
Figure 7.
DNA methylation is involved in telomere homeostasis in Arabidopsis thaliana. (A) Southern blot analysis showing the pattern of DNA methylation of telomeric repeats in cmt3, met1, axe1–5 (left panel), hdt4–1 and attrb2–1 mutants (right panel). DNA blots of methylation-sensitive restriction enzyme-digested gDNA probed with [TTTAGGG]70 repeat sequences. M, MspI; H, HpaII; B, BstNI; E, EcoRII. Arrowheads represent the different signals detected by probe compared to wild-type. Small and large white bar indicate the high and low molecular weight ranges of the gDNAs, respectively, affected by restriction enzyme digestion. (B) Chop-PCR in subtelomeric and ITSs regions using McrBC endonuclease. 4R-300, -600, -800 and -2100 indicate the different regions on the subtelomeric regions closed to telomere in the right arm of chromosome IV. The 60S RP gene was used as a reference for quantitative comparisons. −, +; McrBC-untreated/ -treated.
Because the gDNA bands in this Southern blot presumably contains DNA fragments originated from extratelomeric region, in order to further distinguish the region for methylation and to verify the function of epigenetic regulators in DNA methylation at subtelomeric regions, we treated gDNA from these mutants with McrBC endonuclease, which gDNAs are the same materials used in TRF assay above, and then performed semi-quantitative PCR amplification of subtelomeric regions. McrBC is an endonuclease that recognizes and cleaves DNA containing methyl cytosine, but not unmethylated DNA. Thus, McrBC was used to detect methyl cytosine in certain DNA sequences. Subtelomeric regions in the right arm of chromosome IV (4R) were amplified by PCR regardless of the ecotype background (Col, Ws and Ler, data not shown). As shown in Figure 7A, it is likely that the change in DNA methylation pattern in cmt3 and met1 mutants occurred in the subtelomeric region; however we cannot exclude the possibility of change in ITSs because we used the telomeric repeat sequence as a probe. Thus, we also amplified the 300-bp long ITS in McrBC-treated gDNA from these mutants in order to analyze the DNA methylation on ITSs (37).
In the wild-type, the different regions on 4R close to telomeres (4R-300, -600 and -800 bp) were cleaved by McrBC endonuclease; however, there was no effect of McrBC in the subtelomeric region distant from telomeres (4R-2100 bp) in both the Col and Ws wild-types. In addition, 300-bp ITS was also cleaved by McrBC (Figure 7B). These results indicate the existence of methyl cytosine in subtelomeric regions adjacent to telomeres and ITSs.
Among the mutants, only cmt3 showed amplified PCR products at subtelomeric regions (4R-300, -600, -800 and -2100 bp) regardless of McrBC reaction (Figure 7B, lanes 5–6). On the contrary, q from the met1 mutant was digested by McrBC endonuclease at 4R, similar to that of the wild-type. However, met1 showed the presence of methyl cytosine at 300-bp ITS instead (Figure 7B, lanes 3–4). These results suggest that CMT3 and MET1 mainly regulate DNA methylation in the 4R subtelomeric region and in 300-bp ITSs, respectively. We did not observe the direct effects of HDA6, HDT4 and AtTRB2 on cytosine methylation at subtelomeric regions and ITSs tested in this assay (Figure 7B, lanes 7–8 and 11–14).
DISCUSSION
As previously shown, telomere-binding proteins mainly function in the protection of telomeres. Our study demonstrates the relationship between AtTRB2 and histone deacetylases, providing a broader perspective on the function of telomere-binding proteins in telomeres. Although involvement of epigenetic regulators in telomeres has been demonstrated in previous studies, their mode of action has not yet been fully elucidated. In addition, we characterize HDT4 as a putative H3K27 deacetylase in Arabidopsis.
Molecules involved in the AtTRB2 complex
It has previously been demonstrated that AtTRB proteins interact with each other to form homo- and heteromeric complexes, and also interact with the Arabidopsis G-overhang binding protein POT1b (58,59), and plant telomerase catalytic subunits (TERT) (60). These results suggest that there is functional redundancy among AtTRB proteins. However, our pull-down assay results revealed that AtTRB1, 2 and 3 have distinct binding preferences for different HDACs, indicating that each AtTRB protein may play a specific role in the formation of telomeric complexes and may be further involved in different telomere regulation events (Figure 5A).
Our study reveals that two different types of HDAC proteins can cooperate as a complex. Besides plants studies, this relationship between HDACs has also been reported in yeast (40,41). Rpd3 is necessary to restrict SIR proteins to telomeres, and thus modulates a barrier to the spread of the SIR-dependent telomere position effect (TPE). We hypothesize that this kind of coordination between HDACs is a universal regulatory mechanism in animals and plants.
Our T-ChIP and TRF assay results strongly suggest that histone deacetylases play a role in regulating the chromatin state of telomeric repeat sequences. We hypothesize that the AtTRB2/HDT4/HDA6 complex is involved in histone modification of telomeric chromatin, followed by additional histone methylation and eventually DNA methylation of the telomere. Telomere-binding proteins and epigenetic modifiers are also likely to be involved in the process of the length regulation and chromatin organization in telomeres, respectively. This hypothesis is supported by several previous studies that have reported the involvement of histone methyltransferases in telomere regulation in vivo (37,61,62). However, we cannot exclude the possibility that DNA methyltransferases and HDACs can affect the structure and function of telomere indirectly, for example by affecting the expression of genes encoding the proteins directly regulating the telomere length.
AtTRB2 can interact with HDACs and histone H3 (Supplementary Figure S5). In mammals, TRF2 was shown to be involved in nucleosome organization of heterochromatic marks in telomeric region and associations with other proteins and TERRA RNAs (63–65). We therefore hypothesize that like TRF2 in mammals, AtTRB2 functions as a scaffold protein for epigenetic modifiers of telomeric chromatin.
Regulation of telomeric chromatin state by epigenetic molecules
In Arabidopsis, telomeric regions contain telomeric repeats composed of perfect TTTAGGG sequences, and maintenance of these regions is regulated by telomere-binding proteins, including AtTRFL and AtTRB proteins (33).
Subtelomeric regions are known to contain degenerate telomeric repeat sequences (66). Moreover, subtelomeric regions have a high density of methylated CpG DNA sequences in human (67). The heterochromatic nature of subtelomeric regions is important to prevent spreading of the TPE and abnormal chromosome recombination (40,41,68). In addition, TERRA generation occurs in subtelomeric regions. TERRA expression is regulated by chromatin state, and in turn, telomere length is regulated by the expression level of TERRA (64,69–71).
Recent analyses show that the epigenetic status of subtelomeric regions is heterochromatic, formed by the RdDM pathway and histone methyltransferases (SUVH4 to 6) (37,38). In Arabidopsis, methyl cytosine, H3K9me2 and AGO4-associated siRNA (TERRA/ARRET) are distributed in the same pattern up to 2 kb from the chromosome end, although their distribution patterns differ in each chromosome arm (http://epigenomics.mcdb.ucla.edu/DNAmeth/) (37,38). Our Chop-PCR data also correlates with these previous studies (Figure 7B). The correlations between epigenetic events again suggest that the RdDM pathway, histone modification and DNA methylation processes are involved in possibly common events in telomeric heterochromatin formation. A previous study (38) noted that Arabidopsis RdDM mutants have no alteration in telomere length compared to the wild-type. Meanwhile, our TRF assay using mutants for epigenetic regulators including HDAC proteins and DNA methyltransferases displayed increased telomere length (Figure 6), consistent with the role of DNA methyltransferases in mice (72). From these findings, it is reasonable to infer that the regulation of telomere length mediated by histone modifiers/DNA methyltransferases is distinct from that by the RdDM pathway.
Two previous studies (37,38) have shown that both CpG- and non-CpG DNA methylation processes occur in subtelomeric regions, and non-CpG methylation has a major role in Arabidopsis. Our results (Figure 7) confirm that non-CpG DNA methylation comprises the majority of DNA methylation in chromosome termini. Specifically, CMT3 plays an essential role in the maintenance of telomere/subtelomere non-CpG DNA methylation (Figure 7). These results demonstrate a previously unappreciated role for DNA methylation in maintaining telomere integrity in plants.
Although HDA6 interacts with MET1 directly and further functions in heterochromatic regions (6), our Southern blot analysis shows a similar pattern of DNA methylation in telomeric repeat sequence detected DNA fragments between hda6 and cmt3, but different than the met1 mutant (Figure 7A). The results of Chop-PCR using McrBC endonuclease indicate that the acting loci of CMT3 are separate from those of MET1 (Figure 7B). We assume that HDA6 acts as an HDAC for H3K9Ac and contributes to non-CpG DNA methylation by collaborating with CMT3 in telomeres/sub-telomeres; otherwise, HDA6 acts as an MET1-interacting protein to maintain CpG DNA methylation in ITSs.
Several reports support the regulation of chromatin states by the interplay between the histone- and DNA methylation in eukaryotes (73–75). Specifically, histone modifications assist the function of DNA methyltransferases; for example, H3K9/H3K27 methylation provides a recognition site for CMT3 and further catalyzes methylation at CpNpG sequences in Arabidopsis (57,76). On the other hand, there is evidence that H3K27 modifications regulate chromatin in a separate working pathway distinct from this interplay (77). Furthermore, mutations in the ATXR5 and ATXR6 genes, which encode the H3K27 monomethyltransferases, result in chromatin decondensation by causing the over-replication of heterochromatin without any effects on DNA methylation in Arabidopsis (78–80). Our study shows that the hdt4–1 mutant does not alter DNA methylation of telomeric chromatin despite telomere elongation in contrast with the hda6 mutant (axe1–5) (Figure 7), possibly implying that the different effects of these two HDAC proteins on DNA methylation are due to distinct pathway described above.
In a previous study, we demonstrated that AtTRB2 binds to human-type double-stranded telomere repeats (5′-TTAGGG-3′) with higher affinity than to Arabidopsis-type repeats (5′-TTTAGGG-3′) with unique dual binding activity (35,36). This result raises the possibility that AtTRB2 can associate not only with telomeric regions but also with subtelomeric regions containing degenerated telomeric repeat sequences.
H3K27 modification in telomere regulation
H3K27 methylation is a well-known repressive mark and is thought to play a crucial role in the dynamic regulation of gene expression in plant development (81).
In Arabidopsis, several proteins are involved in H3K27 methylation and demethylation (49,78,82). However, the proteins involved in deacetylation have not been identified in plants. Our results strongly suggest that HDT4 is a deacetylase for H3K27Ac in A. thaliana.
In our genetic study, hdt4–1 mutants did not display any noticeable visible phenotypic differences compared to wild-type plant under normal growth conditions. We attribute this to very low level of expression of HDT4 mRNA, only detected in the stems and flowers with young siliques (54). However, although we cannot exclude the possibility that there are other functionally redundant proteins to HDT4, we hypothesize that HDT4 is involved in specific aspects of plant growth and development, such as telomeric chromatin organization, than in global processes.
Methylated H3K9 and H3K27 marks are enriched in both human and Arabidopsis telomeres (38,83,84). With these previous studies, our data support the notion that HDT4 is a putative H3K27 deacetylase that interacts with AtTRB2 to regulate the chromatin state of Arabidopsis telomeres.
Supplementary Material
Acknowledgments
We thank Dr Steven E. Jacobsen for providing us with seeds of met1 and cmt3 mutants, and Dr Keqiang Wu for providing seeds of met1 and axe1–5 mutants. We thank the INRA-FLAG databases for providing the T-DNA knockout resource. We also thank Dr H.-S. Pai for providing modified pCAMBIA1390 vectors and Dr W.T. Kim for providing the plant telomeric repeat sequences used as a probe. We thank Dr Y.S. Noh for technical help with the ChIP assay.
FUNDING
Basic Science Research Program from the Korean National Research Foundation [NRF-2013R1A12009383 to M.H.C.]; Cooperative Research Program for Agriculture Science & Technology Development, Rural Development Administration, Republic of Korea [PJ01108201 to M.H.C]; Initiative for Biological Function and Systems, Brain Korea 21(BK21) PLUS program of Yonsei University (in part to M.H.C.). Funding for open access charge: Basic Science Research Program from the Korean National Research Foundation [NRF-2013R1A12009383 to M.H.C.]; Cooperative Research Program for Agriculture Science & Technology Development, Rural Development Administration, Republic of Korea [PJ01108201 to M.H.C]; Initiative for Biological Function and Systems, Brain Korea 21(BK21) PLUS program of Yonsei University (in part to M.H.C.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Feng S.H., Jacobsen S.E. Epigenetic modifications in plants: an evolutionary perspective. Curr. Opin. Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Pandey R., Muller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo M., Wang Y.Y., Liu X.C., Yang S.G., Lu Q., Cui Y.H., Wu K.Q. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012;63:3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X.C., Yu C.W., Duan J., Luo M., Wang K.C., Tian G., Cui Y.H., Wu K.Q. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in arabidopsis. Plant Physiol. 2012;158:119–129. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo M., Wang Y.Y., Liu X., Yang S., Wu K. HD2 proteins interact with RPD3-type histone deacetylases. Plant Signal. Behav. 2012;7:608–610. doi: 10.4161/psb.20044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.To T.K., Kim J.M., Matsui A., Kurihara Y., Morosawa T., Ishida J., Tanaka M., Endo T., Kakutani T., Toyoda T. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To T.K., Nakaminami K., Kim J.M., Morosawa T., Ishida J., Tanaka M., Yokoyama S., Shinozaki K., Seki M. Arabidopsis HDA6 is required for freezing tolerance. Biochem. Biophys. Res. Commun. 2011;406:414–419. doi: 10.1016/j.bbrc.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 10.Chen L.T., Luo M., Wang Y.Y., Wu K.Q. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earley K.W., Pontvianne F., Wierzbicki A.T., Blevins T., Tucker S., Costa-Nunes P., Pontes O., Pikaard C.S. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010;24:1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- 13.Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aufsatz W., Mette M.F., van der Winden J., Matzke M., Matzke A.J.M. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murfett J., Wang X.J., Hagen G., Guilfoyle T.J. Identification of arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dangl M., Brosch G., Haas H., Loidl P., Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta. 2001;213:280–285. doi: 10.1007/s004250000506. [DOI] [PubMed] [Google Scholar]
- 18.Lusser A., Brosch G., Loidl A., Haas H., Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 19.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 20.Finnegan E.J., Dennis E.S. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis-thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartee L., Malagnac F., Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindroth A.M., Cao X.F., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 24.Cao X., Springer N.M., Muszynski M.G., Phillips R.L., Kaeppler S., Jacobsen S.E. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4979–4984. doi: 10.1073/pnas.97.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao X., Jacobsen S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. U.S.A. 2002;99(Suppl. 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao X., Jacobsen S.E. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 27.Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 28.McKnight T.D., Shippen D.E. Plant telomere biology. Plant Cell. 2004;16:794–803. doi: 10.1105/tpc.HistPersp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slijepcevic P., Al-Wahiby S. Telomere biology: integrating protection with DNA damage chromosomal end response. Chromosoma. 2005;114:275–285. doi: 10.1007/s00412-005-0338-4. [DOI] [PubMed] [Google Scholar]
- 30.Gallardo F., Chartrand P. Telomerase biogenesis. RNA Biol. 2008;5:212–215. doi: 10.4161/rna.7115. [DOI] [PubMed] [Google Scholar]
- 31.Hug N., Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 32.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 33.Zellinger B., Riha K. Composition of plant telomeres. Biochim. Biophys. Acta. 2007;1769:399–409. doi: 10.1016/j.bbaexp.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Schrumpfova P., Kuchar M., Mikova G., Skrisovska L., Kubicarova T., Fajkus J. Characterization of two Arabidopsis thaliana myb-like proteins showing affinity to telomeric DNA sequence. Genome. 2004;47:316–324. doi: 10.1139/g03-136. [DOI] [PubMed] [Google Scholar]
- 35.Lee W.K., Yun J.H., Lee W., Cho M.H. DNA-binding domain of AtTRB2 reveals unique features of a single Myb histone protein family that binds to both Arabidopsis- and human-type telomeric DNA sequences. Mol. Plant. 2012;5:1406–1408. doi: 10.1093/mp/sss063. [DOI] [PubMed] [Google Scholar]
- 36.Yun J.H., Lee W.K., Kim H., Kim E., Cheong C., Cho M.H., Lee W. Solution structure of telomere binding domain of AtTRB2 derived from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2014;452:436–442. doi: 10.1016/j.bbrc.2014.08.095. [DOI] [PubMed] [Google Scholar]
- 37.Vaquero-Sedas M.I., Gamez-Arjona F.M., Vega-Palas M.A. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2011;39:2007–2017. doi: 10.1093/nar/gkq1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K. siRNA-mediated methylation of Arabidopsis telomeres. Plos Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowen A.J., Gonzalez D., Mullins J.G.L., Bhatt A.M., Martinez A., Conlan R.S. PAH-domain-specific interactions of the Arabidopsis transcription coregulator SIN3-LIKE1 (SNL1) with telomere-binding protein 1 and ALWAYS EARLY2 Myb-DNA binding factors. J. Mol. Biol. 2010;395:937–949. doi: 10.1016/j.jmb.2009.11.065. [DOI] [PubMed] [Google Scholar]
- 40.Noma K., Allis C.D., Grewal S.I.S. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 41.Ehrentraut S., Weber J.M., Dybowski J.N., Hoffmann D., Ehrenhofer-Murray A.E. Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5522–5527. doi: 10.1073/pnas.0909169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tennen R.I., Bua D.J., Wright W.E., Chua K.F. SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L.A., Barrett J.C., et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:U492–U416. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 2004;561:127–131. doi: 10.1016/S0014-5793(04)00148-6. [DOI] [PubMed] [Google Scholar]
- 46.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J., Russell D.W. Molecular Cloning: a Laboratory Manual. 3rd edn. NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 48.Shi X.B., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Pena P., Lan F., Kaadige M.R., et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu F., Cui X., Zhang S., Jenuwein T., Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011;43:715–719. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- 50.Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 51.Hwang M.G., Chung I.K., Kang B.G., Cho M.H. Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1) FEBS Lett. 2001;503:35–40. doi: 10.1016/s0014-5793(01)02685-0. [DOI] [PubMed] [Google Scholar]
- 52.Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- 53.Wu K.Q., Tian L.N., Malik K., Brown D., Miki B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 2000;22:19–27. doi: 10.1046/j.1365-313x.2000.00711.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhou C.H., Labbe H., Sridha S., Wang L., Tian L., Latoszek-Green M., Yang Z., Brown D., Miki B., Wu K.Q. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004;38:715–724. doi: 10.1111/j.1365-313X.2004.02083.x. [DOI] [PubMed] [Google Scholar]
- 55.Loayza D., de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 56.Shakirov E.V., Shippen D.E. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindroth A.M., Shultis D., Jasencakova Z., Fuchs J., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4146–4155. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schrumpfova P.P., Kuchar M., Palecek J., Fajkus J. Mapping of interaction domains of putative telomere-binding proteins AtTRB1 and AtPOT1b from Arabidopsis thaliana. FEBS Lett. 2008;582:1400–1406. doi: 10.1016/j.febslet.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 59.Kuchar M., Fajkus J. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants. FEBS Lett. 2004;578:311–315. doi: 10.1016/j.febslet.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 60.Prochazkova Schrumpfova P., Vychodilova I., Dvorackova M., Majerska J., Dokladal L., Schorova S., Fajkus J. Telomere repeat binding proteins are functional components of Arabidopsis telomeres and interact with telomerase. Plant J. 2014;77:770–781. doi: 10.1111/tpj.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Cao M., O'Sullivan R., Peters A., Jenuwein T., Blasco M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 62.Grafi G., Ben-Meir H., Avivi Y., Moshe M., Dahan Y., Zemach A. Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev. Biol. 2007;306:838–846. doi: 10.1016/j.ydbio.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 63.Galati A., Magdinier F., Colasanti V., Bauwens S., Pinte S., Ricordy R., Giraud-Panis M.J., Pusch M.C., Savino M., Cacchione S., et al. TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS One. 2012;7:e34386. doi: 10.1371/journal.pone.0034386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benetti R., Schoeftner S., Munoz P., Blasco M.A. Role of TRF2 in the assembly of telomeric chromatin. Cell Cycle. 2008;7:3461–3468. doi: 10.4161/cc.7.21.7013. [DOI] [PubMed] [Google Scholar]
- 66.Mizuno H., Wu J.Z., Kanamori H., Fujisawa M., Namiki N., Saji S., Katagiri S., Katayose Y., Sasaki T., Matsumoto T. Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J. 2006;46:206–217. doi: 10.1111/j.1365-313X.2006.02684.x. [DOI] [PubMed] [Google Scholar]
- 67.Brock G.J.R., Charlton J., Bird A. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999;240:269–277. doi: 10.1016/s0378-1119(99)00442-4. [DOI] [PubMed] [Google Scholar]
- 68.Hecht A., Strahl-Bolsinger S., Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 69.Cusanelli E., Romero C.A., Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell. 2013;51:780–791. doi: 10.1016/j.molcel.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 70.Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K., Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 71.Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalo S., Jaco I., Fraga M.F., Chen T.P., Li E., Esteller M., Blasco M.A. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 73.Johnson L.M., Cao X.F., Jacobsen S.E. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 74.Tamaru H., Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 75.Soppe W.J.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jackson J.P., Lindroth A.M., Cao X.F., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 77.Mathieu O., Probst A.V., Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacob Y., Feng S., LeBlanc C.A., Bernatavichute Y.V., Stroud H., Cokus S., Johnson L.M., Pellegrini M., Jacobsen S.E., Michaels S.D. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacob Y., Stroud H., Leblanc C., Feng S., Zhuo L., Caro E., Hassel C., Gutierrez C., Michaels S.D., Jacobsen S.E. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010;466:987–991. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raynaud C., Sozzani R., Glab N., Domenichini S., Perennes C., Cella R., Kondorosi E., Bergounioux C. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006;47:395–407. doi: 10.1111/j.1365-313X.2006.02799.x. [DOI] [PubMed] [Google Scholar]
- 81.Zheng B.L., Chen X.M. Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 2011;14:123–129. doi: 10.1016/j.pbi.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thorstensen T., Grini P.E., Aalen R.B. SET domain proteins in plant development. Biochim. Biophys. Acta. 2011;1809:407–420. doi: 10.1016/j.bbagrm.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 83.Rosenfeld J.A., Wang Z., Schones D.E., Zhao K., DeSalle R., Zhang M.Q. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arnoult N., Van Beneden A., Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat. Struct. Mol. Biol. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.